Abstract
Legionella pneumophila is the predominant cause of Legionnaires' disease in the United States and Europe, while Legionella longbeachae is the common cause of the disease in Western Australia. Although clinical manifestations by both intracellular pathogens are very similar, recent studies have shown that phagosome biogeneses of both species within human macrophages are distinct (R. Asare and Y. Abu Kwaik, Cell. Microbiol., in press). Most inbred mouse strains are resistant to infection by L. pneumophila, with the exception of the A/J mouse strain, and this genetic susceptibility is associated with polymorphism in the naip5 allele and flagellin-mediated early activation of caspase 1 and pyropoptosis in nonpermissive mouse macrophages. Here, we show that genetic susceptibility of mice to infection by L. longbeachae is independent of allelic polymorphism of naip5. L. longbeachae replicates within bone marrow-derived macrophages and in the lungs of A/J, C57BL/6, and BALB/c mice, while L. pneumophila replicates in macrophages in vitro and in the lungs of the A/J mouse strain only. Quantitative real-time PCR studies on infected A/J and C57BL/6 mouse bone marrow-derived macrophages show that both L. longbeachae and L. pneumophila trigger similar levels of naip5 expression, but the levels are higher in infected C57BL/6 mouse macrophages. In contrast to L. pneumophila, L. longbeachae has no detectable pore-forming activity and does not activate caspase 1 in A/J and C57BL/6 mouse or human macrophages, despite flagellation. Unlike L. pneumophila, L. longbeachae triggers only a modest activation of caspase 3 and low levels of apoptosis in human and murine macrophages in vitro and in the lungs of infected mice at late stages of infection. We conclude that despite flagellation, infection by L. longbeachae is independent of polymorphism in the naip5 allele and L. longbeachae does not trigger the activation of caspase 1, caspase 3, or late-stage apoptosis in mouse and human macrophages. Neither species triggers caspase 1 activation in human macrophages.
Legionella longbeachae belongs to the family Legionellaceae, which causes a severe and fatal pneumonia known as Legionnaires' disease. In the United States, more than 90% of cases of Legionnaires' disease are caused by Legionella pneumophila (6). Interestingly, the most predominant species responsible for Legionnaires' disease in Western Australia is L. longbeachae (15). In addition, infection due to L. longbeachae has been reported in New Zealand, Germany, Japan, Denmark, Sweden, Canada, and The Netherlands. Unlike L. pneumophila, which inhabits mostly aquatic environments, L. longbeachae is commonly isolated from moist potting soil (29). In aquatic environments, amoeba serves as a reservoir for the amplification and dissemination of L. pneumophila and is considered the natural host for the bacterium (34). In addition, amoeba has been shown to resuscitate viable nonculturable L. pneumophila after disinfection by biocides, which may account for the reemergence of Legionella in water systems after disinfection (24).
L. pneumophila replicates in alveolar macrophages, which is necessary for the manifestation of Legionnaires' disease. After phagocytosis, L. pneumophila is localized in a unique phagosome that is isolated from the endocytic pathway (26, 41, 43). The L. pneumophila-containing phagosome excludes endocytic markers, including the lysosome-associated membrane glycoproteins lysosome-associated membrane protein 1 (LAMP-1) and LAMP-2 as well as the lysosomal acid protease cathepsin D (10). While the L. pneumophila-containing phagosome does not interact with the dynamic endocytic traffic, the L. longbeachae-containing phagosome interacts with the endocytic traffic and its biogenesis exhibits some maturation within the endocytic pathway (5). Recent studies have shown that within human macrophages, the L. longbeachae-containing phagosome is trafficked into a nonacidified late endosome-like phagosome that acquires the LAMPs and the mannose-6-phosphate receptor late endosomal markers but excludes the vacuolar ATPase proton pump and lysosomal markers (5). In addition, the L. longbeachae-containing phagosome is remodeled by the rough endoplasmic reticulum and bacterial replication occurs within the rough endoplasmic reticulum-remodeled late endosome-like phagosomes (5). Thus, there is a divergence in the mechanisms of pathogenesis of L. longbeachae and L. pneumophila in human macrophages (5). Further studies are needed to dissect further the host-parasite interaction of L. longbeachae, which is lagging behind that of most other intracellular pathogens, including the closely related species L. pneumophila.
Many intracellular pathogens, including L. pneumophila, have been shown to modulate the intrinsic and extrinsic apoptotic pathways of apoptosis that converge on the activation of caspase 3, resulting in apoptosis/programmed cell death (20). L. pneumophila induces the activation of caspase 3 in human macrophages during early stages of infection, which is thought to be essential for evasion of vesicle traffic, since inhibition of caspase 3 in human macrophages results in fusion of the phagosomes to lysosomes (18, 35). The activation of caspase 3 and the subsequent isolation of the phagosome from the endocytic pathway are mediated by the Dot/Icm type IV secretion system (47). Although caspase 3 is induced robustly during early stages of infection in human macrophages, apoptosis is not triggered until late stages of infection, concomitant with the termination of intracellular replication (2, 3, 35). The delay in apoptosis is associated with the induction of antiapoptotic signaling through the activation of NF-κB-dependent and -independent pathways (3, 30). In contrast, caspase 3 is not activated and is not required for the intracellular infection of mouse-derived macrophages (36, 45). Whether L. longbeachae also triggers caspase 3 and subsequent apoptosis in human macrophages is not known.
Among inbred mouse strains, A/J is the only inbred mouse strain susceptible to infection by L. pneumophila, while all the other strains are resistant (31). In contrast, many inbred strains of mice are susceptible to infection by many Legionella species (31). Only one study of permissiveness of mouse macrophages in vitro to L. longbeachae has been reported using a single isolate and indicated that the isolate replicates in both A/J and C57BL/6 thioglycolate-elicited mouse peritoneal macrophages, but whether the growth kinetics are similar to those of L. pneumophila is not known (27). Whether L. longbeachae can replicate in mouse lungs in vivo and whether mice are a suitable animal model for L. longbeachae are not known. The genetic susceptibility of mice has been attributed to a polymorphism in the neuronal apoptosis inhibitory protein 5 (naip5)-birc1e gene (13). At least eight murine homologues of naip genes have been identified (25), and naip5 has been identified as the gene responsible for the differential susceptibilities of A/J mice to L. pneumophila infection (13). The family of Naips is expressed abundantly in macrophage-rich tissues in mice, and their collective expression is increased after phagocytosis by murine macrophages (14), but whether Naip5 is one of the induced Naips is not known. The differential susceptibilities of different inbred mouse strains to infection by L. pneumophila are due to the rapid activation of caspase 1 in C57BL/6 versus A/J mice, resulting in early macrophage pyropoptosis-mediated cell death in C57BL/6 mice (36, 45). The L. pneumophila product that is responsible for the activation of caspase 1 is flagellin, but it is not known how Naip5 contributes to the process (36). Whether L. pneumophila triggers caspase 1 activation in human macrophages is not known, and whether L. longbeachae is capable of activating caspase 1 in mouse or human macrophages is also not known.
Some Naips have been shown to possess antiapoptotic activity (40) due to inhibition of caspase 3, caspase 7, and caspase 9 (17). The role of Naip5 in the activation of caspase 3 and apoptosis has not been determined, although it has been shown that the differential susceptibilities of mice to L. pneumophila are not related to the activation of caspase 3 (36, 45).
Here, we show that polymorphism of the naip5 allele does not play a role in the susceptibility of inbred mouse strains to infection by L. longbeachae. Both in vitro and in vivo studies show that L. longbeachae replicates efficiently in bone marrow-derived macrophages and in the lungs of A/J, C57BL/6, and BALB/c mice. In addition, we show that the induction of naip5 transcription in both L. pneumophila- and L. longbeachae-infected A/J mouse macrophages is less compared to that in C57BL/6 mice. We show that unlike what was observed with L. pneumophila, caspase 3 activation and late-stage apoptosis are triggered only at very low levels in both mouse and human macrophages infected by L. longbeachae. Flagellated L. longbeachae does not trigger caspase 1-mediated pyropoptosis in mouse macrophages, which correlates with the lack of detectable pore-forming activity in this species. Neither species activates caspase 1 in human macrophages.
MATERIALS AND METHODS
Bacterial strains.
The virulent strain of L. pneumophila (AA100) is a clinical isolate that has been described previously and was maintained at −80°C until use (1). The dotA mutant is an isogenic mutant generated from L. pneumophila AA100 (7, 11). L. longbeachae D4968, D4969, and D4973 are clinical isolates obtained from outbreaks in Oregon, Washington, and California, respectively, and were kindly provided by Barry Fields (CDC, Atlanta, GA). The L. longbeachae ATCC strain 33462 was purchased from the American Type Culture Collection (Manassas, VA). The GN229 spontaneous mutant of L. pneumophila strain AA100 is defective in pore-forming activity and has been described previously (32). The green fluorescent protein (GFP) expressed from a plasmid was introduced into some of the strains for analyses by confocal microscopy. The Francisella tularensis subsp. novicida strain U112 was described previously (38, 39). Bacteria were grown from frozen stocks on buffered charcoal-yeast extract (BCYE) agar supplemented with the appropriate antibiotic for 72 h at 37°C (16).
Macrophages.
The U937 macrophage-like cell line was maintained as described previously (22). Isolation and preparation of the human monocyte-derived macrophages (hMDMs) were carried out as described previously (42). Murine bone marrow-derived macrophages (mBDMs) were isolated as described before (46). At 48 h prior to infection, the U937 cells were differentiated with phorbol myristate acetate in either a 96-well plate for the fluorometric caspase 3 assay or a 24-well plate for confocal microscopy. hMDMs and mBDMs were seeded in 24-well plates for confocal analysis. During the preparation of the monolayers, 105 and 5 × 105 cells were seeded onto the 96-well plate and the 24-well plate, respectively. For infection of the monolayers, the bacterial strains were resuspended in RPMI 1640. The infection was carried out as described for each experiment.
Immortalized macrophage cell lines were established by infecting bone marrow-derived macrophages from A/J, C57BL/6, and BALB/c mice with the murine recombinant J2 retrovirus, as previously described (7, 11).
Intracellular growth kinetics.
Monolayers of immortalized bone marrow-derived mouse macrophages were infected with the three different clinical isolates of L. longbeachae grown for 72 h on BCYE at a multiplicity of infection (MOI) of 10 in triplicate. To synchronize the infection, the plates were centrifuged for 5 min at 1,000 rpm using a Thermo IEC Centra GP8R centrifuge. After 1 h of incubation in 5% CO2 at 37°C, the infected macrophages were washed three times with the culture medium to remove extracellular bacteria and incubated with 50 μg/ml gentamicin for 1 h to kill the remaining extracellular bacteria. This was considered the zero time point. The infected mouse macrophages were subsequently incubated for 8, 24, and 48 h. At the end of each time interval, the culture supernatant was removed and the cells were lysed hypotonically by the addition of 200 μl of sterile water for 10 min. The supernatant and the lysate were combined, serial dilutions were prepared, and aliquots were plated on BCYE plates to enumerate the bacteria.
Infection of A/J, C57BL/6, and BALB/c mice with L. longbeachae and L. pneumophila.
Male and female pathogen-free, 6- to 8-week-old A/J, C57BL/6, and BALB/c mice were purchased from Jackson Laboratories. For the preparation of the intratracheal inoculation, two L. longbeachae clinical isolates, D4968 and D4973, and L. pneumophila strain AA100 were grown on BCYE agar plates for 72 h. The mice were inoculated intratracheally with 50 μl of 106 bacteria as described previously (23). At 2 h, 24 h, and 48 h postinoculation, mice were humanely euthanized, the lungs were removed, and the bacteria were cultured on BCYE agar for 72 to 96 h as described previously (23). For the survival assay, the mice were observed at 14 days postinoculation. Our animal protocols comply with NIH guidelines for the humane care and use of animals and are approved by our IACUC.
Determination of naip5 expression by real-time PCR.
Monolayers of immortalized bone marrow-derived macrophages were infected with the L. longbeachae clinical isolate D4968 and L. pneumophila strain AA100 at an MOI of 10. To synchronize the infection, the plates were centrifuged for 5 min at 1,000 rpm using a Thermo IEC Centra GP8R centrifuge. After 1 h of incubation in 5% CO2 at 37°C, the infected macrophages were washed three times with the culture medium to remove extracellular bacteria and incubated with 50 μg/ml gentamicin for 1 h to kill the remaining extracellular bacteria. This was considered the zero time point. The infected mouse macrophages were subsequently incubated for 2, 6, and 12 h. At the end of each time interval, total RNA was isolated using a QIAGEN RNeasy mini kit (QIAGEN, Valencia, CA) and digested with DNase I (Ambione, Austin, TX) to remove chromosomal DNA. The remaining DNase I was inactivated by heating at 70°C for 5 min. Five micrograms of total RNA was reverse transcribed into cDNA with SuperScript III RNase H reverse transcriptase (Invitrogen, Carlsbad, CA). The real-time quantitative PCR was performed in an Opticon continuous fluorescence detector (MJ Research, San Francisco, CA). The primer sequences for Naip5 were 5′-CTCCAGCCACTCTTCCTCAA-3′ (forward primer) and 5′-ACCAGCCACAACCCTCTAAC-3′ (reverse primer), and the primer sequences for the β-actin control were 5′-GATCTGGCACCACACCTTCT-3′ (forward primer) and 5′-GGGGTGTTGAAGGTCTCAAA-3′ (reverse primer). The cDNA sample was amplified with DyNAmo SYBR green quantitative PCR kits containing a modified Thermus brockianus DNA polymerase according to the manufacturer's instructions (New England Biolabs, Beverly, MA). PCR conditions were 5 min at 94°C, 15 s at 96°C, and 15 s at 72°C for 30 cycles. The concentration of Naip5 was determined using the comparative cycle threshold (threshold cycle number at the cross point between amplification plot and threshold) method according to the manufacturer's software. Relative quantitation by quantitative reverse transcription-PCR was validated by the equivalent and linear amplifications of β-actin and naip5 at the assay concentrations.
Caspase 1 activation.
To assess active caspase 1 staining by confocal microscopy, 2.5 × 105 hMDMs or mBDMs on glass coverslips were infected with L. pneumophila and L. longbeachae strains at an MOI of 10 for 18, 24, and 36 h. Macrophages were stained for 1 h with 6-carboxyfluorescein-YVAD-fluoromethylketone (Immunochemistry Technologies) as recommended by the manufacturer. As a positive control for the hMDMs, macrophages were treated with 10 mM simvastatin (Calbiochem) (12). After infection, cultures were then washed extensively, fixed for 1 h with 3.7% formaldehyde solution, washed again, and then mounted with ProLong Gold antifade reagent (Molecular Probes Inc., Eugene, OR).
Caspase 3 activation.
For the detection of caspase 3 activity by artificial substrates, U937 cells in 96-well plates were infected with L. longbeachae clinical isolate D4968, L. pneumophila strain AA100, and its isogenic dotA mutant grown on BCYE plates for 72 h at an MOI of 50. At 6 h postinfection, the activity of caspase 3 in the infected U937 cells was measured by a fluorometric caspase 3 assay kit (BioVision, Inc., CA) as described previously (35). The level of caspase 3 enzymatic activity was measured in arbitrary fluorescence units by using a fluorescent plate reader (PerkinElmer) with an excitation at 400 nm and an emission at 505 nm.
For the detection of active caspase 3 by confocal microscopy, U937 cells attached to glass coverslips in 24-well plates were infected with L. pneumophila strain AA100 (GFP) and L. longbeachae strain ATCC 33462 (GFP) at an MOI of 10 for 6 h and 24 h. The bacteria were grown on BCYE plates containing 5 μg/ml chloramphenicol for 72 h prior to infection. For labeling of the bacteria, cells were fixed with 4% paraformaldehyde (Sigma) for 30 min, permeabilized with 0.1% Triton X-100 (Sigma) on ice for 15 min, blocked with 3% bovine serum albumin (Sigma) for 1 h, incubated with rabbit polyclonal antiactive caspase 3 antiserum (BD, San Diego, CA) for 1 h, and then incubated for 1 h with a goat anti-rabbit immunoglobulin G secondary antibody conjugated to Alexa red (Molecular Probes, Inc., Eugene, OR).
TUNEL assay.
U937 cells attached to glass coverslips in 24-well plates were infected with L. pneumophila strain AA100 and L. longbeachae clinical isolate D4968 at an MOI of 10 for 6 h and 24 h. Bacteria were grown for 72 h on BCYE plates before infection. For labeling of the bacteria, cells were fixed with 4% paraformaldehyde (Sigma) for 30 min, permeabilized with 0.1% Triton X-100 (Sigma) on ice for 15 min, blocked with 3% bovine serum albumin (Sigma) for 1 h, incubated with rabbit polyclonal antiserum (raised against L. pneumophila or L. longbeachae) for 1 h, and then incubated for 1 h with a donkey anti-rabbit immunoglobulin G secondary antibody conjugated to Alexa red (Molecular Probes, Inc., Eugene, OR). For labeling of apoptotic nuclei, the cells were then subjected to fluorescein isothiocyanate-conjugated terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) using an apoptosis detection kit according to the manufacturer's instructions (Boehringer Mannheim Corporation, Indianapolis, IN). Cells were examined with a Zeiss Axiophot photomicroscope and a Leica TCS NT confocal laser scanning microscope. A minimum of 100 cells per sample was counted, and apoptosis was quantified as the percentage of apoptotic cells (TUNEL-positive nuclei) among all of the cells counted. Multiple independent samples were examined.
Histopathological analysis.
Apoptosis in the lungs of A/J mice in response to L. longbeachae and L. pneumophila was assessed by confocal microscopy. L. longbeachae clinical isolate D4968 and L. pneumophila strain AA100 were grown on BCYE plates for 72 h, and 106 bacteria were inoculated intratracheally into the lungs of A/J mice. At 24, 48, and 72 h after inoculation, the mice were humanely sacrificed. Before lung removal, the pulmonary vasculature was perfused with 10 ml of saline containing 5 mM EDTA, via the right ventricle. The excised lungs were inflated and fixed in 10% neutral formalin for 24 h, dehydrated, and embedded in paraffin. Sections (5 μm) were cut, and labeling of apoptotic cells was carried out by TUNEL using an in situ cell death detection kit, as recommended by the manufacturer (Roche, Indianapolis, IN). An analysis of apoptotic cells (TUNEL positive) was carried out using laser scanning confocal microscopy. On average, 10 0.2-μm-thick serial sections of each image were captured and stored for further analyses using Adobe Photoshop (Adobe Photoshop, Inc.).
Statistical analyses.
All experiments were performed at least three times, and the data shown are representative of one experiment. To analyze statistically significant differences between different sets of data, two-tailed Student's t test was used and the P value was obtained.
RESULTS
Efficient replication of L. longbeachae in macrophages of inbred mouse strains.
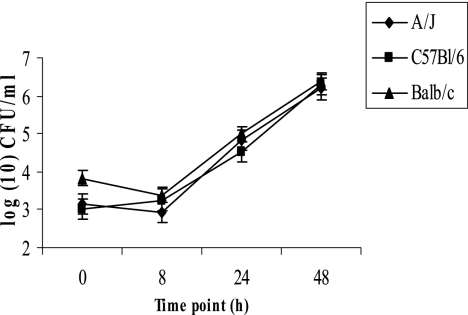
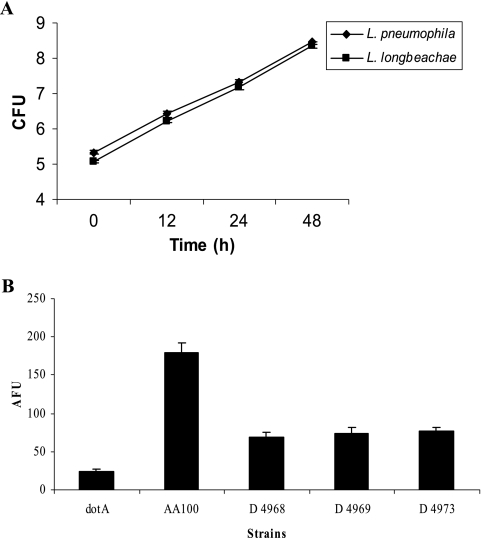
Growth of L. longbeachae in vitro was examined in immortalized bone marrow-derived macrophages from A/J, C57BL/6, and BALB/c mice by using an MOI of 10 for 1-h infection. The data showed that by 48 h postinfection, there was an ∼104-fold increase in the number of CFU of L. longbeachae in macrophages from all three strains of mice examined (Fig. 1). In contrast to L. longbeachae, L. pneumophila strain AA100 replicated only within A/J mouse-derived macrophages (data not shown), consistent with previous observations (8, 9).
FIG. 1.
L. longbeachae strains replicate equally in immortalized bone marrow-derived macrophages from A/J, C57BL/6, and BALB/c mice. Immortalized murine macrophages were infected with L. longbeachae strain D4968 at an MOI of 10 for 1 h, followed by gentamicin treatment for 1 h. The number of CFU of L. longbeachae was enumerated over 48 h. The results are representative of three independent experiments, and error bars represent standard deviations.
L. longbeachae strains are equally lethal to several inbred mouse strains.
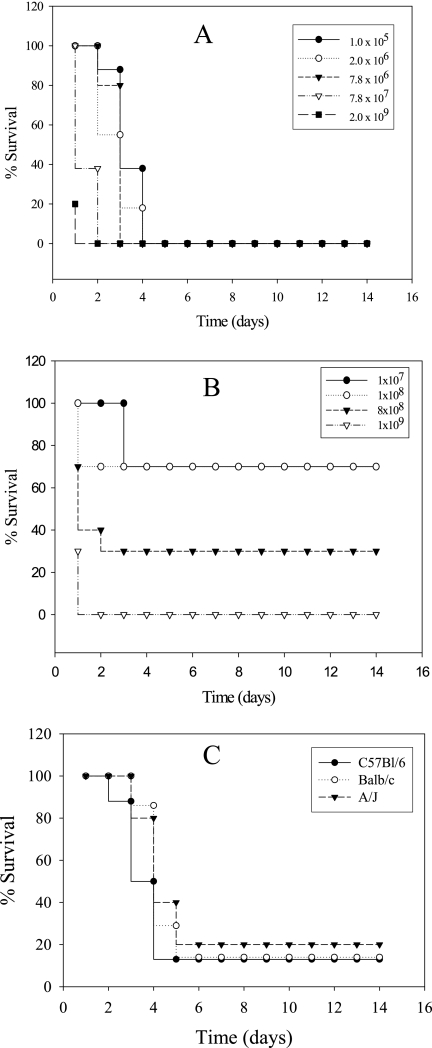
Whether L. longbeachae can replicate in mouse lungs in vivo is not known, and therefore, it would be useful to determine whether mice can be utilized as an animal model for the infection. We determined whether L. longbeachae caused pulmonary infection in the three inbred mouse strains. First, we performed a lethality study of A/J mice to determine the survival of mice after inoculation with L. longbeachae and L. pneumophila strain AA100. Different doses of the L. longbeachae clinical isolate D4968 and L. pneumophila were inoculated intratracheally into 10 A/J mice to determine the number of bacteria needed to establish a lethal infection. As few as 1 × 105 CFU of L. longbeachae were lethal to mice, and most of the mice (7/10) infected by 105 bacteria died by 5 days postinoculation (Fig. 2 A). Unlike infection of A/J mice by L. longbeachae, infection by 108 CFU of L. pneumophila caused only 30% lethality (Fig. 2B). We conclude that the L. longbeachae clinical isolate D4968 causes a more severe infection in A/J mice than L. pneumophila strain AA100.
FIG. 2.
Infection of A/J, C57BL/6, and BALB/c mice with L. longbeachae and L. pneumophila. (A) A/J mice were infected intratracheally with different doses of the L. longbeachae clinical isolate D4968 or (B) L. pneumophila. (C) A/J, C57BL/6, and BALB/c mice were infected with 1 × 105 CFU of L. longbeachae strain D4968, and lethality was monitored over 14 days.
Next, the lethalities of L. longbeachae and L. pneumophila to the three strains of mice (A/J, C57BL/6, and BALB/c) were determined by infecting mice with 105 CFU of the L. longbeachae clinical isolate D4968. The lethalities of L. longbeachae were similar for all mouse strains (Fig. 2C). In contrast, L. pneumophila was not lethal to BALB/c or C57BL/6 mice even when 109 CFU was inoculated, and all 10/10 animals survived (data not shown). As expected, L. pneumophila strain AA100 was lethal to A/J mice in a dose-dependent manner (Fig. 2B).
Intrapulmonary replication of L. longbeachae and L. pneumophila in inbred mouse strains.
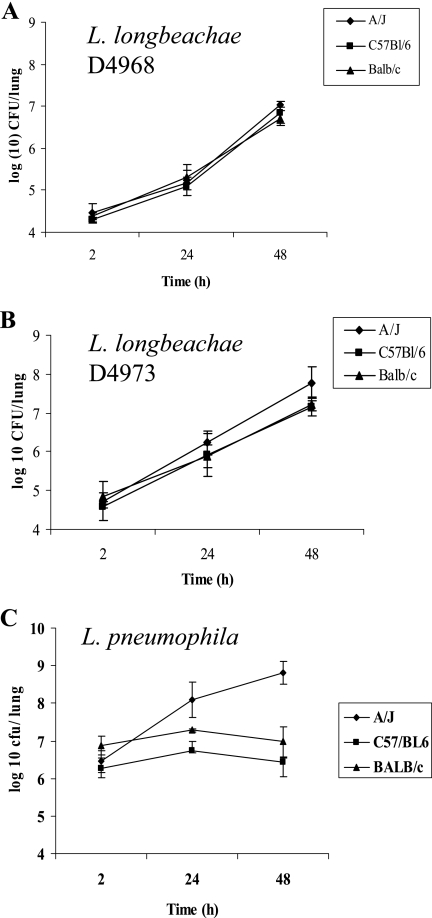
To determine that the lethality of mice was due to replication of L. longbeachae in the lungs and not to acute cytotoxicity, A/J mice were infected with three different doses (105 to 106 CFU) of the L. longbeachae clinical isolate D4968 and L. pneumophila and the number of CFU was determined in the lungs over a 48-h period. All doses of L. longbeachae caused a productive pulmonary infection in A/J mice, while an inoculum of 106 or more of L. pneumophila was essential for a productive infection (data not shown and see below). Next, A/J, C57BL/6, and BALB/c mice were inoculated with ∼105 CFU of the L. longbeachae clinical isolate D4968 and 106 of L. pneumophila and the number of CFU in the lungs was determined over a 48-h period. A total of 106 CFU of L. pneumophila was used, since this is the minimal dose that results in intrapulmonary proliferation compared to L. longbeachae. The results indicated that L. longbeachae replicated equally well in the lungs of all three strains of mice. There was an ∼1,000-fold increase in the number of CFU over the 48-h period in all strains of mice (Fig. 3 A). In contrast to L. longbeachae, L. pneumophila replicated only in A/J mice (Fig. 3C). Unlike growth in A/J mice, in which the CFU of L. pneumophila showed about a 1,000-fold increase over a 48-h period, the CFU in C57BL/6 and BALB/c mice increased only 2- to 4-fold between 2 and 24 h and dropped slightly by 48 h postinfection (Fig. 3C).
FIG. 3.
Inbred mouse strains are equally susceptible to infection by different strains of L. longbeachae. Intrapulmonary replication of L. longbeachae and L. pneumophila in A/J, C57BL/6, and BALB/c mice was determined. The three strains of mice (n = 12 each) were inoculated with (A) 1 × 105 CFU of the L. longbeachae clinical isolate D4968, (B) 1 × 105 CFU of the L. longbeachae clinical isolate D4973, and (C) 106 CFU of L. pneumophila. Lungs were harvested, and the number of CFU in the lungs was determined at the indicated time points. The results are representative of three independent experiments, and error bars represent standard deviations.
Susceptibility of inbred mouse strains to L. longbeachae is not strain dependent.
To confirm that the genetic susceptibility of the three mouse strains was not limited to the D4968 clinical isolate, infection by another clinical isolate, D4973, was examined. A/J, C57BL/6, and BALB/c mice were infected with ∼1 × 105 CFU of clinical isolate D4973, and lethality to mice was monitored over 14 days. Both strains were similarly lethal to all mouse strains (data not shown). In addition, replication of the L. longbeachae clinical isolate D4973 in the lungs of A/J, C57BL/6, and BALB/c mice was determined. The mouse strains were inoculated intratracheally with ∼1 × 105 bacteria, and bacterial replication was determined over a 48-h period. The clinical isolate D4973 replicated in all three strains of mice (Fig. 3B). Taken together, we conclude that in contrast to what was observed with L. pneumophila, polymorphism in the naip5 allele has no effect on the genetic susceptibility of mice to infection by L. longbeachae strains.
Expression of naip5 in C57BL/6 and A/J mice.
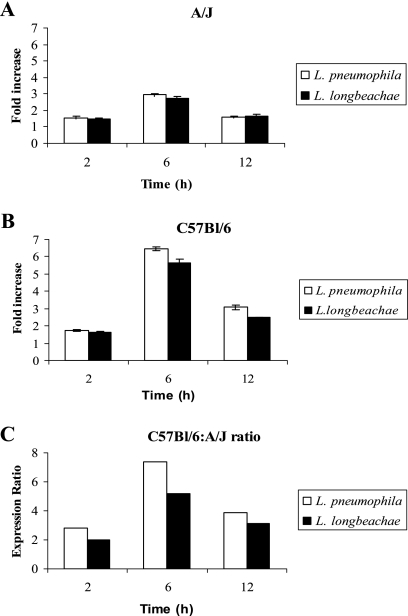
The collective expression of the naip family of eight genes as a group is induced after phagocytosis of L. pneumophila and Salmonella enterica serovar Typhimurium as well as latex beads, but whether naip5 is one of the induced eight naip genes is not known (14). Since naip5 is the only naip gene among the eight naip genes that confers susceptibility to L. pneumophila, we focused our analysis exclusively on naip5 expression. Bone marrow-derived macrophages from both mouse strains were infected with the L. longbeachae clinical isolate D4968 or L. pneumophila for 2, 6, and 12 h. The mRNA was isolated from infected A/J and C57BL/6 mouse macrophages, and the level of naip5 expression was determined by quantitative real-time PCR. We determined the ratio of expression of naip5 in infected macrophages to that in uninfected macrophages (Fig. 4A and B) as well as the ratio of infected C57BL/6 macrophages to infected A/J macrophages (Fig. 4C). The results showed that there was a higher level of expression of naip5 in C57BL/6 mouse macrophages than in A/J mouse macrophages (Fig. 4). At all the time points, there was a slightly higher C57BL/6-to-A/J ratio of naip5 expression in L. pneumophila-infected macrophages than in L. longbeachae-infected macrophages (P < 0.0071 to 0.014, t test). Consistent with that result, L. longbeachae triggered significantly, but modestly, less naip5 expression in C57BL/6 mice than L. pneumophila (P < 0.05, Student's t test).
FIG. 4.
High level of expression of naip5 in C57BL/6 macrophages compared to that in A/J mouse macrophages. Immortalized bone marrow-derived macrophages from A/J and C57BL/6 mice were infected with L. longbeachae or L. pneumophila at an MOI of 10, and total RNA was isolated from macrophages at 2, 6, and 12 h postinfection. The levels of naip5 expression in infected as well as uninfected cells were determined by quantitative reverse transcriptase PCR. Levels of expression of naip5 were determined relative to those in uninfected cells (A and B) and are also expressed as ratios of C57BL/6 to A/J mouse macrophages (C). The results are representative of three independent experiments, and the error bars represent standard deviations.
L. longbeachae expresses flagella but does not induce caspase 1 activation in mouse macrophages.
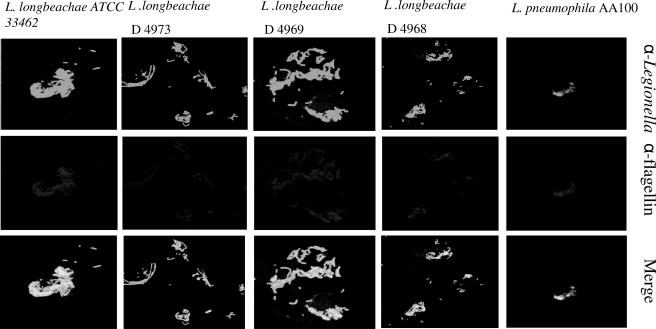
To determine whether the ability of L. longbeachae to grow in resistant mice was related to its failure to express flagella/and or to activate caspase 1, we determined whether the L. longbeachae strains expressed flagella at 36 h (post-exponential phase) after the infection of U937 macrophages. All the L. longbeachae strains expressed flagella at 36 h (Fig. 5). However, we have recently shown that all L. longbeachae strains did not express flagella during the exponential phase of growth (5).
FIG. 5.
Flagellar expression by L. longbeachae during the intracellular infection. Representative images of U937 human macrophages infected with the L. longbeachae clinical isolate D4968 and ATCC strain 33462 at 36 h postinfection. The expression of flagella was determined using anti (α)-flagellin antibody against flagellin and compared to that in L. pneumophila-infected cells. The results are representative of two independent experiments.
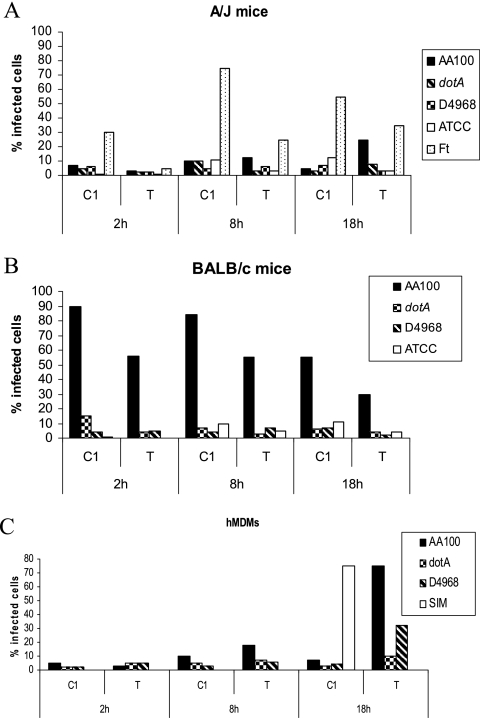
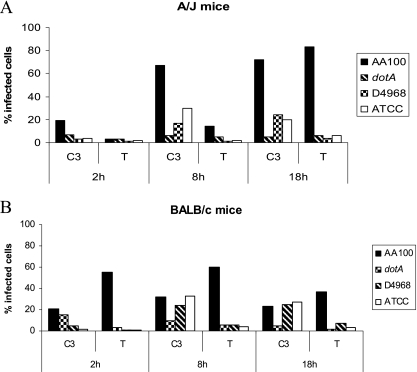
Whether L. longbeachae triggers caspase 1 activation within mouse macrophages has not been reported. Therefore, we determined the activation of caspase 1 by L. longbeachae strains D4968 and ATCC 33462 at different time intervals postinfection of A/J and BALB/c mouse macrophages. Our results showed that there is no activation of caspase 1 by either strain of L. longbeachae in A/J or BALB/c mice at all the time points determined (Fig. 6A and B). In contrast to L. longbeachae, the L. pneumophila control triggered caspase 1 activation in BALB/c mouse macrophages but not in A/J mouse macrophages (Fig. 6A and B) (36, 37, 45). The dotA mutant negative control did not activate caspase 1, while the Francisella tularensis subsp. novicida U112 strain positive control triggered caspase 1 activation (Fig. 6). Results similar to those obtained with BALB/c mouse macrophages were also obtained with C57BL/6 mouse macrophages (data not shown).
FIG. 6.
L. longbeachae does not induce caspase 1 activation in mBDMs or hMDMs. A/J and BALB/c mouse macrophages and hMDMs were infected with the L. longbeachae clinical isolate D4968 and ATCC 33462, and the activation of caspase 1 (C1) and apoptosis (TUNEL [T]) were determined at 2, 8, and 18 h postinfection. L. pneumophila and its isogenic dotA mutant were used as positive and negative controls, respectively, and simvastatin was used as a positive control for caspase 1 in human macrophages. Francisella tularensis (Ft) was used as a positive control for hMDMs. The data are representative of three independent experiments.
Caspase 1 is not triggered by either species within human macrophages.
We have recently shown that L. pneumophila does not trigger caspase 1 activation in hMDMs (M. Santic, R. Asare, M. Doric, and Y. Abu Kwaik, submitted for publication). Therefore, we determined whether L. longbeachae triggered caspase 1 activation in hMDMs. Our data showed that caspase 1 activation was not detected in hMDMs infected by L. longbeachae at all the time points examined up to 36 h postinfection (Fig. 6C and data not shown), similar to that found for the L. pneumophila control (Santic et al., submitted). The hMDMs treated with simvastatin, as a positive control, exhibited caspase 1 activation.
Failure to trigger caspase 1 activation by L. longbeachae within mouse macrophages correlates with the lack of detectable pore-forming activity.
Since the activation of caspase 1 by L. pneumophila requires pore-forming activity, which is detectable by contact-dependent hemolysis of sheep red blood cells (SRBCs) (28), we examined whether the three clinical isolates of L. longbeachae expressed pore-forming activity by using contact-dependent hemolysis of SRBCs (32, 33). The data showed that unlike L. pneumophila, none of the three clinical isolates of L. longbeachae possessed any detectable pore-forming activity, similar to the GN229 mutant negative control (Table 1) (32, 33). Therefore, the lack of detectable pore-forming activity by L. longbeachae correlates with the lack of its activation of caspase 1.
TABLE 1.
Contact-dependent hemolysis of SRBCs as an assay for pore-forming activitya
| Strain | Pore-forming activity (%) |
|---|---|
| AA100 | 100 |
| D4968 | 100 |
| D4969 | 18 |
| D4973 | 20 |
| GN229 | 17 |
| None | 21 |
See reference 28. The results are representative of five independent experiments.
L. longbeachae induces lower levels of caspase 3 activation in both human and murine macrophages.
We determined the ability of the three clinical isolates of L. longbeachae to activate caspase 3 and to trigger subsequent apoptosis in human macrophages. Our data showed that unlike L. pneumophila, all three strains of L. longbeachae triggered very low levels of caspase 3 activation (P < 0.0001, t test) (Fig. 7). Compared to the dotA mutant negative control, the L. pneumophila strain AA100 positive control showed a high level of caspase 3 activation (P < 0.0001, t test) (Fig. 7).
FIG. 7.
Detection of caspase 3 activation by its cleavage of a fluorescent substrate. (A) Human U937 macrophages were infected with the L. longbeachae clinical isolate D4968 and L. pneumophila, and the number of CFU was monitored over 48 h. (B) Human U937 macrophages were infected with the L. longbeachae clinical isolate D4968, D4969, or D4973, L. pneumophila, or its dotA isogenic mutant. The caspase 3 activity to cleave the fluorescent substrate was measured at 6 h postinfection and expressed as arbitrary fluorescence units (AFU). The data are representative of three independent experiments, and the error bars represent standard deviations.
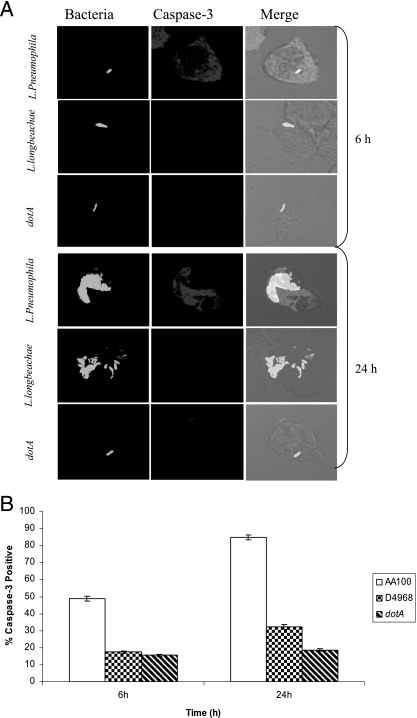
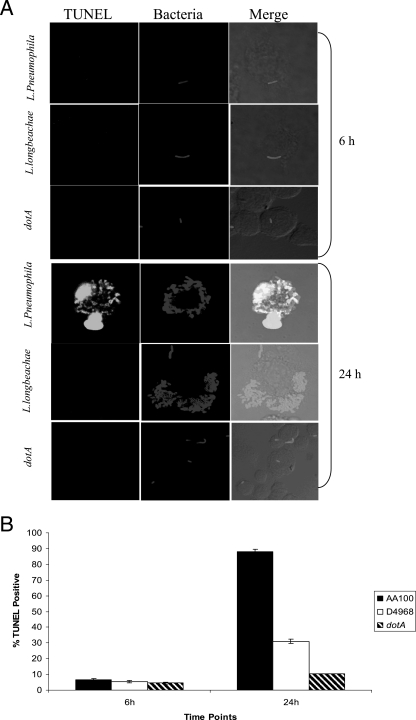
To confirm the low level of activation of caspase 3 by L. longbeachae, we performed single-cell analysis by confocal microscopy to determine the kinetics of caspase 3 activation at 6 h and 24 h postinfection of hMDMs. The L. pneumophila isogenic dotA mutant was used as the negative control. At 6 h postinfection, 50% of L. pneumophila-infected hMDMs showed caspase 3 activation (Fig. 8). In contrast, the level of caspase 3 activation by L. longbeachae-infected macrophages was not significantly different from that by dotA mutant-infected or uninfected macrophages (P > 0.1, t test) (Fig. 8). The number of infected macrophages that exhibited caspase 3 activation at 24 h was significantly less in cells infected with L. longbeachae than in cells infected with the L. pneumophila strain AA100 control (P < 0.0007, t test) (Fig. 8). This may be due to proliferation of L. longbeachae compared to the dotA mutant, which does not replicate. However, bacterial numbers alone could not account for the differential activation of caspase 3 in macrophages, since L. longbeachae and L. pneumophila replicated to the same level (Fig. 7A).
FIG. 8.
Single-cell analysis of caspase 3 activation in human macrophages. hMDMs were infected with the L. longbeachae clinical isolate D4968, L. pneumophila, or its dotA isogenic mutant at an MOI of 10, and caspase 3 activation was analyzed at 6 h and 24 h postinfection. Bacteria were detected by a specific antibody, while active caspase 3 was detected by an antiactive caspase 3 antibody. (A) Representative confocal microscopy images at several time points are shown. (B) Quantification of the percentage of cells with active caspase 3 was determined by analysis of 100 infected cells from three different coverslips in each experiment. There were equivalent numbers of bacteria per phagosome for both bacterial species. The data are representative of three independent experiments, and the error bars represent standard deviations.
We have recently shown that L. pneumophila triggers caspase 3 activation in bone marrow-derived macrophages in A/J mice (Santic et al., submitted). Therefore, we determined whether there was caspase 3 activation in murine macrophages infected by L. longbeachae. We infected bone marrow-derived macrophages from A/J and BALB/c mice with the L. longbeachae clinical isolate D4968 and ATCC strain 33462 and determined the activation of caspase 3 at 2, 8, and 18 h postinfection by single-cell analyses. We used L. pneumophila and its isogenic dotA mutant as our positive and negative controls, respectively. Our data showed that similar to what was observed with human macrophages, L. longbeachae showed low activation of caspase 3 in BALB/c and A/J murine macrophages for all the time points determined (Fig. 9). By 2 h postinfection, less than 5% of L. longbeachae-infected macrophages from both strains of mice exhibited caspase 3 activation, which was not significantly different from what was observed with uninfected cells (P > 0.2, t test). The percentage of caspase 3-positive infected cells increased by 8 to 18 h postinfection by the two L. longbeachae strains (Fig. 9), and no increase was detected by 24 to 36 h (data not shown). In contrast, the L. pneumophila positive control activated caspase 3 in ∼70% of infected A/J mouse macrophages by 8 h postinfection (Fig. 9) (Santic et al., submitted). Minimal activation of caspase 3 was exhibited by L. pneumophila in macrophages from BALB/c mice, similar to what was observed with L. longbeachae-infected BALB/c macrophages (Fig. 9). The dotA mutant control triggered caspase 3 activation in less than 5% of infected macrophages from both strains of mice at all the time points examined (Fig. 9). Staurosporin-treated control cells exhibited apoptosis in ∼85% of BALB/c macrophages (data not shown).
FIG. 9.
L. longbeachae triggers low caspase 3 activation in murine macrophages. A/J and BALB/c mouse macrophages were infected with the L. longbeachae clinical isolate D4968 and ATCC 33462, and levels of caspase 3 activation (C3) and apoptosis (TUNEL [T]) were determined at 2, 8, and 18 h postinfection. L. pneumophila and its isogenic dotA mutant were used as positive and negative controls, respectively. The data are representative of three experiments.
L. longbeachae induces lower levels of apoptosis in human but not murine macrophages.
We used a TUNEL assay to examine the ability of L. longbeachae to induce apoptosis. Human macrophages were infected by the L. longbeachae clinical isolate D4968 or L. pneumophila as a positive control (Santic et al., submitted) at an MOI of 10, and the induction of apoptosis was determined at 6 h and 24 h postinfection. There was no significant difference in apoptosis upon infection by either species at 6 h postinfection compared to that upon infection by the dotA mutant (P > 0.10 to 0.30, t test) (Fig. 10). At 24 h postinfection, ∼80% of the L. pneumophila-infected cells (positive control; Santic et al., submitted) became apoptotic compared to ∼25% of L. longbeachae-infected cells (P < 0.0005, t test). Only ∼5% of the cells infected by the dotA mutant negative control of L. pneumophila (47) became apoptotic by 24 h postinfection (Fig. 10), which was significantly less than the level of apoptosis induced by L. longbeachae (P < 0.02, t test) (Fig. 10).
FIG. 10.
L. longbeachae induces low levels of late-stage apoptosis in human macrophages. hMDMs were infected with L. longbeachae strain D4968, L. pneumophila, or its dotA isogenic mutant at an MOI of 10, and the level of apoptosis in infected cells was analyzed at 6 and 24 h postinfection. The bacteria were labeled with an antibody, while apoptotic nuclei were detected by TUNEL. Representative images at the indicated time points are shown in panel A. (B) Quantification of the percentage of apoptotic cells was determined by analysis of 100 infected cells from three different coverslips in each experiment. The data are representative of three independent experiments, and the error bars represent standard deviations.
Next, we determined whether L. longbeachae induced apoptosis in murine macrophages. Bone marrow-derived macrophages from A/J and BALB/c mice were infected with the L. longbeachae clinical isolate D4968 and the ATCC strain 33462 at an MOI of 10, and apoptosis was determined by single-cell analyses of TUNEL assays at 2, 8, and 18 h postinfection. The results showed that both strains of L. longbeachae triggered very low levels of apoptosis in either strain of mice at all time points examined (Fig. 9). In contrast, the wild-type L. pneumophila control triggered apoptosis in infected A/J macrophages only during late stages of infection (18 h), consistent with our recent observations (M. Santic et al., submitted for publication). Unlike A/J mouse macrophages, L. pneumophila-infected BALB/c macrophages showed high levels of apoptosis at 2 to 8 h postinfection, which is due to the robust activation of caspase 1 (Fig. 6B) (36, 45) that cannot be differentiated from caspase 3-mediated apoptosis by the TUNEL assays.
L. longbeachae induces low levels of pulmonary apoptosis in A/J mice.
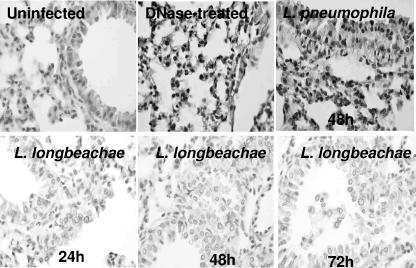
We have recently shown that L. pneumophila triggers pulmonary apoptosis in permissive A/J mice but not in nonpermissive C57BL/6 mice (Santic et al., submitted). We examined pulmonary apoptosis in A/J mice infected with L. longbeachae by in situ cell analyses using laser scanning confocal microscopy. L. pneumophila-infected mice were used as a positive control, sham-infected mice were used as a negative control, and DNase-treated sections were used as a control for the TUNEL assays. Pulmonary apoptosis was observed at 24 to 72 h after infection by L. pneumophila strain AA100 control in the permissive A/J mouse strain (Fig. 11 and data not shown). Unlike that in L. pneumophila-infected lung tissue, there was very a low level of apoptosis in L. longbeachae-infected lung tissue up to 7 days postinfection, consistent with our observations in vitro for mouse-derived macrophages (data not shown). Taken together, these results show that in contrast to L. pneumophila, L. longbeachae induces low levels of apoptosis in vitro and in the lungs of experimental animals.
FIG. 11.
L. longbeachae induces low levels of pulmonary apoptosis in A/J mice. A/J mice were infected with 106 CFU of L. longbeachae strain D4968 or L. pneumophila. Lungs from the infected mice were harvested at 24, 48, and 72 h and stained for TUNEL to detect apoptotic nuclei. Only the 48-h time point is shown for L. pneumophila because the results from all the time points are similar. The images are representative of 20 different microscopic fields from the lungs of three animals for each time point. The results are representative of three independent experiments.
DISCUSSION
All inbred mouse strains are resistant to L. pneumophila infection with the exception of A/J mice (31). Many other Legionella species can replicate within macrophages derived from other mouse strains such as the C57BL/6 mouse strain, including one L. longbeachae isolate from Japan (27), but whether L. longbeachae can replicate within pulmonary cells of mice and whether the mice can be utilized as an animal model for L. longbeachae have never been reported. In this report, we have shown efficient intracellular replication within various mouse macrophages in vitro and intrapulmonary replication of various L. longbeachae clinical isolates in A/J, C57BL/6, and BALB/c mice. Our results show that in contrast to that by L. pneumophila, susceptibility of mice to infection by L. longbeachae is independent of polymorphism in the naip5 allele in vivo and in mouse macrophage cell lines in vitro.
The differences in genetic susceptibilities of inbred mouse strains have been mapped to a polymorphism in the naip5 allele (13). The collective expression of the naip family of genes (approximately eight) as a group is induced after phagocytosis of L. pneumophila and Salmonella enterica serovar Typhimurium as well as latex beads, but whether naip5 is one of the induced naip genes is not known (14). Our current studies have focused exclusively on the expression of the naip5 gene, since it is the only gene in the naip family of eight genes that determines susceptibility to L. pneumophila (13, 44). Our analyses show that upon infection with L. pneumophila and L. longbeachae, there is an increase in the levels of naip5 expression in A/J and C57BL/6 macrophages. However, L. pneumophila-infected C57BL/6 macrophages express higher levels of naip5 than L. longbeachae-infected C57BL/6 macrophages. There are differences in 14 amino acids between Naip5 from C57BL/6 and A/J mice. Therefore, it is possible that structural differences and/or the expression level of Naip5 may contribute to the differential susceptibility to infection by L. pneumophila.
Differential susceptibilities to L. pneumophila in mouse macrophages are mediated by the early rapid triggering of caspase 1 by bacterial flagellin, resulting in early proinflammatory cell death mediated by pyropoptosis (36, 45). Our data show that although L. longbeachae expresses flagella, it does not activate caspase 1 and it does not trigger early rapid pyropoptosis in BALB/c or A/J mouse macrophages. The pore-forming activity of L. pneumophila is essential for the flagellin-mediated activation of caspase 1 (36, 45). Since pore-forming activity is not detectable for L. longbeachae, the flagellin of L. longbeachae may not have access to the macrophage cytoplasm, where it is thought to be involved in caspase 1 activation by L. pneumophila (36, 45). Alternatively, the FlaA of L. longbeachae is not recognized by the Naip5 systems or it does not activate caspase 1.
Activation of caspase 3 by L. pneumophila within human macrophages during early stages of infection is a Dot/Icm-dependent process (47) that is essential for the isolation of the phagosome from the endocytic pathway (18, 19, 21, 35). Interestingly, L. longbeachae does not block endocytic fusion and its phagosome matures to a late endosome-like phagosome (5). Thus, there is a correlation between the maturation of the L. longbeachae phagosome within the endocytic pathway in human macrophages and its inability to activate caspase 3, which is required for the degradation of the early endosome regulator rabaptin-5 upon infection by L. pneumophila (35). In contrast to its function in human macrophages, caspase 3 is not required for the intracellular proliferation of L. pneumophila within mouse macrophages (45). Although caspase 3 is activated early during infection of human macrophages by L. pneumophila, apoptosis is not triggered until ∼18 h postinfection (2, 4). The delay of apoptosis in L. pneumophila-infected macrophages despite robust activation of caspase 3, which is considered to be the executioner of apoptosis, is associated with a Dot/Icm-dependent potent trigger of multiple NF-κB-dependent and -independent antiapoptotic pathways (3, 30). Our results show that unlike L. pneumophila, L. longbeachae induces only a low level of apoptosis in human macrophages during late stages of infection. Similarly, the lungs of A/J mice infected with L. longbeachae exhibit a low level of apoptosis up to 72 h postinfection. The severities of Legionnaires' disease caused by L. pneumophila and L. longbeachae are not different despite the dramatic differences in the abilities of both species to trigger apoptosis in vitro and within the lungs of the mouse model. What role L. pneumophila-triggered apoptosis plays in the pathogenesis of Legionnaires' disease remains to be elucidated.
In summary, we have shown the feasibility of utilizing mice as a model system for L. longbeachae and we have shown that polymorphism of the naip5 allele does not play a role in the susceptibility of inbred mouse strains to L. longbeachae infection, which is quite in contrast to what was observed for infection by L. pneumophila. The induction of naip5 transcription in both L. pneumophila- and L. longbeachae-infected A/J mouse macrophages is less than that in C57BL/6 mice. Despite flagellation of L. longbeachae, this species does not trigger early and rapid activation of caspase 1, which correlates with the lack of detectable pore-forming activity by this species. We show that unlike what was observed with L. pneumophila, caspase 3 activation and late-stage apoptosis are triggered at low levels by L. longbeachae within human and mouse macrophages. However, neither species triggers caspase 1 activation within human macrophages.
Acknowledgments
Y.A.K. is supported by Public Health Service award R01AI065974 and the Commonwealth of Kentucky Research Challenge Trust Fund.
Editor: W. A. Petri, Jr.
Footnotes
Published ahead of print on 29 January 2007.
REFERENCES
- 1.Abu Kwaik, Y., B. I. Eisenstein, and N. C. Engleberg. 1993. Phenotypic modulation by Legionella pneumophila upon infection of macrophages. Infect. Immun. 61:1320-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abu-Zant, A., R. Asare, J. E. Graham, and Y. Abu Kwaik. 2006. Role for RpoS but not RelA of Legionella pneumophila in modulation of phagosome biogenesis and adaptation to the phagosomal microenvironment. Infect. Immun. 74:3021-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abu-Zant, A., S. Jones, R. Asare, J. Suttles, C. Price, J. Graham, and Y. A. Kwaik. 2007. Anti-apoptotic signalling by the Dot/Icm secretion system of L. pneumophila. Cell. Microbiol. 9:246-264. [DOI] [PubMed] [Google Scholar]
- 4.Abu-Zant, A., M. Santic, M. Molmeret, S. Jones, J. Helbig, and Y. Abu Kwaik. 2005. Incomplete activation of macrophage apoptosis during intracellular replication of Legionella pneumophila. Infect. Immun. 73:5339-5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asare, R., and Y. Abu Kwaik. 2007.. Early trafficking and intracellular replication of Legionella longbeachae within an ER-derived late endosome-like phagosome. Cell. Microbiol. [Online.] doi: 10.1111/j.1462-5822.2007.00894.x. [DOI] [PubMed]
- 6.Benin, A. L., R. F. Benson, and R. E. Besser. 2002. Trends in legionnaires disease, 1980-1998: declining mortality and new patterns of diagnosis. Clin. Infect. Dis. 35:1039-1046. [DOI] [PubMed] [Google Scholar]
- 7.Blasi, E., B. J. Mathieson, L. Varesio, J. L. Cleveland, P. A. Borchert, and U. R. Rapp. 1985. Selective immortalization of murine macrophages from fresh bone marrow by a raf/myc recombinant murine retrovirus. Nature 318:667-670. [DOI] [PubMed] [Google Scholar]
- 8.Brieland, J., P. Freeman, R. Kunkel, C. Chrisp, M. Hurley, J. Fantone, and N. C. Engleberg. 1994. Replicative Legionella pneumophila lung infection in intratracheally inoculated A/J mice. A murine model of human Legionnaires' disease. Am. J. Pathol. 145:1537-1546. [PMC free article] [PubMed] [Google Scholar]
- 9.Brieland, J. K., D. G. Remick, P. T. Freeman, M. C. Hurley, J. C. Fantone, and N. C. Engleberg. 1995. In vivo regulation of replicative Legionella pneumophila lung infection by endogenous tumor necrosis factor alpha and nitric oxide. Infect. Immun. 63:3253-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clemens, D. L., and M. A. Horwitz. 1995. Characterization of the Mycobacterium tuberculosis phagosome and evidence that phagolysosomal maturation is inhibited. J. Exp. Med. 181:257-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clemons-Miller, A. R., G. W. Cox, J. Suttles, and R. D. Stout. 2000. LPS stimulation of TNF-receptor deficient macrophages: a differential role for TNF-alpha autocrine signaling in the induction of cytokine and nitric oxide production. Immunobiology 202:477-492. [DOI] [PubMed] [Google Scholar]
- 12.Coward, W. R., A. Marei, A. Yang, M. M. Vasa-Nicotera, and S. C. Chow. 2006. Statin-induced proinflammatory response in mitogen-activated peripheral blood mononuclear cells through the activation of caspase-1 and IL-18 secretion in monocytes. J. Immunol. 176:5284-5292. [DOI] [PubMed] [Google Scholar]
- 13.Diez, E., S. H. Lee, S. Gauthier, Z. Yaraghi, M. Tremblay, S. Vidal, and P. Gros. 2003. Birc1e is the gene within the Lgn1 locus associated with resistance to Legionella pneumophila. Nat. Genet. 33:55-60. [DOI] [PubMed] [Google Scholar]
- 14.Diez, E., Z. Yaraghi, A. MacKenzie, and P. Gros. 2000. The neuronal apoptosis inhibitory protein (Naip) is expressed in macrophages and is modulated after phagocytosis and during intracellular infection with Legionella pneumophila. J. Immunol. 164:1470-1477. [DOI] [PubMed] [Google Scholar]
- 15.Doyle, R. M., T. W. Steele, A. M. McLennan, I. H. Parkinson, P. A. Manning, and M. W. Heuzenroeder. 1998. Sequence analysis of the mip gene of the soilborne pathogen Legionella longbeachae. Infect. Immun. 66:1492-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feeley, J. C., R. J. Gibson, G. W. Gorman, N. C. Langford, J. K. Rasheed, D. C. Mackel, and W. B. Baine. 1979. Charcoal-yeast extract agar: primary isolation medium for Legionella pneumophila. J. Clin. Microbiol. 10:437-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fortier, A., E. Diez, and P. Gros. 2005. Naip5/Birc1e and susceptibility to Legionella pneumophila. Trends Microbiol. 13:328-335. [DOI] [PubMed] [Google Scholar]
- 18.Gao, L.-Y., and Y. Abu Kwaik. 1999. Activation of caspase 3 in Legionella pneumophila-induced apoptosis in macrophages. Infect. Immun. 67:4886-4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao, L.-Y., and Y. Abu Kwaik. 1999. Apoptosis in macrophages and alveolar epithelial cells during early stages of infection by Legionella pneumophila and its role in cytopathogenicity. Infect. Immun. 67:862-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao, L.-Y., and Y. Abu Kwaik. 2000. Hijacking the apoptotic pathways of the host cell by bacterial pathogens. Microbes Infect. 2:1705-1719. [DOI] [PubMed] [Google Scholar]
- 21.Gao, L.-Y., and Y. Abu Kwaik. 2000. The modulation of host cell apoptosis by intracellular bacterial pathogens. Trends Microbiol. 8:306-313. [DOI] [PubMed] [Google Scholar]
- 22.Gao, L.-Y., O. S. Harb, and Y. Abu Kwaik. 1997. Utilization of similar mechanisms by Legionella pneumophila to parasitize two evolutionarily distant hosts, mammalian and protozoan cells. Infect. Immun. 65:4738-4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao, L.-Y., B. J. Stone, J. K. Brieland, and Y. Abu Kwaik. 1998. Different fates of Legionella pneumophila pmi and mil mutants within human-derived macrophages and alveolar epithelial cells. Microb. Pathog. 25:291-306. [DOI] [PubMed] [Google Scholar]
- 24.Garcia, M. T., S. Jones, C. Pelaz, R. D. Millar, and Y. Abu Kwaik. 2007.. Acanthamoeba polyphaga resuscitates viable non-culturable Legionella pneumophila after disinfection. Environ. Microbiol. [Online.] doi: 10.1111/j.1462-2920.2007.01245.x. [DOI] [PubMed]
- 25.Growney, J. D., J. M. Scharf, L. M. Kunkel, and W. F. Dietrich. 2000. Evolutionary divergence of the mouse and human Lgn1/SMA repeat structures. Genomics 64:62-81. [DOI] [PubMed] [Google Scholar]
- 26.Horwitz, M. A. 1983. The Legionnaires' disease bacterium (Legionella pneumophila) inhibits phagosome-lysosome fusion in human monocytes. J. Exp. Med. 158:2108-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Izu, K., S. Yoshida, H. Miyamoto, B. Chang, M. Ogawa, H. Yamamoto, Y. Goto, and H. Taniguchi. 1999. Grouping of 20 reference strains of Legionella species by the growth ability within mouse and guinea pig macrophages. FEMS Immunol. Med. Microbiol. 26:61-68. [DOI] [PubMed] [Google Scholar]
- 28.Kirby, J. E., J. P. Vogel, H. L. Andrews, and R. R. Isberg. 1998. Evidence for pore-forming ability by Legionella pneumophila. Mol. Microbiol. 27:323-336. [DOI] [PubMed] [Google Scholar]
- 29.Koide, M., A. Saito, M. Okazaki, B. Umeda, and R. F. Benson. 1999. Isolation of Legionella longbeachae serogroup 1 from potting soils in Japan. Clin. Infect. Dis. 29:943-944. [DOI] [PubMed] [Google Scholar]
- 30.Losick, V. P., and R. R. Isberg. 2006. NF-kappaB translocation prevents host cell death after low-dose challenge by Legionella pneumophila. J. Exp. Med. 203:2177-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miyamoto, H., K. Maruta, M. Ogawa, M.-C. Beckers, P. Gros, and S.-I. Yoshida. 1996. Spectrum of Legionella species whose intracellular multiplication in murine macrophages is genetically controlled by Lgn1. Infect. Immun. 64:1842-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Molmeret, M., O. A. T. Alli, M. Radulic, M. Susa, M. Doric, and Y. Abu Kwaik. 2002. The C-terminus of IcmT is essential for pore formation and for intracellular trafficking of Legionella pneumophila within Acanthamoeba polyphaga. Mol. Microbiol. 43:1139-1150. [DOI] [PubMed] [Google Scholar]
- 33.Molmeret, M., O. A. T. Alli, S. Zink, A. Flieger, N. P. Cianciotto, and Y. Abu Kwaik. 2002. icmT is essential for pore formation-mediated egress of Legionella pneumophila from mammalian and protozoan cells. Infect. Immun. 70:69-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Molmeret, M., D. M. Bitar, L. Han, and Y. A. Kwaik. 2004. Cell biology of the intracellular infection by Legionella pneumophila. Microbes Infect. 6:129-139. [DOI] [PubMed] [Google Scholar]
- 35.Molmeret, M., S. D. Zink, L. Han, A. Abu-Zant, R. Asari, D. M. Bitar, and Y. Abu Kwaik. 2004. Activation of caspase-3 by the Dot/Icm virulence system is essential for arrested biogenesis of the Legionella-containing phagosome. Cell. Microbiol. 6:33-48. [DOI] [PubMed] [Google Scholar]
- 36.Molofsky, A. B., B. G. Byrne, N. N. Whitfield, C. A. Madigan, E. T. Fuse, K. Tateda, and M. S. Swanson. 2006. Cytosolic recognition of flagellin by mouse macrophages restricts Legionella pneumophila infection. J. Exp. Med. 203:1093-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ren, T., D. S. Zamboni, C. R. Roy, W. F. Dietrich, and R. E. Vance. 2006. Flagellin-deficient Legionella mutants evade caspase-1- and Naip5-mediated macrophage immunity. PLoS Pathogens 2:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Santic, M., M. Molmeret, and Y. Abu Kwaik. 2005. Modulation of biogenesis of the Francisella tularensis subsp. novicida-containing phagosome in quiescent human macrophages and its maturation into a phagolysosome upon activation by IFN-gamma. Cell. Microbiol. 7:957-967. [DOI] [PubMed] [Google Scholar]
- 39.Santic, M., M. Molmeret, K. E. Klose, S. Jones, and Y. A. Kwaik. 2005. The Francisella tularensis pathogenicity island protein IglC and its regulator MglA are essential for modulating phagosome biogenesis and subsequent bacterial escape into the cytoplasm. Cell. Microbiol. 7:969-979. [DOI] [PubMed] [Google Scholar]
- 40.Uren, A. G., M. Pakusch, C. J. Hawkins, K. L. Puls, and D. L. Vaux. 1996. Cloning and expression of apoptosis inhibitory protein homologs that function to inhibit apoptosis and/or bind tumor necrosis factor receptor-associated factors. Proc. Natl. Acad. Sci. USA 93:4974-4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vogel, J. P., H. L. Andrews, S. K. Wong, and R. R. Isberg. 1998. Conjugative transfer by the virulence system of Legionella pneumophila. Science 279:873-876. [DOI] [PubMed] [Google Scholar]
- 42.Welsh, C. T., J. T. Summersgill, and R. D. Miller. 2004. Increases in c-Jun N-terminal kinase/stress-activated protein kinase and p38 activity in monocyte-derived macrophages following the uptake of Legionella pneumophila. Infect. Immun. 72:1512-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wiater, L. A., K. Dunn, F. R. Maxfield, and H. A. Shuman. 1998. Early events in phagosome establishment are required for intracellular survival of Legionella pneumophila. Infect. Immun. 66:4450-4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wright, E. K., S. A. Goodart, J. D. Growney, V. Hadinoto, M. G. Endrizzi, E. M. Long, K. Sadigh, A. L. Abney, I. Bernstein-Hanley, and W. F. Dietrich. 2003. Naip5 affects host susceptibility to the intracellular pathogen Legionella pneumophila. Curr. Biol. 13:27-36. [DOI] [PubMed] [Google Scholar]
- 45.Zamboni, D. S., K. S. Kobayashi, T. Kohlsdorf, Y. Ogura, E. M. Long, R. E. Vance, K. Kuida, S. Mariathasan, V. M. Dixit, R. A. Flavell, W. F. Dietrich, and C. R. Roy. 2006. The Birc1e cytosolic pattern-recognition receptor contributes to the detection and control of Legionella pneumophila infection. Nat. Immunol. 7:318-325. [DOI] [PubMed] [Google Scholar]
- 46.Zamboni, D. S., R. A. Mortara, E. Freymuller, and M. Rabinovitch. 2002. Mouse resident peritoneal macrophages partially control in vitro infection with Coxiella burnetii phase II. Microbes Infect. 4:591-598. [DOI] [PubMed] [Google Scholar]
- 47.Zink, S. D., L. Pedersen, N. P. Cianciotto, and Y. Abu Kwaik. 2002. The Dot/Icm type IV secretion system of Legionella pneumophila is essential for the induction of apoptosis in human macrophages. Infect. Immun. 70:1657-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]