Abstract
Neutralizing effects of antibodies targeting the C‐terminal stalk (S2) subunit of the spike protein of severe acute respiratory syndrome coronavirus have previously been reported, although its mechanism remained elusive. In this study, high titered mouse antisera against the N‐terminal globular (S1) and S2 subunits of the S protein were generated and total immunoglobulin G (IgG) was purified from these antisera. The efficiency of these purified IgGs in virus neutralization and blocking of receptor binding were compared quantitatively using virus neutralization assay and a previously developed cell‐based receptor binding assay, respectively. We demonstrated that anti‐S1 IgG neutralizes the virus and binds to the membrane associated S protein more efficiently than anti‐S2 IgG does. Moreover, both anti‐S1 and anti‐S2 IgGs were able to abolish the binding between S protein and its cellular receptor(s), although anti‐S1 IgG showed a significantly higher blocking efficiency. The unexpected blocking ability of anti‐S2 IgG towards the receptor binding implied a possible role of the S2 subunit in virus docking process and argues against the current hypothesis of viral entry. On the other hand, the functional roles of the previously reported neutralizing epitopes within S2 subunit were investigated using an antigen specific antibody depletion assay. Depletion of antibodies against these regions significantly diminished, though not completely abolished, the neutralizing effects of anti‐S2 IgG. It suggests the absence of a major neutralizing domain on S2 protein. The possible ways of anti‐S2 IgGs to abolish the receptor binding and the factors restricting anti‐S2 IgGs to neutralize the virus are discussed.
Keywords: SARS, severe acute respiratory syndrome, SARS-CoV, SARS coronavirus, S, spike, S1, N-terminal domain of S protein, S2, C-terminal domain of S protein, IgG, immunoglobulin G, HR, heptad repeats, C-HR, C-terminal HR, HIV, human immunodeficiency virus, rAds, recombinant adenoviruses, m-S, membrane associated S, ATCC, American type culture collection, GFP, green fluorescent protein, N, nucleocapsid, rMVA, recombinant modified vaccinia virus Ankara, MFI, mean fluorescent intensity, His-tag, histidine tag, SARS, Spike protein, Antibodies, Virus neutralization, Cell-based receptor binding assay
1. Introduction
Severe acute respiratory syndrome (SARS) is an emerging infectious disease caused by a zoonotic coronavirus (CoV) named SARS‐CoV [1]. The pandemic and re‐emerging potential of the disease urged the development of effective vaccines against the virus. Based on the experiences in animal coronaviral vaccines [2, 3, 4], the development of recombinant vaccines against SARS‐CoV was mainly focused on its spike (S) glycoprotein [4, 5]. The S protein of CoVs, which is classified as a class I viral fusion protein, exists as radially protruded trimers on the viral envelope and can be structurally or functionally divided into two subunits, namely S1 and S2 subunits, representing the N‐terminal globular head and the C‐terminal membrane‐bound stalk, respectively [6, 7]. The S1 subunit is responsible for virus binding to cellular receptor(s) and contains neutralizing epitopes. The S2 subunit mediates membrane fusion during viral infection and is believed to be unable to induce neutralizing antibody in a number of CoVs [8, 9, 10, 11].
On the contrary, several reports suggested that the S2 subunit of SARS‐CoV contains neutralizing epitopes of the virus. Immunization of either Escherichia coli expressed or chemically synthesized S2 peptides elicited neutralizing antibody [12, 13, 14] or specific antibody responses [15, 16] in animal models. Moreover, neutralizing epitopes were mapped on the S2 subunit by biopanning of phage display dodecapeptide library with antisera from convalescent SARS patients [17] or by screening antibody‐phage display library constructed with B‐cells of convalescent SARS patients [18]. Immunization of plasmids encoding the S2 subunit, which may better mimic its native antigenic properties, was reported to induce neutralizing antibodies in rabbits [19]. Our earlier study [20] also demonstrated the cooperative neutralizing effect of anti‐S1 and anti‐S2 antibodies from mice immunized with plasmids encoding S1 and S2 subunits. These results suggest the presence of neutralizing epitopes within the S2 subunit of SARS‐CoV, which was not commonly observed in a number of other CoVs [8].
The mechanism of the efficient neutralizing effect of anti‐S1 antibodies is logically proposed as prevention of spike‐to‐receptor engagement event by blocking of its receptor binding domain [5]. However, the mechanisms involved in the neutralization mediated by anti‐S2 antibodies remain to be clarified. As a class I viral fusion protein, the S2 subunit of coronaviruses has its typical functional domains [21], including a fusion peptide [22], heptad repeats (HR), aromatic amino acids cluster [23] and transmembrane domain. Blocking the actions involving these domains may prevent the virus from entering its host cell, although this concept remains to be proven experimentally. Neutralizing antibody against the S2 subunit of mouse hepatitis virus was reported to bind to a determinant of nine amino acids near the fusion peptide, preventing fusion activity during virus entry [24]. Lip and colleagues [25] demonstrated that monoclonal antibodies targeting the regions both within and immediately after the C‐terminal HR (C‐HR) domain are able to neutralize SARS‐CoV and inhibit cell–cell membrane fusion, implying blocking of membrane fusion may be a possible neutralizing mechanism of anti‐S2 antibodies. Recent advances in epitope mapping of gp41 of human immunodeficiency virus (HIV), which is a membrane anchoring subunit of class I viral fusion proteins and the counterpart of SARS‐CoV S2 subunit, identified several neutralizing epitopes that are proximal to the viral membrane but not near fusion peptide [26, 27, 28]. Although the mechanism of neutralization through these epitopes on gp41 of HIV is not well understood, these results suggest that antibodies targeting the membrane anchoring subunit of a class I viral fusion protein, e.g. S2 subunit of SARS‐CoV, may also efficiently interfere the virus entry into host cells.
Although the neutralizing effects of anti‐S1 and anti‐S2 antibodies to coronaviruses have been widely characterized, they were not compared quantitatively and the neutralizing mechanisms of anti‐S2 antibodies are still unclear. In this study, we immunized mice with plasmids and recombinant adenoviruses (rAds) encoding the S1 and S2 subunits of SARS‐CoV. Virus neutralization efficiency of purified anti‐S1 and anti‐S2 IgGs, as well as their binding ability towards, membrane associated S (m‐S) protein, i.e. S protein anchored on cell membrane that resembles its native structure, were compared quantitatively. In addition, using a cell‐based receptor binding assay, we also showed that the anti‐S2 antibodies are able to abolish the binding between m‐S and the cellular receptors, providing possible new clues towards the understanding of the neutralizing mechanisms of anti‐S2 antibodies.
2. Materials and methods
2.1. Cell culture, plasmids and viral vectors
Mammalian cell lines, including AD293 (Stratagene), Vero E6 [American Type Culture Collection (ATCC), CRL‐1586], FRhK4 (ATCC, CRL‐1688) and BHK21 (ATCC, CCL‐10), and a chicken embryonic fibroblast cell line, DF1 (ATCC, CRL‐12203), were propagated and maintained according to the manufacturers’ instructions. Recombinant adenoviruses encoding the full length S protein (aa 1‐1255) of SARS‐CoV and green fluorescent protein (GFP), designated as rAd‐S and rAd‐GFP respectively, were constructed as described previously [29] and used for in vitro expression. Mammalian expression plasmids and rAds encoding the S1 subunit (aa 18–683), S2 subunit (aa 684–1255) and nucleocapsid (N) protein (aa 1–422) of SARS‐CoV, designated as pCI‐sp‐S1, pCI‐sp‐S2 and pCI‐N for the plasmids or rAd‐S1, rAd‐S2 and rAd‐N for the rAds, respectively, were constructed as described previously [20, 29]. A recombinant modified vaccinia virus Ankara (rMVA) encoding both the full length S (aa 1–1255) of SARS‐CoV and a GFP marker (rMVA‐S/GFP), which was used for in vitro expression, was kindly provided by professor B. Moss [30] at National Institute of Allergy and Infectious Diseases, NIH, USA. It is noted that the amino acid residues of viral genes in this study are numbered with reference to SARS‐CoV strain HKU‐39849 [31].
2.2. Immunization of mice and total IgG purification
Two groups of 20 BALB/c mice aged 6–8 weeks were immunized intramuscularly with plasmid DNA (100 μg per dose), including pCI‐sp‐S1 and pCI‐sp‐S2 respectively, for three doses at interval of three weeks. Sixth week after the last dose of DNA immunization, these two groups of mice were boosted with rAds encoding the corresponding antigens intraperitoneally, i.e. rAd‐S1 and rAd‐S2, at 5 × 108 plaque forming units per dose. A control group was immunized at the same dose and schedule except pCI‐N and rAd‐N were used. Serum samples of all mice groups were collected at one, two, three and four weeks after the rAd booster. The antisera collected from different time points of each mice group were pooled and stored at −20 °C until use. The SARS‐CoV specific antibody titers of the collected antisera were evaluated by using human anti‐SARS‐CoV antibody (IgG) ELISA kit (Beijing Huada GBI Biotechnology, Beijing China) according to the manufacturer's instructions, except that HRP‐conjugated anti‐mouse IgG (H + L) (Zymed) was used as the secondary antibody. Total IgG was purified from the collected antisera by mouse IgG purification kit (Affi‐Gel Protein A MAPS II Kit, BIO‐RAD) according to the manufacturer's instructions. The purified IgGs were quantified with anti‐mouse IgG assay plates (Becton Dickinson) according to the manufacturer's instructions, with mouse IgG (PharMingen) as the standard and HRP‐conjugated anti‐mouse IgG (H + L) as the secondary antibody.
2.3. Virus neutralization assay
The purified IgG samples were diluted using normal mouse serum to the indicated concentrations (2–20 mg/ml). The neutralizing effect of each diluted IgG sample was evaluated using virus neutralization assay as described previously [20], with 100 TCID50 of SARS‐CoV strain HKU‐39849 [31] were used for each well in a 96‐well plate.
2.4. Binding efficiency of anti‐S1 and anti‐S2 antibodies to m‐S protein
AD293 cells expressing m‐S protein (AD293‐S cells) were prepared by transducing AD293 cells with rAd‐S according to the procedures described by Chow et al. [29]. The AD293‐S cells were harvested and washed with FACS buffer (phosphate‐buffered saline (PBS) containing 4% fetal bovine serum (FBS)). To demonstrate the m‐S protein binding ability of anti‐S1 and anti‐S2 antibodies, 50 μl of FACS buffer containing the purified IgG samples was used to resuspend 2 × 105 of AD293‐S and kept on ice for 30 min. After washing three times with ice‐cold FACS buffer, the IgG‐incubated AD293‐S was resuspended in goat anti‐mouse IgG (H + L)‐FITC (Zymed) and kept on ice for 30 min. The mean fluorescent intensity (MFI) of these AD293‐S cells, which is positively related to the amount of bound mouse IgG, is evaluated by counting 1 × 104 cells in each of the samples using Coulter Epics Elite Flow Cytometer. The experiment was performed in triplicate and the data was analyzed with the WinMDI 2.81 (Scripps Research Institute). The average of MFI of the triplicates was presented with standard deviations (S.D.).
To further compare the m‐S binding efficiency of anti‐S1 and anti‐S2 IgGs, a serial antibody binding and depleting assay was carried out. In this assay, approximately 5 × 105 AD293‐S cells were incubated with 100 μl of FACS buffer containing 1.6 mg/ml of the purified IgG samples on ice for 20 min to allow antibody binding. The anti‐S1, anti‐S2 and anti‐N IgG bound AD293‐S cells, designated as anti‐S1/, anti‐S2/ and anti‐N/AD293‐S respectively, were spun down at 1000 × g for 2 min to deplete the bound antibody from the liquid phase and the supernatant, i.e. FACS buffer containing the unbound IgG, was transferred to a new tube containing 5 × 105 AD293‐S cells for the next round of binding and depleting under the same condition. The concentration of the unbound IgG after each depletion round was monitored by analyzing 0.5 μl of the supernatant using the anti‐mouse IgG quantification assay as described. The IgG bound AD293‐S cell pellet was washed three times with FACS buffer and analyzed using flow cytometer after each depletion round. This antibody binding and depleting assay was repeated 20 times in triplicate and the average MFI of the triplicates for each depletion round were recorded.
2.5. Cell‐based receptor binding assay
To evaluate the ability of purified IgG samples to block the m‐S protein from binding with its cellular receptor, a cell‐based receptor binding assay was carried out as described by Chou et al. [32] with modifications. Briefly, three types of ligand cells were prepared, including AD293 transduced with both rAd‐S and rAd‐GFP (AD293‐S/GFP), and two other line cells, BHK21 and DF1, infected with rMVA‐S/GFP (BHK21‐S/GFP and DF1‐S/GFP). A control ligand cell was also prepared from AD293 transduced with rAd‐GFP only (AD293‐GFP). The receptor‐expressing cells, i.e. Vero E6 monolayer, were prepared in six‐well plates with 100% confluence. The purified IgG samples were diluted with FACS buffer to the indicated concentrations and 2 × 105 ligand cells were pre‐incubated with 50 μl of each diluted IgG at 4 °C for 30 min. The ligand cells together with the IgG samples were laid on Vero E6 monolayer for binding and the unbound ligand cells were washed away according to the procedures described by Chou et al. [32]. The bound ligand and Vero E6 cells were detached by trypsin and fixed in 2% of paraformaldehyde in PBS. The number of bound ligand cells in each sample was then counted with flow cytometer and the mean values from triplicates were recorded.
To compare the blocking efficiency between anti‐S1 and anti‐S2 IgGs, only AD293‐S/GFP was used as the ligand cell and the blocking efficiency was defined as (N unblock − N block)/(N unblock − N background) × 100%, where N block and N unblock represent the number of bound ligand cells, i.e. AD293‐S/GFP, with and without IgG pre‐incubation respectively, and N background is the number of bound control ligand cells, i.e. AD293‐GFP.
2.6. Antigen specific antibody depletion assay
Regions within or proximal to the fusion peptide and the C‐HR of the S2 subunit have been reported to contain neutralizing epitopes (Fig. 1 ). To investigate the functional roles of these epitopes, E. coli expressed peptides covering these regions were used to deplete antibodies against the linear epitopes within these regions in the purified anti‐S2 IgG. Two regions covering aa 787–828 and aa 1023–1131, with reference to strain HKU‐39849 [31], were PCR amplified and fused using overlapping extension PCR. The fused coding sequence, designated as S2‐Ept, was cloned into pRSET‐A (Invitrogen), sequenced and then expressed in E. coli strain BL‐21 (Invitrogen) according to the manufacturer's instructions. The expressed peptide was affinity purified through its N‐terminal fused histidine tag (His‐tag) with Ni2+ charged agarose (Milli‐Pore) and its identity was confirmed by Western blots with AP‐conjugated anti‐His‐tag antibody (Invitrogen) according to the manufacturer's instructions. The purified peptide was incubated with 50 μl of anti‐S2 IgG (3.2 mg/ml) at 4 °C for 2 h. The IgG bound peptide complexes and unbound peptides were then removed using His‐tag affinity purification. The peptide–IgG mixture was allowed to pass through the Ni2+‐agarose column three times and the flowthrough, which represents the S2‐Ept bound IgG depleted anti‐S2 IgG, designated as dep‐anti‐S2 IgG, were collected and subjected to virus neutralization and IgG quantification ELISA as described. Three control depletions, including anti‐S1 IgG with or without addition of S2‐Ept peptide (designated as dep‐anti‐S1 IgG and mock depleted anti‐S1 IgG, respectively), and anti‐S2 without addition of S2‐Ept peptide (i.e. mock depletion), were performed to evaluate the specificity the depletion process and its possible artifacts in virus neutralization. The specific binding between anti‐S2 IgG and S2‐Ept peptide was confirmed by Western blots.
Figure 1.
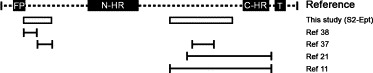
Schematic diagram of S2 functional domains and regions containing neutralizing epitopes. The black boxes, white boxes and black lines represent the S2 functional domains, regions chosen for this study (designated as S2‐Ept) and regions containing previously reported neutralizing epitopes, respectively. The number of the references of the reported neutralizing regions is listed on the right column. FP, N‐HR, C‐HR and T refer to fusion peptide, N‐terminal and C‐terminal heptad repeat, and transmembrane domain respectively.
2.7. Statistical analysis
A student's t‐test was used to compare the significance between specified groups, with P ⩽ 0.05, or 0.01 be defined as statistically significant.
3. Results
3.1. Neutralizing efficiency and additive neutralizing effect of anti‐S1 and anti‐S2 antibodies
With plasmid DNA prime and viral vector boost immunization regimen, SARS‐CoV specific antibodies of much higher titer were observed when compared to that of our previous study, which employed plasmid DNA immunization only [20]. Mice immunized with recombinant vaccines encoding S1 subunit generally showed higher neutralizing titer against SARS‐CoV in their antisera than those immunized with S2 antigens (data not shown). In order to quantitatively compare the neutralizing efficiency of anti‐S1 and anti‐S2 antibodies, purified anti‐S1 or anti‐S2 total IgGs with the same concentration range were subjected to virus neutralization assay. The negative control, i.e. anti‐N IgG, was also purified and subjected to virus neutralization assay, which showed no neutralizing effect in all tested concentrations. The neutralizing efficiency of anti‐S1 and anti‐S2 IgGs, which is generally defined as the increment of neutralizing titer with increasing concentration of IgG, or simply as the slopes of the plots in Fig. 2 A, was compared. Fig. 2A demonstrates that both anti‐S1 and anti‐S2 IgG neutralized SARS‐CoV in a wide range of concentrations and the neutralizing efficiency of anti‐S1 IgG is approximately 2.3 times higher than that of anti‐S2 IgG (Slopanti‐S1/Slopanti‐S2) in the tested IgG concentrations.
Figure 2.
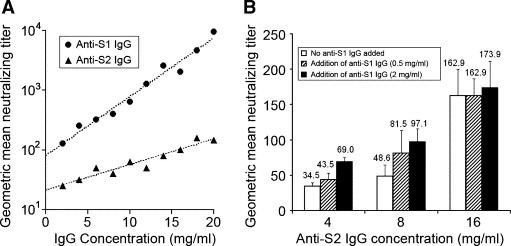
The neutralizing efficiency and additive neutralizing effect of anti‐S1 and anti‐S2 IgG. (A) The geometric mean neutralizing titers of anti‐S1 and anti‐S2 IgGs of the same concentration range, i.e. 2–20 mg/ml, were compared. The values of y‐axis are showed in logarithmic scale and the dotted lines represent the best‐fit line obtained from linear regression of the datasets. Data of Anti‐N IgG is not shown since it did not show any neutralizing effect in all tested concentrations. (B) The geometric mean neutralizing titers of anti‐S2 IgG of different concentrations mixed with 0.5 or 2 mg/ml of anti‐S1 IgG. The average values of the triplicates were indicated and the error bars represent the SD of the triplicates. The geometric mean neutralizing titers of 0.5 and 2 mg/ml of anti‐S1 IgG are 29.6 (S.D. = 3.8) and 51.3 (S.D. = 11.8), respectively.
Our previous results suggested that anti‐S1 and anti‐S2 antibodies cooperatively neutralize SARS‐CoV [20]. To further investigate this phenomenon, anti‐S2 IgG at final concentration of 4, 8 and 16 mg/ml were subjected to neutralization assays alone or mixed with anti‐S1 IgG at a final concentration of 2 or 0.5 mg/ml. Additive neutralizing effects, i.e. enhancement of neutralizing titer when anti‐S1 IgG is added, were observed in anti‐S2 IgG at concentration of 4 and 8 mg/ml, although at high concentration of anti‐S2 IgG (16 mg/ml), the addition of anti‐S1 IgG only provided little further increase in virus neutralizing titer (Fig. 2B). Present results demonstrated that both anti‐S1 and anti‐S2 IgGs were able to neutralize SARS‐CoV with different efficiency and additive neutralizing effects of these antibodies were observed at a relatively low concentration.
3.2. The binding efficiency of anti‐S1 and anti‐S2 antibodies to m‐S protein
Fig. 3 A demonstrates that both anti‐S1 and anti‐S2 IgGs were able to bind specifically with AD293‐S cells, while the anti‐N control IgG shows no significant binding. All IgGs showed no significant binding toward non‐transduced AD293 (data not shown), demonstrating the specificity of the binding between anti‐S1 or anti‐S2 IgG and m‐S protein. Moreover, the binding capacity of m‐S protein towards anti‐S1 IgG, i.e. the amount of anti‐S1 IgG bound to m‐S protein, is substantially larger than that of anti‐S2 IgG under the same tested concentrations, i.e. 3.2 and 0.8 mg/ml (Fig. 3A). The differential binding of both IgGs to m‐S was further demonstrated with different IgG dosages. The binding efficiency to m‐S, i.e. the increment of MFI with increasing concentration of IgG, between anti‐S1 and anti‐S2 IgG was very different in the tested range of IgG concentrations (Fig. 3B).
Figure 3.
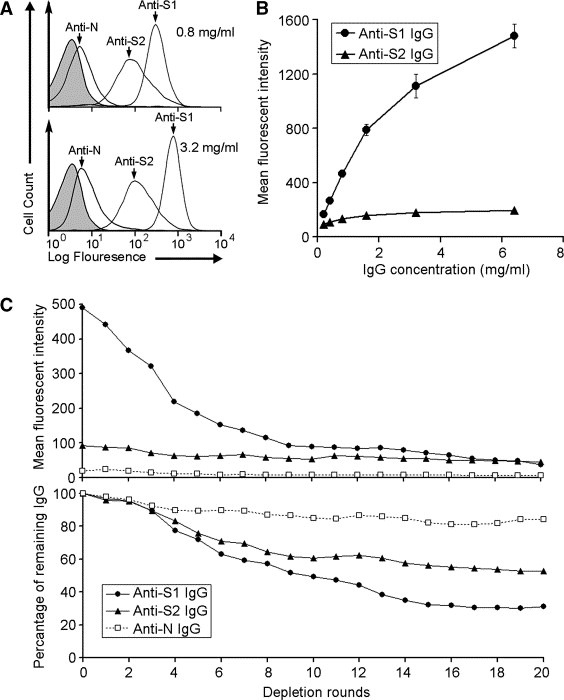
The binding efficiency of anti‐S1 and anti‐S2 IgGs towards m‐S protein. (A) Flow cytometry showing that anti‐S1 and anti‐S2 IgGs specifically bind to m‐S protein on AD293 cells. The shaded peaks represent the background signal, i.e. cells without IgG incubation, and the non‐shaded peaks represent cells incubated with 0.8 (upper panel) or 3.2 (lower panel) mg/ml of anti‐N, anti‐S1 and anti‐S2 IgG as indicated. (B) Anti‐S1 IgG binds more efficiently to m‐S protein than anti‐S2 IgG does. The error bars represent the SD of the triplicates. (C) The results of the serial antibody binding and depleting assay. The upper and lower panels show the decrement of MFI and percentage of remaining IgG after each round of depletion, respectively. Each data point represents the average value of three triplicates.
To investigate the observed differences between the binding capacities of m‐S protein towards anti‐S1 and anti‐S2 IgGs, a serial antibody binding and depleting assay, was performed and the results are shown in Fig. 3C. The behaviors of anti‐S1 and anti‐S2 IgGs are strikingly different in this assay. Firstly, the MFI of anti‐S1/AD293‐S cells drops more than 50% of its starting MFI after the first 5 depletion rounds and slowly decreases after each of the last 15 depletion rounds, while no considerable decrement is observed in the MFI of anti‐S2/AD293‐S cells even after 20 rounds of depletion (Fig. 3C, upper panel). Secondly, the absolute amounts of both the unbound anti‐S1 and anti‐S2 IgGs decrease during the depletion rounds but the decrement of the unbound anti‐S1 is substantially larger than that of the unbound anti‐S2 IgG after each depletion round (Fig. 3C, lower panel). It is also noted that the MFI of anti‐N/AD293‐S cells is significantly lower than that of anti‐S1/ and anti‐S2/AD293‐S cells during the depletion rounds, and the absolute amount of anti‐S1 and anit‐S2 IgG is depleted at a considerably higher rate than that of anti‐N IgG, demonstrating the depletion of IgG is S protein specific. These observations imply that: (1) anti‐S1 IgG has a good binding capacity to m‐S and therefore can be efficiently removed from the IgG pool after every binding‐depleting cycle; and (2) binding of anti‐S2 IgG to m‐S protein, however, is restricted and hence the dynamic change of free anti‐S2 IgG is different from that of anti‐S1 and anti‐N IgG in the process of depletion, characterized by a limited amount of anti‐S2 IgG bound and removed in each depletion cycle and an abundant free IgG sustained a close value of MFI even after many round of depletion.
3.3. Abolition of receptor binding by anti‐S2 IgG in cell‐based receptor binding assay
In the cell‐based receptor binding assay, we found that not only anti‐S1 IgG, but also anti‐S2 IgG abolished the binding between the ligand cells, i.e. AD293‐S/GFP and the receptor expressing cells, i.e. Vero E6, while the control anti‐N IgG did not (Fig. 4 A). Since the abolition of receptor binding to m‐S protein by anti‐S2 IgG is not reported previously, we further confirmed this phenomenon by repeating the binding assay with other ligand cells. Fig. 4B demonstrates that anti‐S2 IgG is able to abolish the binding of BHK21‐S/GFP or DF1‐S/GFP to Vero E6 cells with different efficiencies, indicating that the abolition of receptor binding is specific to S protein and independent of ligand cell types.
Figure 4.
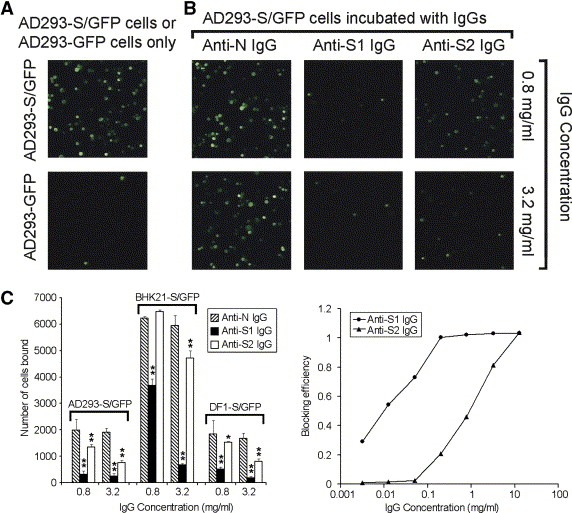
Abolition of receptor binding by anti‐S1 and anti‐S2 IgGs in cell‐based receptor binding assay. (A) The abolition of receptor binding by anti‐S1 and anti‐S2 IgGs is specific. Ligand cells, i.e. AD293‐S/GFP, incubated with anti‐S1 and anti‐S2 IgGs showed a substantially smaller number of bound cells (green fluorescent spots) when compared with those incubated with anti‐N IgG or no IgG incubation. The images shown are representative views (magnification of 100×) of each treatment as indicated. (B) Abolition of receptor binding of different types of ligand cells by anti‐S2 IgGs. The significance of differences between the numbers of bound cells of the control IgG, i.e. anti‐N IgG, and anti‐S1 or anti‐S2 IgG is represented by asterisks (P < 0.01, ∗∗ or P < 0.05, ∗). The results are presented in average of the triplicates with S.D. The types of ligand cells used are indicated at the top of each data group. (C) The blocking efficiency of anti‐S1 IgG is higher than that of anti‐S2 IgG under the same concentrations tested. Each data point represents the average value of three replicates. The x‐axis, i.e. IgG concentration, is presented in logarithmic scale.
In order to quantitatively compare the performance of anti‐S1 and anti‐S2 IgGs in abolition of receptor binding, their blocking efficiency were evaluated in a wide range of IgG concentration (Fig. 4C). Remarkably, anti‐S1 IgG was able to abolish the receptor binding at a relatively low concentration (<0.01 mg/ml) while anti‐S2 IgG did not. Moreover, the blocking efficiency of anti‐S1 IgG was generally higher than that of anti‐S2 IgG under the same tested concentration, although both IgGs showed complete blocking of receptor binding at about 10 mg/ml (Fig. 4C). The above data indicated that both anti‐S1 and anti‐S2 IgGs are able to specifically abolish the binding between m‐S protein and the receptor(s) on Vero E6 cells, while anti‐S1 IgG abolishes the binding more efficiently than anti‐S2 IgG does under the same tested conditions.
3.4. The role of S2‐Ept in virus neutralization
To investigate the contribution of two specific regions on the S2 subunit, i.e. S2‐Ept (Fig. 1), to the elicitation of neutralizing antibody by S2 subunit expressed in vivo, antibodies against the epitopes of S2‐Ept in anti‐S2 IgG were depleted by E. coli expressed peptide covering these regions. The specific binding between the S2‐Ept peptide and anti‐S2 IgG is demonstrated in Fig. 5 A. The neutralizing titer of dep‐anti‐S2 IgG was found to decrease significantly (P < 0.05, ∗) at various peptide concentrations. Such IgG depletion is demonstrated to be S2 specific as no significant decrease of neutralizing titer was observed in dep‐anti‐S1 IgG and the mock depleted anti‐S2 IgG (data not shown). It is noted that antibodies against epitopes in S2‐Ept was undetectable in dep‐anti‐S2 IgG but detectable in the mock depleted anti‐S2 IgG using Western blots (data not shown), demonstrating that only a negligible amount of anti‐S2‐Ept antibodies is present in dep‐anti‐S2 IgG. In addition, according to the IgG quantification ELISA, no significant loss of total IgG was observed between the dep‐anti‐S2 IgG and the mock depleted anti‐S2 IgG (data not shown) and indicating that the IgG against S2‐Ept only contributes a negligible fraction of the total anti‐S2 IgG population although it contributes significantly to virus neutralization. On the other hand, the blocking efficiency of mock depleted anti‐S2 IgG and dep‐anti‐S2 IgG in the cell‐based receptor binding assay were not significantly different (data not shown), implying anti‐S2‐Ept IgG may not play a major role in the blocking of receptor binding.
Figure 5.
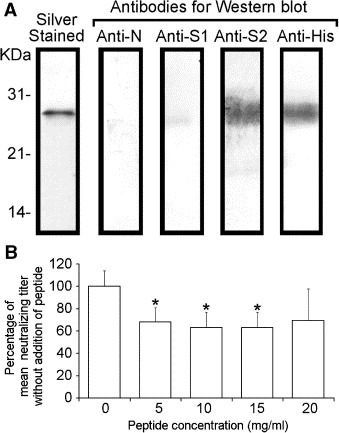
The role of S2‐Ept in virus neutralization. (A) Identification of S2‐Ept peptide using Western blot. (B) The neutralizing titer of S2‐Ept depleted anti‐S2 IgG is lower than the mock depleted anti‐S2 IgG (0 mg/ml). The results were presented as the average of geometric mean of neutralizing titer of three replicates and the error bar represents its SD. The statistical significance of differences between the geometric mean of neutralizing titer of dep‐anti‐S2 IgG and mock depleted anti‐S2 IgG is represented by an asterisk (P < 0.05, ∗). It is noted that no significant decrease of neutralizing titer was observed between dep‐anti‐S1 IgG and mock depleted anti‐S1 IgG (data not shown).
4. Discussion
While the neutralizing effect of anti‐S1 antibodies in most of the human and animal CoVs is well established, the contribution of anti‐S2 antibodies to virus neutralization remains controversial. This was mainly reported for SARS‐CoV and even in our earlier studies [20] we failed to prove that anti‐S2 antibodies have a neutralizing effect independent of anti‐S1 antibodies. This was probably due to the low titer of the antisera used. In this study, high titered anti‐S1 and anti‐S2 antibodies were generated with DNA‐prime‐viral‐booster immunization regimen in mice, allowing us to characterize the roles of the S1 and S2 subunits in virus docking and virus neutralization. Here, we demonstrated both anti‐S1 and anti‐S2 IgGs are able to neutralize the virus with significant difference in efficacy (Fig. 2A), which were not compared quantitatively in previous studies of other CoVs or other class I virus fusion proteins. In addition, the observed differences of neutralizing efficacies may be accounted, at least partially, by the antibody binding to m‐S assay (Fig. 3), in which we showed that the binding capacities of m‐S protein for anti‐S1 IgG is significantly larger than that of anti‐S2 IgG. The low antibody binding capacity of the S2 subunit (Fig. 3C) may be explained by the limited accessible surface for anti‐S2 IgG, as it has a coiled‐coil trimeric structure forming a compact short stalk which is hided under the globular S1 subunit and radially covered by oligosaccharides [33]. A similar finding on the steric constraints for antibody to access gp41, another class I virus fusion protein of HIV, was reported and discussed recently [34, 35].
Despite the widely reported neutralizing epitopes within the S2 subunit, the role of anti‐S2 antibodies in virus neutralization remains unknown. Based on the existing viral entry model of CoVs [7], it is proposed that the virus docks to host cell surface through the binding between the globular S1 subunit and cellular receptor(s), then the stalk‐like S2 subunit undergoes structural changes to initiate the fusion between viral envelope and host cell membrane, allowing the migration of the viral nucleocapsid into the cytosol. According to the previous studies of class I virus fusion proteins in other viruses [21, 36, 37], immediately after the receptor binding of their globular domains, the six‐helix bundled HR regions of their membrane fusion subunits, i.e. S2 subunit in the case SARS‐CoV, undergo dramatic conformational transition to expose its hydrophobic fusion peptide and mediates the subsequent virus–cell fusion. However, no evidence was reported to relate these structural changes to the attachment and docking of these viruses. Interestingly, in our study, anti‐S2 antibodies were demonstrated to be able to abolish the binding between S protein and its host cell in the cell‐based receptor binding assay, implying the possible involvement of the S2 subunit in virus docking process. The unanticipated blocking ability of anti‐S2 antibodies argues the current hypothesis that the S2 subunit only participates in membrane fusion but not virus docking. Therefore, we propose that successful docking or attachment of SARS‐CoV may not solely rely on the receptor binding of S1 subunit but may also depend on actions involving the S2 subunit, which therefore can be blocked by anti‐S2 IgG. To this end, we hypothesize two possible mechanisms to explain this observation. Firstly, we propose that the S2 subunit may participate in a global structural change of the spike which strengthens the binding or enhance the surface complementariness [38] between the receptor binding domain and its receptor. Alternatively speaking, the bound anti‐S2 antibodies may stress the S1 subunit and prevent the receptor binding domain from forming a complementary surface with its receptor [38]. Secondly, binding between the S1 subunit and host receptor(s) may be reversible and the virus may fail to stably dock to the host cell surface without subsequent insertion of the fusion peptide into the host cell membrane. One other possible explanation is simply the bulkiness of the bound anti‐S2 antibodies may hinder the spike from binding with its receptor.
Currently, the neutralizing epitopes of S2 subunit are mainly mapped to regions proximal to the fusion peptide and the C‐HR (Fig. 1). In addition, neutralizing antibodies targeting one of these regions have been found in sera of recovered SARS patients [17]. Using antigen specific antibody depletion assay, we demonstrated that the existence of neutralizing determinants within these regions as depletion of antibodies against these regions significantly diminished the neutralizing effects of anti‐S2 IgG (Fig. 5B). However, the residual neutralizing effect of the dep‐anti‐S2 IgG implies these regions may not be the sole neutralizing determinants and suggests the presence of neutralizing epitopes within other regions of the S2 subunit, such as the recent report on monoclonal neutralizing antibody targeting the C‐HR [13, 25]. It is noted that a similar depletion assay has been used to prove the existence neutralizing epitopes beyond the receptor binding domain of the SARS S1 subunit [39]. Furthermore, the blocking efficiency of dep‐anti‐S2 IgG was comparable to that of the mock depleted anti‐S2 IgG, indicating these regions may not be involved in virus docking, and alternatively speaking, the neutralizing mechanism of these epitopes may not involve blocking of receptor binding. On the other hand, some neutralizing epitopes may only be exposed during the conformational change of S2 protein triggered by receptor binding since binding of anti‐S2 IgG to m‐S is nearly saturated in low range of IgG concentrations (Fig. 3B) but such saturation was not observed in neutralizing assay (Fig. 2A) and cell‐based receptor binding assays (Fig. 4C). Although currently no neutralizing epitope was mapped to the C‐terminal tail of the S2 subunit of SARS‐CoV, neutralizing epitopes were mapped on the corresponding region of gp41 [40], implying the presence of possible functional roles of the regions other than the defined functional domains (e.g. HR and fusion peptide) in class I virus fusion proteins. The present experimental data suggests that there is no major neutralizing domain, like receptor binding domain of S1, on S2 protein.
In conclusion, the efficiency of anti‐S1 and anti‐S2 IgGs in virus neutralizing, S protein binding and blocking of receptor binding were compared quantitatively in this study. Although anti‐S2 IgG neutralizes the virus less efficiently as compared with the anti‐S1 IgG, the sequence conservation of S2 subunit across various strains, including the SARS‐like CoV in civet [41] and bats [42, 43], and its additive neutralizing effects with anti‐S1 antibody still make it a candidate for development of recombinant vaccines if the neutralizing efficiency can be greatly improved. Taken together with the results of the quantitative comparisons and the antigen specific antibody depletion assay, this study puts toward new understanding of the functional roles of the S2 subunit, hence the neutralizing mechanisms of anti‐S2 antibodies, which would certainly provide valuable information for rational design of vaccines or antibody therapeutics against the virus.
Conflict of interest statement
No conflicts declared.
Acknowledgements
This work was supported by HWF CR/3/6/3921/03 Research Grant funded by the Hong Kong Government. We thank Mr. Ken Y.C. Chow from Molecular Viral Pathogenesis Unit, Institute Pasteur, Paris, France, for critical reading of this manuscript.
Zeng Fanya,Hon Chung Chau,Yip Chi Wai,Law Ka Man,Yeung Yin Shan,Chan Kwok Hung,Malik Peiris Joseph S. and Leung Frederick Chi Ching(2006), Quantitative comparison of the efficiency of antibodies against S1 and S2 subunit of SARS coronavirus spike protein in virus neutralization and blocking of receptor binding: Implications for the functional roles of S2 subunit, FEBS Letters, 580, doi: 10.1016/j.febslet.2006.08.085
Author contributions: F.Z., J.S.M.P. and F.C.C.L. designed research; F.Z., C.C.H. and C.W.Y. wrote the paper; F.Z., K.M.L., Y.S.Y. performed animal test and adenovirus propagation; K.H.C. performed virus neutralization assay; C.C.H. and C.W.Y. performed molecular cloning; F.Z. performed all other experiments.
References
- 1. Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W., Nicholls J., Yee W.K., Yan W.W., Cheung M.T., Cheng V.C., Chan K.H., Tsang D.N., Yung R.W., Ng T.K., Yuen K.Y., Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet, 361, (2003), 1319– 1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cavanagh D., Severe acute respiratory syndrome vaccine development: experiences of vaccination against avian infectious bronchitis coronavirus. Avian Pathol., 32, (2003), 567– 582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Saif L.J., Animal coronavirus vaccines: lessons for SARS. Dev. Biol. (Basel), 119, (2004), 129– 140. [PubMed] [Google Scholar]
- 4. Taylor D.R., Obstacles and advances in SARS vaccine development. Vaccine, 24, (2006), 863– 871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. He Y., Jiang S., Vaccine design for severe acute respiratory syndrome coronavirus. Viral Immunol., 18, (2005), 327– 332. [DOI] [PubMed] [Google Scholar]
- 6. Dimitrov D.S., Virus entry: molecular mechanisms and biomedical applications. Nat. Rev. Microbiol., 2, (2004), 109– 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hofmann H., Pohlmann S., Cellular entry of the SARS coronavirus. Trends Microbiol., 12, (2004), 466– 472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cavanagh D., Davis P.J., Darbyshire J.H., Peters R.W., Coronavirus IBV: virus retaining spike glycopolypeptide S2 but not S1 is unable to induce virus-neutralizing or haemagglutination-inhibiting antibody, or induce chicken tracheal protection. J. Gen. Virol., 67, Pt 7 (1986), 1435– 1442. [DOI] [PubMed] [Google Scholar]
- 9. Delmas B., Rasschaert D., Godet M., Gelfi J., Laude H., Four major antigenic sites of the coronavirus transmissible gastroenteritis virus are located on the amino-terminal half of spike glycoprotein S. J. Gen. Virol., 71, Pt 6 (1990), 1313– 1323. [DOI] [PubMed] [Google Scholar]
- 10. Godet M., Grosclaude J., Delmas B., Laude H., Major receptor-binding and neutralization determinants are located within the same domain of the transmissible gastroenteritis virus (coronavirus) spike protein. J. Virol., 68, (1994), 8008– 8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vautherot J.F., Madelaine M.F., Boireau P., Laporte J., Bovine coronavirus peplomer glycoproteins: detailed antigenic analyses of S1, S2 and HE. J. Gen. Virol., 73, Pt 7 (1992), 1725– 1737. [DOI] [PubMed] [Google Scholar]
- 12. Keng C.T., Zhang A., Shen S., Lip K.M., Fielding B.C., Tan T.H., Chou C.F., Loh C.B., Wang S., Fu J., Yang X., Lim S.G., Hong W., Tan Y.J., Amino acids 1055 to 1192 in the S2 region of severe acute respiratory syndrome coronavirus S protein induce neutralizing antibodies: implications for the development of vaccines and antiviral agents. J. Virol., 79, (2005), 3289– 3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lai S.C., Chong P.C., Yeh C.T., Liu L.S., Jan J.T., Chi H.Y., Liu H.W., Chen A., Wang Y.C., Characterization of neutralizing monoclonal antibodies recognizing a 15-residues epitope on the spike protein HR2 region of severe acute respiratory syndrome coronavirus (SARS-CoV). J. Biomed. Sci., 12, (2005), 711– 727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang H., Wang G., Li J., Nie Y., Shi X., Lian G., Wang W., Yin X., Zhao Y., Qu X., Ding M., Deng H., Identification of an antigenic determinant on the S2 domain of the severe acute respiratory syndrome coronavirus spike glycoprotein capable of inducing neutralizing antibodies. J. Virol., 78, (2004), 6938– 6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Choy W.Y., Lin S.G., Chan P.K., Tam J.S., Lo Y.M., Chu I.M., Tsai S.N., Zhong M.Q., Fung K.P., Waye M.M., Tsui S.K., Ng K.O., Shan Z.X., Yang M., Wu Y.L., Lin Z.Y., Ngai S.M., Synthetic peptide studies on the severe acute respiratory syndrome (SARS) coronavirus spike glycoprotein: perspective for SARS vaccine development. Clin. Chem., 50, (2004), 1036– 1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hua R., Zhou Y., Wang Y., Hua Y., Tong G., Identification of two antigenic epitopes on SARS-CoV spike protein. Biochem. Biophys. Res. Commun., 319, (2004), 929– 935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhong X., Yang H., Guo Z.F., Sin W.Y., Chen W., Xu J., Fu L., Wu J., Mak C.K., Cheng C.S., Yang Y., Cao S., Wong T.Y., Lai S.T., Xie Y., Guo Z., B-cell responses in patients who have recovered from severe acute respiratory syndrome target a dominant site in the S2 domain of the surface spike glycoprotein. J. Virol., 79, (2005), 3401– 3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Duan J., Yan X., Guo X., Cao W., Han W., Qi C., Feng J., Yang D., Gao G., Jin G., A human SARS-CoV neutralizing antibody against epitope on S2 protein. Biochem. Biophys. Res. Commun., 333, (2005), 186– 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang S., Chou T.H., Sakhatskyy P.V., Huang S., Lawrence J.M., Cao H., Huang X., Lu S., Identification of two neutralizing regions on the severe acute respiratory syndrome coronavirus spike glycoprotein produced from the mammalian expression system. J. Virol., 79, (2005), 1906– 1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zeng F., Chow K.Y., Hon C.C., Law K.M., Yip C.W., Chan K.H., Peiris J.S., Leung F.C., Characterization of humoral responses in mice immunized with plasmid DNAs encoding SARS-CoV spike gene fragments. Biochem. Biophys. Res. Commun., 315, (2004), 1134– 1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Earp L.J., Delos S.E., Park H.E., White J.M., The many mechanisms of viral membrane fusion proteins. Curr. Top. Microbiol. Immunol., 285, (2005), 25– 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sainz B. Jr., Rausch J.M., Gallaher W.R., Garry R.F., Wimley W.C., Identification and characterization of the putative fusion peptide of the severe acute respiratory syndrome-associated coronavirus spike protein. J. Virol., 79, (2005), 7195– 7206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sainz B. Jr., Rausch J.M., Gallaher W.R., Garry R.F., Wimley W.C., The aromatic domain of the coronavirus class I viral fusion protein induces membrane permeabilization: putative role during viral entry. Biochemistry, 44, (2005), 947– 958. [DOI] [PubMed] [Google Scholar]
- 24. Taguchi F., Shimazaki Y.K., Functional analysis of an epitope in the S2 subunit of the murine coronavirus spike protein: involvement in fusion activity. J. Gen. Virol., 81, (2000), 2867– 2871. [DOI] [PubMed] [Google Scholar]
- 25. Lip K.M., Shen S., Yang X., Keng C.T., Zhang A., Oh H.L., Li Z.H., Hwang L.A., Chou C.F., Fielding B.C., Tan T.H., Mayrhofer J., Falkner F.G., Fu J., Lim S.G., Hong W., Tan Y.J., Monoclonal antibodies targeting the HR2 domain and the region immediately upstream of the HR2 of the S protein neutralize in vitro infection of severe acute respiratory syndrome coronavirus. J. Virol., 80, (2006), 941– 950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McGaughey G.B., Barbato G., Bianchi E., Freidinger R.M., Garsky V.M., Hurni W.M., Joyce J.G., Liang X., Miller M.D., Pessi A., Shiver J.W., Bogusky M.J., Progress towards the development of a HIV-1 gp41-directed vaccine. Curr. HIV Res., 2, (2004), 193– 204. [DOI] [PubMed] [Google Scholar]
- 27. Zwick M.B., Jensen R., Church S., Wang M., Stiegler G., Kunert R., Katinger H., Burton D.R., Anti-human immunodeficiency virus type 1 (HIV-1) antibodies 2F5 and 4E10 require surprisingly few crucial residues in the membrane-proximal external region of glycoprotein gp41 to neutralize HIV-1. J. Virol., 79, (2005), 1252– 1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zwick M.B., Labrijn A.F., Wang M., Spenlehauer C., Saphire E.O., Binley J.M., Moore J.P., Stiegler G., Katinger H., Burton D.R., Parren P.W., Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J. Virol., 75, (2001), 10892– 10905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chow K.Y., Yeung Y.S., Hon C.C., Zeng F., Law K.M., Leung F.C., Adenovirus-mediated expression of the C-terminal domain of SARS-CoV spike protein is sufficient to induce apoptosis in Vero E6 cells. FEBS Lett., 579, (2005), 6699– 6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bisht H., Roberts A., Vogel L., Bukreyev A., Collins P.L., Murphy B.R., Subbarao K., Moss B., Severe acute respiratory syndrome coronavirus spike protein expressed by attenuated vaccinia virus protectively immunizes mice. Proc. Natl. Acad. Sci. USA, 101, (2004), 6641– 6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zeng F.Y., Chan C.W., Chan M.N., Chen J.D., Chow K.Y., Hon C.C., Hui K.H., Li J., Li V.Y., Wang C.Y., Wang P.Y., Guan Y., Zheng B., Poon L.L., Chan K.H., Yuen K.Y., Peiris J.S., Leung F.C., The complete genome sequence of severe acute respiratory syndrome coronavirus strain HKU-39849 (HK-39). Exp. Biol. Med. (Maywood), 228, (2003), 866– 873. [DOI] [PubMed] [Google Scholar]
- 32. Chou C.F., Shen S., Tan Y.J., Fielding B.C., Tan T.H., Fu J., Xu Q., Lim S.G., Hong W., A novel cell-based binding assay system reconstituting interaction between SARS-CoV S protein and its cellular receptor. J. Virol. Meth., 123, (2005), 41– 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Krokhin O., Li Y., Andonov A., Feldmann H., Flick R., Jones S., Stroeher U., Bastien N., Dasuri K.V., Cheng K., Simonsen J.N., Perreault H., Wilkins J., Ens W., Plummer F., Standing K.G., Mass spectrometric characterization of proteins from the SARS virus: a preliminary report. Mol. Cell. Proteomics, 2, (2003), 346– 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Burton D.R., Desrosiers R.C., Doms R.W., Koff W.C., Kwong P.D., Moore J.P., Nabel G.J., Sodroski J., Wilson I.A., Wyatt R.T., HIV vaccine design and the neutralizing antibody problem. Nat. Immunol., 5, (2004), 233– 236. [DOI] [PubMed] [Google Scholar]
- 35. Hamburger A.E., Kim S., Welch B.D., Kay M.S., Steric accessibility of the HIV-1 gp41 N-trimer region. J. Biol. Chem., 280, (2005), 12567– 12572. [DOI] [PubMed] [Google Scholar]
- 36. Eckert D.M., Kim P.S., Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem., 70, (2001), 777– 810. [DOI] [PubMed] [Google Scholar]
- 37. Zelus B.D., Schickli J.H., Blau D.M., Weiss S.R., Holmes K.V., Conformational changes in the spike glycoprotein of murine coronavirus are induced at 37 °C either by soluble murine CEACAM1 receptors or by pH 8. J. Virol., 77, (2003), 830– 840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li F., Li W., Farzan M., Harrison S.C., Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science, 309, (2005), 1864– 1868. [DOI] [PubMed] [Google Scholar]
- 39. Chen Z., Zhang L., Qin C., Ba L., Yi C.E., Zhang F., Wei Q., He T., Yu W., Yu J., Gao H., Tu X., Gettie A., Farzan M., Yuen K.Y., Ho D.D., Recombinant modified vaccinia virus Ankara expressing the spike glycoprotein of severe acute respiratory syndrome coronavirus induces protective neutralizing antibodies primarily targeting the receptor binding region. J. Virol., 79, (2005), 2678– 2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Heap C.J., Reading S.A., Dimmock N.J., An antibody specific for the C-terminal tail of the gp41 transmembrane protein of human immunodeficiency virus type 1 mediates post-attachment neutralization, probably through inhibition of virus-cell fusion. J. Gen. Virol., 86, (2005), 1499– 1507. [DOI] [PubMed] [Google Scholar]
- 41. Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., Luo S.W., Li P.H., Zhang L.J., Guan Y.J., Butt K.M., Wong K.L., Chan K.W., Lim W., Shortridge K.F., Yuen K.Y., Peiris J.S., Poon L.L., Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science, 302, (2003), 276– 278. [DOI] [PubMed] [Google Scholar]
- 42. Lau S.K., Woo P.C., Li K.S., Huang Y., Tsoi H.W., Wong B.H., Wong S.S., Leung S.Y., Chan K.H., Yuen K.Y., Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. USA, 102, (2005), 14040– 14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., Zhang J., McEachern J., Field H., Daszak P., Eaton B.T., Zhang S., Wang L.F., Bats are natural reservoirs of SARS-like coronaviruses. Science, 310, (2005), 676– 679. [DOI] [PubMed] [Google Scholar]


