Abstract
We have developed array Comparative Genomic Hybridization for Caenorhabditis elegans as a means of screening for novel induced deletions in this organism. We designed three microarrays consisting of overlapping 50-mer probes to annotated exons and micro-RNAs, the first with probes to chromosomes X and II, the second with probes to chromosome II alone, and a third to the entire genome. These arrays were used to reliably detect both a large (50 kb) multigene deletion and a small (1 kb) single-gene deletion in homozygous and heterozygous samples. In one case, a deletion breakpoint was resolved to fewer than 50 bp. In an experiment designed to identify new mutations we used the X:II and II arrays to detect deletions associated with lethal mutants on chromosome II. One is an 8-kb deletion targeting the ast-1 gene on chromosome II and another is a 141-bp deletion in the gene C06A8.1. Others span large sections of the chromosome, up to >750 kb. As a further application of array Comparative Genomic Hybridization in C. elegans we used the whole-genome array to detect the extensive natural gene content variation (almost 2%) between the N2 Bristol strain and the strain CB4856, a strain isolated in Hawaii and JU258, a strain isolated in Madeira.
Comparative Genomic Hybridization (CGH) allows the detection of copy number differences between two DNA samples (Kallioniemi et al. 1992; Mantripragada et al. 2004). The two DNA samples, one a reference and the other a test sample, are differentially labeled and hybridized to a representative genome arrayed on a matrix. Over the past several years a number of different array platforms have been utilized for CGH from bacteria artificial chromosomes (BACs) and cosmids to cDNA clones and oligonucleotides (Solinas-Toldo et al. 1997; Pinkel et al. 1998; Mantripragada et al. 2004). As the ability to detect small alterations is limited by the spacing and size of the probes on the matrix, there has been a move away from BAC clones to oligonucleotide arrays for experiments where high resolution is required (Carvalho et al. 2004; Ishkanian et al. 2004; Sebat et al. 2004; Selzer et al. 2005). For example, oligonucleotide array-based CGH (oaCGH) was recently used to measure copy number variation at specific exons in several human genes with a resolution between 50 and 500 bp (Dhami et al. 2005; Selzer et al. 2005).
We were interested in determining whether oaCGH could be used to detect copy number alterations (insertions and deletions, or “indels”) among different DNA samples of the nematode Caenorhabditis elegans. Specifically, we wished to determine whether CGH has the required sensitivity and resolving power to detect single-gene knockouts, where the deletions may be small and the animals may be heterozygous. Our laboratory is a member of the C. elegans Knockout Consortium (http://celeganskoconsortium.omrf.org/) and we are interested in examining techniques that might help us identify and clone single-gene knockouts more efficiently. Array CGH, if efficient, has a number of potential advantages over our current PCR-based method (Barstead 1999) of screening for deletions, including the ability to screen thousands of genes in a single experiment, no constraint on the maximum detectable deletion size, and identification of copy number alterations at other loci in the mutant genome. As an example, the ability to detect large deletions would be useful for screening for tandem gene family knockouts, such as the Serpentine Receptor class AB (srab) family of seven-transmembrane chemoreceptors and integral membrane proteins, where consecutive genes may share functional redundancies (Chen et al. 2005; Thomas 2006b).
We have designed three exon-tiled oligonucleotide arrays, one for two chromosomes (II and X), one for a single chromosome (II), and one for the entire genome. Using these arrays, we have detected both previously characterized deletions of 1–50 kb in control experiments and new deletion alleles of genes with no known mutations. The sensitivity of CGH is such that we can detect small deletions even in heterozygous animals. The ability to detect single-copy single-gene deletions at this resolution will allow us to use oaCGH to screen for novel induced deletions in mutagenized populations. This will greatly aid our efforts to generate knockout strains for the research community. The resolution of oaCGH may also make it an attractive tool for those studying the population biology and evolution of C. elegans. The large number of indel differences observed among the Bristol, Hawaiian, and Madeiran nematode strains points to the dynamic nature of genomes and the flux of many of the gene families within this organism.
Results
Oligonucleotide probe quality and detection of homozygous 50-kb and 1-kb deletions
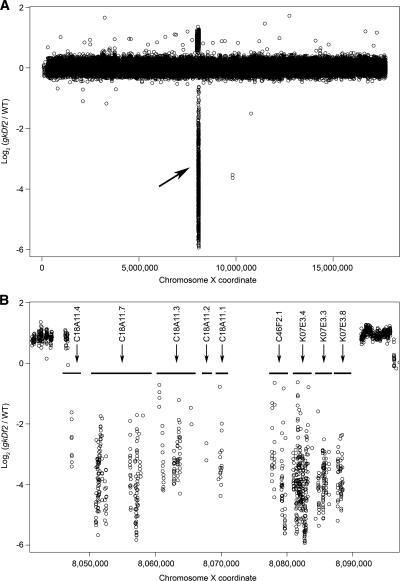
We designed a pilot microarray composed of a tiled set of oligonucleotide probes to nearly 90% of the exons and 94% of the genes on chromosomes X and II. This set revealed a remarkable consistency in signal-to-noise ratios over all of the experiments. Our initial oaCGH experiment was designed to determine whether a large (50 kb) homozygous deletion could be distinguished reliably from wild-type DNA. For this experiment we used gkDf2, a homozygous-viable deletion of the dim-1 locus on chromosome X. PCR analysis indicated that the deletion breakpoints lay between 8,046,205 and 8,046,422 on the left and 8,088,676 and 8,108,916 on the right, a physical interval of ∼50 kb that potentially included up to 12 genes. For this large deletion experiment, fluorescence intensities were collected for all probes, and we calculated log2 fluorescence intensity ratios for the mutant test sample with the reference wild-type sample (gkDf2/WT). After normalizing with a LOESS regression, the average log2 intensity ratios for the probe pairs gave a SD of 0.13 with very few outliers (see Fig. 1A). Note that these few outliers are from a plot of >92,000 forward and reverse complement probe pairs. The gkDf2 deletion was identified unambiguously in this plot as a prominent peak of negative log2 ratios for probe pairs targeting the chromosome X region around dim-1. An enlarged view of this region is shown in Figure 1B, showing that nine genes are affected by this deletion. The deletion breakpoints are clearly defined at the resolution of individual exons. These results indicated that we could certainly identify deletions smaller than 50 kb. Interestingly, probes adjacent to the breakpoints exhibit a positive log2 ratio (Fig. 1B), possibly indicating previously unknown duplications of the flanking sequences, which have been periodically observed for deletions caused by this type of mutagenesis (data not shown).
Figure 1.
Detection of a 50-kb homozygous viable deletion in gkDf2. (A) Normalized log2 ratios (gkDf2/WT) of the average fluorescent intensities for each of the 92,209 forward and reverse pairs of probes to the X chromosome are represented by circles. The deletion is identified by negative log2 ratios and indicated by an arrow. (B) A higher resolution view of fluorescence ratios for probe pairs targeting the 50-kb deletion. Horizontal bars indicate the positions of the nine genes targeted by the deletion. Duplications of sequences flanking the deletion are indicated by positive log2 ratios. Adjacent 50-mer probes in this region overlap by as much as 44 bp.
Using the same X:II array design, we next examined whether oaCGH could detect a smaller homozygous deletion elsewhere on the X chromosome. The mutation gk329 is a 1047-bp deletion in the gene ceh-39. A hybridization plot comparing gk329 with wild-type DNA (Fig. 2A) showed a deleted region in the chromosome X region around ceh-39, the site of the gk329 deletion. This region is enlarged in Figure 2B and aligned with a diagram of the coding regions for ceh-41, ceh-21, and ceh-39. The 30 probe pairs representing exons 1, 2, and 3 of ceh-39 (T26C11.7) showed strong negative fluorescence log2 ratios, while the nine probe pairs representing exon 4 of ceh-39 and the probe pairs targeting the five nearest exons of ceh-21 (T26C11.6) yielded lower amplitudes, but still statistically significant non-zero log2 ratios (with P-values of 3 × 10−8 and 7 × 10−5, respectively).
Figure 2.
Detection of a 1047-bp homozygous viable deletion in ceh-39 (gk329). (A) Normalized log2 ratios (gk329/WT) for the average fluorescence intensities for all probe pairs to the X chromosome are shown. The arrow indicates the deletion. (B) Intensity ratios for probes to ceh-39, ceh-21, and ceh-41 are shown with WormBase gene models to illustrate probe coverage in exons near the deletion. Sequenced deletion breakpoints are indicated by dotted lines. CGH accurately identified the left breakpoint between exons 3 and 4 of ceh-39.
Probe pairs targeting ceh-41 (T26C11.5) had log2 ratios closer to zero. The negative ratios for probes targeting exons 1–3 of gk329 corresponded exactly with deletion breakpoints determined by DNA sequencing (chromosome X coordinates 1,854,827/1,855,875). The next gene to the right of the deletion, T26C11.t1 (encoding a tRNA-Glu), was not represented among the probes on the array. Fluorescence ratios for probes to the next closest gene, tbx-41 (T26C11.1), lying 9365 bp beyond the distal deletion breakpoint, showed no evidence of reduced signal intensity in the gk329 sample (data not shown). From this experiment it was clear that the X:II chip design permits detection of deletion breakpoints at the resolution of individual exons.
Detection of single-copy number differences between hermaphrodite and male X chromosomes and in a balanced chromosome II deficiency
Broader application of oaCGH in C. elegans research and other model systems would be feasible if its sensitivity extended to detecting heterozygous (single-copy) deletions. We performed two experiments to determine whether log2 fluorescence ratios from a heterozygote are sufficient to give unambiguous identification of deletions. In the first experiment we compared the hybridization signal from wild-type C. elegans hermaphrodite DNA (two X chromosomes) to male DNA (one X chromosome) for all probes on chromosomes II and X (Fig. 3). The median log2 fluorescence ratios for probe pairs to chromosome II (which should be equally represented in the two samples) was set to zero, and these exhibited a SD of 0.23. Setting the log2 ratios for chromosome II to zero led to a median log2 ratio for forward and reverse complement probe pairs to chromosome X of −0.82, with a SD of 0.22. These distributions for chromosomes II and X overlapped by only 4% (Fig. 3).
Figure 3.
Comparison of the normalized average fluorescence ratios (XO male/XX hermaphrodite) for all probe pairs to chromosomes II and X. The graph in the top right corner plots the probe density versus the log2 fluorescence ratio for probes to chromosomes II and X. The curve peaking on the left is for the X chromosome and the curve peaking on the right is for chromosome II. The distributions overlap by ∼4%.
In a second experiment we compared wild-type DNA with that from a heterozygous 1202-bp deletion on chromosome II, using a balanced strain of genotype dab-1(gk291)/mIn1[mIs14 dpy-10(e128)]. Heterozygous animals are wild type with a pharyngeal green fluorescent protein (GFP) signal conferred by the mIn1 balancer chromosome, and they segregate ∼50% heterozygotes, 25% gk291 homozygotes (viable and fertile but slow-growing), and 25% mIn1 homozygotes (viable and fertile Dpy with small broods and a strong pharyngeal GFP signal). Initially, we compared the hybridization signal from wild-type DNA to that from DNA made from confirmed gk291/mIn1 heterozygotes. A separate hybridization compared the wild-type signal with that from DNA made from a population containing all progeny genotypes in their normal proportions. To obtain this latter sample we simply washed animals off a plate and isolated DNA from the mixed population of animals. The data plots from these two hybridizations were virtually indistinguishable, and both yielded reliable detection of the gk291 deletion (P = 4 × 10−13 [data not shown] and P = 8 × 10−14 [Fig. 4], respectively). These experiments demonstrate that single-copy deletions within a single gene can be reliably detected using oaCGH.
Figure 4.
Detection of the 1202-bp deletion in dab-1 (gk291) in a wash-sampled balanced heterozygous population. (A) The normalized log2 ratios [(dab-1(-)/mIn1)/WT] of the average fluorescence intensities for probe pairs to chromosome II are plotted. The arrow indicates the dab-1 deletion (other features are discussed in the text). (B) Normalized fluorescence ratios for probe pairs targeting dab-1 are shown. Sequenced deletion breakpoints are indicated by dotted lines and were accurately predicted by CGH. The left breakpoint lies within the second intron. Overlapping probes targeting the right breakpoint span just 73 bp, allowing resolution of the right breakpoint to within fewer than 50 bp.
Figure 4B shows a fluorescence ratio plot for probe pairs to the dab-1 locus aligned with a diagram of the dab-1 gene model from WormBase WS120. The sequenced deletion breakpoints lie at chromosome II coordinates 8,226,388 and 8,226,391, and agree perfectly with breakpoints predicted by the log2 fluorescence ratios. The log2 fluorescence ratios for probes to the deleted region were similar to those observed for probes to the X chromosome in the male/hermaphrodite experiment. The proximal deletion breakpoint is within an intron, while the distal deletion breakpoint is within an exon. The oligo probes around these breakpoints serve to illustrate the high resolution of oaCGH. Since we targeted all oaCGH probes to exons rather than using a tiling-path approach, the resolution of the proximal breakpoint was only about 400 nucleotides due to the first intron causing a 400-plus gap between adjacent oligos in that region. However, the distal deletion breakpoint, which lies within an exon, was more accurately resolved since it is targeted by two overlapping oligos (Fig. 4B). Together, these two probes span just 73 base pairs, thus resolving the distal deletion breakpoint to <50 nucleotides. In this experiment, probes flanking the deletion on either side yielded significant positive log2 fluorescence ratios.
The mIn1 balancer chromosome
Figure 4A reveals several copy number alterations in addition to the gk291 deletion, including a deletion (roughly chromosome II coordinates 1,020,000–1,050,000) and four amplifications (roughly chromosome II coordinates 1,561,000–1,567,000, 11,698,000–11,701,000, 12,847,000–12,848,000, and 13,482,000– 13,490,000). Since these experiments were the first to include the mIn1 balancer chromosome, we speculated that these additional features might represent deletions or amplifications on the balancer chromosome, or elsewhere in the balancer strain genome. Formally, the additional chromosome features could be linked to the gk291-bearing chromosome, the balancer, or distributed between them or other chromosomes (this latter possibility applies to amplifications only). We suspected that these were features of mIn1, as the construction of this balancer chromosome required two rounds of mutagenesis (Edgley and Riddle 2001). Comparison of N2 DNA with DNA from mIn1 homozygotes showed that all of the additional features observed in the dab-1/mIn1 heterozygote (Fig. 4A) were indeed derived from the mIn1 strain (data not shown). These alterations included the deletion on the left arm plus the various amplified regions throughout the chromosome. We can only be certain that the deletion involves the mIn1 chromosome II, since the amplifications of chromosome II sequences in the mIn1 strain do not necessarily reside on chromosome II.
Novel balanced lethal deletion on chromosome II
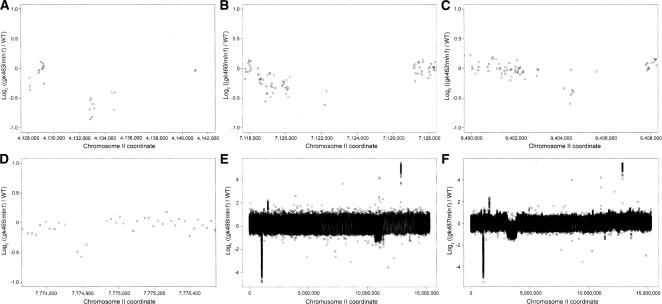
To demonstrate that oaCGH can be used to reliably detect novel deletions, we conducted a screen for lethal mutations on chromosome II balanced by the mIn1 inversion, using TMP/UV as a mutagen. We mutagenized a predominantly L4 population of DR2078 [bli-2(e768) unc-4(e120)/mIn1[mIs14 dpy-10(e128)]], set up clonal populations from the F1 progeny, and screened the F2 for absence of viable, fertile Bli-2 Unc-4 adults. Such lines indicated the presence of new recessive lethal mutations linked to bli-2 unc-4 and balanced by mIn1. Approximately 200 balanced lethal lines were obtained. We analyzed 30 of these strains by array CGH, using DNA prepared from mixed populations washed off plates, and detected 25 new deletions (0–3 deletions per strain). We describe six of these new deletions here (see Fig. 5). For one of the candidates, gk463, PCR using primers to the regions flanking the deletion confirmed the presence of an 8-kb deletion; sequencing this PCR product demonstrated that gk463 deletes 8063 bp between chromosome II coordinates 4,131,236/4,139,298 encoding the ast-1 (T08H4.3) gene (Fig. 5A). Similar experiments confirmed three other deletions. The gk460 lesion is 4.5-kb deletion encompassing the genes F10E7.4 (spon-1), F10E7.11, and F10E7.2 (breakpoints at chromosome II coordinates 7,118,909 and 7,123,417; Fig. 5B); gk462 is a 2.2-kb deletion encompassing Y51B9A.5 and an internal tRNA (Fig. 5C); and gk465 is a 141-bp deletion affecting the gene C06A8.1 (breakpoints at chromosome II coordinates 7,774,796 and 7,774,938; Fig. 5D).
Figure 5.
Deletions detected in a screen for homozygous lethal mutations in six wash-sampled balanced heterozygous populations. The following normalized log2 ratios are shown: (A) (gk463/mIn1)/WT; (B) (gk460/mIn1)/WT; (C) (gk462/mIn1)/WT; (D) (gk465/mIn1)/WT; (E) (gk488/mIn1)/WT; and (F) (gk487/mIn1)/WT.
In addition to these single-gene deletions, we identified several larger deletions spanning several genes. The gk488 deletion is nearly 500 kb in size, spanning chromosome II coordinates 10,662,230 to 11,160,425, affecting 93 genes (Fig. 5E). An even larger deletion affecting 274 genes is identified in gk487. The deletion spans chromosome II coordinates 3057,725 through 3841,090, completely deleting over 783 kb with the exception of ∼4.5 kb (from chromosome II coordinates 3,131,948 through 3,136,511) (Fig. 5F). From these experiments, we conclude that oaCGH is a powerful and efficient method for discovery of knockout mutations and for characterizing large deletions in this organism.
Whole-genome array CGH: Comparing N2 Bristol to Hawaiian and Madeiran wild isolates
The amount of gene content variability within natural populations of animal species is largely unknown. High-resolution array CGH appears to be an excellent method for exploring this variability. To examine this variability we designed a whole-genome oligonucleotide CGH array to detect alterations among wild-type nematode strains. This array targets the entire C. elegans genome, with 94% coverage of the exons and 98% coverage of the genes. Using our selection criteria it was not possible to obtain 100% coverage with unique probes (see Methods). Here we compared the N2 Bristol strain, isolated in England, to the wild-type strain CB4856 that was isolated on one of the Hawaiian Islands and to a strain isolated on the island of Madeira (JU258). We chose the Hawaiian strain because it is a popular strain for single nucleotide polymorphism (SNP) mapping, as it has many sequence variations compared with N2 (Wicks et al. 2001). Comparisons among wild isolates are potentially complicated by the presence of single nucleotide changes relative to N2, which might cause reduced log2 fluorescence ratios that do not reflect deletions. To minimize this possibility, we used a conservative analysis for copy number changes and counted regions as deleted only if they had a consistently low log2 fluorescence ratio over a substantial distance covering many probes (see Methods).
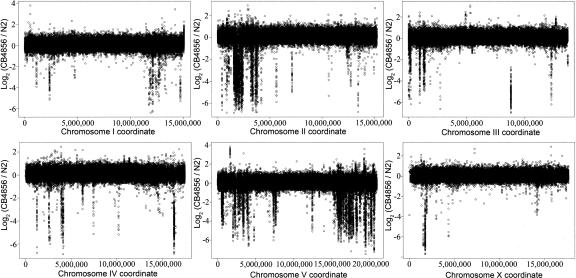
Using these conservative criteria we were able to detect many indel differences between N2 and the Hawaiian strain (Fig. 6), illustrating that natural large-scale gene content variation exists between populations. We observed similar differences between N2 and the Madeiran strain (see Supplemental Fig. 1). The Hawaiian strain exhibited 141 deletions relative to N2, with a total length of 1.54 Mb of DNA deleted (1.54% of the genome). These deletions removed 483 predicted genes and 48 predicted pseudogenes (2.54% of all genes) (Table 1). The Madeiran isolate had 122 deletions relative to N2, deleting 1.94 Mb (1.94% of the genome), removing 670 loci (39 of which are pseudogenes) (Table 1). Supplemental Tables 1 and 2 show chromosomal coordinates and interpretations for every deleted gene for pairwise genome comparisons between N2 and the Hawaiian and Madeiran strains, respectively.
Figure 6.
Whole-genome array CGH comparing Hawaiian (CB4856) and Bristol N2 (VC196) hermaphrodites. Large-scale copy number polymorphism is evident between these two wild-type isolates. Normalized log2 fluorescence ratios (CB4856/N2) for all probes on the chip are shown.
Table 1.
Gene family members deleted in natural isolates from Hawaii (CB4856) and Madeira (JU258)
P-values were computed only for families with potentially higher rates of deletions, and only values <0.05 are shown. P-values are relative to all genes and are one-sided and computed by a 2 × 2 chi-square test with Yates correction. P-values are not corrected for multiple testing, and those with marginal values after Bonferroni correction are enclosed in parentheses. NA, not applicable; NS, not significantly different.
Alterations in the Hawaiian and Madeiran strains relative to N2 Bristol are unevenly distributed both within and between chromosomes, appearing more often on the chromosome arms than in the centers, and a large number of changes on chromosomes II and V, but relatively few changes on chromosome X (Fig. 6; Supplemental Fig. 1). Most of the copy number alterations detected appear to be deletions in the Hawaiian and Madeiran strains relative to N2, but a few amplifications are also evident. The genome regions deleted in the Hawaiian and Madeiran strains are not gene poor or enriched in known pseudogenes, indicating that there are major differences in the functional gene content among these isolates. Among many gene families analyzed, a few were overrepresented among deleted genes (Table 1). The frequency of deletions was particularly high for the MATH-BTB, F-box, C-type lectin, and Srz chemoreceptor families.
It was impractical in this study to validate all copy number changes detected by array CGH between these strains, but we did test one representative deletion extending over several probes. We identified a 2942-bp deletion on chromosome V in the Hawaiian strain, CB4856, that affects two adjacent genes, C49G7.1 and D1065.3. Both are uncharacterized genes containing ankyrin repeats as well as BRCT and WSN domains. We designed primers flanking the deletion, amplified the affected region using PCR, and sequenced the region to determine the deletion breakpoints. The deletion falls between chromosome V coordinates 4,057,455 and 4,057,457 for the proximal breakpoint and 4,060,396 and 4,060,398 for the distal breakpoint, confirming a deletion for these two genes in the Hawaiian strain relative to N2 Bristol (Fig. 7). We also examined a gene, gst-38, that has been sequenced from the Hawaiian strain and is known to have several SNPs relative to the Bristol strain (Denver et al. 2003). Probe targets in the Hawaiian genome contain 0–3 SNPs each, which resulted in a significantly negative log2 ratio in that region of the genome (−1.6), but not of sufficient amplitude to pass our conservative criteria for identifying deletions (see Methods).
Figure 7.
A homozygous viable deletion identified on chromosome V in the Hawaiian strain (CB4856). Normalized log2 ratios (CB4856/N2) indicate that the deletion targets the genes C49G7.1 and D1065.3.
Discussion
The utility of oaCGH in screening for novel deletions
We have demonstrated that oaCGH is a viable platform for detecting heterozygous deletions as small as 141 bp in size in C. elegans. By targeting exons it is more likely that any detected deletion alters the structure of the gene product. Depending on the overlap of oligonucleotides on the array, the resolution of a deletion breakpoint can be <50 bp. To increase resolution, chromosome-specific arrays can be manufactured as we did for chromosome II, which may be desirable depending on the experiment being undertaken. For identifying lethal mutations this may be the most fruitful approach, as the lethal mutation will already be balanced (as described above). PCR amplification and DNA sequencing of the deleted region in the mutant genome can be utilized to precisely identify the breakpoints after oaCGH has made the initial identification.
The ability to detect deletion and amplification events in heterozygous animals is a testament to the sensitivity of oaCGH. This is particularly important when screening for lethal mutations, as it means one can use DNA samples from balanced heterozygous populations that are simply washed from a plate. The added convenience of not having to separate out mutant animals should make this type of analysis more amenable as a high-throughput method.
An important feature of oaCGH is that it yields a high-resolution view of a whole chromosome, or even a whole genome, without the size limitation of ∼100 kb when using a BAC-based platform. Combining an oligonucleotide-based approach with a high-density array format (∼385,000 unique probes) is already leading to widespread adoption of this method for high-resolution mapping of DNA breakpoints for larger sized chromosomal rearrangements in tumors and microdeletion syndromes in humans (Pollack et al. 2002; Selzer et al. 2005; Stallings et al. 2006; Strefford et al. 2006; Urban et al. 2006), as well as the detection of amplifications and deletions such as copy number polymorphisms <0.1 Mb in size (Lucito et al. 2003; Sebat et al. 2004; Conrad et al. 2006). The power of screening a whole chromosome or whole genome for gains and losses of genomic DNA was amply illustrated when we tested for mutations balanced by the inversion mIn1. Besides the known inversion, the mIn1 strain contained several previously unknown deletions and amplifications, some linked to the inversion, but others possibly resident elsewhere in the genome (CGH identifies only the presence of a sequence in a genome, not its location). We also found a previously undetected deletion of exons 4 and 5 of the gene K05F6.2 (floxb-50) in our N2 strain. Curiously, this deletion must have occurred relatively recently, as all of the mutations studied here were isolated from N2 in this laboratory. Without whole-genome testing by array CGH, these novel features present in the genomes of N2 and the balancer strain would have remained undetected.
Natural gene content variation in wild populations
The results of our whole-genome experiment comparing N2 to the Hawaiian and Madeiran wild-type strains revealed a large amount of gene-content variation among these natural isolates. Most of these differences are deletions in the Hawaiian or Madeiran strains relative to N2. Obviously, there is a bias in favor of detecting deletions, because all probes target sequences that are present in the N2 genome. Probe targets containing several SNPs could potentially cause the identification of spurious deletions, so we have used very conservative criteria to ameliorate this possibility and observed that even a gene as divergent as gst-38 is not mistakenly identified as a deletion. Our exon-centric probe selection should also help to reduce the impact of SNPs on hybridization, since SNPs are less common in coding sequences. To identify N2 deletions we will need to compare N2 to a sequenced Hawaiian or Madeiran strain. Previous work in nematodes has shown that chromosomal rearrangements, repeat elements, and transposons are all more common on chromosome arms than in the central region of the chromosomes (Stein et al. 2003). Homologous gene clusters are also more common on the chromosome arms, particularly on the proximal arm of chromosome II and both arms of chromosome V (Thomas 2006b), where we observe the largest number of deletions in the Hawaiian and Madeiran strains. This result suggests that non-allelic homologous recombination (Lupski 1998) on chromosome arms between repeat sequences and/or homologous gene clusters could be responsible for many of the deletions observed in these strains relative to N2. This could also explain the smaller number of gene content alterations observed between N2 and the Hawaiian and Madeiran strains on the X chromosome, where chromosomal rearrangements are less common (Stein et al. 2003). Our array designs target only annotated exons in the sequenced N2 genome, but the large number of deletions observed in the Hawaiian and Madeiran strains relative to N2 implies the likelihood that N2 has also lost novel genes present in the other wild-type isolates.
The frequency of deletions was particularly high for the MATH-BTB, F-box, C-type lectin, and Srz chemoreceptor families. These four gene families are among those with the highest rates of birth–death evolution among Caenorhabditis species (J.H. Thomas, unpubl.). The correlation indicates that indel population diversity within the C. elegans species is related to long-term evolutionary stability in gene families. The nature and level of deletion polymorphisms that we find in the nematode is mirrored in human populations (Conrad et al. 2006; Hinds et al. 2006; Locke et al. 2006; McCarroll et al. 2006). In the study by Conrad et al. (2006), they reported that genes involved in immunity and defense, sensory perception, cell adhesion, and signal transduction were especially prone to deletion, categories that overlap the gene families highlighted as prone to deletion in C. elegans (Thomas et al. 2005; Thomas 2006a). Array studies in nematodes and humans are the first experiments to view wholesale gene-content variation of large numbers of genes in many diverse gene families between populations. These observations from humans and nematodes offer strong support for the “less-is-more” hypothesis of evolutionary change (Olson 1999). In his review, Olson argued “loss of gene function may represent a common evolutionary response of populations undergoing a shift in environment and, consequently, a change in the pattern of selective pressures.” He went on to suggest that “adaptive loss of function may occur regularly and may spread rapidly through small populations.” With their small genome size, rapid life cycle, and self-fertilizing mode of reproduction, dispersed wild populations of nematodes are perhaps ideally suited to monitor genomic responses to environmental selective pressures.
Similar to others, we observe that the Hawaiian and Madeiran strains are more similar to each other than either are to the Bristol (N2) strain (Haber et al. 2005; Stewart et al. 2005). At first this seems surprising; why should nematodes from the Hawaiian Islands located in the middle of the Pacific Ocean and nematodes from Madeira, an island in the Atlantic off the coast of the African continent, be so similar? As previously suggested (Stewart et al. 2005), we think there may be a simple explanation based on the migration of human populations. During the last half of the nineteenth and first half of the twentieth centuries, planting and harvesting sugar cane was a major crop in Hawaii. The workers in the cane fields came from many countries including China, Japan, the Philippines, and after 1878, from Portugal (Bartholomew and Bailey 1994). Almost all of the new immigrants from Portugal came from either the Azores or the island of Madeira (Bartholomew and Bailey 1994), and these immigrants may have inadvertently brought C. elegans with them. If this is true, we have a fairly precise timeline for the introduction of a new strain of C. elegans, or possibly C. elegans itself, to Hawaii.
In the experiments described here we have demonstrated that oaCGH is a robust technology with many possible applications. These include experiments as diverse as screens for novel induced deletions to population genetic studies comparing evolutionary differences among natural isolates. The protocols and chips described here for the C. elegans genomes can similarly be made for other organisms as is already evident in human, mouse, and yeast studies. The high-resolution genome-wide investigation of DNA copy number changes reported here for C. elegans will likely prove to be a powerful tool in genome-wide studies of other model organisms, such as the fly and zebrafish genomes, and the more recently sequenced chicken and dog genomes.
Methods
Probe selection, microarray design, and microarray manufacture
The pilot project focused initially on chromosomes II and X. DNA oligonucleotides, 50 nucleotides in length, were selected to tile open reading frames from both chromosomes. Several types of filters were applied in the selection process in order to maximize the sensitivity and specificity of the oligonucleotides and the signal-to-noise ratio. The applied filters were intentionally relatively mild in order to produce data that would reveal the most important characteristics of oligonucleotides for future chip designs. As a result, ∼90% of the exons and 94% of the genes from both chromosomes are represented on the array. Our oligonucleotide selection can be arbitrarily divided into eight sequential phases. Unless stated otherwise, all of the computer programs have been developed as part of the current work and are freely available from one of the authors (S. Flibotte). (1) The sequences of all curated exons and RNA transcripts on chromosomes II and X were extracted from WormBase (data freeze WS120). Sequences smaller than 50 bases were extended to 50 bases and overlapping sequences were merged. (2) All of the repeats annotated in WormBase were masked. All non-masked subsequences <50 bases in length were then masked (this was also done after phases 3 and 4). (3) All of the 20-mers occurring more than once in the genome were masked. (4) Homopolymers >5 bases in length were masked. (5) All possible 50-mers were extracted from the non-masked subsequences and only those with GC content between 30% and 56% were kept, which corresponds to a melting temperature range of Tm = 72.6 ± 5°C. (6) All of the 50-mers with folding energy larger than –1 kcal/mol according to a hybrid-ss-min calculation (Markham 2003) were kept. (7) Following a MegaBLAST (Zhang et al. 2000) calculation, 50-mers without significant homology with other locations in the genome were kept. (8) For all remaining subsequences, the 50-mers with the lowest overall 15-mer counts were selected using a greedy algorithm and probe spacing parameter, insuring that the distance between the starting positions of two neighboring oligonucleotides is at least 22 bases for chromosome II and 21 bases for chromosome X, except for the region around dim-1, where the distance was set to 6 bases. For each subsequence, the selection continued until no further oligonucleotides could be selected while respecting the overlap constraint. The overall 15-mer count of an oligonucleotide is defined as the sum of the genomic frequencies of all constituent 15-mers. The application of all of these filters resulted in the selection of 97,481 oligonucleotides for chromosome II and 92,209 oligonucleotides for chromosome X. Microarrays were manufactured by NimbleGen Systems, with each oligonucleotide and its corresponding reverse complement synthesized at random positions on the array.
A similar procedure was used to design a chip targeting the whole C. elegans genome (using release WS139) and a chip targeting chromosome II alone (using data freeze WS150). The only differences were that no reverse complement probes were synthesized, the probe spacing parameter was adjusted, and a procedure was introduced to rescue exons targeted by fewer than two oligonucleotides. For the whole-genome chip we tried to select one probe upstream and one probe downstream as close as possible to the underrepresented exon following the filters 2–7 described in the previous paragraph. With a probe spacing parameter of 39, this resulted in 61,910 probes for chromosome I, 64,165 for chromosome II, 56,856 for chromosome III, 59,422 for chromosome IV, 82,944 for chromosome V, and 59,564 for chromosome X. For the chromosome II chip we selected 332,334 probes targeting annotated exons with a probe spacing parameter of 6, and 47,853 probes targeting noncoding sequences with a spacing parameter of 85.
Nematode culture, harvest, and DNA preparation
Nematodes were generally grown as previously described (Brenner 1974) on 60- or 150-mm NGM agar plates seeded with Escherichia coli strain OP50 or χ1666. Strains used were N2 (VC196, a hermaphrodite subculture of N2 received from the Caenorhabditis Genetics Center in 2002); N2 males (male stock of CGC N2 received in 1998); mIn1[mIs14 dpy-10(e128)] homozygotes derived from a single Dpy animal selected from CGC strain DR2078 (strain not kept); VC100 (unc-112(r367) V; gkDf2 X); VC615 (dab-1(gk291)/mIn1[mIs14 dpy-10(e128)] II); VC766 (ceh-39(gk329) X); CB4856 (a subculture of the Hawaiian C. elegans wild isolate HA-8); and JU258 (a wild C. elegans isolate from Madeira). All mutant strains (excluding mIn1) were generated by mutagenesis with trimethylpsoralen (TMP) and UV-irradiation. For DNAs prepared from plate cultures, populations were grown to starvation and harvested by washing into 15-mL centrifuge tubes with 10 mL of M9 buffer containing 0.01% Triton X-100. Each population was washed seven times by centrifugation, removal of supernatant by aspiration, and resuspension and vortexing in fresh M9/Triton X-100. After the final wash, populations were plated on unseeded agar plates and left overnight at 20°C to digest any bacteria remaining in their guts, then reharvested by washing and centrifugation. For DNA from N2 males and confirmed dab-1(gk291)/mIn1[mIs14 dpy-10(e128)] II balanced heterozygotes, worms were picked directly into M9/Triton X-100 in labeled 1.8-mL microcentrifuge tubes, and washed free of bacteria in seven rounds of dilution/centrifugation/aspiration. Aliquots of pelleted worms were transferred to 1.5-mL microcentrifuge tubes containing lysis buffer (50 mM KCl, 10 mM Tris-HCl at pH 8.3, 2.5 mM MgCl2, 0.45% NP-40 [Igepal], 0.45% Tween-20, 0.01% gelatin, 300 μg/mL Proteinase K), frozen at −20°C, and incubated at 55°C–60°C for 3 h. DNA was prepared either by standard phenol-chloroform extraction followed by ethanol precipitation or with the Puregene DNA Purification Kit (D-7000A, Gentra Systems) using the solid tissue protocol. Purified DNAs were resuspended in nuclease-free sterile dH2O or TE (10 mM Tris-HCl, 1 mM EDTA at pH 7.0–8.0). DNA concentrations were determined with a spectrophotometer (Biomate3, Thermo Spectronic) and adjusted to 500 ng/μL for submission to NimbleGen Systems, Inc. for further processing.
DNA fragmentation and labeling
Samples were fragmented and labeled in the NimbleGen Service Laboratory as follows. Two micrograms of each genomic DNA sample were diluted to 80 μL with deionized (DI) water and fragmented by sonication. A portion (0.3 μg) of each sonicated sample was run on a 1% agarose gel to confirm that most of the DNA fragments were between 500 and 2000 bp in length.
Cy3 and Cy5 dye-labeled random 9-mers (TriLink BioTechnologies, Inc.) were diluted to 1 O.D./42 μL of buffer containing 0.125 M Tris-HCl (pH 8.0), 0.125 M MgCl2, 1.75 μL/mL β-mercaptoethanol. Mutant DNA samples were labeled with Cy3 and the wild-type DNA sample (VC196) was labeled with Cy5. One microgram of genomic DNA was added to each random 9-mer buffer solution, denatured at 95°C, and then chilled on ice in 0.2 mL PCR tubes. A total of 10 μL of 50× dNTP mixture (1× TE buffer, 10 mM each of dATP, dCTP, dGTP, and dTTP), 8 μL of DI water, and 100 U of Klenow fragment (exo-) was added to each tube and mixed well with a pipet. Samples were centrifuged and incubated at 37°C for 2 h and 10 μL of 0.5 M EDTA was added and mixed well to stop the labeling reaction. DNA was precipitated by adding 11.5 μL of 0.5 M NaCl and 110 μL of isopropanol, vortexing, incubating in the dark for 10 min at room temperature, and centrifuging at 12,000g for 10 min. The supernatant was removed and the DNA pellet was washed with 500 μL of 80% ethanol. After centrifugation at 12,000g for 2 min, the supernatant was removed, and the pellet was dried in a SpeedVac on low heat for 5 min before being rehydrated in 25 μL of DI water. DNA concentration was measured using a spectrophotometer.
Sample hybridization and imaging
Samples were hybridized in the NimbleGen Service Facility using standard operating procedures, as previously described (Selzer et al. 2005). Briefly, 15 μg of each labeled test and reference DNA sample were added to a single 1.5 mL tube and dried down in the dark in a SpeedVac on low heat. The DNA was resuspended in 3.5 μL of DI water and vortexed; 41.5 μL of NimbleGen hybridization buffer was added to the tube, mixed well, and heated at 95°C for 5 min in the dark. Samples were hybridized at the NimbleGen Service Facility for 16–20 h at 42°C. and then washed with NimbleGen wash buffers and scanned on an Axon scanner (Model # 4000B).
Data analysis
The fluorescence intensity of each feature on the array was extracted with the NimbleScan 2.1 software for the sample and reference images. The intensity ratios were normalized with the help of the robust LOESS regression on the so-called M-A plot, where M = log2 I1/I2 and A = log2 sqrt(I1*I2), I1, and I2 being the intensities of the feature in the two images, similar to the procedure described in Yang et al. (2002). The LOESS regression was implemented with the library from Cleveland et al. (1992). The log2 ratios, M, corresponding to the probes targeting the forward and reverse strands at the same genomic location, were averaged. No outliers were excluded from the subsequent analysis. Copy number aberrations were detected both by careful visual inspection and with a segmentation algorithm developed and currently being tested by one of the authors (S. Flibotte). This segmentation algorithm is a very efficient implementation of a bottom-up approach. The P-value for each aberration was calculated with a one-sample t-test (however, with the total number of non-aberrant data points being very large, one-sample and Welch two-sample t-tests give essentially the same P-values). Also synthesized at random locations on the chip were 9500 50-mer oligonucleotides of random sequence but with the same GC content distribution as our probes. Use of data from these probes as an estimate of background tends to increase the overall standard deviation of the data, and therefore our analysis includes no background subtraction.
For indel comparisons between wild-type strains, genes were from the WormBase release WS150 and were classified into families using a combination of the blastclust clustering algorithm and protein alignments and trees, performed using clustalw and phyml (Thompson et al. 1994; Guindon and Gascuel 2003). We set conservative cutoff values for identifying indels, requiring log2 ratios of 1 or more for amplification segments and −2 or less for deletions. Chromosomal start and end coordinates for each gene were used to determine whether the gene was entirely contained with an assigned deletion; genes that spanned the end of a deletion were not included.
Acknowledgments
We thank Marco Marra and Wan Lam for their enthusiasm, support, and advice concerning this project. We also thank Peggy Eis of NimbleGen for her enthusiasm for this project and her comments on the manuscript. This work was supported by grants from Genome Canada, Genome British Columbia, the Michael Smith Health Research Foundation, the Canadian Institute of Health Research, and the Natural Sciences and Engineering Research Council of Canada to D.G.M.
Footnotes
[Supplemental material is available online at www.genome.org. All data reported in this manuscript is available at GEO through accession number GSE6294.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5690307
References
- Barstead R.J. Reverse genetics. In: Hope I.A., editor. C. elegans: A practical approach. Oxford University Press; Oxford, UK: 1999. pp. 97–118. [Google Scholar]
- Bartholomew G., Bailey B., Bailey B. Maui remembers: A local history. Mutual Publishing; Honolulu, HI: 1994. [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho B., Ouwerkerk E., Meijer G.A., Ylstra B., Ouwerkerk E., Meijer G.A., Ylstra B., Meijer G.A., Ylstra B., Ylstra B. High resolution microarray comparative genomic hybridisation analysis using spotted oligonucleotides. J. Clin. Pathol. 2004;57:644–646. doi: 10.1136/jcp.2003.013029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Pai S., Zhao Z., Mah A., Newbury R., Johnsen R.C., Altun Z., Moerman D.G., Baillie D.L., Stein L.D., Pai S., Zhao Z., Mah A., Newbury R., Johnsen R.C., Altun Z., Moerman D.G., Baillie D.L., Stein L.D., Zhao Z., Mah A., Newbury R., Johnsen R.C., Altun Z., Moerman D.G., Baillie D.L., Stein L.D., Mah A., Newbury R., Johnsen R.C., Altun Z., Moerman D.G., Baillie D.L., Stein L.D., Newbury R., Johnsen R.C., Altun Z., Moerman D.G., Baillie D.L., Stein L.D., Johnsen R.C., Altun Z., Moerman D.G., Baillie D.L., Stein L.D., Altun Z., Moerman D.G., Baillie D.L., Stein L.D., Moerman D.G., Baillie D.L., Stein L.D., Baillie D.L., Stein L.D., Stein L.D. Identification of a nematode chemosensory gene family. Proc. Natl. Acad. Sci. 2005;102:146–151. doi: 10.1073/pnas.0408307102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland W.S., Grosse E., Shyu M.J., Grosse E., Shyu M.J., Shyu M.J. A package of C and fortran routines for fitting local regression models. In: Chambers J.M., et al., editors. Statistical methods in S. Chapman and Hall Ltd.; London, UK: 1992. [Google Scholar]
- Conrad D.F., Andrews T.D., Carter N.P., Hurles M.E., Pritchard J.K., Andrews T.D., Carter N.P., Hurles M.E., Pritchard J.K., Carter N.P., Hurles M.E., Pritchard J.K., Hurles M.E., Pritchard J.K., Pritchard J.K. A high-resolution survey of deletion polymorphism in the human genome. Nat. Genet. 2006;38:75–81. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- Denver D.R., Morris K., Thomas W.K., Morris K., Thomas W.K., Thomas W.K. Phylogenetics in Caenorhabditis elegans: An analysis of divergence and outcrossing. Mol. Biol. Evol. 2003;20:393–400. doi: 10.1093/molbev/msg044. [DOI] [PubMed] [Google Scholar]
- Dhami P., Coffey A.J., Abbs S., Vermeesch J.R., Dumanski J.P., Woodward K.J., Andrews R.M., Langford C., Vetrie D., Coffey A.J., Abbs S., Vermeesch J.R., Dumanski J.P., Woodward K.J., Andrews R.M., Langford C., Vetrie D., Abbs S., Vermeesch J.R., Dumanski J.P., Woodward K.J., Andrews R.M., Langford C., Vetrie D., Vermeesch J.R., Dumanski J.P., Woodward K.J., Andrews R.M., Langford C., Vetrie D., Dumanski J.P., Woodward K.J., Andrews R.M., Langford C., Vetrie D., Woodward K.J., Andrews R.M., Langford C., Vetrie D., Andrews R.M., Langford C., Vetrie D., Langford C., Vetrie D., Vetrie D. Exon array CGH: Detection of copy number changes at the resolution of individual exons in the human genome. Am. J. Hum. Genet. 2005;76:750–762. doi: 10.1086/429588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgley M.L., Riddle D.L., Riddle D.L. LG II balancer chromosomes in Caenorhabditis elegans: mT1(II;III) and the mIn1 set of dominantly and recessively marked inversions. Mol. Genet. Genomics. 2001;266:385–395. doi: 10.1007/s004380100523. [DOI] [PubMed] [Google Scholar]
- Guindon S., Gascuel O., Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Haber M., Schungel M., Putz A., Muller S., Hasert B., Schulenburg H., Schungel M., Putz A., Muller S., Hasert B., Schulenburg H., Putz A., Muller S., Hasert B., Schulenburg H., Muller S., Hasert B., Schulenburg H., Hasert B., Schulenburg H., Schulenburg H. Evolutionary history of Caenorhabditis elegans inferred from microsatellites: Evidence for spatial and temporal genetic differentiation and the occurrence of outbreeding. Mol. Biol. Evol. 2005;22:160–173. doi: 10.1093/molbev/msh264. [DOI] [PubMed] [Google Scholar]
- Hinds D.A., Kloek A.P., Jen M., Chen X., Frazer K.A., Kloek A.P., Jen M., Chen X., Frazer K.A., Jen M., Chen X., Frazer K.A., Chen X., Frazer K.A., Frazer K.A. Common deletions and SNPs are in linkage disequilibrium in the human genome. Nat. Genet. 2006;38:82–85. doi: 10.1038/ng1695. [DOI] [PubMed] [Google Scholar]
- Ishkanian A.S., Malloff C.A., Watson S.K., DeLeeuw R.J., Chi B., Coe B.P., Snijders A., Albertson D.G., Pinkel D., Marra M.A., Malloff C.A., Watson S.K., DeLeeuw R.J., Chi B., Coe B.P., Snijders A., Albertson D.G., Pinkel D., Marra M.A., Watson S.K., DeLeeuw R.J., Chi B., Coe B.P., Snijders A., Albertson D.G., Pinkel D., Marra M.A., DeLeeuw R.J., Chi B., Coe B.P., Snijders A., Albertson D.G., Pinkel D., Marra M.A., Chi B., Coe B.P., Snijders A., Albertson D.G., Pinkel D., Marra M.A., Coe B.P., Snijders A., Albertson D.G., Pinkel D., Marra M.A., Snijders A., Albertson D.G., Pinkel D., Marra M.A., Albertson D.G., Pinkel D., Marra M.A., Pinkel D., Marra M.A., Marra M.A., et al. A tiling resolution DNA microarray with complete coverage of the human genome. Nat. Genet. 2004;36:299–303. doi: 10.1038/ng1307. [DOI] [PubMed] [Google Scholar]
- Kallioniemi A., Kallioniemi O.P., Sudar D., Rutovitz D., Gray J.W., Waldman F., Pinkel D., Kallioniemi O.P., Sudar D., Rutovitz D., Gray J.W., Waldman F., Pinkel D., Sudar D., Rutovitz D., Gray J.W., Waldman F., Pinkel D., Rutovitz D., Gray J.W., Waldman F., Pinkel D., Gray J.W., Waldman F., Pinkel D., Waldman F., Pinkel D., Pinkel D. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258:818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- Locke D.P., Sharp A.J., McCarroll S.A., McGrath S.D., Newman T.L., Cheng Z., Schwartz S., Albertson D.G., Pinkel D., Altshuler D.M., Sharp A.J., McCarroll S.A., McGrath S.D., Newman T.L., Cheng Z., Schwartz S., Albertson D.G., Pinkel D., Altshuler D.M., McCarroll S.A., McGrath S.D., Newman T.L., Cheng Z., Schwartz S., Albertson D.G., Pinkel D., Altshuler D.M., McGrath S.D., Newman T.L., Cheng Z., Schwartz S., Albertson D.G., Pinkel D., Altshuler D.M., Newman T.L., Cheng Z., Schwartz S., Albertson D.G., Pinkel D., Altshuler D.M., Cheng Z., Schwartz S., Albertson D.G., Pinkel D., Altshuler D.M., Schwartz S., Albertson D.G., Pinkel D., Altshuler D.M., Albertson D.G., Pinkel D., Altshuler D.M., Pinkel D., Altshuler D.M., Altshuler D.M., et al. Linkage disequilibrium and heritability of copy number polymorphisms within duplicated regions of the human genome. Am. J. Hum. Genet. 2006;79:275–290. doi: 10.1086/505653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucito R., Healy J., Alexander J., Reiner A., Esposito D., Chi M., Rodgers L., Brady A., Sebat J., Troge J., Healy J., Alexander J., Reiner A., Esposito D., Chi M., Rodgers L., Brady A., Sebat J., Troge J., Alexander J., Reiner A., Esposito D., Chi M., Rodgers L., Brady A., Sebat J., Troge J., Reiner A., Esposito D., Chi M., Rodgers L., Brady A., Sebat J., Troge J., Esposito D., Chi M., Rodgers L., Brady A., Sebat J., Troge J., Chi M., Rodgers L., Brady A., Sebat J., Troge J., Rodgers L., Brady A., Sebat J., Troge J., Brady A., Sebat J., Troge J., Sebat J., Troge J., Troge J., et al. Representational oligonucleotide microarray analysis: A high-resolution method to detect genome copy number variation. Genome Res. 2003;13:2291–2305. doi: 10.1101/gr.1349003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski J.R. Genomic disorders: Structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14:417–422. doi: 10.1016/s0168-9525(98)01555-8. [DOI] [PubMed] [Google Scholar]
- Mantripragada K.K., Buckley P.G., de Stahl T.D., Dumanski J.P., Buckley P.G., de Stahl T.D., Dumanski J.P., de Stahl T.D., Dumanski J.P., Dumanski J.P. Genomic microarrays in the spotlight. Trends Genet. 2004;20:87–94. doi: 10.1016/j.tig.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Markham N.R. “Hybrid: A software system for nucleic acid folding, hybridizing and melting predictions.” Masters thesis, Rensselaer Polytechnic Institute, Troy, NY. 2003. [Google Scholar]
- McCarroll S.A., Hadnott T.N., Perry G.H., Sabeti P.C., Zody M.C., Barrett J.C., Dallaire S., Gabriel S.B., Lee C., Daly M.J., Hadnott T.N., Perry G.H., Sabeti P.C., Zody M.C., Barrett J.C., Dallaire S., Gabriel S.B., Lee C., Daly M.J., Perry G.H., Sabeti P.C., Zody M.C., Barrett J.C., Dallaire S., Gabriel S.B., Lee C., Daly M.J., Sabeti P.C., Zody M.C., Barrett J.C., Dallaire S., Gabriel S.B., Lee C., Daly M.J., Zody M.C., Barrett J.C., Dallaire S., Gabriel S.B., Lee C., Daly M.J., Barrett J.C., Dallaire S., Gabriel S.B., Lee C., Daly M.J., Dallaire S., Gabriel S.B., Lee C., Daly M.J., Gabriel S.B., Lee C., Daly M.J., Lee C., Daly M.J., Daly M.J., et al. Common deletion polymorphisms in the human genome. Nat. Genet. 2006;38:86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- Olson M.V. When less is more: Gene loss as an engine of evolutionary change. Am. J. Hum. Genet. 1999;64:18–23. doi: 10.1086/302219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkel D., Segraves R., Sudar D., Clark S., Poole I., Kowbel D., Collins C., Kuo W.L., Chen C., Zhai Y., Segraves R., Sudar D., Clark S., Poole I., Kowbel D., Collins C., Kuo W.L., Chen C., Zhai Y., Sudar D., Clark S., Poole I., Kowbel D., Collins C., Kuo W.L., Chen C., Zhai Y., Clark S., Poole I., Kowbel D., Collins C., Kuo W.L., Chen C., Zhai Y., Poole I., Kowbel D., Collins C., Kuo W.L., Chen C., Zhai Y., Kowbel D., Collins C., Kuo W.L., Chen C., Zhai Y., Collins C., Kuo W.L., Chen C., Zhai Y., Kuo W.L., Chen C., Zhai Y., Chen C., Zhai Y., Zhai Y., et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat. Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- Pollack J.R., Sorlie T., Perou C.M., Rees C.A., Jeffrey S.S., Lonning P.E., Tibshirani R., Botstein D., Borresen-Dale A.L., Brown P.O., Sorlie T., Perou C.M., Rees C.A., Jeffrey S.S., Lonning P.E., Tibshirani R., Botstein D., Borresen-Dale A.L., Brown P.O., Perou C.M., Rees C.A., Jeffrey S.S., Lonning P.E., Tibshirani R., Botstein D., Borresen-Dale A.L., Brown P.O., Rees C.A., Jeffrey S.S., Lonning P.E., Tibshirani R., Botstein D., Borresen-Dale A.L., Brown P.O., Jeffrey S.S., Lonning P.E., Tibshirani R., Botstein D., Borresen-Dale A.L., Brown P.O., Lonning P.E., Tibshirani R., Botstein D., Borresen-Dale A.L., Brown P.O., Tibshirani R., Botstein D., Borresen-Dale A.L., Brown P.O., Botstein D., Borresen-Dale A.L., Brown P.O., Borresen-Dale A.L., Brown P.O., Brown P.O. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc. Natl. Acad. Sci. 2002;99:12963–12968. doi: 10.1073/pnas.162471999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J., Lakshmi B., Troge J., Alexander J., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., Lakshmi B., Troge J., Alexander J., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., Troge J., Alexander J., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., Alexander J., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., Lundin P., Maner S., Massa H., Walker M., Chi M., Maner S., Massa H., Walker M., Chi M., Massa H., Walker M., Chi M., Walker M., Chi M., Chi M., et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- Selzer R.R., Richmond T.A., Pofahl N.J., Green R.D., Eis P.S., Nair P., Brothman A.R., Stallings R.L., Richmond T.A., Pofahl N.J., Green R.D., Eis P.S., Nair P., Brothman A.R., Stallings R.L., Pofahl N.J., Green R.D., Eis P.S., Nair P., Brothman A.R., Stallings R.L., Green R.D., Eis P.S., Nair P., Brothman A.R., Stallings R.L., Eis P.S., Nair P., Brothman A.R., Stallings R.L., Nair P., Brothman A.R., Stallings R.L., Brothman A.R., Stallings R.L., Stallings R.L. Analysis of chromosome breakpoints in neuroblastoma at sub-kilobase resolution using fine-tiling oligonucleotide array CGH. Genes Chromosomes Cancer. 2005;44:305–319. doi: 10.1002/gcc.20243. [DOI] [PubMed] [Google Scholar]
- Solinas-Toldo S., Lampel S., Stilgenbauer S., Nickolenko J., Benner A., Dohner H., Cremer T., Lichter P., Lampel S., Stilgenbauer S., Nickolenko J., Benner A., Dohner H., Cremer T., Lichter P., Stilgenbauer S., Nickolenko J., Benner A., Dohner H., Cremer T., Lichter P., Nickolenko J., Benner A., Dohner H., Cremer T., Lichter P., Benner A., Dohner H., Cremer T., Lichter P., Dohner H., Cremer T., Lichter P., Cremer T., Lichter P., Lichter P. Matrix-based comparative genomic hybridization: Biochips to screen for genomic imbalances. Genes Chromosomes Cancer. 1997;20:399–407. [PubMed] [Google Scholar]
- Stallings R.L., Nair P., Maris J.M., Catchpoole D., McDermott M., O'Meara A., Breatnach F., Nair P., Maris J.M., Catchpoole D., McDermott M., O'Meara A., Breatnach F., Maris J.M., Catchpoole D., McDermott M., O'Meara A., Breatnach F., Catchpoole D., McDermott M., O'Meara A., Breatnach F., McDermott M., O'Meara A., Breatnach F., O'Meara A., Breatnach F., Breatnach F. High-resolution analysis of chromosomal breakpoints and genomic instability identifies PTPRD as a candidate tumor suppressor gene in neuroblastoma. Cancer Res. 2006;66:3673–3680. doi: 10.1158/0008-5472.CAN-05-4154. [DOI] [PubMed] [Google Scholar]
- Stein L.D., Bao Z., Blasiar D., Blumenthal T., Brent M.R., Chen N., Chinwalla A., Clarke L., Clee C., Coghlan A., Bao Z., Blasiar D., Blumenthal T., Brent M.R., Chen N., Chinwalla A., Clarke L., Clee C., Coghlan A., Blasiar D., Blumenthal T., Brent M.R., Chen N., Chinwalla A., Clarke L., Clee C., Coghlan A., Blumenthal T., Brent M.R., Chen N., Chinwalla A., Clarke L., Clee C., Coghlan A., Brent M.R., Chen N., Chinwalla A., Clarke L., Clee C., Coghlan A., Chen N., Chinwalla A., Clarke L., Clee C., Coghlan A., Chinwalla A., Clarke L., Clee C., Coghlan A., Clarke L., Clee C., Coghlan A., Clee C., Coghlan A., Coghlan A., et al. The genome sequence of Caenorhabditis briggsae: A platform for comparative genomics. PLoS Biol. 2003;1:e45. doi: 10.1371/journal.pbio.0000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M.K., Clark N.L., Merrihew G., Galloway E.M., Thomas J.H., Clark N.L., Merrihew G., Galloway E.M., Thomas J.H., Merrihew G., Galloway E.M., Thomas J.H., Galloway E.M., Thomas J.H., Thomas J.H. High genetic diversity in the chemoreceptor superfamily of Caenorhabditis elegans. Genetics. 2005;169:1985–1996. doi: 10.1534/genetics.104.035329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strefford J.C., van Delft F.W., Robinson H.M., Worley H., Yiannikouris O., Selzer R., Richmond T., Hann I., Bellotti T., Raghavan M., van Delft F.W., Robinson H.M., Worley H., Yiannikouris O., Selzer R., Richmond T., Hann I., Bellotti T., Raghavan M., Robinson H.M., Worley H., Yiannikouris O., Selzer R., Richmond T., Hann I., Bellotti T., Raghavan M., Worley H., Yiannikouris O., Selzer R., Richmond T., Hann I., Bellotti T., Raghavan M., Yiannikouris O., Selzer R., Richmond T., Hann I., Bellotti T., Raghavan M., Selzer R., Richmond T., Hann I., Bellotti T., Raghavan M., Richmond T., Hann I., Bellotti T., Raghavan M., Hann I., Bellotti T., Raghavan M., Bellotti T., Raghavan M., Raghavan M., et al. Complex genomic alterations and gene expression in acute lymphoblastic leukemia with intrachromosomal amplification of chromosome 21. Proc. Natl. Acad. Sci. 2006;103:8167–8172. doi: 10.1073/pnas.0602360103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J.H. Adaptive evolution in two large families of ubiquitin-ligase adapters in nematodes and plants. Genome Res. 2006a;16:1017–1030. doi: 10.1101/gr.5089806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J.H. Analysis of homologous gene clusters in Caenorhabditis elegans reveals striking regional cluster domains. Genetics. 2006b;172:127–143. doi: 10.1534/genetics.104.040030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J.H., Kelley J.L., Robertson H.M., Ly K., Swanson W.J., Kelley J.L., Robertson H.M., Ly K., Swanson W.J., Robertson H.M., Ly K., Swanson W.J., Ly K., Swanson W.J., Swanson W.J. Adaptive evolution in the SRZ chemoreceptor families of Caenorhabditis elegansand Caenorhabditis briggsae. Proc. Natl. Acad. Sci. 2005;102:4476–4481. doi: 10.1073/pnas.0406469102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J., Higgins D.G., Gibson T.J., Gibson T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban A.E., Korbel J.O., Selzer R., Richmond T., Hacker A., Popescu G.V., Cubells J.F., Green R., Emanuel B.S., Gerstein M.B., Korbel J.O., Selzer R., Richmond T., Hacker A., Popescu G.V., Cubells J.F., Green R., Emanuel B.S., Gerstein M.B., Selzer R., Richmond T., Hacker A., Popescu G.V., Cubells J.F., Green R., Emanuel B.S., Gerstein M.B., Richmond T., Hacker A., Popescu G.V., Cubells J.F., Green R., Emanuel B.S., Gerstein M.B., Hacker A., Popescu G.V., Cubells J.F., Green R., Emanuel B.S., Gerstein M.B., Popescu G.V., Cubells J.F., Green R., Emanuel B.S., Gerstein M.B., Cubells J.F., Green R., Emanuel B.S., Gerstein M.B., Green R., Emanuel B.S., Gerstein M.B., Emanuel B.S., Gerstein M.B., Gerstein M.B., et al. High-resolution mapping of DNA copy alterations in human chromosome 22 using high-density tiling oligonucleotide arrays. Proc. Natl. Acad. Sci. 2006;103:4534–4539. doi: 10.1073/pnas.0511340103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicks S.R., Yeh R.T., Gish W.R., Waterston R.H., Plasterk R.H., Yeh R.T., Gish W.R., Waterston R.H., Plasterk R.H., Gish W.R., Waterston R.H., Plasterk R.H., Waterston R.H., Plasterk R.H., Plasterk R.H. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 2001;28:160–164. doi: 10.1038/88878. [DOI] [PubMed] [Google Scholar]
- Yang Y.H., Dudoit S., Luu P., Lin D.M., Peng V., Ngai J., Speed T.P., Dudoit S., Luu P., Lin D.M., Peng V., Ngai J., Speed T.P., Luu P., Lin D.M., Peng V., Ngai J., Speed T.P., Lin D.M., Peng V., Ngai J., Speed T.P., Peng V., Ngai J., Speed T.P., Ngai J., Speed T.P., Speed T.P. Normalization for cDNA microarray data: A robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Schwartz S., Wagner L., Miller W., Schwartz S., Wagner L., Miller W., Wagner L., Miller W., Miller W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]