Abstract
Nontypeable Haemophilus influenzae is an important respiratory pathogen, causing otitis media in children and lower respiratory tract infection in adults with chronic obstructive pulmonary disease (COPD). Immunoglobulin A1 (IgA1) protease is a well-described protein and potential virulence factor in this organism as well as other respiratory pathogens. IgA1 proteases cleave human IgA1, are involved in invasion, and display immunomodulatory effects. We have identified a second IgA1 protease gene, igaB, in H. influenzae that is present in addition to the previously described IgA1 protease gene, iga. Reverse transcriptase PCR and IgA1 protease assays indicated that the gene is transcribed, expressed, and enzymatically active in H. influenzae. The product of this gene is a type 2 IgA1 protease with homology to the iga gene of Neisseria species. Mutants that were deficient in iga, igaB, and both genes were constructed in H. influenzae strain 11P6H, a strain isolated from a patient with COPD who was experiencing an exacerbation. Analysis of these mutants indicated that igaB is the primary mediator of IgA1 protease activity in this strain. IgA1 protease activity assays on 20 clinical isolates indicated that the igaB gene is associated with increased levels of IgA1 protease activity. Approximately one-third of 297 strains of H. influenzae of diverse clinical and geographic origin contained igaB. Significant differences in the prevalence of igaB were observed among isolates from different sites of isolation (sputum > middle ear > nasopharynx). These data support the hypothesis that the newly discovered igaB gene is a potential virulence factor in nontypeable H. influenzae.
Nontypeable Haemophilus influenzae, a gram-negative coccobacillus, is an important respiratory pathogen (36). This organism is the second most common cause of bacterial otitis media in children and the leading cause of recurrent otitis media (24, 36). Nontypeable H. influenzae also plays an important role in the course and pathogenesis of chronic obstructive pulmonary disease (COPD). It is the most commonly isolated organism from the lower airways of adults with COPD and is the most common bacterial cause of exacerbations or periodic worsening of the disease (57). Nontypeable H. influenzae is also a contributing etiologic agent in sinusitis in children and adults as well as in adult pneumonia (36).
A number of virulence factors have been identified in nontypeable H. influenzae, including (i) several adhesin molecules, which promote attachment to host structures; (ii) lipooligosaccharide, which promotes inflammation at the site of infection; and (iii) proteins involved in iron acquisition (49, 59, 60). Additionally, an enzyme which cleaves immunoglobulin A1 (IgA1) has been identified in H. influenzae (22, 32). This capability is advantageous to this organism because IgA is the predominant mucosal immunoglobulin, and most of the IgA in the respiratory mucosa is of the IgA1 subclass (34).
IgA1 proteases have been identified in several human mucosal pathogens, including H. influenzae, Streptococcus pneumoniae, Neisseria meningitidis, and Neisseria gonorrhoeae (22, 32, 41). The IgA1 protease genes of Haemophilus and Neisseria show a high degree of nucleotide homology by Southern blotting (25) and encode serine-type IgA1-specific endopeptidases which share approximately 50% amino acid homology with each other (40). These proteins cleave the IgA1 molecule at specific locations within the hinge region, releasing Fc and Fab fragments (34). The gram-negative IgA1 proteases are released from the bacterial cell by way of a type V secretion (autosecretory) mechanism (44) in an Omp85-dependent manner (64). The C-terminal autotransporter (β-core) domain is placed in the membrane and mediates the translocation of the proteolytic domain, which is cleaved from the precursor protein by an autoproteolytic mechanism. Previous studies have determined that almost all IgA1 protease activity from H. influenzae, N. meningitidis, and N. gonorrhoeae is found in the supernatant of broth cultures, indicating the secretion of these proteins (51).
All or nearly all strains of H. influenzae contain the iga gene that encodes an IgA1 protease (47, 54, 62). Sequence analysis of IgA1 protease genes from H. influenzae have demonstrated long stretches of amino acid homology punctuated by short spans of variable regions in the protease domain of the gene. The C-terminal autotransporter (β-core) domain is highly conserved (48). The H. influenzae IgA1 proteases are classified as type 1 or type 2 based on the specific site of cleavage in the hinge region of the IgA1 molecule (34, 35, 40). Vitovski et al. (62) demonstrated that the level of IgA1 protease activity was higher in invasive disease-causing strains than in strains isolated from throat swabs of asymptomatic people. However, the precise role of IgA1 protease in the pathogenesis of H. influenzae infection has not yet been elucidated.
In previous studies, we identified a second H. influenzae IgA1 protease gene, igaB, that is separate and distinct from the previously identified H. influenzae IgA1 protease gene, iga (12). The igaB gene displays a high degree of homology to neissserial IgA1 protease genes. In the present study, we characterize the structure and function of this second gene as well as its distribution among strains of H. influenzae isolated from patients with a variety of clinical infections.
MATERIALS AND METHODS
Bacterial strains and growth.
H. influenzae strain 11P6H was initially isolated from the sputum of an adult during an acute exacerbation of COPD (56, 58, 65). Sources and clinical characteristics of other strains are listed in Table 1.
TABLE 1.
Sources of Haemophilus influenzae strains
| Strain collection | Clinical source of isolates | No. of isolates | Reference(s) |
|---|---|---|---|
| National | Blood, cerebrospinal fluid, middle ear fluid | 54 | 33, 37 |
| Buffalo, NY | Sputum of adults with COPD | 132 | 56 |
| Michigan day care centers | Nasopharynx of children | 33 | 11 |
| Otitis media (various centersa) | Middle ear fluid obtained by tympanocentesis | 78 | 7, 10 |
Buffalo, NY; Rochester, NY; Seattle, WA; Dallas, TX; St. Louis, MO; and Columbus, OH.
Strains of H. influenzae were grown on chocolate agar, in brain heart infusion (BHI) broth supplemented with hemin and isovitalex, or in chemically defined media (CDM) as described previously (53).
Sequence analysis.
DNA sequences, obtained by standard methods, were aligned using Sequencher (Version 4.5; Gene Code Company). Comparisons to known genes were performed with BLASTN, BLASTP, and BLASTX analysis (http://www.ncbi.nlm.nih.gov/BLAST/).
Reverse transcriptase PCR (RT-PCR).
All experiments were carried out with RNA isolated from strain 11P6H or the mutant strains using the QIAGEN RNeasy kit and the Qiashredder column, followed by 30 min of treatment with DNase I (Promega, Madison, WI) at 37°C and 10 min of DNase stop solution (Promega) at 65°C. RT-PCR was performed on these samples using the QIAGEN OneStep RT-PCR kit, using the optional Q solution and RNaseOut RNase inhibitor (Invitrogen, Carlsbad, Calif.) according to the manufacturer's instructions. Primers were designed based on strain 11P6H sequence to amplify fragments approximately 500 bp in size. For every amplification, additional reactions were performed using primers to amplify a fragment of the constitutively expressed P2 outer membrane protein as an RNA control. Every reaction performed had a parallel reaction performed using the same primers and template but with HotMasterMix (Eppendorf, Hamburg, Germany) instead of the RT-PCR mix to test for contaminating DNA in the samples. Following amplification, samples were electrophoresed on 1.5% agarose gels.
Construction of mutants.
Two 1-kb regions were amplified by PCR using primers (Table 2) specific to the areas immediately upstream and downstream of the igaB gene in strain 11P6H. The primers also contained H. influenzae uptake sequences and appropriate restriction enzyme sites. These fragments were sequentially cloned into pGEM-3Zf (Promega). Similar amplifications and cloning of the 5′ 1 kb and 3′ 1 kb of the iga gene (the previously described IgA1 protease gene [6, 48]) were performed. The chloramphenicol resistance cassette from pACYC184 (New England BioLabs, Beverly, MA) was amplified by PCR with specific primers containing appropriate restriction enzyme sites. The two plasmids were linearized by restriction enzyme digestion between the two genomic fragments, and the resistance cassette was ligated to the upstream and downstream regions. These constructs were then linearized by appropriate restriction endonuclease digestion and transformed individually into strain 11P6H, which was made competent by the method of Herriott et al. (17) using the transformation protocol of Poje and Redfield (46). This resulted in two mutant strains, Δiga and ΔigaB. To construct a double mutant, the procedure for the ΔigaB mutant was repeated using a spectinomycin cassette from pSpec1 (2). The Δiga strain was then transformed with this construct to create strain Δiga ΔigaB. Confirmation of homologous recombination was verified by PCR and sequencing of the regions surrounding the antibiotic resistance cassettes in all three mutants.
TABLE 2.
Oligonucleotide primersa
| Primer set | 5′ Primer sequenceb | 3′ Primer sequence | Purpose of primers |
|---|---|---|---|
| igaBscreen | TGCCCGTTACCGGATAAATG | AACCACGGGCACAAACAGCC | Amplify 1,650-bp region of igaB in screen of 298 strains |
| igaB2screen | TTCTTCGCCAAAGAAACCGC | ATCTATAAAAAAGAATTTGC | Amplify 1,011-bp region of igaB in screen of 298 strains |
| igaup | AAAGAATTCAAGTGCGGTAATTTTATTGCACTTACTGTC | AAAGGTACCGCCCAGTAATCATAAGAACC | Amplify and clone 5′ 1 kb of iga gene |
| igadown | AAAGGTACCAATAACTGATAATTCAGAAAG | AAATCTAGAAAGTGCGGTCTGCAGTTTTTTGTTTTTCCG | Amplify and clone 3′ 1 kb of iga gene |
| igaBup | AAAGAATTCAAGTGCGGTGCGGGCTTTTATCTTAATGC | AAAGGATCCTTATACCATTATTATCTTAAC | Amplify and clone 1 kb upstream of igaB gene |
| igaBdown | AAAGGATCCATGATTACAATTAAGAAAGG | AAACTGCAGAAGTGCGGTCTTTTTCATTACCTTCAGCG | Amplify and clone 1 kb downstream of igaB gene |
| igaprobe | AACGCCACCAATGTATCTGG | TATTTAAATGAATATGCCCC | Amplify iga gene probe for Southern blots |
| igaBprobe | AACACGATAACGCCGGCACC | AAAAACAGACCGCCCGCACC | Amplify igaB gene probe for Southern blots |
| specprobe | AAAAAATAGGAGATAAAAGC | TTTTCGTTTTGCTTGGTAAAGC | Amplify spectinomycin cassette probe for Southern blots |
| catprobe | AGTAAGTTGGCAGCATCACCC | CATTGAGCAACTGACTGAAATGCC | Amplify chloramphenicol cassette probe for Southern blots |
The sequence of other oligonucleotide primers referred to in the text are available upon request.
Restriction enzyme sites are underlined; H. influenzae uptake sequences are italicized.
Southern blot analysis of mutants.
Genomic DNA was purified from strain 11P6H and the three mutant strains using the Wizard genomic DNA purification kit (Promega). Approximately 20 μg of DNA was digested with BglII and EcoRI simultaneously. Five-microgram samples of each digest were subjected to electrophoresis on a 0.8% agarose gel. The DNA was transferred to a Hybond-XL membrane (Amersham Biosciences, Buckinghamshire, United Kingdom) using a Hoefer TransVac vacuum blotting unit (San Francisco, CA) following the manufacturer's instructions. The membrane was cross-linked by UV light exposure and cut into four pieces, each containing one sample from each of the four strains. Probes ranging from 200 bp to 350 bp in size, corresponding to the iga and igaB genes as well as the chloramphenicol and spectinomycin resistance cassettes, were amplified by PCR from strain 11P6H, pACYC184, or pSpec1. Probes were labeled with the NEBlot Phototope kit (New England Biolabs) according to the manufacturer's instructions so as to produce approximately 1,400 ng of labeled probe. The membranes were incubated for 90 min in prehybridization buffer followed by overnight hybridization with approximately 50 ng/ml of the probes at 50°C. The signal was detected using the NEBlot Phototope Detection kit (New England Biolabs) according to the manufacturer's instructions.
Direct visualization of IgaB.
H. influenzae strain 11P6H and the mutant strains were grown in 22-ml CDM cultures to an optical density at 600 nm of 0.65 to 0.7. Cells were removed by centrifugation at 2,440 × g. Complete protease inhibitor cocktail (Roche, Basel, Switzerland) was added according to the manufacturer's instructions, and culture supernatants were concentrated approximately 100-fold using the Amicon ultrafiltration system with a molecular mass cutoff of 10 kDa (Millipore, Billerica, MA). Concentrated supernatants were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in an 8% gel and Coomassie stained. Extended destaining with 40% methanol and 10% acetic acid (30 min), oxidizer reagent (5 min; Bio-Rad, Hercules, CA), and water (15 min; changed multiple times) was performed.
Determination of IgA cleavage specificity.
H. influenzae strain 11P6H and the mutant strains, as well as KW20 Rd (cleavage type 1 control) (5) and H. influenzae biotype aegyptius BPF strain F3030 (cleavage type 2 control) (4, 29), were grown in supplemented BHI to an optical density at 600 nm of 0.8. Chloramphenicol was added to a concentration of 30 μg/ml to arrest production of bacterial proteins as described previously (51). Cells were removed by centrifugation, and culture supernatants were incubated with an equal volume of 20 μg/ml myeloma IgA1 or myeloma IgA2 (Calbiochem/EMD Biosciences, San Diego, CA) in phosphate-buffered saline (PBS) in a total reaction volume of 100 μl. Reactions were incubated at 37°C overnight and then subjected to SDS-PAGE and transferred to nitrocellulose. The membranes were probed overnight with 1:1,000 anti-IgA horseradish peroxidase conjugate (Dako, Carpinteria, CA) and developed.
IgA1 protease activity assays.
IgA1 protease assays were performed as described previously (50, 51). Briefly, to prepare protease reaction solutions, serial twofold dilutions (50 μl) of culture supernatants, obtained as described above, were made with supplemented BHI broth. These were arrayed in a sterile 96-well plate which had been blocked with 0.15% Tween 20. To each sample, 50 μl myeloma IgA1 diluted to 20 μg/ml in PBS was added. For each sample, controls were culture supernatant with no IgA1 and sterile BHI broth with IgA1. The plates were incubated for 6 h at 37°C with gentle agitation. Following incubation, IgA1 protease reactions were stopped by adding 200 μl 0.01 M bathocuproindisulfonic acid (Sigma-Aldrich, St. Louis, MO) in PBS.
Enzyme-linked immunosorbent assay (ELISA) plates were prepared by coating 1:2,000 rabbit anti-mouse Ig (Z0259; Dakocode) to Immulon 4HBX plates (ThermoLabsystems, Franklin, MA) for 2 h at 37°C, followed by washing with PBS and blocking with 2% nonfat dry milk for 90 min at room temperature. After washing with PBS, wells were incubated with 70 ng/ml mouse monoclonal anti-IgA1 (Fc specific) (code MAhu/IgA1/MAb; Nordic, Tilburg, The Netherlands) for 2 h at room temperature and washed with PBS again. The protease reaction solutions, prepared as described above (100 μl), were added to wells and incubated at room temperature for 2 h. After washing, 100 μl of a 1:1,000 anti-lambda light-chain horseradish peroxidase conjugate (code P0130; Dako) was added to the wells and incubated at room temperature for 2 h. After washing, color development was accomplished using tetramethyl benzidine. Absorbance readings were normalized to controls for that supernatant sample to allow for comparisons between assays.
Screening of strains for igaB.
The sequence of the igaB gene was used in a PCR-based screening method as previously described (12). Whole bacterial cell lysates were made from strains of H. influenzae using previously described methods (19). Briefly, bacteria grown overnight on chocolate agar plates were harvested with a sterile loop and suspended in 100 μl sterile water by vortexing. The bacteria were incubated at 100°C in a heat block for 5 min, resuspended by vortexing, and incubated at 100°C for an additional 5 min. The samples were centrifuged for 1 min at 16,000 × g, and the supernatants were saved for use as the templates in PCR. Two sets of internal primers (Table 2) were designed for igaB based on the nucleotide sequence of strain 11P6H in this region. PCRs were carried out under the following conditions: 10 min at 94°C; 30 cycles of 30 s at 94°C, 30 s at 55°C, and 90 s at 72°C; followed by 3 min at 72°C. Samples prepared from the following strains were used as positive and negative controls in the PCR screening experiments: H. influenzae strain 11P6H (positive control), the sequenced H. influenzae strain KW20 Rd (negative control), and Moraxella catarrhalis strain 43617 (negative control). Water controls were also included in the analysis. Univariate analyses to identify associations between the presence of igaB and a variety of clinical syndromes were performed with chi-square analysis.
RESULTS
Sequence analysis of the igaB locus.
In previous work, we identified igaB, a gene in H. influenzae strain 11P6H whose product has high homology to neisserial IgA1 proteases, that is separate and distinct from the previously identified IgA1 protease encoded by iga (6, 48). To confirm the sequence of the igaB locus, a series of overlapping fragments that span the entire region was amplified by PCR from the genomic DNA of strain 11P6H, and the sequences of the fragments were determined (GenBank accession no. DQ423203). The igaB open reading frame is 5,664 nucleotides (1,887 amino acids) in length (Fig. 1). Blastx analysis determined that the igaB gene is homologous to the IgA1 protease genes of Neisseria gonorrhoeae and Neisseria meningitidis. The degree of homology of igaB to neisserial iga genes is greater than the homology of igaB to Haemophilus iga genes. The majority of the IgaB protein homology to neisserial Iga proteins occurred in amino acids 1 to 1020 (92% identity, 95% similarity; corresponding to the secreted protease domain) and amino acids 1529 to 1887 (53% identity, 71% similarity; corresponding primarily to the autotransporter [β-core] domain) (Fig. 1).
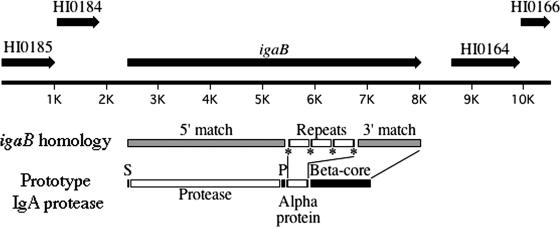
FIG. 1.
Diagram of the igaB locus. The upstream and downstream genes are labeled based on designations in the sequenced strain KW20 Rd. Shaded boxes of igaB denote regions of high homology to neisserial iga genes. Open boxes labeled “Repeats” indicate the positions of 423-bp unique tandem repeats in the igaB gene. Asterisks indicate positions of four direct repeats 13 bp in length found in the context of the larger tandem repeats. Also shown are the conserved domains of neisserial IgA1 protease proteins as they relate to igaB. S, 25-amino-acid signal sequence; protease, 961-amino-acid secreted IgA1 protease domain; P, 32-amino-acid short peptide conserved among IgA1 proteases; alpha protein, 476-amino-acid region, including repeated sequence, potentially corresponding to the alpha protein of neisserial IgA1 proteases; beta-core, 393-amino-acid β-core (autosecretory domain).
Both the igaB gene and the previously identified IgA1 protease gene (known as iga or HI0990 in strain KW20 Rd) are present in strain 11P6H. The iga gene was amplified by PCR from strain 11P6H using previously described primers (62), and the sequence was determined. The sequence was highly similar to H. influenzae iga genes in the National Center for Biotechnology Information database. Whereas iga is present in the genomes of all four sequenced strains of H. influenzae (9, 13, 15), igaB is absent from the four strains. Furthermore, igaB is distinct from other known variants of the iga gene in this organism (48).
The igaB gene is 885 to 1,068 nucleotides longer than IgA1 protease genes with the highest BLAST matches. This size differential and the low level of sequence match in the third quarter of the gene are due to the presence of a repeated element in this section of the gene. A 423-bp section is present three times in succession in this region (Fig. 1, Repeats). The first 13 bp of these repeats (noted by asterisks in Fig. 1) are repeated a fourth time at the 3′ end of this region. Blast analysis of the 423-bp sequence showed small stretches of nucleotide match to Neisseria IgA1 protease genes but yielded no amino acid match.
Analysis of the deduced amino acid sequence of the igaB gene reveals a primary structure similar to that of known IgA1 proteases. Based on homology to other IgA1 proteases, the first 25 amino acids are a signal sequence capable of targeting translocation to the periplasm via the Sec pathway (38). Autocatalytic cleavage sites that produce the 961-amino-acid mature secreted enzyme and a short peptide (∼32 amino acids) 3′ to the secreted domain are present in the N-terminal half of the igaB gene (Fig. 1). Conserved sequences predicted to encode the catalytically active site of IgA1 proteases (30, 44, 48) are found in the igaB gene. Further analysis of the amino acid sequence of the region that determines cleavage specificity (14) of the igaB gene indicates that the gene likely encodes a type 2 IgA1 protease. There is no apparent cleavage site in IgaB that would yield an α-protein as described for N. gonorrhoeae strain MS11 (44); however, there is a proline-rich region closer to the conserved β-core that, if cleaved, would yield a protein approximately 476 amino acids in length. This proline-rich region is homologous to a similar region in N. gonorrhoeae strain FA 1090 (GenBank accession no. YP_207437). The C-terminal 414 amino acids of IgaB, primarily corresponding to the β-core autotransporter domain, are conserved among neisserial IgA1 proteases. This region of the gene encodes the portion of IgA1 proteases that forms a β-barrel in the outer membrane, facilitating export of the mature protein.
Transcription of IgA1 proteases in strain 11P6H.
To determine whether the iga and igaB genes are transcribed in H. influenzae strain 11P6H, reverse transcriptase PCR (RT-PCR) was performed. RNA was isolated from this strain and treated with DNase I to eliminate contaminating DNA. RT-PCR was performed using primers designed to amplify 500-bp fragments of iga, igaB, and the constitutively expressed OMP P2 gene as a positive control. RT-PCR analysis demonstrated transcription of both the iga and igaB genes in strain 11P6H (Fig. 2). Simultaneous parallel reactions performed using the same primers and template but with Taq polymerase and with no reverse transcriptase were all negative, indicating the absence of contaminating DNA.
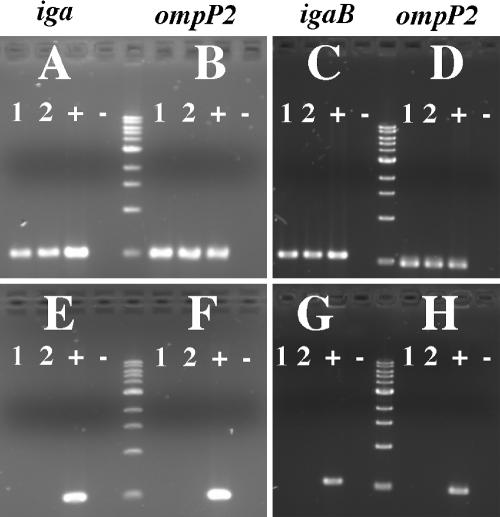
FIG. 2.
Results of RT-PCR of IgA1 protease genes from H. influenzae strain 11P6H. (A) RT-PCR with iga primers; (C) RT-PCR with igaB primers; (B and D) RT-PCR with ompP2 primers for positive control. Lanes 1 and 2, purified 11P6H RNA template; lanes +, H. influenzae strain 11P6H genomic DNA template for positive control; lanes −, water template for negative control. The bottom panels (E to H) depict the same templates and primers corresponding to upper panels amplified with Taq polymerase only, confirming the absence of DNA contamination.
Construction and characterization of IgA1 protease mutants.
To analyze the role of the two IgA1 proteases in the phenotype of strain 11P6H, three mutants were constructed. A Δiga mutant was constructed by replacing ∼3 kb of the gene with a chloramphenicol resistance cassette. A ΔigaB mutant was constructed by replacing the entire gene with the chloramphenicol resistance cassette. A Δiga ΔigaB double mutant was constructed by replacing the igaB gene with a spectinomycin resistance cassette in the Δiga mutant. To confirm putative mutants, transformants that grew on appropriate selective media were tested by PCR for the presence and size of the construct, the presence of the resistance cassette, and the absence of the deleted gene. The three mutant strains were further confirmed by PCR and sequencing using one primer from the sequence within the resistance cassettes and one primer from the genomic sequence located outside of the sequence contained on the plasmid used to create the mutant. These reactions showed amplification of the appropriate-sized amplicons (data not shown). Sequencing across the regions of homologous recombination confirmed the presence of the cassette in the proper location.
Southern blots were also performed to confirm the absence of the IgA1 protease genes and the presence of the resistance cassettes in the appropriate knockouts. Genomic DNA from the parent strain and the three mutants was digested, electrophoresed, transferred, and hybridized with Phototope-Star-labeled probes that were amplified from the iga gene, the igaB gene, the chloramphenicol resistance cassette, and the spectinomycin resistance cassette. The patterns of signal confirmed the presence of iga in the parent and the ΔigaB mutant, the presence of igaB in the parent and the Δiga mutant, the presence of the chloramphenicol cassette in all three mutants, and the presence of the spectinomycin cassette in the double mutant only (Fig. 3).
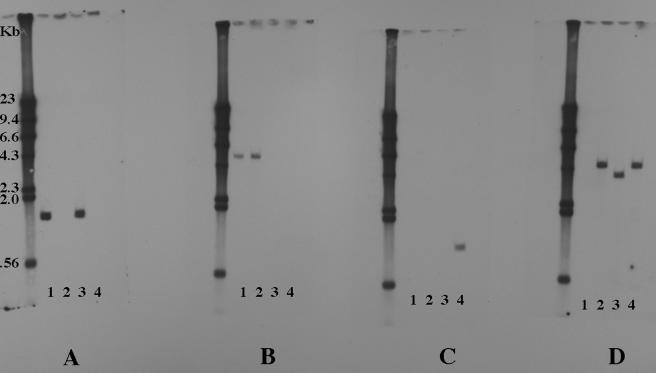
FIG. 3.
Southern blot assays of genomic DNA of H. influenzae strain 11P6H and mutants. Lanes 1, strain 11P6H; lanes 2, Δiga mutant; lanes 3, ΔigaB mutant; lanes 4, Δiga ΔigaB double mutant. (A) iga probe; (B) igaB probe; (C) spectinomycin cassette probe; (D) chloramphenicol cassette probe. Molecular size markers are labeled in kilobases.
To further confirm that the iga and igaB genes were not transcribed in the mutants, RT-PCR was performed (Fig. 4). As expected, the Δiga mutant showed only igaB transcription, the ΔigaB mutant showed only iga transcription, and the Δiga ΔigaB mutant showed transcription of neither.
FIG. 4.
Results of RT-PCR of IgA1 protease mutants. Lanes 1, Δiga mutant RNA template; lanes 2, ΔigaB mutant RNA template; lanes 3, Δiga ΔigaB double mutant RNA template; lanes +, H. influenzae strain 11P6H genomic DNA template for positive control; lanes −, water template for negative control. (A) RT-PCR with iga primers; (B) RT-PCR with igaB primers; (C) RT-PCR with ompP2 primers for positive control. The bottom panels (D to F) depict the same templates and primers corresponding to upper panels amplified with Taq polymerase only, confirming the absence of DNA contamination.
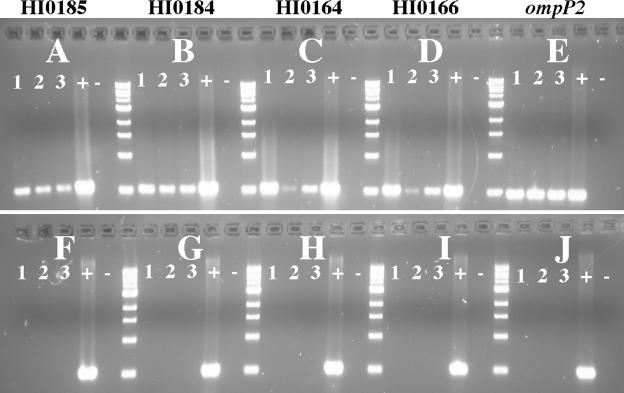
The ΔigaB and Δiga ΔigaB mutants were further analyzed for transcription of the upstream (HI0185 and HI0184) and downstream (HI0164 and HI0166) genes. Primers were designed to amplify approximately 500-bp fragments from each of these genes. RT-PCRs were performed as described above, with strain 11P6H RNA used as a positive control. Both mutants showed transcription of the upstream and downstream genes (Fig. 5). The RT-PCR amplification reactions of the downstream genes in the ΔigaB strain showed less product than those from the parent strain. This is likely due to encroachment on the promoter region of those genes by the chloramphenicol resistance cassette in the construct that was used to create the ΔigaB mutant. These genes encode subunits of a Na+-translocating NADH-quinone reductase, thus, reduction in transcription of these genes is not expected to affect IgA1 protease activity of this strain. The gene downstream of the iga gene is convergently expressed, so polar effects of mutations in the iga gene are not expected. Further, this mutant was constructed so as to homologously recombine within the 5′ 1 kb and 3′ 1 kb of the iga gene, thus, adverse effects on regulatory regions of adjacent genes are not expected.
FIG. 5.
RT-PCR of genes upstream and downstream of igaB. Lanes 1, H. influenzae strain 11P6H RNA template; lanes 2, ΔigaB mutant RNA template; lanes 3, Δiga ΔigaB double mutant RNA template; lanes +, H. influenzae strain 11P6H genomic DNA template for positive control; lanes −, water template for negative control. (A) RT-PCR with HI0185 primers; (B) RT-PCR with HI0184 primers; (C) RT-PCR with HI0164 primers; (D) RT-PCR with HI0166 primers; (E) RT-PCR with ompP2 primers for positive control. The bottom panels (F to J) depict the same templates and primers corresponding to upper panels amplified with Taq polymerase only, confirming the absence of DNA contamination.
Outer membrane proteins were isolated from strain 11P6H and the three mutant strains to analyze any effects of deleting IgA1 protease genes on other membrane protein patterns. All strains had the same outer membrane protein pattern on SDS-PAGE (data not shown).
As seen in other IgA1 protease mutants in H. influenzae (5, 42), growth curves of the parent and the mutants were not different from one another, indicating the growth rate of this strain is unaffected by the deletions of either or both IgA1 protease genes.
Identification of secreted IgA1 protease encoded by igaB.
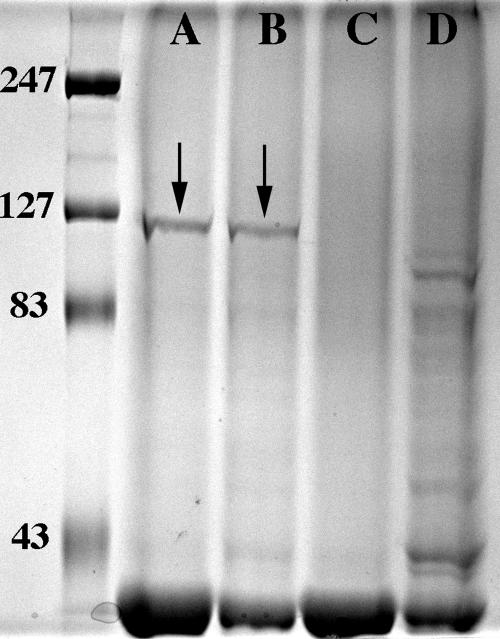
Strain 11P6H and the three mutant strains were grown in chemically defined media to late logarithmic phase. The cells were removed by centrifugation, and the supernatants were concentrated approximately 100-fold using the Amicon ultrafiltration system and electrophoresed on SDS-PAGE (Fig. 6). A band was present in both strain 11P6H (lane A) and the Δiga mutant (lane B) at the expected position of ∼106 kDa, which corresponds to the predicted secreted portion of IgaB protein. If the iga gene expressed a stable product in this experiment, we would expect to see a second band in strain 11P6H (lane A) and a corresponding band in the ΔigaB mutant (lane C) only, or a band at the same level as IgaB in the ΔigaB mutant that was still not seen in the double mutant (lane D). No such band that would correspond to the product of the iga gene was noted. This result indicates that (i) strain 11P6H secretes IgaB, and (ii) although transcription of the iga gene was noted by RT-PCR, final expression of the gene was significantly lower than that of igaB, or the protein is labile in the culture supernatant.
FIG. 6.
Coomassie-stained SDS-polyacrylamide gel of concentrated culture supernatants from cultures in chemically defined media. Lane A, H. influenzae strain 11P6H; lane B, Δiga mutant; lane C, ΔigaB mutant; lane D, Δiga ΔigaB double mutant. Molecular mass markers are labeled in kilodaltons. Arrows denote IgaB-secreted protease bands.
Cleavage specificity of IgA1 proteases in strain 11P6H.
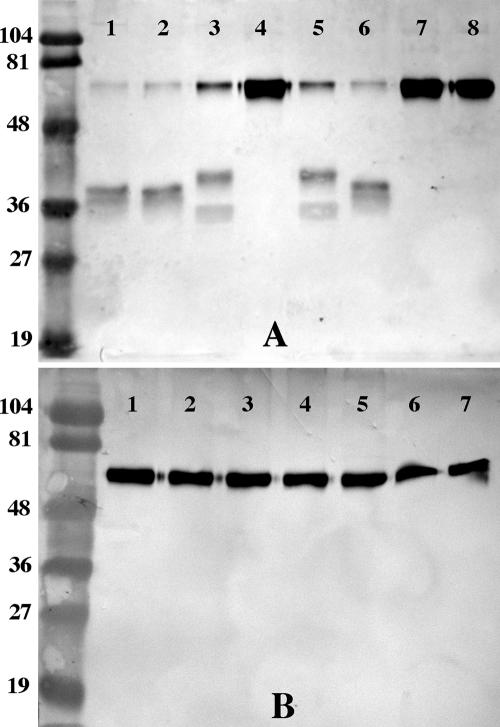
IgA1 proteases are classified as type 1 or type 2 based on the specific site of enzymatic cleavage in the hinge region of the IgA1 molecule. To characterize the enzymatic specificity of the IgA1 proteases of strain 11P6H, digests were performed with IgA and culture supernatants of the parent and mutant strains. Culture supernatants were incubated with myeloma IgA1 overnight. The reaction mixtures were then separated by SDS-PAGE, transferred to nitrocellulose, and probed with anti-IgA peroxidase conjugate that binds to the heavy chain of IgA. Control reactions were performed using culture supernatants of H. influenzae strain KW20 Rd, which produces a type 1 IgA1 protease (5), and H. influenzae biogroup aegyptius strain BPF F3030, which produces a type 2 IgA1 protease (29). Negative controls included IgA1 incubated in BHI broth and PBS. The heavy-chain cleavage in this assay yields two fragments, the Fc fragment (larger) and the Fd fragment (smaller). The pattern of IgA1 digestion (Fig. 7) of the parent (lane 1) and the Δiga strain (lane 2) correspond to the type 2 control (lane 6), while the ΔigaB strain (lane 3) corresponds to the type 1 control (lane 5). These results indicate that the igaB gene encodes a type 2 IgA1 protease, while the iga gene encodes a type 1 IgA1 protease. Further, these results suggest that the dominant activity in strain 11P6H is derived from IgaB, not Iga. The double mutant showed no IgA1 protease activity (lane 4).
FIG. 7.
Western blot assays of IgA digests. (A) Lanes contain purified human IgA1 digested with culture supernatants of H. influenzae or PBS in place of the supernatant, as follows: strain 11P6H (lane 1); Δiga mutant (lane 2); ΔigaB mutant (lane 3); Δiga ΔigaB double mutant (lane 4); type 1 IgA1 protease control, H. influenzae strain KW20 Rd (lane 5); type 2 IgA1 protease control, H. influenzae biogroup aegyptius strain BPF 17 (lane 6); sterile BHI broth (lane 7); and PBS (lane 8). (B) Lanes contain purified human IgA2 digested with culture supernatants of H. influenzae or broth or PBS in place of the supernatant, as follows: strain 11P6H (lane 1); Δiga mutant (lane 2); ΔigaB mutant (lane 3); Δiga ΔigaB double mutant (lane 4); sterile BHI broth (lane 5); PBS (lane 6); and PBS (unincubated) (lane 7). Molecular mass markers are labeled in kilodaltons.
A similar experiment performed with myeloma IgA2 showed no digestion by strain 11P6H or the mutant strains, indicating that neither IgA1 protease in this strain is active against IgA2 (Fig. 7). This is consistent with previous reports that most IgA1 proteases do not digest IgA2 (34, 40, 43).
IgA1 protease activity in strain 11P6H.
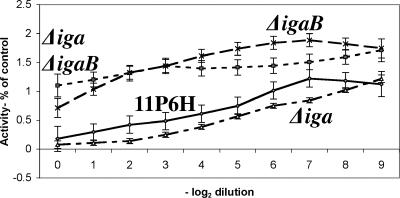
To quantify the relative contributions of the products of iga and igaB in the IgA1 protease activity of strain 11P6H, assays were performed as previously described on the parent strain and the three mutant strains (50, 51). In this assay, serial dilutions of broth culture supernatants are incubated with IgA1, and the resulting reactions are analyzed for intact IgA1 in ELISA. Therefore, a greater signal indicates more intact IgA, thus, less IgA1 protease activity. Data from the dilutions of supernatant yield roughly sigmoidal curves (Fig. 8). More concentrated samples have more IgA1 protease activity, thus, less intact IgA and lower values. More dilute samples have less IgA1 protease activity, thus, more intact IgA and higher values. Taken as a whole, curves shifted to the left indicate that the strains in question have less IgA1 protease activity, while curves shifted to the right indicate strains with more activity.
FIG. 8.
IgA1 protease activity assays. Lines indicate LOESS plots of the IgA1 protease activity as a percentage of control of culture supernatants of H. influenzae strain 11P6H and the mutant strains as noted. x axis, the degree of twofold dilution of the original samples; y axis, activity as a percentage of maximal activity based on positive (IgA with sterile BHI broth) and negative (undiluted culture supernatants absent IgA) controls. Each data point is an average of eight replicates. Error bars represent the standard error for each dilution.
IgA1 protease activity ELISAs were performed on strain 11P6H and the three mutant strains using a total of four aliquots of culture supernatant from three different cultures (with each aliquot assayed in duplicate, this resulted in eight data points for each dilution for each strain). Each data point was normalized by converting the absolute value to a percentage of the controls for that strain in that assay (absent IgA means 0% signal, thus, maximum protease activity; IgA and sterile broth means 100% signal, thus, zero protease activity). Curves for each strain were generated using the LOESS (locally weighted scatterplot smoothing; SAS v9.1; SAS Institute, Inc.) technique. The curve for strain 11P6H indicates that this strain has IgA1 protease activity. The curves for 11P6H and the Δiga mutant were very similar, indicating that the majority of the IgA1 protease activity for this strain comes from igaB. The curve for the double mutant shows no degradation of IgA1, confirming previous observations that this strain has no detectable IgA1 protease activity. The curve for the ΔigaB mutant shows a low level of activity at maximal concentration that is quickly diluted out to form a curve that is similar to that of the double mutant, indicating that iga contributes minimally to the IgA1 protease activity of this strain.
Distribution of igaB among clinical isolates of H. influenzae.
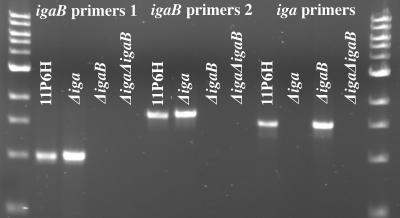
A total of 297 well-characterized clinical isolates of nontypeable H. influenzae collected from a variety of sources (Table 1) were screened by PCR for the presence of igaB as described previously (12). Two sets of internal primers were designed to amplify fragments of the secreted portion of the igaB gene. Each assay contained control amplification reactions using template DNA from strain 11P6H (positive), the sequenced strain KW20 Rd (negative), Moraxella catarrhalis strain 43617 (negative), and water (negative). PCRs were performed in duplicate on separate days, with all strains scored for presence or absence of amplification of the predicted size fragments detected by agarose gel electrophoresis. When replicates of a strain-primer combination did not agree (less than 1% of all reactions), additional reactions were carried out with either purified genomic DNA or a second lysate preparation. The final determination was made based on these repeated reactions. The specificity of the igaB primers was confirmed using the IgA1 protease mutant strains. PCRs were performed with both igaB primer sets on strain 11P6H as well as the mutant strains. These reactions showed amplification from the parent and the Δiga mutant but not the ΔigaB mutant or the double mutant (Fig. 9). These results indicate that the igaB primers amplify a portion of the igaB gene but not the iga gene.
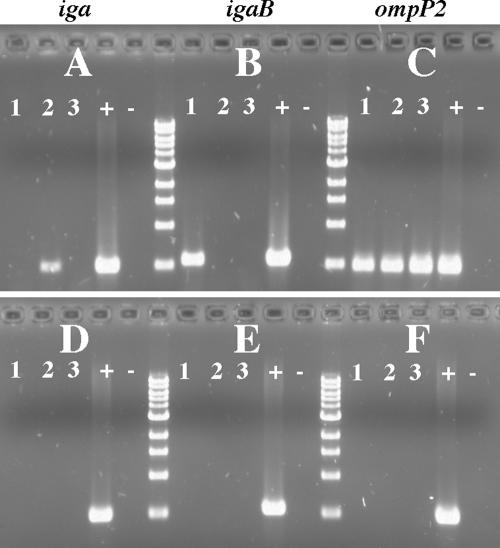
FIG. 9.
Agarose gel showing specificity of primers for PCR amplification of IgA1 protease genes. DNA from strain 11P6H and the mutant strains were used as the templates for PCRs as noted. Primers used in each PCR are noted at the top of the gel. Primers used for the amplification of iga were the “P” primers used by Vitovski et al. (62).
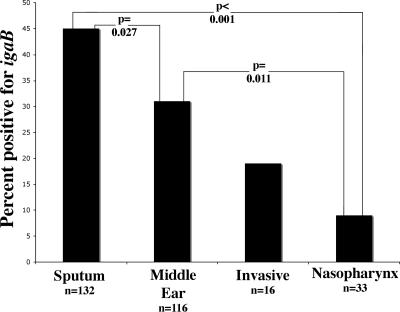
Among the 297 strains of nontypeable H. influenzae, 101 (or 34%) were positive for amplification of the igaB gene (Fig. 10). Analysis of the presence of igaB related to the site of isolation (sputum, middle ear, nasopharynx, and invasive disease [blood and cerebrospinal fluid {CSF}]) demonstrated a significant distribution (P < 0.001; chi square). Significant differences between individual sites of infection were also seen by chi square (Fig. 10). The proportion of sputum isolates from adults with COPD with igaB was significantly greater than the proportion of middle ear isolates from children with igaB (P = 0.027; chi square). Further, nasopharyngeal colonizing strains had significantly lower prevalence of igaB than sputum isolates, middle ear isolates, and all disease isolates combined (P < 0.001, P = 0.011, and P = 0.0014, respectively).
FIG. 10.
Percentage of strains containing igaB by site of infection. Chi-square P values are shown.
IgA1 protease activity of strains of H. influenzae with and without igaB.
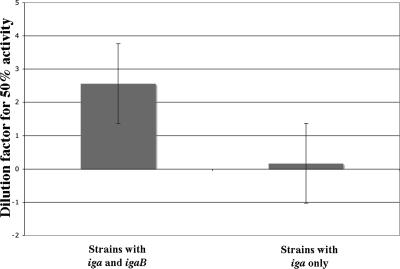
To assess the IgA1 protease activity in strains of H. influenzae that contain igaB compared with those that do not, the IgA1 protease activity assay was performed on 10 clinical isolates that contain the igaB gene and 10 that do not. All strains were isolated from the sputum of adults with COPD. The assays were carried out on two aliquots of culture supernatant, each from different cultures (each aliquot was assayed in duplicate for a total of four data points at each dilution). The data were normalized relative to the absent IgA and sterile broth controls as previously described, and LOESS plots were generated. Based on these plots, the dilution that resulted in 50% IgA1 protease activity for each strain was determined. A higher value indicates more IgA1 protease activity, while lower values indicate less IgA1 protease activity. For those strains that had undiluted supernatants that did not produce 50% activity, theoretical values were determined based on extrapolation of the LOESS curves into negative values. The results of this analysis are summarized in Fig. 11. The mean value of the 10 strains that contain the igaB gene (2.57) was significantly greater than the mean value of the 10 strains that lacked the igaB gene (0.17; P = 0.0411; unpaired t test). These data indicate that those strains that contain igaB exhibit more IgA1 protease activity than those that do not. We conclude that the total IgA1 protease activity of strains of H. influenzae results from the combination of the activities of the IgA1 proteases encoded by iga and igaB.
FIG. 11.
Results of IgA1 protease activity assays on 20 strains of H. influenzae from adults with COPD. y axis, the dilution factor required to produce 50% IgA1 protease activity from culture supernatants as determined by four replicates. The bars are the averages of two groups of 10 strains each with iga and igaB genes as noted. The difference between groups is significant (P = 0.04; t test). Error bars represent the standard errors for each group.
DISCUSSION
In this study, we have characterized a second IgA1 protease gene, igaB, and have demonstrated that the gene is present in one-third of strains of H. influenzae isolated from a wide range of clinical and geographic sources. The igaB gene is separate and distinct from the well-characterized IgA1 protease encoded by iga that is present in all strains of H. influenzae studied thus far. The IgA1 protease encoded by igaB shows greater homology to neisserial IgA1 proteases than to the IgA1 protease encoded by the H. influenzae iga gene. Detailed analysis of a strain of H. influenzae that was isolated from an adult with COPD at the time of an exacerbation revealed that the igaB gene is transcribed, and the protein is expressed and secreted in its active form with type 2 IgA1 protease activity.
IgA1 proteases cleave the hinge region of IgA1, which is the predominant immunoglobulin on the human respiratory tract mucosal surface. The potential significance of IgA1 protease in the pathogenesis of H. influenzae infection has been difficult to elucidate in part because the IgA1 subclass molecule is unique to humans and some higher primates, limiting the use of animal models to study IgA1 protease. However, several lines of evidence suggest that IgA1 protease is a virulence factor. Among gram-negative bacteria, IgA1 protease is present predominantly in pathogenic species. Neisseria meningitidis, N. gonorrhoeae, and H. influenzae have IgA1 proteases, while the commensals N. mucosa, N. lactamica, N. flavescens, N. cinerea, N. subflava, N. flava, N. perflava, N. sicca, N. elongata, H. parainfluenzae, H. aphrophilus, H. paraphrophilus, H. segnis, H. paraphrohaemolyticus, H. hemoglobinophilus, and H. haemolyticus do not (20, 39, 54). In addition, studies of N. meningitidis showed significantly increased levels of IgA1 protease activity among invasive isolates compared to isolates from asymptomatic carriers (63). A similar finding was reported in a study of nontypeable H. influenzae. Strains isolated from sputum, blood, CSF, and other disease-associated sites had higher IgA1 protease levels than nasopharyngeal isolates from healthy volunteers (62), suggesting that IgA1 protease is a virulence factor. The present study contributes another indirect line of evidence suggesting that IgA1 protease is a potential virulence factor in human infection. The proportion of H. influenzae strains isolated from middle ear fluid of children with otitis media with the igaB gene (31%) was significantly greater than the proportion of nasopharyngeal strains with igaB (9%; P = 0.013). Further studies of the prevalence of igaB in well-characterized clinical isolates are needed to test this hypothesis further.
The present study indicates that genes iga and igaB are transcribed in strain 11P6H. The study further showed that igaB is directly responsible for a substantial amount of IgA1 protease activity in strain 11P6H. The observation that strains of H. influenzae that have both IgA1 protease genes express higher levels of total IgA1 protease activity than strains that have iga exclusively suggests that igaB contributes to the IgA1 protease activity in other strains as well (Fig. 11). However, additional study is needed to establish this observation with certainty in other strains. Overall, these observations support the conclusion that the IgA1 protease activity measured in assays represents the cumulative IgA1 protease activity of both iga and igaB genes. The observation of the contribution of a second IgA1 protease in H. influenzae has implications in the interpretation of previous studies involving measurements of IgA1 protease activities of clinical isolates. Several groups have identified strains that have two protease activity specificity types (23, 28, 29, 35, 39, 54). Lomholt and Kilian (28) identified a strain of nontypeable H. influenzae with two different protease types that had inhibition of IgA1 protease activity by using antisera raised to neisserial IgA1 proteases. Also, antiserum raised against secreted proteases from that strain inhibited IgA1 proteases from N. meningitidis and N. gonorrhoeae, suggesting that this strain encodes a neisserial iga-like IgA1 protease (i.e., igaB). Further, this strain showed Southern hybridization of probes derived from iga to two portions of the genome (47). We speculate that these strains have both iga and igaB genes expressing IgA1 proteases.
In addition, the varying levels of IgA1 protease activity among clinical isolates observed by Vitovski et al. (62) is likely a result of the IgA1 proteases encoded by iga in all strains and igaB in some strains. In that study, PCR was used to confirm the presence of the iga gene in virtually all strains of H. influenzae isolates examined. We confirmed that the primers used in the Vitovski study (62) did not amplify the igaB gene. Further, the primers we used to screen for igaB did not amplify the iga gene (Fig. 9). These data confirm the results of epidemiological studies performed by Vitovski et al. that showed iga in all strains of H. influenzae, while we showed that igaB is present in approximately one-third of strains.
In view of the high degree of homology between igaB and the iga genes from both N. meningitidis and N. gonorrhoeae, we speculate that igaB was acquired by horizontal transfer from Neisseria. The major difference between igaB and the neisserial iga genes is the presence of 423-bp direct repeats between base pairs 5498 and 6779. It appears that this sequence originally appeared as a single copy that was expanded by a mechanism possibly mediated by 13-bp direct repeats on both ends of the sequence. In addition, the G+C content of this gene is greater than the average for sequenced H. influenzae strains, approaching the G+C contents of neisserial iga genes and neisserial genomes (12). Transfer from Haemophilus to Neisseria has been suggested in a number of studies (8, 26, 52, 55), but evidence of transfer from Neisseria to Haemophilus has not been established. Analysis of the igaB locus suggests that this gene was acquired and integrated into the H. influenzae genome as part of a large genomic inversion event, as the genes upstream and downstream of igaB in strain 11P6H are not present in such close proximity nor in the same orientation in the genomes of the four sequenced strains of this organism.
IgA1 proteases may play a role in pathogenesis by a mechanism independent of their ability to cleave IgA1. IgA1 was the only known substrate of IgA1 protease until recently. LAMP1, a membrane component of late endosomes and lysosomes, is also cleaved by IgA1 proteases, resulting in destabilized lysosomes and increased invasion of human cells by the bacterium (1, 16, 18, 27). Further, the α-protein, which may be a component of the igaB protein, induces human T-cell responses and translocates to human nuclei, potentially altering host gene transcription (21, 45). IgA1 proteases also induce a number of proinflammatory cytokines, including tumor necrosis factor alpha (TNF-α), interleukin-1β (IL-1β), IL-6, and IL-8 independent of proteolytic activity (31), as well as a specific T-cell response in humans (61). In addition, IgA1 proteases inhibit TNF-α-mediated apoptosis of human monocytes by cleavage of the TNF receptor II (3). These examples illustrate contributions that one or both of the IgA1 proteases may make to the pathogenesis of H. influenzae, thus, the relative importance of each of these genes goes well beyond a discussion of IgA1 protease activity levels. It will be important to determine the relative contributions of both IgA1 proteases in these other functions.
In summary, this study characterized the second IgA1 protease gene, igaB, which is separate and distinct from the well-characterized IgA1 protease encoded by iga, which is present in all strains of H. influenzae. The igaB gene is transcribed and expressed in H. influenzae strain 11P6H and is present in approximately one-third of strains. The IgA1 protease activity of strains of H. influenzae results from IgA1 proteases encoded by the iga and igaB genes. Future studies will examine the significance of these genes in the pathogenesis of H. influenzae in the context of human infection.
Acknowledgments
We thank Janet Gilsdorf, May Patel, Terence Stull, Daniel Morton, Howard Faden, Diane Dryja, Anne Vacca-Smith, Michael Pichichero, Eric Hansen, Steven Barenkamp, and Arnold Smith for providing isolates; Lauren Bakaletz and Robert Munson for providing isolates and plasmid pSpec1; Mogens Kilian for advice regarding IgA1 protease activity assays; Charmaine Kirkham for expert technical assistance; and Anthony Campagnari for reviewing the manuscript.
This work was supported by NIH grant AI19641 and by the Department of Veterans Affairs.
Editor: J. N. Weiser
REFERENCES
- 1.Ayala, B. P., B. Vasquez, S. Clary, J. A. Tainer, K. Rodland, and M. So. 2001. The pilus-induced Ca2+ flux triggers lysosome exocytosis and increases the amount of Lamp1 accessible to Neisseria IgA1 protease. Cell Microbiol. 3:265-275. [DOI] [PubMed] [Google Scholar]
- 2.Bakaletz, L. O., B. D. Baker, J. A. Jurcisek, A. Harrison, L. A. Novotny, J. E. Bookwalter, R. Mungur, and R. S. Munson, Jr. 2005. Demonstration of type IV pilus expression and a twitching phenotype by Haemophilus influenzae. Infect. Immun. 73:1635-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beck, S. C., and T. F. Meyer. 2000. IgA1 protease from Neisseria gonorrhoeae inhibits TNF-α-mediated apoptosis of human monocytic cells. FEBS Lett. 472:287-292. [DOI] [PubMed] [Google Scholar]
- 4.Brenner, D. J., L. W. Mayer, G. M. Carlone, L. H. Harrison, W. F. Bibb, M. C. Brandileone, F. O. Sottnek, K. Irino, M. W. Reeves, J. M. Swenson, et al. 1988. Biochemical, genetic, and epidemiologic characterization of Haemophilus influenzae biogroup aegyptius (Haemophilus aegyptius) strains associated with Brazilian purpuric fever. J. Clin. Microbiol. 26:1524-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bricker, J., M. Mulks, E. R. Moxon, A. G. Plaut, and A. Wright. 1985. Physical and genetic analysis of DNA regions encoding the immunoglobulin A proteases of different specificities produced by Haemophilus influenzae. Infect. Immun. 47:370-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bricker, J., M. H. Mulks, A. G. Plaut, E. R. Moxon, and A. Wright. 1983. IgA1 proteases of Haemophilus influenzae: cloning and characterization in Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 80:2681-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casey, J. R., and M. E. Pichichero. 2004. Changes in frequency and pathogens causing acute otitis media in 1995-2003. Pediatr. Infect. Dis. J. 23:824-828. [DOI] [PubMed] [Google Scholar]
- 8.Davis, J., A. L. Smith, W. R. Hughes, and M. Golomb. 2001. Evolution of an autotransporter: domain shuffling and lateral transfer from pathogenic Haemophilus to Neisseria. J. Bacteriol. 183:4626-4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erwin, A. L., K. L. Nelson, T. Mhlanga-Mutangadura, P. J. Bonthuis, J. L. Geelhood, G. Morlin, W. C. Unrath, J. Campos, D. W. Crook, M. M. Farley, F. W. Henderson, R. F. Jacobs, K. Muhlemann, S. W. Satola, L. van Alphen, M. Golomb, and A. L. Smith. 2005. Characterization of genetic and phenotypic diversity of invasive nontypeable Haemophilus influenzae. Infect. Immun. 73:5853-5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faden, H., J. Bernstein, L. Brodsky, J. Stanievich, D. Krystofik, C. Shuff, J. J. Hong, and P. L. Ogra. 1989. Otitis media in children. I. The systemic immune response to nontypable Haemophilus influenzae. J. Infect. Dis. 160:999-1004. [DOI] [PubMed] [Google Scholar]
- 11.Farjo, R. S., B. Foxman, M. J. Patel, L. Zhang, M. M. Pettigrew, S. I. McCoy, C. F. Marrs, and J. R. Gilsdorf. 2004. Diversity and sharing of Haemophilus influenzae strains colonizing healthy children attending day-care centers. Pediatr. Infect. Dis. J. 23:41-46. [DOI] [PubMed] [Google Scholar]
- 12.Fernaays, M. M., A. J. Lesse, S. Sethi, X. Cai, and T. F. Murphy. 2006. Differential genome contents of nontypeable Haemophilus influenzae strains from adults with chronic obstructive pulmonary disease. Infect. Immun. 74:3366-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. F. Tomb, B. A. Dougherty, J. M. Merrick, et al. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 14.Grundy, F. J., A. G. Plaut, and A. Wright. 1990. Localization of the cleavage site specificity determinant of Haemophilus influenzae immunoglobulin A1 protease genes. Infect. Immun. 58:320-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrison, A., D. W. Dyer, A. Gillaspy, W. C. Ray, R. Mungur, M. B. Carson, H. Zhong, J. Gipson, M. Gipson, L. S. Johnson, L. Lewis, L. O. Bakaletz, and R. S. Munson, Jr. 2005. Genomic sequence of an otitis media isolate of nontypeable Haemophilus influenzae: comparative study with H. influenzae serotype d, strain KW20. J. Bacteriol. 187:4627-4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hauck, C. R., and T. F. Meyer. 1997. The lysosomal/phagosomal membrane protein h-lamp-1 is a target of the IgA1 protease of Neisseria gonorrhoeae. FEBS Lett. 405:86-90. [DOI] [PubMed] [Google Scholar]
- 17.Herriott, R. M., E. M. Meyer, and M. Vogt. 1970. Defined nongrowth media for stage II development of competence in Haemophilus influenzae. J. Bacteriol. 101:517-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hopper, S., B. Vasquez, A. Merz, S. Clary, J. S. Wilbur, and M. So. 2000. Effects of the immunoglobulin A1 protease on Neisseria gonorrhoeae trafficking across polarized T84 epithelial monolayers. Infect. Immun. 68:906-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson, J. R., and J. J. Brown. 1996. A novel multiply primed polymerase chain reaction assay for identification of variant papG genes encoding the Gal(alpha 1-4)Gal-binding PapG adhesins of Escherichia coli. J. Infect. Dis. 173:920-926. [DOI] [PubMed] [Google Scholar]
- 20.Jose, J., G. W. Otto, and T. F. Meyer. 2003. The integration site of the iga gene in commensal Neisseria sp. Mol. Genet. Genomics 269:197-204. [DOI] [PubMed] [Google Scholar]
- 21.Jose, J., U. Wolk, D. Lorenzen, H. Wenschuh, and T. F. Meyer. 2000. Human T-cell response to meningococcal immunoglobulin A1 protease associated alpha-proteins. Scand. J. Immunol. 51:176-185. [DOI] [PubMed] [Google Scholar]
- 22.Kilian, M., J. Mestecky, and R. E. Schrohenloher. 1979. Pathogenic species of the genus Haemophilus and Streptococcus pneumoniae produce immunoglobulin A1 protease. Infect. Immun. 26:143-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kilian, M., and B. Thomsen. 1983. Antigenic heterogeneity of immunoglobulin A1 proteases from encapsulated and non-encapsulated Haemophilus influenzae. Infect. Immun. 42:126-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klein, J. O. 1994. Otitis media. Clin. Infect. Dis. 19:823-833. [DOI] [PubMed] [Google Scholar]
- 25.Koomey, J. M., and S. Falkow. 1984. Nucleotide sequence homology between the immunoglobulin A1 protease genes of Neisseria gonorrhoeae, Neisseria meningitidis, and Haemophilus influenzae. Infect. Immun. 43:101-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kroll, J. S., K. E. Wilks, J. L. Farrant, and P. R. Langford. 1998. Natural genetic exchange between Haemophilus and Neisseria: intergeneric transfer of chromosomal genes between major human pathogens. Proc. Natl. Acad. Sci. USA 95:12381-12385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin, L., P. Ayala, J. Larson, M. Mulks, M. Fukuda, S. R. Carlsson, C. Enns, and M. So. 1997. The Neisseria type 2 IgA1 protease cleaves LAMP1 and promotes survival of bacteria within epithelial cells. Mol. Microbiol. 24:1083-1094. [DOI] [PubMed] [Google Scholar]
- 28.Lomholt, H., and M. Kilian. 1994. Antigenic relationships among immunoglobulin A1 proteases from Haemophilus, Neisseria, and Streptococcus species. Infect. Immun. 62:3178-3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lomholt, H., and M. Kilian. 1995. Distinct antigenic and genetic properties of the immunoglobulin A1 protease produced by Haemophilus influenzae biogroup aegyptius associated with Brazilian purpuric fever in Brazil. Infect. Immun. 63:4389-4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lomholt, H., K. Poulsen, and M. Kilian. 1995. Comparative characterization of the iga gene encoding IgA1 protease in Neisseria meningitidis, Neisseria gonorrhoeae, and Haemophilus influenzae. Mol. Microbiol. 15:495-506. [DOI] [PubMed] [Google Scholar]
- 31.Lorenzen, D. R., F. Dux, U. Wolk, A. Tsirpouchtsidis, G. Haas, and T. F. Meyer. 1999. Immunoglobulin A1 protease, an exoenzyme of pathogenic Neisseriae, is a potent inducer of proinflammatory cytokines. J. Exp. Med. 190:1049-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Male, C. J. 1979. Immunoglobulin A1 protease production by Haemophilus influenzae and Streptococcus pneumoniae. Infect. Immun. 26:254-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meats, E., E. J. Feil, S. Stringer, A. J. Cody, R. Goldstein, J. S. Kroll, T. Popovic, and B. G. Spratt. 2003. Characterization of encapsulated and noncapsulated Haemophilus influenzae and determination of phylogenetic relationships by multilocus sequence typing. J. Clin. Microbiol. 41:1623-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mestecky, J., and M. Kilian. 1985. Immunoglobulin A (IgA). Methods Enzymol. 116:37-75. [DOI] [PubMed] [Google Scholar]
- 35.Mulks, M. H., S. J. Kornfeld, B. Frangione, and A. G. Plaut. 1982. Relationship between the specificity of IgA proteases and serotypes in Haemophilus influenzae. J. Infect. Dis. 146:266-274. [DOI] [PubMed] [Google Scholar]
- 36.Murphy, T. F. 2003. Respiratory infections caused by non-typeable Haemophilus influenzae. Curr. Opin. Infect. Dis. 16:129-134. [DOI] [PubMed] [Google Scholar]
- 37.Musser, J. M., S. J. Barenkamp, D. M. Granoff, and R. K. Selander. 1986. Genetic relationships of serologically nontypeable and serotype b strains of Haemophilus influenzae. Infect. Immun. 52:183-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parsons, H. K., S. Vitovski, and J. R. Sayers. 2004. Immunoglobulin A1 proteases: a structure-function update. Biochem. Soc Trans. 32:1130-1132. [DOI] [PubMed] [Google Scholar]
- 39.Plaut, A. G. 1983. The IgA1 proteases of pathogenic bacteria. Annu. Rev. Microbiol. 37:603-622. [DOI] [PubMed] [Google Scholar]
- 40.Plaut, A. G., and W. W. Bachovchin. 1994. IgA-specific prolyl endopeptidases: serine type. Methods Enzymol. 244:137-151. [DOI] [PubMed] [Google Scholar]
- 41.Plaut, A. G., J. V. Gilbert, M. S. Artenstein, and J. D. Capra. 1975. Neisseria gonorrhoeae and Neisseria meningitidis: extracellular enzyme cleaves human immunoglobulin A. Science 190:1103-1105. [DOI] [PubMed] [Google Scholar]
- 42.Plaut, A. G., J. Qiu, F. Grundy, and A. Wright. 1992. Growth of Haemophilus influenzae in human milk: synthesis, distribution, and activity of IgA protease as determined by study of iga+ and mutant iga- cells. J. Infect. Dis. 166:43-52. [DOI] [PubMed] [Google Scholar]
- 43.Plaut, A. G., and A. Wright. 1995. Immunoglobulin A-metallo-type specific prolyl endopeptidases. Methods Enzymol. 248:634-642. [DOI] [PubMed] [Google Scholar]
- 44.Pohlner, J., R. Halter, K. Beyreuther, and T. F. Meyer. 1987. Gene structure and extracellular secretion of Neisseria gonorrhoeae IgA protease. Nature 325:458-462. [DOI] [PubMed] [Google Scholar]
- 45.Pohlner, J., U. Langenberg, U. Wolk, S. C. Beck, and T. F. Meyer. 1995. Uptake and nuclear transport of Neisseria IgA1 protease-associated alpha-proteins in human cells. Mol. Microbiol. 17:1073-1083. [DOI] [PubMed] [Google Scholar]
- 46.Poje, G., and R. J. Redfield. 2003. Transformation of Haemophilus influenzae, p. 57-70. In M. Herbert, D. Wood, and E. Moxon (ed.), Haemophilus influenzae protocols. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 47.Poulsen, K., J. P. Hjorth, and M. Kilian. 1988. Limited diversity of the immunoglobulin A1 protease gene (iga) among Haemophilus influenzae serotype b strains. Infect. Immun. 56:987-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poulsen, K., J. Reinholdt, and M. Kilian. 1992. A comparative genetic study of serologically distinct Haemophilus influenzae type 1 immunoglobulin A1 proteases. J. Bacteriol. 174:2913-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rao, V. K., G. P. Krasan, D. R. Hendrixson, S. Dawid, and J. W. St Geme, III. 1999. Molecular determinants of the pathogenesis of disease due to non-typable Haemophilus influenzae. FEMS Microbiol. Rev. 23:99-129. [DOI] [PubMed] [Google Scholar]
- 50.Reinholdt, J. 1996. A method for titration of inhibiting antibodies to bacterial immunoglobulin A1 proteases in human serum and secretions. J. Immunol. Methods 191:39-48. [DOI] [PubMed] [Google Scholar]
- 51.Reinholdt, J., and M. Kilian. 1997. Comparative analysis of immunoglobulin A1 protease activity among bacteria representing different genera, species, and strains. Infect. Immun. 65:4452-4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saunders, N. J., D. W. Hood, and E. R. Moxon. 1999. Bacterial evolution: bacteria play pass the gene. Curr. Biol. 9:R180-R183. [DOI] [PubMed] [Google Scholar]
- 53.Segada, L. M., G. M. Carlone, L. L. Gheesling, and A. J. Lesse. 2000. Characterization of P1-deficient isogenic mutant of Haemophilus influenzae biogroup aegyptius associated with Brazilian purpuric fever. Microb. Pathog. 28:145-155. [DOI] [PubMed] [Google Scholar]
- 54.Senior, B. W., and C. L. Ip. 1999. An investigation into the influences of species and biotype on the type of IgA1 protease produced by isolates of Haemophilus. J. Med. Microbiol. 48:389-394. [DOI] [PubMed] [Google Scholar]
- 55.Serino, L., and M. Virji. 2002. Genetic and functional analysis of the phosphorylcholine moiety of commensal Neisseria lipopolysaccharide. Mol. Microbiol. 43:437-448. [DOI] [PubMed] [Google Scholar]
- 56.Sethi, S., N. Evans, B. J. Grant, and T. F. Murphy. 2002. New strains of bacteria and exacerbations of chronic obstructive pulmonary disease. N. Engl. J. Med. 347:465-471. [DOI] [PubMed] [Google Scholar]
- 57.Sethi, S., and T. F. Murphy. 2001. Bacterial infection in chronic obstructive pulmonary disease in 2000: a state-of-the-art review. Clin. Microbiol. Rev. 14:336-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sethi, S., C. Wrona, B. J. Grant, and T. F. Murphy. 2004. Strain-specific immune response to Haemophilus influenzae in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 169:448-453. [DOI] [PubMed] [Google Scholar]
- 59.St. Geme, J. W., III. 2002. Molecular and cellular determinants of non-typeable Haemophilus influenzae adherence and invasion. Cell Microbiol. 4:191-200. [DOI] [PubMed] [Google Scholar]
- 60.St. Geme, J. W., III. 2000. The pathogenesis of nontypable Haemophilus influenzae otitis media. Vaccine 19(Suppl. 1):S41-S50. [DOI] [PubMed] [Google Scholar]
- 61.Tsirpouchtsidis, A., R. Hurwitz, V. Brinkmann, T. F. Meyer, and G. Haas. 2002. Neisserial immunoglobulin A1 protease induces specific T-cell responses in humans. Infect. Immun. 70:335-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vitovski, S., K. T. Dunkin, A. J. Howard, and J. R. Sayers. 2002. Nontypeable Haemophilus influenzae in carriage and disease: a difference in IgA1 protease activity levels. JAMA 287:1699-1705. [DOI] [PubMed] [Google Scholar]
- 63.Vitovski, S., R. C. Read, and J. R. Sayers. 1999. Invasive isolates of Neisseria meningitidis possess enhanced immunoglobulin A1 protease activity compared to colonizing strains. FASEB J. 13:331-337. [DOI] [PubMed] [Google Scholar]
- 64.Voulhoux, R., M. P. Bos, J. Geurtsen, M. Mols, and J. Tommassen. 2003. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 299:262-265. [DOI] [PubMed] [Google Scholar]
- 65.Yi, K., S. Sethi, and T. F. Murphy. 1997. Human immune response to nontypeable Haemophilus influenzae in chronic bronchitis. J. Infect. Dis. 176:1247-1252. [DOI] [PubMed] [Google Scholar]