Abstract
We evaluated a new real-time PCR-based prototype assay for the detection of Chlamydia trachomatis and Neisseria gonorrhoeae developed by Abbott Molecular Inc. This assay is designed to be performed on an Abbott m2000 real-time instrument system, which consists of an m2000sp instrument for sample preparation and an m2000rt instrument for real-time PCR amplification and detection. The limit of detection of this prototype assay was determined to be 20 copies of target DNA for both C. trachomatis and N. gonorrhoeae, using serially diluted linearized plasmids. No cross-reactivity could be detected when 55 nongonococcal Neisseria isolates and 3 non-C. trachomatis Chlamydia isolates were tested at 1 million genome equivalents per reaction. Concordance with the Roche Amplicor, BDProbeTec ET, and Gen-Probe APTIMA Combo 2 tests was assessed using unlinked/deidentified surplus clinical specimens previously analyzed with these tests. For C. trachomatis, concordance for positive results ranged from 93.7% to 100%, while concordance for negative results ranged from 98.2% to 100%. For N. gonorrhoeae, concordance for positive and negative results ranged from 91.4% to 100% and 99.3% to 100%, respectively. A workflow analysis of the prototype assay was conducted to obtain information on throughput under laboratory conditions. At 48 samples/run, the time to first result for both C. trachomatis and N. gonorrhoeae was 4.5 h. A total of 135 patient specimens could be analyzed in 8.9 h, with 75 min of hands-on time. This study demonstrated the technical and clinical feasibility of the new Abbott real-time PCR C. trachomatis/N. gonorrhoeae assay.
Chlamydia trachomatis and Neisseria gonorrhoeae are common sexually transmitted microorganisms. These bacteria infect epithelial cells of the cervixes and urethras of women and the urethras of men, but symptoms of lower genital tract infection are expressed in only a small percentage of patients (1). Most infections remain undiagnosed and untreated, increasing the risk of ascending infections, especially in women, who may experience pelvic inflammatory disease, ectopic pregnancy, or infertility. The consequences of this “silent epidemic” are so significant that public health authorities are initiating screening programs to identify and treat infected individuals and their partners. Traditional samples, such as cervical and urethral swabs, and less invasive clinical samples, such as first-void urines (FVUs) and vaginal swabs, have been shown to be effective indicators of infection when tested by nucleic acid amplification tests (NAATs) (2, 3, 8, 11, 14-19). For NAATs to be successfully implemented in screening programs, assays need to possess appropriate analytical performance characteristics and be sufficiently automated to enable large numbers of specimens to be analyzed rapidly with minimal intervention. An automated magnetic sample preparation platform combined with homogeneous real-time multiplexed PCR has been developed for the diagnosis of urogenital infection with C. trachomatis and N. gonorrhoeae, with the dual goals of improving the workflow and improving the diagnostic sensitivity and specificity of NAATs for these organisms. This report describes the characteristics of a prototype assay, including its analytical sensitivity and specificity. Residual samples that had been tested using a variety of other assays were analyzed to examine the concordance of results across assay platforms. In addition, an analysis of the workflow, throughput, and labor requirements of the m2000 assay was performed.
MATERIALS AND METHODS
Clinical samples.
Cervical, vaginal, and urethral specimens that had been analyzed for C. trachomatis, using either LCx from Abbott Laboratories (n = 64), BDProbeTec ET from Becton Dickinson (n = 496), or Amplicor C. trachomatis from Roche (n = 67), and FVUs previously tested for C. trachomatis and N. gonorrhoeae, using either APTIMA Combo 2 from Gen-Probe (n = 496) or BDProbeTec ET (n = 156), were shipped from several laboratories to Abbott Laboratories for m2000 testing. All swab samples were collected in the proprietary transport devices provided by the various assay manufacturers and were stored and shipped per the manufacturers' instructions. Swab specimens containing sufficient volume (minimum of 500 μl) were tested directly on the m2000 system. For swabs with no or not enough liquid medium remaining, the volume was brought up to 1 ml using Abbott specimen transport buffer, allowing for further testing. FVU samples were collected in either the manufacturer-supplied device (APTIMA Combo 2) or DNA/RNA Protect (Sierra Diagnostics, Sonora, CA) (for BDProbeTec ET testing) and were stored and shipped to Abbott per each manufacturer's instructions. Prior to shipment, all samples were coded and patient identifiers removed. After blinded testing of samples in the m2000 system, concordance of positive and negative test results was determined by comparing m2000 results with those obtained using the FDA-cleared tests. Samples yielding discordant results for C. trachomatis were subsequently retested using either the LCx C. trachomatis assay or a Real Art C. trachomatis PCR kit (artus, Hamburg, Germany).
Magnetic sample preparations.
The m2000sp sample preparation technology (Abbott Molecular Inc.) captured DNAs from 400-μl specimens and an internal control sequence (linearized plasmid DNA containing the hydroxypyruvate reductase gene from the pumpkin plant) added to each specimen, using silica-based magnetic particles. The microparticles were washed three times, allowing for the removal of PCR amplification inhibitors. Each sample was eluted in 100 μl of elution buffer, and 25 μl was added to an equal volume of PCR master mix. In this study, 48 specimens were processed per run.
PCR amplification and real-time sequence detection.
The assay amplifies a region of the C. trachomatis cryptic plasmid and a region in the N. gonorrhoeae opacity (Opa) gene, as well as an internal control. The master mix contained a C. trachomatis probe labeled with 6-carboxyfluorescein (5′) and BHQ1 (3′) for the detection of a 102-base C. trachomatis cryptic plasmid DNA amplicon, an N. gonorrhoeae probe labeled with VIC (5′) and BHQ2 (3′) for the detection of a 122-base DNA amplicon of the N. gonorrhoeae Opa gene, and an internal control probe labeled with NED (5′) and BHQ1 (3′) for the detection of a 136-base amplicon of the hydroxypyruvate reductase gene from the pumpkin plant. The homogenous assay format allowed for the detection of all three DNA sequences without the necessity of opening the reaction vessels. Amplification and detection were performed in an m2000rt PCR instrument.
Analytical sensitivity and specificity.
Serial dilutions of linearized plasmids containing the C. trachomatis and N. gonorrhoeae target sequences were analyzed in the system to determine the limits of detection of the assay for each analyte. The C. trachomatis dilution panel consisted of members at 20, 75, 300, 3 × 103, 4 × 104, and 8 × 105 copies/assay. The N. gonorrhoeae dilution panel consisted of members at 20, 75, 300, 1.5 × 103, 1.5 × 104, and 3 × 105 copies/assay. For each concentration, 16 replicates were analyzed. To assess the analytical specificity, genomic DNAs were prepared from a panel of 55 nongonococcal Neisseria organisms and 3 non-C. trachomatis Chlamydia organisms (Table 1) at 1 million copies/assay and analyzed in the m2000 system.
TABLE 1.
Composition of the specificity panel
| Organism | No. of strains tested |
|---|---|
| Chlamydia pneumoniae | 2 |
| Chlamydia psittaci | 1 |
| Neisseria animalis | 1 |
| Neisseria caniae | 2 |
| Neisseria canis | 1 |
| Neisseria cinerea | 2 |
| Neisseria cuniculi | 1 |
| Neisseria denitrificans | 1 |
| Neisseria elongata subsp. elongata | 1 |
| N. elongata subsp. glycolytica | 1 |
| Neisseria flavescens | 1 |
| Neisseria iguanae | 1 |
| Neisseria lactamica | 6 |
| Neisseria macacae | 1 |
| Neisseria meningitidis | 2 |
| N. meningitidis B | 1 |
| N. meningitidis C | 2 |
| N. meningitidis D | 2 |
| Neisseria mucosa | 3 |
| Neisseria polysaccharea | 1 |
| Neisseria siccaa | 3 |
| Neisseria subflavaa | 21 |
| Neisseria weaveri | 1 |
One of the 3 N. sicca isolates and 5 of the 21 N. subflava isolates in the specificity panel had previously been demonstrated to cross-react with the Roche Amplicor N. gonorrhoeae assay (7).
Throughput experiments.
During this study, a prototype m2000sp instrument capable of preparing and analyzing a maximum of 48 samples was utilized. Runs were conducted using various numbers of samples to determine both the time to first result and the time to completion (total run time and hands-on time) for both the sample preparation and real-time PCR components of the assay.
RESULTS
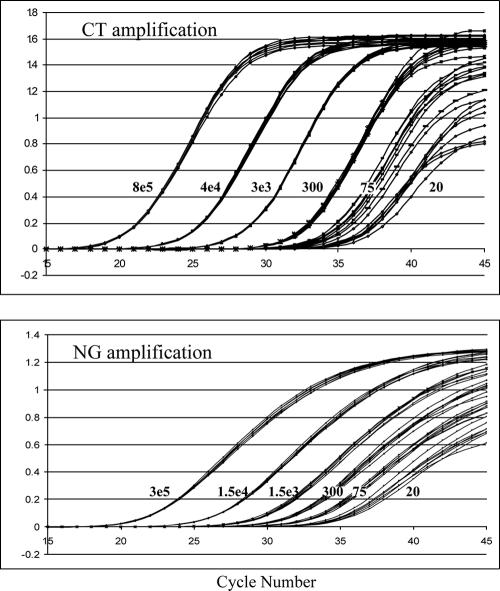
The analytical sensitivity of the m2000 C. trachomatis/N. gonorrhoeae real-time PCR prototype assay was assessed by analyzing a dilution series of 16 replicates at each concentration level (8 × 105 copies/reaction to 20 copies/reaction) of linearized plasmid DNA. Representative amplification curves for the C. trachomatis and N. gonorrhoeae components of the assay are shown in Fig. 1. Earlier detection of amplification corresponds to higher target levels. The amplification curves for the 16 replicate samples at each concentration were tightly grouped, particularly at or above 300 plasmid copies. A target concentration of 20 copies per reaction yielded robust amplification curves among all 16 replicates for each target, indicating that both the C. trachomatis and N. gonorrhoeae targets can be detected reproducibly at a level of 20 copies of DNA per PCR.
FIG. 1.
Fluorescent detection during 45 amplification cycles of Chlamydia trachomatis (CT) and Neisseria gonorrhoeae (NG) target DNAs in cloned and quantified linearized plasmid dilution series. The C. trachomatis dilution panel consisted of members at 20, 75, 300, 3 × 103, 4 × 104, and 8 × 105 copies/assay, and the N. gonorrhoeae dilution panel consisted of members at 20, 75, 300, 1.5 × 103, 1.5 × 104, and 3 × 105 copies/assay, as labeled on the curves.
All of the organisms included in the specificity panel (Table 1) gave negative results in the m2000 test when tested at 1 million genome equivalents per reaction (data not shown), a concentration of target significantly above what would be expected in a typical biological specimen. Six of the nongonococcal Neisseria isolates (indicated in the table) used in the specificity panel had previously been demonstrated to cross-react with the Amplicor N. gonorrhoeae assay (7).
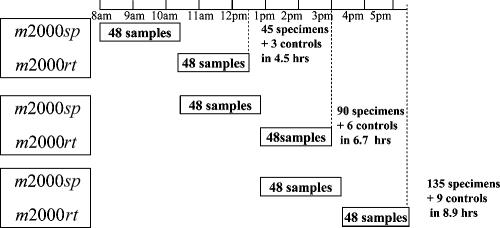
The workflow of a typical assay run of 48 samples is shown in Fig. 2 and includes the following steps: (i) 15 min of hands-on reagent setup and sample loading on the m2000sp instrument, (ii) 100 min of walk-away automated DNA extraction (m2000sp), (iii) 3 min of hands-on PCR master mix reagent loading on the m2000sp instrument, (iv) 16 min of walk-away automated PCR plate preparation (m2000sp), (v) 5 min of hands-on PCR plate sealing and transfer to the m2000rt instrument, (vi) 126 min of walk-away automated amplification and detection (m2000rt), and (vii) 2 min of hands-on result review and printout (m2000rt). Since the sample preparation and real-time PCR components of the assay can be overlapped, preparation of the m2000sp instrument for the second batch of samples was carried out during step iv of processing of the first batch of samples, and the automated extraction for the second sample set was performed while the first batch of samples were undergoing simultaneous PCR amplification and product detection on the m2000rt instrument (step vi), as shown in Fig. 2. This enabled a total of 45 specimens to be analyzed in 4.5 h, with the analysis of an additional 45 specimens being completed in 6.7 h. Analysis of a third set of 45 specimens could be commenced once the samples from the second sample batch reached step 4 of the process, enabling a maximum of 135 specimens to be analyzed and reported in 8.9 h. For convenience, the final set of specimens could be left in the m2000rt machine at the conclusion of a shift to complete the analytical process, and the postanalytical steps could be completed the following day. The total hands-on time for completing three runs (135 specimens) was approximately 75 min. In addition to the actual assay time, approximately 15 min of daily maintenance was required for the m2000sp instrument and could be completed either prior to or at the conclusion of testing. No daily maintenance was required for the m2000rt instrument.
FIG. 2.
Throughput with one m2000sp instrument and one m2000rt instrument. The total hands-on time (for three runs) is 75 min.
The concordance of the results obtained with the m2000 assay and other currently available assays is shown in Table 2. For C. trachomatis results for swab specimens, 99.1% (325/328) concordance was obtained with m2000 for BDProbeTec ET-positive samples, 96.3% (26/27) concordance was obtained for Amplicor-positive samples, and 100% (32/32) concordance was obtained for LCx-positive samples. The levels of concordance of m2000 for C. trachomatis-negative swab samples were 98.2% (165/168) with BDProbeTec ET, 100% (40/40) with Amplicor, and 100% (32/32) with LCx. Overall, the agreement for C. trachomatis results for swab samples ranged from 98.5% to 100%. For FVU specimens tested for C. trachomatis, the m2000 assay results were concordant with 93.7% (59/63) of APTIMA Combo 2-positive results and 96.8% (60/62) of BDProbeTec ET-positive samples. The concordance of negative samples was 99.3% (430/433) with APTIMA Combo 2 and 98.9% (93/94) with BDProbeTec ET. The overall agreement of m2000 C. trachomatis results for FVUs with both APTIMA Combo 2 and BDProbeTec ET was 98.6%. All FVU specimens (n = 6) that were negative on initial testing in the m2000 C. trachomatis assay but had been reported as positive in either the APTIMA Combo 2 (n = 4) or BDProbeTec ET (n = 2) assay were negative upon retesting with the LCx C. trachomatis test, and both BDProbeTec ET-positive/m2000-negative samples were also negative in the RealArt C. trachomatis assay. The single urine sample that yielded a BDProbeTec ET-negative/m2000-positive result on initial testing was negative by the LCx test and positive by the RealArt C. trachomatis assay upon repeat testing. The concordance of results for FVU samples tested for N. gonorrhoeae was 91.4% (32/35) for APTIMA Combo 2-positive samples and 100% (35/35) for BDProbeTec ET-positive samples, with concordance for APTIMA Combo 2- and BDProbeTec ET-negative samples being 99.3% (460/463) and 100% (121/121), respectively. The overall agreement of N. gonorrhoeae results for FVUs was 99.2% with APTIMA Combo 2 and 100% with BDProbeTec ET. N. gonorrhoeae testing was not performed on all of the swab samples by using BDProbeTec ET, Amplicor, and LCx. Therefore, no concordance data for N. gonorrhoeae results for swab samples were available.
TABLE 2.
Concordance of m2000 C. trachomatis and N. gonorrhoeae results with those of other amplification assays
| Specimen type | Previous test (n) | % m2000 C. trachomatis concordance (no. of concordant results/total no. of samples)
|
% m2000 N. gonorrhoeae concordance (no. of concordant results/total no. of samples)b
|
||||
|---|---|---|---|---|---|---|---|
| Positive | Negative | Overall | Positive | Negative | Overall | ||
| Swabs | BDProbeTec ET (496) | 99.1 (325/328) | 98.2 (165/168) | 98.8 | ND | ND | ND |
| Amplicor (67) | 96.3 (26/27) | 100 (40/40) | 98.5 | ND | ND | ND | |
| LCx (64) | 100 (32/32) | 100 (32/32) | 100 | ND | ND | ND | |
| Urine | APTIMA Combo 2 (496) | 93.7 (59/63) | 99.3 (430/433) | 98.6 | 91.4 (32/35) | 99.3 (460/463) | 99.2 |
| BDProbeTec ET (156)a | 96.8 (60/62) | 98.9 (93/94) | 98.6 | 100 (35/35) | 100 (121/121) | 100 | |
Urine specimens tested using BDProbeTec ET were collected, transported, and stored in DNA/RNA Protect and validated by the laboratory.
ND, not determined.
DISCUSSION
The new m2000 real-time PCR prototype assay demonstrated an analytical sensitivity of 20 copies of DNA per reaction for both C. trachomatis and N. gonorrhoeae. Since the cryptic plasmid of C. trachomatis is typically present at approximately 7 to 10 copies per elementary body (EB) (13), this corresponds to a detection limit of two EBs. Similarly, approximately 11 copies of the Opa gene are found in the N. gonorrhoeae genome (12), and thus, 20 copies of genomic DNA corresponds to approximately two organisms. Further testing also revealed that the limit of detection for C. trachomatis was maintained when 20 copies of C. trachomatis and 10 million copies of N. gonorrhoeae were copresent in a sample, and vice versa, i.e., the limit of detection for N. gonorrhoeae was maintained when 20 copies of N. gonorrhoeae and 10 million copies of C. trachomatis were copresent in a sample (data not shown). Previous studies have shown a range of analytical sensitivity end points for other commercially available assays. For example, Chong et al. (4) demonstrated end points of 12 EBs for the LCx Chlamydia test and 0.01 EB for the APTIMA Combo 2 assay. For 15 common C. trachomatis serovars, the package inserts report limits of detection of 1 inclusion-forming unit per reaction for COBAS Amplicor, 5 to 200 (median of 35) EBs per reaction for BDProbeTec ET, and 1 inclusion-forming unit per assay for APTIMA Combo 2. For N. gonorrhoeae, the package insert claims are 5 CFU/test for 15 strains of N. gonorrhoeae for COBAS Amplicor, 5 to 25 cells per reaction (median of 10 cells) for 39 strains of N. gonorrhoeae for BDProbeTec ET, and 50 cells/assay for 57 strains of N. gonorrhoeae for APTIMA Combo 2.
Prior to formally evaluating the new m2000 assay in clinical trials, we elected to perform a preliminary assessment of assay performance by testing residual material from swab and urine samples previously collected for and tested by other FDA-cleared assays. This protocol involved shipping of positive and negative samples to Abbott, where they were tested in a blinded fashion. For each specimen type, the agreements were close to 100% for C. trachomatis- or N. gonorrhoeae-positive and -negative samples, suggesting that the m2000 assay may be expected to offer a comparable clinical performance to those reported for currently available tests. Additional testing of the few samples yielding discordant results revealed somewhat inconsistent patterns of positive and negative results in the different assays, suggesting that most, if not all, of these specimens were low-level positive specimens producing inconsistent results near the limits of analytical sensitivity for the various assays. A multicenter evaluation of the BDProbeTec ET system for C. trachomatis and N. gonorrhoeae evaluated swabs and urines from men and women and compared its performance to that of the LCx assay (17). The study found that the sensitivity rates for male urethral swabs and FVUs and female cervical swabs ranged from 92.5 to 98.5% but that sensitivity rates for female FVUs were >10% lower for both C. trachomatis and N. gonorrhoeae. This may be a reflection of a lower analytical sensitivity (3) and of inhibitors in urine (10), which have been reported for most assays, including the BDProbeTec ET test (5, 6). The Roche COBAS Amplicor tests for C. trachomatis and N. gonorrhoeae were evaluated at eight U.S. centers (11, 16, 20). For both organisms, urine sensitivity rates were usually lower than those for swabs, but comparisons were done to cell culture and direct fluorescent-antibody assays and did not employ amplification methods other than PCR. The internal controls for Amplicor revealed that 2.4% of the specimens were inhibitory when initially tested. The transcription-mediated amplification assay (APTIMA Combo 2) also demonstrated excellent sensitivities for C. trachomatis and N. gonorrhoeae compared to those of other commercial NAATs, with a slightly lower value for N. gonorrhoeae detection in urines from asymptomatic women (8).
The m2000 assay was also challenged for specificity by using numerous strains of nongonococcal Neisseria and three non-C. trachomatis strains of Chlamydia, and all yielded negative results at 1 million genome copies per reaction. Analytical specificity data reported in the package inserts of commercially available assays and in the present report are similar, with little or no specific cross-reactivity observed when physiological levels of nucleic acids from related organisms were tested in the assay. Not surprisingly, therefore, none of the multicenter evaluations of current assays reported aberrant numbers of false-positive results (11, 16-18, 20).
Concern about the relative risk of false-positive results increasing to an unacceptable level as NAATs for C. trachomatis and N. gonorrhoeae are expanded to low-prevalence populations has been expressed, including suggestions that all positive results by NAATs be confirmed by retesting samples in the same assay or, ideally, a different one (6). The present study provides an insight into the feasibility of using the new m2000 real-time PCR assay in this capacity, since it is capable of generating reliable results for specimens collected in transport systems not designed for the m2000 system. Although further laboratory validation is needed to fully substantiate the interchangeability of samples between the m2000 and other assay systems, this capability may help establish the optimal role, either confirmatory or screening, for the various testing platforms (8).
Since the establishment of screening programs will undoubtedly increase testing volumes for laboratories, sample processing and testing workflow will become increasingly important, and a preliminary assessment of these parameters was made for the m2000 system. The m2000 system has a maximum capacity of processing 96 samples at a time, matching 96-well PCRs on a thermal cycler. At the time of the study, however, software for handling only 48 samples per run was available for evaluation. Figure 2 demonstrates the workflow of the assay. For both C. trachomatis and N. gonorrhoeae results, the time to first result for 45 specimens was 4.5 h. Ninety patient specimens were tested in 6.7 h, and 135 specimens were tested in 8.9 h. When the 96-samples/run capability becomes available, the time to first result for 93 C. trachomatis and N. gonorrhoeae patient results is estimated to be 5.6 h, and 186 and 279 patient specimens can be tested using this format in 8 h and 11 h, respectively. This would present a significant improvement compared to the manual versions of Amplicor, BDProbeTec ET, and APTIMA Combo 2 and would approach the throughput of automated BDProbeTec ET with the Viper system and APTIMA Combo 2 with the TIGRIS system. Hanson and Cartwright (9) adapted the automated liquid-handling Tecan Genesis RSP 100 system to the LCx Chlamydia test and reported a preservation of assay accuracy while reducing technician pipetting time by 75%. Vincelette et al. (20) reported that the COBAS Amplicor test for C. trachomatis allowed automation of amplification and detection. One technician performed 3 to 4 h of hands-on work while testing 96 specimens in a workday. Significantly improving the efficiency of C. trachomatis/N. gonorrhoeae NAA testing requires the kind of integrated approach to automating sample preparation, amplification, detection, and reporting afforded by the Abbott m2000 system.
In conclusion, this study reports the first evaluation of the Abbott m2000 prototype real-time PCR C. trachomatis/N. gonorrhoeae assay and demonstrates satisfactory preliminary analytical and clinical performance. The workflow assessment of the prototype assay also offers important throughput information to testing laboratories. More studies with larger patient sample sizes will be conducted to provide further assessments of the clinical performance characteristics of the assay.
Footnotes
Published ahead of print on 3 January 2007.
REFERENCES
- 1.Cates, W., and J. N. Wasserheit. 1991. Genital Chlamydia infections; epidemiology and reproductive sequelae. Am. J. Obstet. Gynecol. 164:1771-1781. [DOI] [PubMed] [Google Scholar]
- 2.Chernesky, M., D. Jang, H. Lee, J. D. Burczak, H. Hu, J. Sellors, S. J. Tomazic-Allen, and J. B. Mahony. 1994. Diagnosis of Chlamydia trachomatis infections in men and women by testing first-void urine by ligase chain reaction. J. Clin. Microbiol. 32:2682-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chernesky, M., D. Jang, K. Luinstra, S. Chong, M. Smieja, W. Cai, B. Hayhoe, E. Portillo, C. MacRitchie, C. Main, and R. Ewert. 2006. High analytical sensitivity and low rates of inhibition may contribute to detection of Chlamydia trachomatis in significantly more women by the APTIMA Combo 2 assay. J. Clin. Microbiol. 44:400-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chong, S., D. Jang, X. Song, J. Mahony, A. Petrich, P. Barriga, and M. Chernesky. 2003. Specimen processing and concentration of Chlamydia trachomatis added can influence false-negative rates in the LCx assay but not in the APTIMA Combo 2 assay when testing for inhibitors. J. Clin. Microbiol. 41:778-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cosentino, L. A., D. V. Landers, and S. L. Hillier. 2003. Detection of Chlamydia trachomatis and Neisseria gonorrhoeae by strand displacement amplification and relevance of the amplification control for use with vaginal swab specimens. J. Clin. Microbiol. 41:3592-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Culler, E. E., A. M. Caliendo, and F. S. Nolte. 2003. Reproducibility of positive test results in the BDProbeTec ET system for detection of Chlamydia trachomatis and Neisseria gonorrhoeae. J. Clin. Microbiol. 41:3911-3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farrell, D. 1999. Evaluation of AMPLICOR Neisseria gonorrhoeae using cppB nested PCR and 16S rRNA PCR. J. Clin. Microbiol. 37:386-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaydos, C. A., T. C. Quinn, D. Willis, A. Weissfeld, E. W. Hook, D. H. Martin, D. C. Ferrero, and J. Schachter. 2003. Performance of the APTIMA Combo 2 assay for detection of Chlamydia trachomatis and Neisseria gonorrhoeae in female urine and endocervical swab specimens. J. Clin. Microbiol. 41:304-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanson, K. L., and C. P. Cartwright. 2001. Evaluation of an automated liquid-handling system (Tecan Genesis RSP 100) in the Abbott LCx assay for Chlamydia trachomatis. J. Clin. Microbiol. 39:1975-1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mahony, J. B., S. Chong, D. Jang, K. Luinstra, M. Faught, D. Dalby, J. Sellors, and M. A. Chernesky. 1998. Urine specimens from pregnant and nonpregnant women inhibitory to amplification of Chlamydia trachomatis nucleic acid by PCR, ligase chain reaction, and transcription-mediated amplifications: identification of urinary substances associated with inhibition and removal of inhibitory activity. J. Clin. Microbiol. 36:3122-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin, D. H., C. Cammarata, B. Van der Pol, R. B. Jones, T. C. Quinn, C. A. Gaydos, K. Crotchfelt, J. Schachter, J. Moncada, D. Jungkind, B. Turner, and C. Peyton. 2000. Multicenter evaluation of AMPLICOR and automated COBAS AMPLICOR CT/NG tests for Neisseria gonorrhoeae J. Clin. Microbiol. 38:3544-3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer, T. F., C. P. Gibbs, and R. Hass. 1990. Variation and control of protein expression in Neisseria. Annu. Rev. Microbiol. 44:451-477. [DOI] [PubMed] [Google Scholar]
- 13.Palmer, L., and S. Falkow. 1986. A common plasmid of Chlamydia trachomatis. Plasmid 16:52-62. [DOI] [PubMed] [Google Scholar]
- 14.Schachter, J., E. W. Hook III, W. M. McCormack, T. C. Quinn, M. Chernesky, S. Chong, J. I. Girdner, P. B. Dixon, L. DeMeo, E. Williams, A. Cullen, and A. Lorincz. 1999. Ability of the Digene hybrid capture II test to identify Chlamydia trachomatis and Neisseria gonorrhoeae in cervical specimens. J. Clin. Microbiol. 37:3668-3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schachter, J., W. M. McCormack, M. A. Chernesky, D. H. Martin, B. Van der Pol, P. A. Rice, E. W. Hook III, W. E. Stamm, T. C. Quinn, and J. M. Chow. 2003. Vaginal swabs are appropriate specimens for diagnosis of genital tract infection with Chlamydia trachomatis. J. Clin. Microbiol. 41:3784-3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van der Pol, B., T. C. Quinn, C. A. Gaydos, K. Crotchfelt, J. Schachter, J. Moncada, D. Jungkind, D. H. Martin, B. Turner, C. Peyton, and R. B. Jones. 2000. Multicenter evaluation of the AMPLICOR and automated COBAS AMPLICOR CT/NG tests for detection of Chlamydia trachomatis. J. Clin. Microbiol. 38:1105-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van der Pol, B., D. V. Ferrero, L. Buck-Barrington, E. Hook III, C. Lenderman, T. Quinn, C. A. Gaydos, J. Lovchik, J. Schacter, J. Moncada, G. Hall, M. J. Tuohy, and R. B. Jones. 2001. Multicenter evaluation of the BDProbeTec ET system for detection of Chlamydia trachomatis and Neisseria gonorrhoeae in urine specimens, female endocervical swabs, and male urethral swabs. J. Clin. Microbiol. 39:1008-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van der Pol, B., J. A. Williams, N. J. Smith, B. E. Batteiger, A. P. Cullen, H. Erdman, T. Edens, K. Davis, H. Salim-Hammad, V. W. Chou, L. Scearce, J. Blutman, and W. J. Payne. 2002. Evaluation of the Digene hybrid capture II assay with the rapid capture system for detection of Chlamydia trachomatis and Neisseria gonorrhoeae. J. Clin. Microbiol. 40:3558-3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Dyck, E., M. Ieven, S. Pattyn, L. van Damme, and M. Laga. 2001. Detection of Chlamydia trachomatis and Neisseria gonorrhoeae by enzyme immunoassay, culture, and three nucleic acid amplification tests. J. Clin. Microbiol. 39:1751-1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vincelette, J., J. Schirm, M. Bogard, A.-M. Bourgault, D. S. Luijt, A. Bianchi, P. C. van Voorst Vader, A. Butcher, and M. Rosenstraus. 1999. Multicenter evaluation of the fully automated COBAS AMPLICOR PCR test for detection of Chlamydia trachomatis in urogenital specimens. J. Clin. Microbiol. 37:74-80. [DOI] [PMC free article] [PubMed] [Google Scholar]