Abstract
Supernumerary or B chromosomes are selfish entities that maintain themselves in populations by accumulation mechanisms. The accumulation mechanism of the B chromosome of maize (Zea mays) involves nondisjunction at the second pollen mitosis, placing two copies of the B chromosome into one of the two sperm. The B chromosome long arm must be present in the same nucleus for the centromere to undergo nondisjunction. A centromere, containing all of the normal DNA elements, translocated from the B chromosome to the short arm of chromosome 9 was recently found to be epigenetically silenced for centromeric function. When intact B chromosomes were added to this genotype, thus supplying the long arm, the inactive centromere regained the property of nondisjunction causing the translocation chromosome 9 to be differentially distributed to the two sperm or resulted in chromosome breaks in 9S, occasionally producing new translocations. Translocation of the inactive B centromere to chromosome 7 transferred the nondisjunction property to this chromosome. The results provide insight into the molecular and evolutionary basis of this B chromosome accumulation mechanism by demonstrating that nondisjunction is caused by a process that does not depend on normal centromere function but that the region of the chromosome required for nondisjunction resides in the centromeric region.
INTRODUCTION
Chromosomes that do not contain genetic information that is vital for the survival of the organism are found in many different species, including more than a thousand in the plant kingdom (Jones and Rees, 1982). Such dispensable chromosomes, called B chromosomes, have a neutral or slightly negative impact on the fitness of individuals that carry them (Puertas, 2002; Jones and Houben, 2003) and are typically highly heterochromatic, reflecting their gene poor nature. Because they are not maintained by fitness selection, successful B chromosomes that have spread through populations have acquired accumulation mechanisms that allow them to be transmitted to progeny at frequencies higher than predicted by Mendelian ratios (Puertas, 2002; Jones and Houben, 2003). B chromosomes are undoubtedly derived from A chromosomes at some point (Jones and Houben, 2003). The genes on the proto-B must be silenced to avoid aneuploid syndromes, and these modified chromosomes must acquire accumulation mechanisms to be maintained in the population. Given the frequency of known B chromosomes, the generation of proto-B chromosomes is probably fairly common. However, the processes that give rise to B chromosomes during evolution have remained a mystery, as have the molecular bases of their accumulation mechanisms (Burt and Trivers, 2006). It is unknown if these mechanisms are caused by subtle alterations to existing centromeric functions or if the B chromosomes acquire novel molecular properties.
In many species, the B chromosomes fail to disjoin properly during a particular mitosis (Puertas, 2002; Jones and Houben, 2003). By doubling the chromosome number in one of the cells, nondisjunction increases the copy number that can be transmitted to progeny. The maize (Zea mays) B chromosome undergoes nondisjunction at the second pollen mitosis, and the sperm with the B chromosomes preferentially fertilizes the egg rather than the polar nuclei in the process of double fertilization (Roman, 1948; Carlson, 1978). At least two B chromosome long arm regions that act in trans must be present in the same cell for nondisjunction of the centromere: the very distal tip (Ward, 1973) and a site in the proximal euchromatin (Lin, 1978).
Previously, a chromosome containing an inactive B centromere was recovered through screening progeny of a plant undergoing the breakage-fusion-bridge (BFB) cycle (Zheng et al., 1999; Han et al., 2006). During the genomic instability resulting from this process, an intact B centromere region became affixed at the end of chromosome 9S. Because the B centromere is inactive, the chromosome is stable in both mitosis and meiosis. To gain insight into the molecular mechanism responsible for nondisjunction, we studied the passage of this chromosome, 9-B inactivated centromere-1 (9Bic-1), through the second pollen mitosis. Our results provide evidence that the mechanism of nondisjunction is independent of centromere function but that the site of action is coincident with the DNA elements typical of the B centromere.
RESULTS
Genetic Test of Nondisjunction
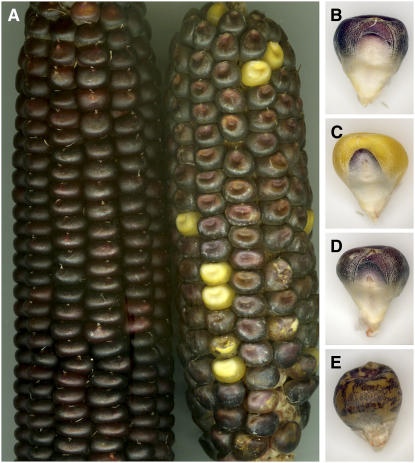
Both 9Bic-1 and the regular chromosome 9 contain the dominant allele, C1, of the c1 gene that is required for anthocyanin pigmentation in the endosperm and embryo of the kernel. The 9Bic-1 chromosome disjoins normally at the second pollen mitosis, as does any A chromosome. This result is expected because the long arm of B is not present in the genotype, which is required for nondisjunction (Roman, 1950; Ward, 1973; Lin, 1978; Carlson and Chou, 1981). Plants containing two intact B chromosomes and 9Bic-1 heterozygous with a normal chromosome 9 [9Bic-1 (C1)/C1 + 2B] were crossed reciprocally with a c1 tester line (c1/c1) (Figure 1). The results from these crosses were distinctly different and suggested that nondisjunction was reinstated to the inactive B centromere sequences. The different outcomes when 9Bic-1 + 2B was the pollen source are illustrated in Figure 2. Nondisjunction of 9Bic-1 would be expected to create a sperm with two copies of C1 and the other sperm with no C1. Fertilization by the deficient sperm is visualized by lack of pigmentation. When 9Bic-1 (C1)/C1 + 2B plants were used as males, ∼71.4% of the kernels containing 9Bic-1 segregants (n = 440) have a phenotype suggesting nondisjunction (Table 1), including kernels with colorless endosperm and colored embryos (Figure 1B) and those with colored endosperm and colorless embryos (Figure 1C). Also, kernels with a mosaic endosperm and colored embryo were observed (Figures 1A and 1E). This color pattern results when the inactive B centromeres fail to separate as the centromeres from chromosome 9 move to opposite poles during the second pollen mitosis, causing a chromosome break and initiating the BFB cycle that continues in the endosperm following fertilization. Because B chromosome nondisjunction does not occur during ovule development, all kernels were entirely colored when the c1 tester was the pollen source (Figure 1A, left ear). Also, because B chromosome nondisjunction requires the transacting factors on the B long arm, reciprocal crosses between c1 testers and plants with 9Bic-1 but no B chromosomes produced ears with fully colored kernels in both cross directions.
Figure 1.
Genetic Test of Nondisjunction of 9Bic-1.
Plants containing two B chromosomes, one standard chromosome 9, and one chromosome 9Bic-1, both with the dominant color marker C1, were crossed reciprocally with c1 tester plants. The left ear in (A) is from a 9Bic-1 (C1)/ 9 (C1) + 2B plant pollinated by the c1 tester. All the resulting kernels are fully colored. The right ear is from a c1 tester plant crossed with pollen from the 9Bic-1 (C1)/9 (C1) + 2B plant. When the 9Bic-1 chromosome nondisjoins, it places two copies of C1 in a single sperm and no copies in the other. Fertilization of the endosperm can occur by either sperm. Kernels with colored endosperm and colorless embryos (B), colorless endosperm and colored embryos (C), fully colored kernels (D), and mosaic endosperms with colored embryos (E), which result from chromosome 9S breakage, are present on the ear to the right. These kernel types are the expected products if 9Bic-1 undergoes nondisjunction in the presence of the normal B chromosomes.
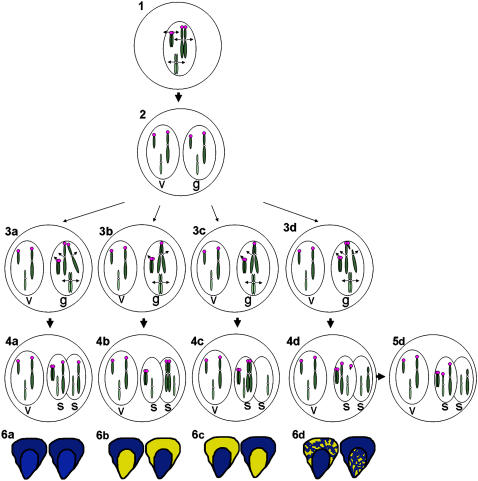
Figure 2.
Possible Outcomes Resulting from Nondisjunction of 9Bic-1 or 7Bic-1.
In this illustration, the B chromosome is depicted as a small telocentric chromosome. The magenta circles indicate tandem arrays of the B-specific element, ZmBs, that are present in and surrounding the B centromere. The larger blue chromosome with ZmBs at the tip of the short arm represents either 9Bic-1 or 7Bic-1. The green chromosome illustrates a representative A chromosome. Centromeres from A chromosomes are illustrated with a white circle. During the first pollen division (1), sister chromatids separate and move to opposite poles, producing two cells: the vegetative (v) and the generative (g). Chromosomes in the generative cell are replicated a second time. During the second pollen division (2), nondisjunction of the B chromosome occurs. 9Bic-1 may disjoin properly (3a), nondisjoin and move to the opposite pole as the B chromosomes (3b), nondisjoin and move to the same pole as the B chromosomes (3c), or break in the short arm as the active A centromeres move to opposite poles but the inactive B centromeres are adhered together. The different outcomes of the second division produce a variety of different sperm (4). Chromosome breakage produces a large fragment containing 9L, the centromere of 9, and a small portion of 9S in one sperm (s) and a small fragment from the short arm of 9Bic-1 in the other (4d). Because the small fragment does not contain an active centromere, it will be lost in the cell divisions that follow fertilization unless it acquires centromere activity (data not shown), or it becomes attached to another chromosome (5). Either of the two sperm depicted in each pollen grain can fertilize the egg (giving rise to the embryo) or the polar nuclei (giving rise to the endosperm). Kernels representing fertilization of the egg by the first sperm or the second sperm are depicted (6). The color of the embryo and endosperm depends on the presence of the C1 allele on 9Bic-1. When chromosome breakage occurs, if the breakpoint is distal to C1, the fragment containing 9L, the centromere, and C1 will enter the chromatid-type BFB cycle, producing a mosaic color pattern in the endosperm (6d). If breakage occurs proximal to C1, the chromatid entering the BFB will not produce any color, so the kernels will appear the same as in 6a.
Table 1.
Frequency of Kernels and Pollen Resulting from Nondisjunction of 9Bic-1
| Kernel Counts for c1/c1 Tester × 2B + 9Bic-1/9 Reciprocal Testcrosses
| ||
|---|---|---|
| Endosperm Color | ||
| Purple | 722 | |
| Yellow | 81 | |
| Mosaic | 76 | |
| Total | 879 | |
| Total with aberrant phenotype | 157 | |
| Total kernels (%) | 17.86% | |
| Adjusted total with nondisjunction phenotype | 314 | |
| Estimated total that contain 9Bic-1a | 439.5 | |
| Estimated frequency of nondisjunction for 9Bic-1b | 71.44% | |
| FISH Data for Pollen from 2B + 9Bic-1(C1)/C1 Plants
| |||||
|---|---|---|---|---|---|
| Number of ZmBs Signals in the Nucleusc
|
|||||
| Vegetative | Sperm 1 | Sperm 2 | No. of Pollen | Total (%) | 9Bic-1 Containing Pollen (%) |
| 1 | 1 | 0 | 142 | 51.64% | |
| 2 | 1 | 1 | 52 | 18.91% | 39.10% |
| 2 | 2 | 0 | 50 | 18.18% | 37.59% |
| 2 | 2 | 1 | 31 | 11.27% | 23.31% |
| Total | 275 | ||||
| Total with 9Bic-1 chromosome | 133 | ||||
| Percentage of 9Bic-1 pollen showing nondisjunctiona | 76.69% | ||||
The endosperm color of kernels from a c1/c1 × 9(C1)/9Bic-1(C1) + 2B testcross was scored as either purple (colored), yellow, or mosaic. Yellow and mosaic endosperm result from failure of the inactive B centromere sister chromatids to separate at the second pollen mitosis. All kernels from the reciprocal crosses were fully colored. Pollen grains with two clearly separated sperm were scored for the number of the B chromosome–specific ZmBs signals in the vegetative and sperm nuclei. Although sperm with a stretched appearance and connected sperm were observed, they were not recorded as such but were assigned to one of the listed categories. Because there are two copies of the B chromosome, every pollen is assured of receiving a single copy. FISH examination of pollen containing only a B chromosome indicates that the B centromere rarely, if ever, separates. Proper disjunction places a ZmBs cluster from 9Bic-1 in both sperm. Failure of the inactive B centromere to separate leads to sperm pairs with two and zero or one and one ZmBs signals. The former results from cases in which the normal B and 9Bic-1 move to the same sperm, while the latter results from their distribution to different poles during the second microspore division. Calculation of ZmBs separation failure using kernel phenotype or pollen FISH produces a similar percentage (pink box).
One-half of the total pollen and seeds should contain 9Bic-1.
Embryo phenotypes were not determined, but assuming no preferential fertilization, they should occur at approximately the same frequency as for the endosperm because each sperm can fertilize either the egg or the central cell. Nonrandom fertilization of the embryo by the sperm containing the B chromosome at rates of ∼70% has been reported in specific genotypes (Gonzalez-Sanchez et al., 2003). It has not been determined if the inactive centromere on 9Bic-1 influences fertilization. If it does, the number of kernels with nonconcordant embryo phenotypes would be less than the endosperm phenotypes. This difference would be minimal because the B chromosome can move independently of 9Bic-1, and only sperm containing both the two B chromosomes and two 9Bic-1 chromosomes would fertilize the embryo at the higher frequency. In any case, the comparison between frequencies of nondisjunction phenotypes in the kernels and pollen is only approximate.
Small ZmBs signals present on the tip of the B chromosome were not counted.
Pollen Fluorescence in Situ Hybridization Visualization of Nondisjunction
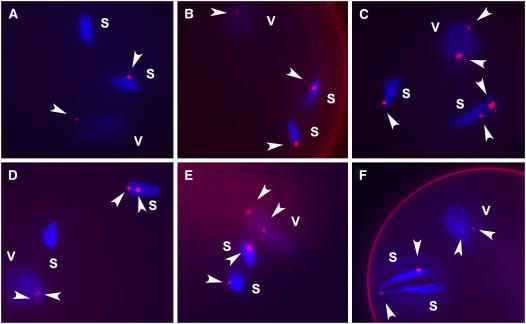
Nondisjunction of 9Bic-1 could also be detected by performing fluorescence in situ hybridization (FISH) on pollen (Shi et al., 1996; Rusche et al., 1997; Lamb et al., 2006) from 9Bic-1(C1)/C1 + 2B plants using a probe for the B chromosome–specific DNA element, ZmBs, found in and around the B centromere and at a distal site on the long arm (Alfenito and Birchler, 1993; Jin et al., 2005; Lamb et al., 2005) (Figure 3; see Supplemental Figure 1 online). Considering only pollen that contained 9Bic-1, the frequency of separation failure of the inactive B centromere was calculated to be 76.69% (n = 133), similar to the approximate percentage determined by examining kernels (Table 1). In immature pollen, pairs of sperm were occasionally observed that were connected by a long thin strand of DNA (Figure 3). Slides were scanned for sperm with this appearance, and 20 out of 20 such pollen contained 9Bic-1. Therefore, connected sperm are likely caused by chromosome bridges that form during the second pollen mitosis as the centromeres of 9Bic-1 move to the two sperm but the short arms remain connected at the inactive B centromere. Connected sperm are not seen in slightly older pollen, suggesting that the situation is quickly resolved, probably through chromosome breakage. In a small number of kernels (2 out of 110), resulting from crosses using pollen from a 9Bic-1(C1)/(C1) + 2B plant, the embryo contained 32 chromosomes consisting of 28 A chromosomes, two 9Bic-1 chromosomes, and two B chromosomes. These individuals contained a single chromosome 9 from the mother plant. This outcome could result from failure of the second pollen mitosis giving rise to a single diploid sperm, which can still function in fertilization and produce a viable kernel if another (haploid) sperm fertilizes the polar nuclei (Kato, 1997).
Figure 3.
Pollen FISH Analysis of Nondisjunction.
Pollen FISH was performed with the ZmBs probe (magenta) on microsporocytes at the three-cell stage and counterstained with 4′,6-diamidino-2-phenylindole (blue). Arrowheads indicate ZmBs signals from the B centromere region. The small ZmBs signal from the B chromosome tip is not indicated. The pollen in (A) was obtained from a plant with two B chromosomes. One signal is observed in the vegetative nucleus (v). Nondisjunction places two B chromosomes in one sperm (s). ZmBs signal is seen in one of the sperm and not the other. Only one ZmBs centromere signal is typically observed in the sperm, although two can occasionally be distinguished, suggesting that the sister centromeres have not separated. The pollen in (B) contains a single 9Bic-1 chromosome. In the absence of the B chromosome long arm, 9Bic-1 disjoins properly and each sperm receives a copy of the chromosome. Because the 9Bic-1 chromosome contains an inactive B centromere region, ZmBs signals are present in all three nuclei. (C) to (F) show observed outcomes for pollen containing a 9Bic-1 chromosome and an intact B chromosome. 9Bic-1 may disjoin properly (C), nondisjoin and both copies move to the same sperm as do the B chromosomes (D), or move to the opposite sperm (E). In immature pollen, pairs of sperm were occasionally observed that were connected by a long thin strand of DNA (F).
Cytological Visualization of Nondisjunction and Chromosome Breakage
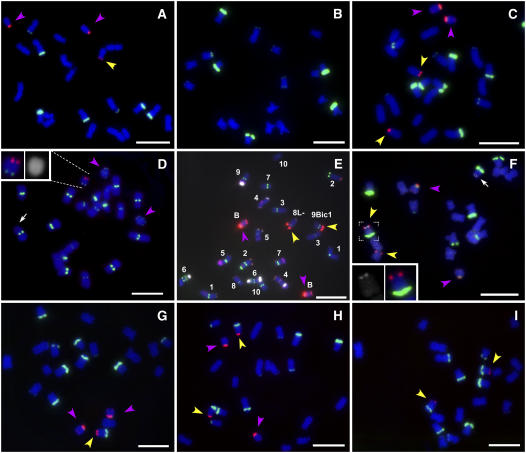
Concordance of kernel phenotype and predicted chromosomal constitutions were tested by examining mitotic chromosome preparations from germinated kernels (Figure 4). Fifty-four kernels with a colorless endosperm and a colored embryo were examined cytologically, and six contained 19 normal A chromosomes, two copies of 9Bic-1, and zero or two intact B chromosomes (Figure 4C, Table 2). Other kernels with this phenotype have one copy of 9Bic-1 and zero or two B chromosomes (Figure 2) and might result from the following scenario (Figure 2). Failure of the inactive B centromere to separate at the second microspore causes breakage of 9Bic-1 in the short arm. If the breakpoint is proximal to C1, a deficient sperm is produced containing a fragment of 9Bic-1 that could fertilize the polar nuclei giving rise to the endosperm. The reciprocal sperm contains an intact 9Bic-1 and a fragment that includes the inactive B centromere and C1. During development, the 9S fragment without a functional centromere would be lost. Among kernels with colored endosperm and apparently colorless embryos were individuals that had only 19 A chromosomes and zero or two Bs (Figure 4B, Table 2). This class results from nondisjunction of 9Bic-1 and fertilization of the egg by the deficient sperm. From kernels of the same phenotype, fragments of 9Bic-1 missing the short arm were recovered (Figure 4D, Table 2). By contrast, from the reciprocal cross of 9Bic-1(C1)/C1 + 2B plants by the tester male line, all 101 screened seedlings had 20 A chromosomes or 19 A chromosomes plus one 9Bic-1 chromosome. More than 200 kernels have been screened from reciprocal crosses of the tester and 9Bic-1(C1)/C1 without B chromosomes, also without evidence of nondisjunction. Thus, nondisjunction of 9Bic-1 only occurs in pollen development and only when an intact B chromosome is present.
Figure 4.
Somatic Chromosome Spreads Demonstrating Nondisjunction.
In all images except (B) and (D), the B-specific sequence, ZmBs, is labeled in magenta and the 180-bp knob repeat is labeled in green. Magenta arrows indicate B chromosomes, and yellow arrows indicate inactive B centromeres from 9Bic-1, 8Bic-1, or 7Bic-1.
(A) to (F) Progeny of ♀ c1/c1 × 9(C1)/9Bic-1(C1) + 2B testcrosses include proper disjunction of 9Bic-1 (A), nondisjunction of 9Bic-1, and movement to the same sperm as the B chromosome followed by fertilization of the egg by a sperm without any chromosome 9 or B chromosomes (B) or by the sperm with two B chromosomes and two 9Bic-1 chromosomes (C). Progeny, in which the nondisjoined 9Bic-1 chromosomes moved to the opposite sperm as did the B chromosomes, were recovered but are not shown. Separation of the chromosome 9 centromeres of 9Bic-1 but failure to separate the inactive B centromere can lead to chromosome breakage, producing fragments containing 9L, the centromere from 9, and a portion of 9S. In (D), a spread containing a fragment is labeled with the White Cap probe (magenta), which detects the Zm CCD1 gene that is present as tandem arrays on 9L. The fluorescence intensity of the White Cap signal in FISH preparations reflects the variation in array size present among different lines (B.C. Tan and D.R. McCarty, personal communication). The 9Bic-1 long arm is strongly labeled by the White Cap probe, making it easy to distinguish from the weakly labeled chromosome 9 of the c1 tester line (white arrow). The centromere is labeled with CentC (green). The reciprocal fragment containing the centromere from 9Bic-1 and a small portion of 9S would also be produced but is usually lost since it lacks a functional centromere. However, it can be recovered if it becomes attached to an A chromosome. A chromosome containing such a translocated fragment was identified as chromosome 8 by application of a cocktail of repetitive elements that allows each chromosome to be identified (E). The cocktail includes CentC (green), 5S ribosomal genes (green), TAG microsatellite (magenta), Cent4 (magenta), TR1 (white), 1.1-kb subtelomeric repeat (magenta), subtelomeric repeat 4-12-1 (green), the 180-bp knob repeat (blue and light magenta), 45S ribosomal genes (green), and ZmBs (magenta). The assignment of chromosome 8 was confirmed by serial application of a number of chromosome-specific probes ruling out the other nine chromosomes. A second translocation chromosome was found that has an inactive B centromere on 7S (F). The spread was hybridized with a 22-kD zein B gene probe that labels 7S (white), ZmBs (magenta), and the 180-bp knob repeat (green). The inset shows an enlarged view of the translocation chromosome, including the gray value image of the 22-kD zein B gene probe. The white arrow indicates the site of the 22-kD zein B gene hybridization on the other chromosome 7 homolog. The 7Bic-1 chromosome also nondisjoins.
(G) to (I) Chromosome spreads from progeny of a tester plant crossed by a pollen parent containing one copy of the 7Bic-1 chromosome and two B chromosomes include individuals in which the 7Bic-1 chromosome disjoins properly (G) or nondisjoins and can move to the same sperm as the B chromosomes (H) or to the opposite sperm (I). Also recovered but not shown were individuals with only 19 chromosomes resulting from fertilization of the polar nuclei by the sperm with two 7Bic-1 and two B chromosomes.
Table 2.
Cytological Screen of Kernels from Crosses Involving 9Bic-1
| Chromosome Constitutiona | Total | 20 A Chromosomes | 19A + 1 9Bic-1 | 19A | 19A + 2 9Bic-1 | 19A+ 1 9Bic-1 + 1 Tiny Fragment | 19A + 1 9L Fragment |
|---|---|---|---|---|---|---|---|
| Kernel phenotype | |||||||
| Cross 1: ♀ c1/c1 × ♂ 9(C1)/9Bic-1(C1) + 2B | |||||||
| Colored embyro, colorless endosperm | 54 | 0 | 45 | 0 | 7 | 2 | 0 |
| Colorless embryo, colored endospermb | 28 | 18 | 2 | 5 | 0 | 0 | 3 |
| Colored embryo, mosiac endosperm | 29 | 0 | 24 | 0 | 2 | 3 | 0 |
| Both colored | 33 | 18 | 14 | 0 | 0 | 1c | 0 |
| Random (mix of all above) | 40 | 16 | 22 | 0 | 0 | 1 | 1 |
| Cross 1: ♀ 9(C1)/9Bic-1(C1) + 2B × ♂ c1/c1 | |||||||
| Both colored | 101 | 50 | 51 | 0 | 0 | 0 | 0 |
A sampling of kernels from reciprocal crosses involving the c1 tester and 9(C1)/9Bic-1(C1) + 2B were germinated and the chromosome constitution examined.
The presence or absence of intact B chromosomes was not scored. When present, there were always two B chromosomes.
It is not possible to identify this kernel type with accuracy in this background. Misclassified kernels explain the recovery of plants with a standard chromosome 9.
The presence of a tiny chromosome in a single kernel of this phenotype is the only case that can not be explained via chromosome breakage.
Recovery and Characterization of Additional Translocations Involving an Inactive B Centromere
Mosaic endosperms result when 9Bic-1 breaks distal to the C1 gene, initiating the BFB cycle in the endosperm and placing an intact 9Bic-1 and a small fragment into the embryo. Although the fragments would usually be lost, they could be recovered in two circumstances: (1) the fragment attaches to another chromosome or (2) the fragment regains centromere activity. Two cases of transposed B centromeres were observed. The first involved translocation to chromosome 8S, but this case was not sexually transmitted, presumably because genetic material essential for gametophytic functions was lost from 8S (Figure 4E). The other, 7Bic-1, placed the inactive B centromere at the tip of 7S (Figure 4F). This chromosome now carries all of the DNA elements typical of the B centromere as does 9Bic-1 (Han et al., 2006). This plant was grown to maturity and self-pollinated. The progeny was screened via root tip chromosome preparations for individuals without 9Bic-1 but still containing 7Bic-1 plus two normal B chromosomes. These 7/7Bic-1 + 2B individuals were tested for nondisjunction via pollen FISH, and reciprocal crosses were made to normal lines without B chromosomes. Examination of ears and pollen and chromosomal preparations of progeny seedlings resulting from these crosses demonstrated nondisjunction of 7Bic-1 in the presence of intact B chromosomes (Figures 4H and 4I, Table 3; see Supplemental Figure 2 online), just as was found with 9Bic-1. Thus, despite the fact that the translocated B centromere is still inactive, the property of nondisjunction is now conferred to chromosome 7.
Table 3.
FISH Examination of Immature Pollen from a 7Bic-1(C1)/Standard Chromosome 7 + 2B Plant
| FISH Data for Immature Pollen from 2B + 7Bic-1/Standard 7
| |||||
|---|---|---|---|---|---|
| Number of ZmBs Signals in the Nucleusa
|
|||||
| Vegetative | Sperm1 | Sperm 2 | No. of Pollen | Total (%) | 7Bic-1 Containing Pollen (%) |
| 1 | 1 | 0 | 104 | 43.15% | |
| 2 | 1 | 1 | 30 | 12.45% | 21.90% |
| 2 | 2 | 0 | 33 | 13.69% | 24.09% |
| 2 | 2 | 1 | 52 | 21.58% | 37.96% |
| Sperm stretched or connected | |||||
| 2 | ZmBs signal split | 7 | 2.90% | 5.11% | |
| 2 | ZmBs signal joined | 15 | 6.22% | 10.95% | |
| Total | 241 | 9.13% | |||
| Total with 7Bic-1 chromosome | 137 | ||||
| Percentage of 7Bic-1 pollen showing nondisjunction | 56.93% | ||||
| Percentage of 7Bic-1 pollen with stretched sperm | 16.06% | ||||
Immature pollen were extruded from nondehisced anthers and examined using FISH. Many sperm (∼9%) had a stretched appearance, or the two sperm were connected by a thin strand of DNA (e.g., Figure 3F). In all cases, pollen with stretched sperm also contained the 7Bic-1 chromosome. When the sperm were connected and the ZmBs signals from the two sister chromatids of 7Bic-1 were connected, these cases were included with those showing nondisjunction.
Small ZmBs signals present near the tip of the B chromosome were not counted.
With regard to the second possible fate of the fragments from 9Bic-1 breakage, a few tiny chromosomes (in 6 out of 110 randomly selected kernels) consisting primarily of the B centromere were recovered among the progeny of testcrosses using 2B with 9-Bic-1 as the pollen parent, but not from crosses in which 9Bic-1 was transmitted through the female (232 kernels) or without an intact B chromosome (>400 kernels). For such fragments to be inherited, they must have centromere activity and could result from reactivation of the inactive B centromere from 9Bic-1. However, because active B centromeres are also present in the same cell, it is not possible to definitively conclude whether the B centromere fragments resulted from centromere reactivation or from fractionation of the normal B.
Localization of the Site in the B Centromere Region Required for Nondisjunction
The 9Bic-1 and 7Bic-1 chromosomes carry small portions of the B chromosome centromeric region. This region includes a heterochromatic knob (Han et al., 2006) adjacent to the centromere (Lamb et al., 2005). Previous work has suggested a possible requirement for this region in efficient nondisjunction (Carlson and Chou, 1981). To narrow the potential sequences involved, a previously described minichromosome derived from the B chromosome was tested for nondisjunction. Minichromosome 9 has been studied over many generations (Kato et al., 2005). It contains no detectable knob sequence, consisting basically of the centromeric region of the B chromosome (Kato et al., 2005), and does not undergo nondisjunction in the absence of the B long arm. We crossed this minichromome to lines containing full-length B chromosomes. A plant containing a copy of minichromosome 9 and one normal B was examined by pollen FISH. Pollen grains were observed that contained one sperm with no B-specific signals and the other sperm with both of them, indicating that nondisjunction occurred at the second pollen mitosis (see Supplemental Figure 3 online). A plant with two 9 minichromosomes and two normal B chromosomes produced progeny with increased numbers of the minichromosomes when crossed as a male in contrast with the material with only minichromosome 9 (Kato et al., 2005). Thus, the knob sequences can be ruled out as being required for nondisjunction, leaving the region spanned by the B-specific sequence array, including the centromere core region (Jin et al., 2005; Lamb et al., 2005) as the part of the chromosome responsible for the nondisjunction property.
DISCUSSION
Our findings show that nondisjunction of the maize B chromosome involves strong attachment of the two sister chromatids at a specific site within, or in close proximity to, the B centromere. This attachment is dependent on the transacting factors on the B chromosome long arm. Because it occurs at the inactive centromere regions of 9Bic-1 and 7Bic-1, attachment does not require centromere function. The strength of attachment is greater than the force required to break a chromatid. The site of attachment being in or immediately surrounding the B centromere would not produce chromosome breakage in intact B chromosomes by not allowing separation of sister centromeres at the second pollen mitosis. However, when the site of chromosome attachment was located distal to the centromere in 9Bic-1 or 7Bic-1, chromosome breakage was more frequent than nondisjunction. Thus, to be an effective mechanism for B chromosome nondisjunction, the site that fails to separate at the second pollen mitosis must reside at the centromere.
Specificity for this function could be caused by targeting of factors to the B centromere region in a sequence-specific fashion. The B-specific DNA element, ZmBs, is present as tandem arrays in and around the B centromere (Alfenito and Birchler, 1993; Kaszas and Birchler, 1996, 1998; Jin et al., 2005; Lamb et al., 2005). The maize B chromosome centromere has been defined extensively by capitalizing on the B-specific repeat to identify this particular centromere (Kaszas and Birchler, 1996, 1998; Jin et al., 2005; Lamb et al., 2005). A core region has been delineated as the site of kinetochore formation, which contains an interspersion of two DNA elements common to all maize centromeres, CentC and CRM, among the B-specific array (Jin et al., 2005; Lamb et al., 2005). Thus, the unique molecular feature that correlates with nondisjunction is the B repeat, ZmBs, although it is not possible to rule out other minor undetected sequences.
Indeed, minor sites of B-specific sequences exist elsewhere on the B chromosome (Alfenito and Birchler, 1993; Lamb et al., 2005). These sites are incapable of causing chromosomal nondisjunction or breakage when translocated to A chromosome centromeres in B-A translocations (Roman, 1947; Beckett, 1991). If the B-specific repeat is responsible for nondisjunction, then either the minor long arm arrays have a sequence variant that does not condition nondisjunction or a sufficiently large array must be necessary to cause failure of the sister chromatids to separate at the second pollen mitosis. The percentage of pollen grains with nondisjunction of the B chromosome is never 100%, so a critical mass of the responsible sequences might be necessary and increasing amounts might foster higher rates of nondisjunction. An interspersion of B repeat, CentC, and CRM at the centromere core might also be a requirement, whether centromere activity is present or not. Yet another possibility is the presence of an epigenetic chromatin state that conditions nondisjunction and that is coincidently correlated with the ZmBs array at the centromere region.
The ZmBs repeat has a small region of homology shared with both the tandemly arrayed 180-bp repeat that composes the prominent heterochromatic blocks, called knobs (Alfenito and Birchler, 1993), found at various locations on maize chromosomes and the Cent4 repeat that is present as tandem arrays in the chromosome 4 centromeric region (Page et al., 2001). In the presence of the B chromosome, increased rates of nondisjunction and chromosome breakage have been observed for A chromosomes in tapetal cells and other terminal tissues where B chromosome nondisjunction can be observed (Chiavarino et al., 2000; Gonzalez-Sanchez et al., 2004). Chromosomes containing knobs are especially susceptible to B chromosome–induced instability. Additionally, for chromosomes containing large knobs, addition of two or more B chromosomes causes an increase in breakage and loss at the second pollen mitosis in a specific line of maize (Rhoades et al., 1967; Rhoades and Dempsey, 1972). Thus, in this line, the behavior of knobbed chromosomes with regard to breakage is similar to 9Bic-1 and 7Bic-1 except that the frequency is much lower. Because the B-specific repeat is related in sequence to the knobs, the increase in chromosome instability may be due to low levels of targeting of the B chromosome nondisjunction mechanisms to the knobs or other similar sequences.
There is a knob close to the B chromosome centromere (Pryor et al., 1980; Peacock et al., 1981). Given the interaction of B chromosomes with knobs as described above, one possible mechanism of nondisjunction might be that the transacting sites on the B long arm interact with the proximal knob to prevent sister chromatid separation at the second pollen mitosis. The knobs in general are the last sites replicated compared with other types of chromatin in the maize genome (Pryor et al., 1980). Their replication is increasingly delayed with increasing numbers of B chromosomes in the genotype (Pryor et al., 1980). If replication is delayed beyond the time of anaphase movement during the second pollen mitosis, such a scenario could account for nondisjunction (Pryor et al., 1980). The 9Bic-1 and 7Bic-1 chromosomes include this proximal knob in the small portion of the B that is translocated in the two events and could potentially provide the site causing nondisjunction. However, there are four observations that argue against this hypothesis in its simplest form.
First, in most genetic backgrounds, the knobs on the A chromosomes are unaffected by the presence of B chromosomes, but nondisjunction of the B chromosomes regularly occurs. Thus, the knob on B would need to be distinct in sequence or other properties from the other knobs to be the responsible agent. Second, the finding that adding normal B chromosomes to the genotype can restore nondisjunction to mini-B chromosome 9, which has no detectable knob sequence, suggests that the knob is not required. Third, a B-A translocation, TB-10Sc, divides the proximal knob (Beckett, 1991). The B-A chromosome carrying the B centromere and part of the knob undergoes nondisjunction at the second pollen mitosis. However, the 10-B chromosome that carries the other part of this knob and the remainder of the B chromosome does not undergo nondisjunction. If late replication of knob sequences was solely responsible for nondisjunction, both chromosomes would be expected to nondisjoin or in the case of the 10-B chromosome to undergo chromosomal breakage. Fourth, it is now known that the knob is displaced from the B centromere functional core (Jin et al., 2005; Lamb et al., 2005). Given our current results indicating that the forces that hold together the two sister chromatids at the second pollen mitosis are usually stronger than the forces involved with anaphase movement resulting in frequent chromosome breakage, the site of adhesion most probably resides within the centromeric region itself; otherwise, the B chromosome would frequently undergo chromosome breakage at the proximal knob site, which is not observed. Thus, the available evidence points to the centromere itself as the site causing delayed sister chromatid separation at the second pollen mitosis. However, the proximal knob is embedded near the edge of the overall array of B-specific repeats (Lamb et al., 2005). Given the sequence relationship between the knob and the B-specific repeats, it is possible that similar replication timing occurs throughout the B-specific array region, which would prevent chromosome rupture and account for nondisjunction.
The specificity of nondisjunction for the B centromere region and its unique mode of action indicate that it is controlled by a novel molecular mechanism that arose independently of normal centromere functions. Centromere specification has an epigenetic component, as illustrated by stable inheritance of an inactive state over several generations (Han et al., 2006). By contrast, the nondisjunction property of the B chromosome is not affected by the epigenetic inactivation of the centromere. The sequences or epigenetic state responsible for nondisjunction apparently invaded and expanded in the centromeric region to provide the currently robust accumulation mechanisms of this selfish nonvital chromosome.
METHODS
FISH
Probe preparation, somatic chromosome spreading, FISH, image capture, and processing were performed as previously described (Kato et al., 2004; Yu et al., 2007). To screen kernels and recover plants with chromosomal constitutions of interest, kernels were germinated in moist vermiculite, the primary root tip excised, and then transplanted to the greenhouse or field. The chromosome identification cocktail components were as described (Kato et al., 2004). The chromosome-specific probes used to confirm the identity of new translocation chromosomes were produced in our lab and previously described (Lamb et al., 2007), except the White Cap probe (B.C. Tan and D.R. McCarty, unpublished data), which was provided by D.R. McCarty (University of Florida).
Pollen FISH was as previously described (Lamb et al., 2006). Mature pollen was collected by shaking tassels over a white piece of paper. Immature pollen grains can be extruded from nondehisced anthers. Pollen was fixed in ethanol:acetic acid (3:1) overnight at −20°C. The next day, the fixative was removed and the pollen was rinsed in 70% ethanol and stored at −20°C. A small amount of pollen was transferred to a 1.5-mL tube, washed in 2× SSC three times, 5 min each, and centrifuged briefly to collect pollen at the bottom of the tube between each wash. Next, the pollen was washed in 10 mM HCl for 10 min. Pollen was treated with pepsin (50 μg/mL in 10 mM HCl) for 10 to 30 min at 37° C and then washed with 2× SSC three times for 5 min each wash. Excess liquid was removed, and 10 μL of hybridization mixture was added (200 ng of each probe in 2× SSC/50% formamide). The mixture was denatured by heating at 100°C for 10 min and then placed on ice for 5 min. The tube was heated to 80°C for 6 min and then incubated at 37°C in the dark for 22 to 24 h. The pollen was washed briefly with 2× SSC, excess liquid was removed, and a drop of Vectashield containing 4′,6-diamidino-2-phenylindole was added (Vector Laboratories; catalog number H-1200) and gently mixed. After incubation for 1 or 2 h at 4°C, the pollen mixture was placed on a glass slide and a cover slip applied.
Supplemental Data
The following materials are available in the online version of this article.
Supplementary Figure 1. Development of Pollen Containing 9Bic-1 and a B Chromosome.
Supplementary Figure 2. Reciprocal Cross between 7Bic-1/+ and a Standard Line.
Supplementary Figure 3. Pollen FISH Analysis of Nondisjunction for Minichromosome 9.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Science Foundation Plant Genome Initiative (DBI0421671 and DBI0423898).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: James A. Birchler (birchlerj@missouri.edu).
Online version contains Web-only data.
References
- Alfenito, M.R., and Birchler, J.A. (1993). Molecular characterization of a maize B chromosome centric sequence. Genetics 135 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckett, J.B. (1991). Cytogenetic, genetic and plant breeding applications of B-A translocation in maize. In Chromosome Engineering in Plants: Genetics, Breeding and Evolution, P.K. Gupta and T. Tsuchiya, eds (New York: Elsevier), pp. 493–529.
- Burt, A., and Trivers, R. (2006). Genes in Conflict. (Cambridge, MA: Belknap Press).
- Carlson, W.R. (1978). The B chromosome of corn. Annu. Rev. Genet. 12 5–23. [DOI] [PubMed] [Google Scholar]
- Carlson, W.R., and Chou, T.S. (1981). B chromosome nondisjunction in corn: Control by factors near the centromere. Genetics 97 379–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiavarino, A.M., Rosato, M., Manzanero, S., Jimenez, G., Gonzalez-Sanchez, M., and Puertas, M.J. (2000). Chromosome nondisjunction and instabilities in tapetal cells are affected by B chromosomes in maize. Genetics 155 889–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Sanchez, M., Gonzalez-Gonzalez, E., Molina, F., Chiavarino, A.M., Rosato, M., and Puertas, M.J. (2003). One gene determines maize B chromosome accumulation by preferential fertilisation; another gene(s) determines their meiotic loss. Heredity 90 122–129. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Sanchez, M., Rosto, M., Chiavarino, M., and Puertas, M.J. (2004). Chromosome instabilities and programmed cell death in tapetal cells of maize with B chromosomes and effects on pollen viability. Genetics 166 999–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, F., Lamb, J.C., and Birchler, J.A. (2006). High frequency of centromere inactivation resulting in stable dicentric chromosomes of maize. Proc. Natl. Acad. Sci. USA 103 3238–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, W., Lamb, J.C., Vega, J.M., Dawe, R.K., Birchler, J.A., and Jiang, J. (2005). Molecular and functional dissection of the maize B chromosome centromere. Plant Cell 17 1412–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, G.H., and Rees, H. (1982). B Chromosomes. (London: Academic Press).
- Jones, N., and Houben, A. (2003). B chromosomes in plants: Escapees from the A chromosome genome? Trends Plant Sci. 8 417–423. [DOI] [PubMed] [Google Scholar]
- Kato, A. (1997). Induced single fertilization in maize. Sex. Plant Reprod. 19 96–100. [Google Scholar]
- Kato, A., Lamb, J.C., and Birchler, J.A. (2004). Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc. Natl. Acad. Sci. USA 101 13554–13559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, A., Zheng, Y.-Z., Auger, D.L., Phelps-Durr, T., Bauer, M.J., Lamb, J.C., and Birchler, J.A. (2005). Minichromosomes derived from the B chromosome of maize. Cytogenet. Genome Res. 109 156–165. [DOI] [PubMed] [Google Scholar]
- Kaszas, E., and Birchler, J.A. (1996). Misdivision analysis of centromere structure in maize. EMBO J. 15 5246–5255. [PMC free article] [PubMed] [Google Scholar]
- Kaszas, E., and Birchler, J.A. (1998). Meiotic transmission rates correlate with physical features of rearranged centromeres in maize. Genetics 150 1683–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb, J.C., Danilova, T., Bauer, M.J., Meyer, J.M., Holland, J.J., Jensen, M.D., and Birchler, J.A. (January 21, 2007). Single loci detection and karyotyping using small target FISH on maize somatic chromosomes. Genetics http://dx.doi.org/10.1534/genetics.106.065573. [DOI] [PMC free article] [PubMed]
- Lamb, J.C., Han, F., Auger, D.L., and Birchler, J.A. (2006). A trans-acting factor required for non-disjunction of the B chromosome is located distal to the TB-4Lb breakpoint on the B chromosome. Maize Genet. Coop. News Lett. 80 51–54. [Google Scholar]
- Lamb, J.C., Kato, A., and Birchler, J.A. (2005). Sequences associated with A chromosome centromeres are present throughout the maize B chromosome. Chromosoma 113 337–349. [DOI] [PubMed] [Google Scholar]
- Lin, B.-Y. (1978). Regional control of nondisjunction of the B chromosome in maize. Genetics 90 613–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, B.T., Wanous, M.K., and Birchler, J.A. (2001). Characterization of a maize chromosome 4 centromeric sequence: Evidence for an evolutionary relationship with the B chromosome centromere. Genetics 159 291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock, W.J., Dennis, E.S., Rhoades, M.M., and Pryor, A.J. (1981). Highly repeated DNA sequence limited to knob heterochromatin in maize. Proc. Natl. Acad. Sci. USA 78 4490–4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryor, A., Faulkner, K., Rhoades, M.M., and Peacock, A.J. (1980). Asynchronous replication of heterochromatin in maize. Proc. Natl. Acad. Sci. USA 77 6705–6709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puertas, M.J. (2002). Nature and evolution of B chromosomes in plants: A non-coding but information-rich part of plant genomes. Cytogenet. Genome Res. 96 198–205. [DOI] [PubMed] [Google Scholar]
- Rhoades, M.M., Dempsey, E., and Ghidoni, A. (1967). Chromosome elimination in maize induced by supernumerary B chromosome. Proc. Natl. Acad. Sci. USA 57 1626–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades, M.M., and Dempsey, E. (1972). On the mechanisms of chromatin loss induced by the B chromosome of maize. Genetics 71 73–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman, H. (1947). Mitotic nondisjunction in the case of interchanges involving the B-type chromosome in maize. Genetics 32 391–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman, H. (1948). Directed fertilization in maize. Proc. Natl. Acad. Sci. USA 34 36–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman, H. (1950). Factors affecting mitotic nondisjunction in maize. Genetics 35 132. [Google Scholar]
- Rusche, M.L., Mogensen, H.L., Shi, L., Keim, P., Rougier, M., Chaboud, A., and Dumas, C. (1997). B chromosome behavior in maize pollen as determined by a molecular probe. Genetics 147 1915–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, L., Zhu, T., Mogensen, H.L., and Keim, P. (1996). Sperm identification in maize by fluorescence in situ hybridization. Plant Cell 8 815–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, E. (1973). Nondisjunction: Localization of the controlling site in the maize B chromosome. Genetics 73 387–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, W., Lamb, J.C., Han, F., and Birchler, J.A. (2007). Cytological visualization of transposable elements and their transposition pattern in somatic cells of maize. Genetics 175 31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, Y.Z., Roseman, R.R., and Carlson, W.R. (1999). Time course study of the chromosome-type breakage-fusion-bridge cycle in maize. Genetics 153 1435–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.