Abstract
The geographic diffusion of highly pathogenic influenza A H5N1 has largely been traced from the perspective of the virus's victims. Birds of a variety of avian orders have been sampled across localities, and their infection has been identified by a general genetic test. Another approach tracks the migration from the perspective of the virus alone, by way of a phylogeography of H5N1 genetic sequences. Although several phylogenies in the literature have labeled H5N1 clades by geographic region, none has analytically inferred the history of the virus's migration. With a statistical phylogeography of 192 hemagglutinin and neuraminidase isolates, we show that the Chinese province of Guangdong is the source of multiple H5N1 strains spreading at both regional and international scales. In contrast, Indochina appears to be a regional sink, at the same time demonstrating bidirectional dispersal among localities within the region. An evolutionary trace of HA1 across the phylogeography suggests a mechanism by which H5N1 is able to infect repeated cycles of host species across localities, regardless of the host species first infected in each locale. The trace also hypothesizes amino acid replacements that preceded the first recorded outbreak of pathogenic H5N1 in Hong Kong, 1997.
Keywords: phylogeny, geography, epidemiology, parallelism, diffusion
With the recent emergence of several highly pathogenic strains of avian influenza, pandemic influenza has attracted new attention. Several waves of outbreaks of influenza A H5N1, the most prevalent of the new strains, have swept across Eurasia, spreading as far as sub-Saharan Africa. Hundreds of millions of domestic and migratory birds have been killed by H5N1 or culled in an effort at control. Disturbingly, H5N1 has demonstrated considerable capacity for xenospecific transmission, including to human hosts. Since 2003, a total of 267 patients across 10 countries are known to have been infected, and 161 have died (January 15, 2007, WHO). In both laboratory and clinical studies, H5N1 has shown itself capable of evolving phenotypes dangerous to humans (1, 2). The previous year's (2006) spate of outbreaks was intermittently marked by lengthening, although still limited, chains of human-to-human transmission (3, 4). The possibility of a pandemic and the clinical course of the most virulent cases have inspired widespread concern and myriad plans of action. Intervention efforts can be better brought to focus when the localities from which H5N1 strains repeatedly evolve are identified. Combinations of interventions, be they vaccines, antivirals, social distancing, or changes to the agricultural industry, can be better targeted once public health officials learn what areas are seeding recurrent outbreaks and are epidemiologically connected.
Genetic phylogenies represent the “gold standard” in characterizing viral genealogy, transmission, and molecular evolution. They provide as good a marker of dispersal available for organisms on which no radio transmitter can be placed. However, no H5N1 phylogeny we reviewed (e.g., refs. 5–8) has analytically traced the migration of the virus through its evolutionary history. We tracked H5N1 migration by using a modification of Slatkin and Maddison's (9) method for inferring migratory events through an intraspecific phylogeny. A gene tree is built from sampled sequences by one of the standard methods: maximum parsimony, maximum likelihood, or distance. The localities from which the sequences were sampled are assigned as a single character to the tips of the resulting tree. The resident locality of any node can be inferred from the tree structure and the tip localities alone with a parsimony algorithm. The resulting assignments minimize the number of migration events in the tree necessary for the localities of the sample sequences to be consistent with the genetic phylogeny. The method as originally formulated omitted tests of significance for the geographic associations inferred. We introduce a nonparametric statistical test for determining whether the frequencies of migration events among localities that occur from node to node through a phylogeny are significantly greater than for phylogenies first characterized by localities randomly distributed across the tips. We used the method to infer migration patterns for influenza A H5N1 phylogenies derived from 192 hemagglutinin and neuraminidase nucleotide sequences sampled from 20 localities across Eurasia.
Results and Discussion
Inferring Migration Events.
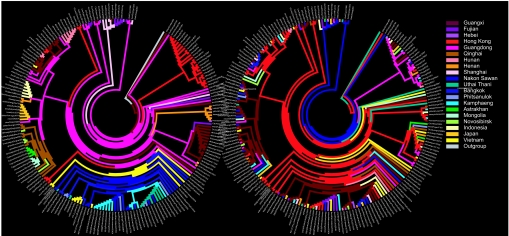
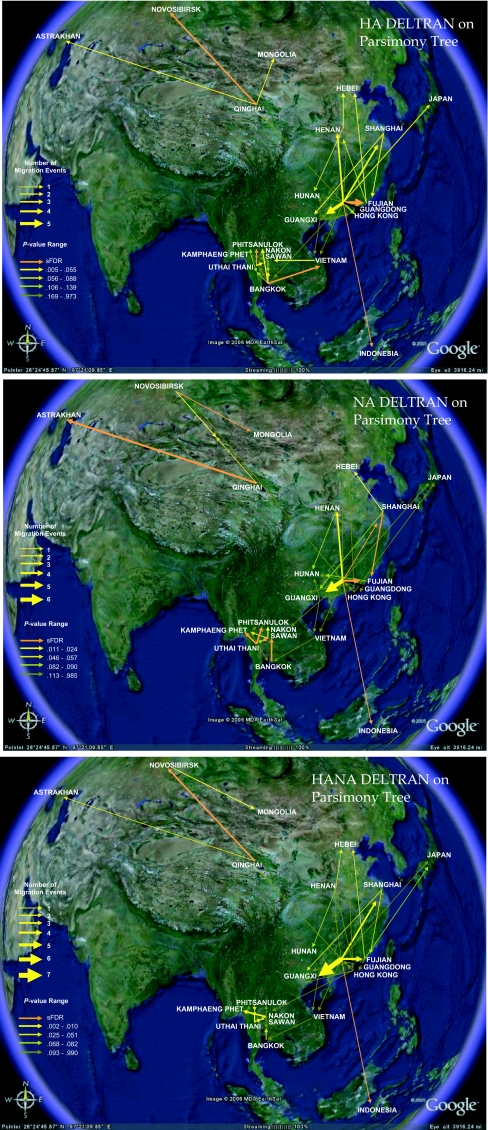
The migration events inferred across the parsimony genetic phylogenies for hemagglutinin, neuraminidase, and their concatenation showed the Chinese province of Guangdong acted as a geographic source for multiple H5N1 variants [Figs. 1A and 2 and supporting information (SI) Tables 1 and 2). In contrast, a migration tree for a typical Monte Carlo random trial hypothesized Hong Kong and Guangxi, which have the greatest sample sizes among the localities tested (n = 29), to be the major sources of migration (Fig. 1B). For the real data, major migration events out of Guangdong (to Hong Kong, Indochina, Qinghai, and Indonesia) were supported by large bootstrap values in the original genetic tree (SI Fig. 4). The Monte Carlo analysis generated frequency distributions for each entry in the resulting migration matrix (SI Table 3). A sparse false discovery rate (sFDR) correction applied to the multiple comparisons test we ran for the hemagglutinin migration matrix identified five pathways with migration significantly greater than expected under the Monte Carlo distributions: Guangdong to Fujian, Guangdong to Indonesia, Bangkok to Vietnam, Uthai Thani to Phitsanulok, and Qinghai to Novosibirsk (Fig. 2). sFDR identified 10 neuraminidase pathways as significant. Many of the other pathways accumulated considerable support even as their P values missed their sFDR cutoff (SI Table 3). The sFDR corrections for the two genes and their concatenation showed optimization by delayed transformation (DELTRAN) to support greater numbers of significant pathways than under accelerated transformation (ACCTRAN) (SI Fig. 5 and SI Table 4). Under DELTRAN, character state changes in the phylogeny (here, migration events) are delayed and concentrated toward the tips of the tree. Under ACCTRAN, state changes are accelerated and concentrated toward the root of the tree.
Fig. 1.
H5N1 migration events through hemagglutinin DELTRAN parsimony. (A) DELTRAN parsimony phylogeny for locality character on a parsimony gene tree for 192 H5N1 hemagglutinin (HA) sequences sampled across 20 Eurasian localities. Branches are colored by locality. Changes in color represent migration events. (B) Typical randomized Monte Carlo trial (no. 9732) for DELTRAN parsimony phylogeny for locality. The tree based on the real sample localities supports 66 migration events without consideration of the outgroup. The Monte Carlo trial supports 147 migration events without the outgroup. The trees were colorized in Mega 3.1 (10).
Fig. 2.
Map of H5N1 migration events DELTRAN parsimony: hemagglutinin, neuraminidase, and concatenation. Migration events in DELTRAN parsimony phylogenies of 192 H5N1 nucleotide sequences sampled across 20 Eurasian localities 1996–2005 (3 ≤ n ≤ 29 isolates per locality). Orange vectors are statistically significant (α = 0.05) under an upper-tail Monte Carlo test of 10,000 trials and a sparse false discovery rate (sFDR) correction: hemagglutinin (HA) (a), neuraminidase (NA) (b), and their concatenation (HANA) (c). Nonsignificant vectors are color-coded by Monte Carlo P value: the brighter the yellow, the greater the support. Quintiles are defined by breaks in ranked P values of > 0.01, except within the final quintile. The maps are based on satellite pictures made available in Google Earth (http://earth.google.com).
Phylogeographic Concordance and Methodological Convergence.
We determined the degree to which the migration matrices across the two H5N1 genes and their concatenation differed under DELTRAN and ACCTRAN optimization by conducting a singular value decomposition on each matrix. We calculated the arccos of the scalar product of the resulting eigenvectors (11), contrasting the three migration matrices in round robin fashion. The resulting theta values, in degrees, quantify the concordance in phylogeographic structure across genes. The theta values showed largely orthogonal differences between the hemagglutinin and neuraminidase matrices (SI Fig. 5), indicating some variance between their phylogeographies. Of the two genes, hemagglutinin contributed the greater effect on the concatenation's migration structure.
Methodological congruence (12) was tested for by rerunning the DELTRAN migration analysis on hemagglutinin and neuraminidase tree topologies derived via (i) a gamma-corrected maximum likelihood (ML) general time-reversible model (13) and (ii) a distance method based on neighbor joining (NJ) and a gamma-corrected Tamura–Nei substitution model (10) (SI Figs. 6–8). A scalar product analysis within each gene showed that for genetic trees generated by the three phylogenetic methods used (parsimony, ML, NJ), parsimony and maximum likelihood produced migration structures of the greatest similarity (SI Table 6). The methods converged best for neuraminidase migration. Despite discrepancies in some of the pathways inferred (SI Figs 7 and 8), the key patterns of migration appear reproduced across phylogenetic methodologies.
Specific Pathways of Dispersal.
The phylogeographies all capture what are known to be major thrusts in H5N1's spatial diffusion since 1996, validating the approach used. What is new here is that the phylogeographies offered a finer distinction than previous work, which defined southern China as an influenza epicenter (5). The province of Guangdong appears the prime source of H5N1's diversity and diffusion, seeding multiple migration events (Figs. 1 and 2). Guangdong, along with much of southern China, hosts a combination of circumstances that apparently promote H5N1 diversification and spread. These include explosive growth in the production of factory farm poultry and free-range ducks (14); extensive use of vaccines on industrial Galloanserae (15); an expanding interface between wildfowl and domestic birds brought about by reduced wetlands and a newfound wildfowl penchant for human agriculture (16, 17); and, with Hong Kong's reintegration into China, greater access to international trade (18).
The phylogeographies also captured H5N1's general thrust west into Europe. However, some of the details appear discordant. The DELTRAN parsimony results showed the H5N1 Qinghai-like sublineage spreading directly from Qinghai to Astrakhan, near the Caspian Sea (Fig. 2). Neither migration by wildfowl nor the poultry trade likely accounts for such a jump. The result may have arisen from limited sample sizes for the more western localities included in the study. Another explanation may be found in the speed of diffusion. H5N1 spread to Western Europe in a matter of months. The short duration may have given the virus's genome little chance to change via isolation by distance, increasing the likelihood that sequences from comparatively distant geographic regions are grouped together in the phylogeny.
The fundamental advance the method used here offers is its capacity to identify, and test the significance of, specific pathways of dispersal across a variety of geographic scales. China's influenza epidemiology, as with those of other countries (19), appeared defined by a hierarchy of diffusion potentials. From Guangdong, H5N1 spreads to a variety of international destinations, including Japan, Vietnam, and Indonesia, as well as to regional localities within China. Guangxi's migratory targets, on the other hand, were more spatially localized. Indochina, as represented here by Thailand and Vietnam, appeared largely isolated in its own endemic disease ecosystem, cut off from the rest of Asia but with extensive bidirectional dispersal within the region, although the latter result may stem in part from the effects of truncated sequencing on H5N1's Thai phylogeny. Many of the Thai sequences included in this study are truncated at both the 5′ and 3′ ends, reducing branch support.
The results suggest that interventions should be directed toward Guangdong and at the socioecological pathways among Indochinese sites (20). The present data set does not, however, address more recent incursions across Indonesia, Western Europe, and Africa, or the recently emergent Fujian-like sublineage in Asia (15). The extent of H5N1's dispersal from Guangdong, including to Japan and Indonesia, conflicts with a recent hypothesis suggesting outbreaks in those two countries originated from South Korea and Thailand, respectively (21).
Phylogeographic Pulses.
The nature of influenza spread may help explain why the migration analysis developed here reproduces known patterns of H5N1 diffusion. Broadly speaking, virulent pathogens such as highly pathogenic influenza must spread across large geographic regions before wiping out local supplies of susceptibles or else risk local extinction (22), although endemic infections have also been documented (19). If we assume widespread geographic spread, influenza outbreaks, as captured by gene trees, represent repeated sampling of the virus's migration structure, regardless of what part of the phylogenetic space each outbreak explores. A caveat, of course, is that migration structures can change over time along with the types of hosts infected and the range of epidemic spread.
The DELTRAN analyses produced maps most comparable to known H5N1 incidence and with greater numbers of significant pathways (SI Table 4). DELTRAN optimization delays character state changes in the phylogeny, favoring parallel state changes across the tree over state reversals. In a phylogeographic context parallel changes in locality represent geographic pulses, in which epidemics repeatedly radiate out from an epicenter, as appears to have been the case for H5N1 this past decade.
The significance of individual pathways likely arises from some function of migration structure, sequence coverage, the number of isolates, the number of localities, phylogenetic dispersion, and the geographic scale surveyed, even as the Monte Carlo test used here helps control for differences in sample size and nonindependence. Nonsignificance may result from limited sampling rather than a lack of effect, and for that reason we advocate that all H5N1 sequences generated be made publicly available for analysis, along with the precise locality where the isolates were sampled.
An Evolutionary Mechanism for Geographic Spread.
Finally, the phylogeographies speak to the evolution of H5N1 transmission. Although in different parts of H5N1's evolutionary space, viral strains can infect the same host species, indicating (i) multiple mechanisms by which to successfully infect any one host species and/or (ii) parallel evolution to successful viral phenotypes. Ducatez et al. (6) found H5N1 independently introduced into Nigerian poultry three separate times, from different regions and host species. In this study, we inferred that each of the three phylogenetically distinct hemagglutinin samples isolated from poultry in the Chinese province of Hebei originated from a different locality: under parsimony, Guangdong, Henan, and Fujian (Fig. 2). Two of the Hebei samples were most related to duck isolates, the third to that of a tree sparrow (SI Fig. 4).
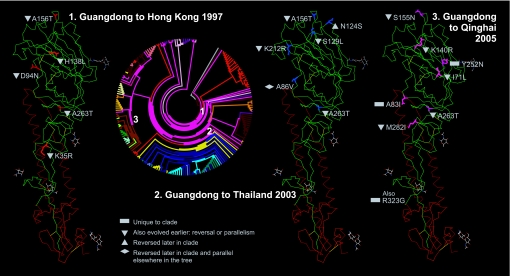
H5N1's phylogeny is littered with parallel evolutionary changes and reversals across a variety of amino acid hotspots, a characteristic of influenzas in general (23, 24). Such conflicts in character state evolution may be a part of real influenza biology rather than the result of a limiting number of character states. In this case, the observed homoplasies may represent geographic gradients of repeated viral ecotypes (Fig. 3). The three sequen-tial migration events we focused on here shared exact changes at two HA1 positions. Of the 19 residue changes depicted across the migration events, all except four occurred elsewhere in the tree.
Fig. 3.
Evolutionary trace for HA1 across three H5N1 migration events: Guangdong to Hong Kong 1997, Guangdong to Thailand 2003, and Guangdong to Qinghai 2005. Each of the three migration events are supported by large bootstrap values in the original gene tree. The three migration events included the same change at residue position 263 (H5 numbering) in antigenic site E. Guangdong to Hong Kong and Guangdong to Thailand also shared a change at position 156, a site for possible N-linked glycosylation (7). The A156T replacement apparent here is associated with increased adaptation to and virulence in poultry (8). Of the 19 amino acid replacements depicted in HA1 across the three migration events depicted in the figure, all except four occurred elsewhere in the tree. All residue changes in the Guangdong-to-Thailand migration were identified as part of the natural variation apparent across the 192 sequences included in the migration analysis. In contrast, only changes at two sites (140, 262) appeared a part of the natural variation for the Guangdong-to-Qinghai migration, emblematic of the differentiation of the QH-like sublineage that has since spread west into Europe and Africa (15). Most of the changes in the globular head represented here appear to arise from escape mutations in antigenic sites near receptor binding site residues (2). One change, at site 129 in the Guangdong-to-Thailand migration, occurred in the receptor binding domain itself. Previous work (e.g., ref. 8) defined H5N1 amino acid replacements by directly comparing field isolates. The trace instead infers replacements through H5N1's evolutionary history, including some of those leading to the first H5N1 outbreak in Hong Kong in 1997. The histidine at site 138 for the Guangdong-to-Hong Kong migration appears a historic artifact now abandoned. The asparagine at site 94 evolves from aspartic acid and is not ancestral, contrary to analyses of 1997 isolates alone.
Correlated evolution across residues could account for the repeated phenotypic parallelisms, allowing H5N1 to toggle between what can be thought of as molecular dialects. Repeatable combinations of amino acid replacements arise from stereochemical relationships among residues and by compensatory evolution (25). The capacity for such combinations, originating in antigenic drift, may represent an exaptation that now permits influenza easier transmission across repeated cycles of host species over geographic space, regardless of the host species initially infected for any given outbreak. Influenza may have long practiced such an epidemiology in its Arctic habitat. Influenza strains adaptively rotate among host reservoirs as different species of migratory birds move on and off the tundra during the breeding season (26).
By cycling among molecular dialects, a more southern H5N1 may circumvent the patches of antibody and cytotoxic lymphocyte memory that host populations sustain. These molecular alternatives permit H5N1 to switch across the sialic acid receptors that define which organs the virus targets and, as previously shown, which host species are infected and modes of transmission used (27, 28). It seems, then, that the effects of the selection pressure under which viral evolution operates extends beyond accelerated mutation rates to differences in the mechanisms by which phenotypic solutions are derived. In the case of influenza A H5N1, one such mechanism appears to involve appropriating evolutionary parallelisms and reversals to circumvent host barriers to geographic spread.
Methods
Samples.
From GenBank (29) and the Influenza Sequence Database (30), we downloaded 192 influenza A H5N1 hemagglutinin and neuraminidase gene sequences isolated from a variety of hosts 1996–2005 across 20 Eurasian localities (Table 1). The hemagglutinin homotrimer binds to the sialic acid receptor of its target host cell and, after endocytosis and a conformational change, induces fusion with the target cell's endosomal compartments. The neuraminidase homotetramer cleaves the sialic acid residues to which newly released influenza virions would otherwise stick. Under recurrent antigenic attack, the two surface proteins accumulate considerable escape variation across viremia, and their sequences have long been used to construct influenza phylogenies. The hemagglutinin sequences used in this study measured 1,698 nucleotides, indels included, covering all of the precursor peptide, HA1 and HA2, except HA2's final six nucleotide sites. The neuraminidase sequences sampled for this study covered the entire monomer, across 1,407 nucleotide sites. Each individual host sampled provided a hemagglutinin and neuraminidase sequence except two neuraminidase sequences from Vietnam, which were sampled from other individuals of the same host type, locality, and year as their hemagglutinin counterparts.
Our analyses used a convenience sample of sequences available from a variety of researchers from across the world, sampling at a variety of geographic scales. The localities sampled were represented by a variable number of isolates (n = 3–29) and included the Chinese city of Shanghai; the Hong Kong Special Administrative Region of China; the Chinese provinces of Hebei, Henan, Hunan, Guangdong, Guangxi, Fujian, and Qinghai; the Thai provinces of Bangkok, Nakhon Sawan, Kamphaeng Phet, Phitsanulok and Uthai Thani; the countries of Vietnam, Japan, Mongolia, and Indonesia; and the southern Russian cities of Novosibirsk and Astrakhan. Duplicate sequences sampled from the same locality add no phylogeographic information to the analysis and were not included among the 192 samples used. On the other hand, duplicate sequences from different localities were included, as they are by definition differentiated by a migration event.
The H5N1 isolates used in this study differed in sequencing coverage across locality (Table 1). Some of the H5N1 samples, particularly from Thailand, are truncated at both the 5′ and 3′ ends for hemagglutinin and neuraminidase, possibly confounding the phylogeny of local clades. Epidemiologists under the pressure of an avian influenza outbreak may sequence field isolates only enough to identify the influenza strains or, with inhibition assays and microarray chips (31), conduct no sequencing at all. Phylogeneticists, on the other hand, often require as much sequence data as possible to resolve intraspecific relationships. Another obstacle, many of the H5N1 isolates included in this study were at best identified by country or province. Much finer influenza phylogeographies could be constructed with samples whose localities are more precisely identified, including by global positioning system coordinates. To take full advantage of the public health applications of phylogeography, we encourage full sequencing of outbreak strains when and where possible and that the exact dates and locales of isolation be provided for each sample.
Phylogeny.
The nucleotide sequences were aligned in ClustalW (32). Gaps arising from truncated sequencing at either end of the sequence were treated as missing data. Gaps within the sequences (e.g., of a length of 19–20 residues in the neuraminidase stalk) were treated as real indels and defined as a fifth character state. Character state changes were ordered by outgroup comparison with isolate A/chicken/Scotland/59(H5N1) and phylogenetic trees constructed with maximum parsimony using PAUP* 4.0b10 (33). An outgroup too distant from its ingroup can generate homoplasies brought about by evolution across limited character states over too much time apart. However, H5N1 phylogenies are constrained by outgroup availability, as the 1959 sample is the sole outgroup isolate with both hemagglutinin and neuraminidase genetic sequences publicly available. A migration analysis using the oldest of the ingroup isolates (A/Goose/Guangdong/1/96) as the outgroup did not significantly affect the outcomes. The most parsimonious trees were heuristically generated by stepwise addition and branch swapping via tree-bisection-recombination. The resulting gene tree used for migration analysis was assumed, and appeared for hemagglutinin and neuraminidase, to be representative of all most parsimonious trees generated.
The question of methodological applicability may arise from the network-like nature of much intraspecific evolution, but traditional phylogenetic methods such as parsimony and maximum likelihood are appropriate here. Influenza is defined by a rapid cladogenesis atypical of many intraspecific phylogenies for which traditional methods perform poorly (34). Influenza taxa are also characterized by rapid extinction and their phylogenies are limited in extant ancestors (35, 36). Finally, the traditional methods can be used to infer ancestral character states with which migration-specific mutations can be catalogued, an important line of research for efforts aimed at developing phylogeographic interventions for influenza.
Statistical Phylogeography.
As in Slatkin and Maddison (9), the localities of the isolates in the gene tree were assigned to the tips as a single character with 20 states. Moving recursively up the tree, PAUP* 4.0 assigned the localities of ancestral nodes by way of the same parsimony optimization algorithm used in generating the original gene tree, such that the tree supported the fewest possible migration events between localities consistent with the gene tree.
For each gene, the total number of migration events occurring through the tree between every possible pair of localities was tallied. A Monte Carlo test of 10,000 trials was conducted to determine the probability that the frequencies of migration events between each pair of localities in the original migration tree are more than expected when the localities are randomly distributed across the tree's tips. The localities of the real data sets were randomized without changing the sample sizes for each locality. The migration events among localities through the gene tree were inferred by DELTRAN optimization and the migration events among localities summed for each randomized trial. Another Monte Carlo test was conducted for ACCTRAN optimization. The Java code used to sum migration events and conduct the Monte Carlo test is available upon request.
To test the significance of the resulting P values across all localities, α, the nominal type 1 error rate, was corrected for multiple tests across locality pairs. Bonferroni and Sidak corrections to α, and their sharpened stepwise equivalents, control the familywise error rate against any wrong rejection of the null hypothesis. These corrections tend to be overly conservative, especially for multiple comparisons numbering in the hundreds and thousands. For greater power, we used the false discovery rate (FDR) correction, another correction for simultaneous tests, which aims to minimize the fraction of wrong rejections among rejected hypotheses (37).
Still greater power can be generated by reducing the number of comparisons (38). Sparse matrices are characterized by numerous cells of zero count whose entries are by definition never greater than produced under the Monte Carlo randomization. Null cells, in effect, carry little information regarding the relative significance among migration pathways other than the triviality that these pathways do not exist. It follows that a sFDR correction can be calculated by dividing α by the greatest number of nonzero entries possible in the migration matrix (66 for DELTRAN hemagglutinin) rather than by the total possible comparisons (20 localities × 19 = 380 comparisons). The Monte Carlo P values of significant pathways are less than their sFDR cutoff: P value rank × (0.05/total migration events). The test is capped at a distribution conservative enough to account for a regular spatial distribution in which each hypothesized migration occupies a different cell in the migration matrix. The sFDR procedure was run for the hemagglutinin, neuraminidase, and concatenation migration trees under both DELTRAN and ACCTRAN optimization.
Evolutionary Trace.
In an effort to observe correlated migration-specific substitutions, we conducted an evolutionary trace of hemagglutinin HA1 amino acid residues for three H5N1 migration events in the DELTRAN parsimony migration tree (Fig. 1A): Guangdong to Hong Kong 1997, Guangdong to Thailand 2003, and Guangdong to Qinghai 2005. The amino acid changes were inferred using the original nucleotide tree topology (Fig. 4A). Not all correlated changes across residue sites need occur at a single branching point or at the migration event alone: such changes can accumulate. For Guangdong to Hong Kong, we included residue replacements occurring in the branch immediately following the migration event. For Guangdong to Thailand and Guangdong to Qinghai, we included replacements in the branch immediately preceding the migration event. The migration tree was colorized in Mega 3.1 (10), the nucleotide sequences translated into amino acid residues in Mega, the evolutionary trace conducted in MacClade 4.06 (39), and the hemagglutinin solution structure annotated in JEvtrace (40).
Supplementary Material
Acknowledgments
We thank John Avise, Mike Davis, Ford Doolittle, Mark Finkelstein, Marius Gilbert, Marcin Joachimiak, Marcella McClure, Tihomir Petrov, William Schopf, Howard Tucker, Rodrick Wallace, and Jianzhi George Zhang for invaluable discussion and correspondence. We also thank the hundreds of scientists and technicians involved in sequencing the nucleotide samples used in this study. The work was made possible by National Institutes of Health National Institute of Allergy and Infectious Diseases Grant 1R21AI063275-01.
Abbreviations
- sFDR
sparse false discovery rate
- DELTRAN
delayed transformation
- ACCTRAN
accelerated transformation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700435104/DC1.
References
- 1.Yuen KY, Wong SS. Hong Kong Med J. 2005;11:189–199. [PubMed] [Google Scholar]
- 2.Stevens J, Blixt O, Tumpey TM, Taubenberger JK, Paulson JC, Wilson IA. Science. 2006;312:404–410. doi: 10.1126/science.1124513. [DOI] [PubMed] [Google Scholar]
- 3.Butler D. Nature. 2006;439:248–249. doi: 10.1038/439248a. [DOI] [PubMed] [Google Scholar]
- 4.Butler D. Nature. 2006;442:37. [Google Scholar]
- 5.Chen H, Smith GJ, Li KS, Wang J, Fan XH, Rayner JM, Vijaykrishna D, Zhang JX, Zhang LJ, Guo CT, et al. Proc Natl Acad Sci USA. 2006;103:2845–2850. doi: 10.1073/pnas.0511120103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ducatez MF, Olinger CM, Owoade AA, De Landtsheer S, Ammerlaan W, Niesters HG, Osterhaus AD, Fouchier RA, Muller CP. Nature. 2006;442:37. doi: 10.1038/442037a. [DOI] [PubMed] [Google Scholar]
- 7.Smith GJ, Naipospos TS, Nguyen T. D., de Jong MD, Vijaykrishna D, Usman TB, Hassan SS, Nguyen TV, Dao TV, Bui NA, et al. Virology. 2006;350:258–268. doi: 10.1016/j.virol.2006.03.048. [DOI] [PubMed] [Google Scholar]
- 8.World Health Organization Global Influenza Program Surveillance Network. Emerg Infect Dis. 2005;11:1515–1521. doi: 10.3201/eid1110.050644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Slatkin M, Maddison W. Genetics. 1989;123:603–613. doi: 10.1093/genetics/123.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar S, Tamura K, Nei M. Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 11.Arfken GB, Weber H.-J. Mathematical Methods for Physicists. Burlington, MA: Elsevier; 2005. [Google Scholar]
- 12.Miyamoto M, Fitch WM. Syst Biol. 1995;44:64–76. [Google Scholar]
- 13.Zwickl DJ. Austin: University of Texas; 2006. PhD dissertation. [Google Scholar]
- 14.Gilbert M, Chaitaweesub P, Parakamawongsa T, Premashthira S, Tiensin T, Kalpravidh W, Wagner H, Slingenbergh J. Emerg Infect Dis. 2006;12:227–234. doi: 10.3201/eid1202.050640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith GJ, Fan XH, Wang J, Li KS, Qin K, Zhang JX, Vijaykrishna D, Cheung CL, Huang K, Rayner JM, et al. Proc Natl Acad Sci USA. 2006;103:16936–16941. doi: 10.1073/pnas.0608157103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu L, Tian H, Liu M, Zhuang D, Melillo JM, Zhang Z. Geophys Res Lett. 2005;32:L02405.1–L02405.5. [Google Scholar]
- 17.Rapport D, Howard J, Maffi L, Mitchell B. Avian Influenza and the Environment: An Ecohealth Perspective. New York: United Nations Environment Programme; 2006. www.unep.org/dewa/products/publications/2006/DRapport_AI_Final_180506_Edit3.doc.pdf. [Google Scholar]
- 18.Sit VFS. Asian Surv. 2004;44:815–835. [Google Scholar]
- 19.Cliff AD, Haggett P, Ord JK. Spatial Aspects of Influenza Epidemics. Pion: London; 1986. [Google Scholar]
- 20.Songserm T, Jam-on R, Sae-Heng N, Meemak N, Hulse-Post DJ, Sturm-Ramirez KM, Webster RG. Emerg Infect Dis. 2006;12:575–581. doi: 10.3201/eid1204.051614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kilpatrick AM, Chmura AA, Gibbons DW, Fleischer RC, Marra PP, Daszak P. Proc Natl Acad Sci USA. 2006;103:19368–19373. doi: 10.1073/pnas.0609227103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boots M, Sasaki A. Proc R Soc London B. 1999;266:1933–1938. doi: 10.1098/rspb.1999.0869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fitch WM, Leiter X, Li Q, Palese P. Proc Natl Acad Sci USA. 1991;88:4270–4274. doi: 10.1073/pnas.88.10.4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolf YI, Viboud C, Holmes EC, Koonin EV, Lipman DJ. Biol Direct. 2006;1:34. doi: 10.1186/1745-6150-1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rimmelzwaan GF, Berkhoff EG, Nieuwkoop NJ, Smith DJ, Fouchier RA, Osterhaus AD. J Gen Virol. 2005;86:1801–1805. doi: 10.1099/vir.0.80867-0. [DOI] [PubMed] [Google Scholar]
- 26.Olsen B, Munster VJ, Wallensten A, Waldenstrom J, Osterhaus A. D., Fouchier R. A. Science. 2006;312:384–388. doi: 10.1126/science.1122438. [DOI] [PubMed] [Google Scholar]
- 27.Stevens J, Blixt O, Glaser L, Taubenberger JK, Palese P, Paulson JC, Wilson IA. J Mol Biol. 2006;355:1143–1155. doi: 10.1016/j.jmb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 28.Yamada S, Suzuki Y, Suzuki T, Le MQ, Nidom CA, Sakai-Tagawa Y, Muramoto Y, Ito M, Kiso M, Horimoto T, et al. Nature. 2006;444:378–382. doi: 10.1038/nature05264. [DOI] [PubMed] [Google Scholar]
- 29.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. Nucleic Acids Res. 2006;34:D16. doi: 10.1093/nar/gkj157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Macken C, Lu H, Goodman J, Boykin L. In: Options for the Control of Influenza IV. Osterhaus ADME, Cox N, Hampson AW, editors. Amsterdam: Elsevier; 2001. pp. 103–106. [Google Scholar]
- 31.Dawson ED, Moore CL, Smagala JA, Dankbar DM, Mehlmann M, Townsend MB, Smith CB, Cox NJ, Kutchta RD, Rowlen KL. Anal Chem. 2006;78:7610–7615. doi: 10.1021/ac061739f. [DOI] [PubMed] [Google Scholar]
- 32.Thompson JD, Higgins DG, Gibson TJ. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swofford D. PAUP*: Phylogenetic Analysis Using Parsimony (And Other Methods) 4.0 beta for Windows/DOS. Sunderland, MA: Sinauer; 2002. [Google Scholar]
- 34.Benjamini Y, Hochberg Y. J Roy Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- 35.McClintock JN, Edenberg HJ. BMC Bioinformatics. 2006;7:49. doi: 10.1186/1471-2105-7-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hillis DM, Huelsenbeck JP, Cunningham CW. Science. 1994;264:671–677. doi: 10.1126/science.8171318. [DOI] [PubMed] [Google Scholar]
- 37.Buonagurio DA, Nakada S, Parvin JD, Krystal M, Palese P, Fitch WM. Science. 1986;232:980–982. doi: 10.1126/science.2939560. [DOI] [PubMed] [Google Scholar]
- 38.Bush RM, Bender CA, Subbarao K, Cox NJ, Fitch WM. Science. 1999;286:1921–1925. doi: 10.1126/science.286.5446.1921. [DOI] [PubMed] [Google Scholar]
- 39.Maddison D, Maddison W. MacClade 4: Analysis of Phylogeny and Character Evolution. Sunderland, MA: Sinauer; 2000. [DOI] [PubMed] [Google Scholar]
- 40.Joachimiak MP, Cohen FE. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-12-research0077. RESEARCH0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.