Abstract
Development of invariant natural killer T (iNKT) cells requires the presentation of lipid ligand(s) by CD1d molecules in the thymus. The glycosphingolipid (GSL) isoglobotrihexosylceramide (iGb3) has been proposed as the natural iNKT cell-selecting ligand in the thymus and to be involved in peripheral activation of iNKT cells by dendritic cells (DCs). However, there is no direct biochemical evidence for the presence of iGb3 in mouse or human thymus or DCs. Using a highly sensitive HPLC assay, the only tissue where iGb3 could be detected in mouse was the dorsal root ganglion (DRG). iGb3 was not detected in other mouse or any human tissues analyzed, including thymus and DCs. Even in mutant mice that store isoglobo-series GSLs in the DRG, we were still unable to detect these GSLs in the thymus. iGb3 is therefore unlikely to be a physiologically relevant iNKT cell-selecting ligand in mouse and humans. A detailed study is now warranted to better understand the nature of iNKT cell-selecting ligand(s) in vivo.
Keywords: glycosphingolipid, thymus, CD1d, lipid presentation
CD1 is a nonpolymorphic MHC class I-like molecule (1). Five isoforms exist in humans (hCD1a–e), which can be subdivided into two groups; group I contains CD1a, -b, -c, and -e, and group II contains CD1d (1). Two homologues of hCD1d exist in mouse (mCD1.1 and mCD1.2) (1). The group II CD1 molecules are believed to present self and exogenous lipid antigens (2–4), whereas group I molecules present exogenous ligands, such as mycobacterial lipids and perhaps endogenous lipids (1). The conserved nature of the CD1 molecules and the T cells that respond to them suggests this system plays an important role in mammalian innate and adaptive immunity (1). Lipids bound to CD1d are recognized by specialized T cells with a restricted T cell receptor repertoire (1). In mouse, these T cells are either CD4 single-positive or CD4/8 double-negative, express some natural killer (NK) cell markers, and are referred to as invariant NK T (iNKT) cells (5).
iNKT cells are selected in the thymus by the presentation of endogenous unknown lipid(s) bound to CD1d on cortical CD4/8 double-positive thymocytes (6). Evidence suggests this lipid ligand is a glycosphingolipid (GSL), because of the inability of a glucosylceramide-deficient cell line to stimulate iNKT cell hybridomas (7). Intracellular trafficking of CD1d to the lysosome is required for lipid loading in vitro and in vivo (8), as are lysosomal proteases (9) and lipid transfer proteins or saposins (10, 11).
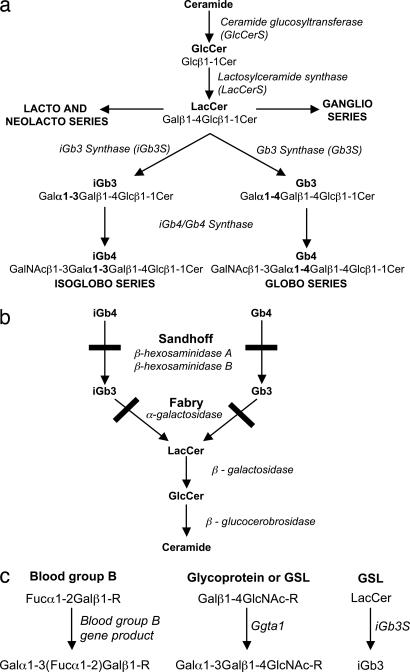
Recently, Zhou et al. (12) suggested that the natural selecting ligand in the thymus is the neutral GSL, isoglobotrihexosylceramide (iGb3)†† (Fig. 1a). This notion was based on the observation that the number of iNKT cells was greatly reduced in a Sandhoff (SH) disease (hexb−/−) mouse model. It was therefore hypothesized that the lysosomal enzymes deficient in SH disease, β-hexosaminidase (β-hex) A and B (Fig. 1b), are responsible for the generation of the natural lipid-ligand in the thymus. This would implicate β-hex-catalyzed degradation products of the globo, isoglobo, ganglio, and lacto series of GSLs as potential iNKT cell natural ligands. The ganglio-series GSLs were eliminated as candidate ligands, because mice deficient in either of two key biosynthetic enzymes required to generate complex gangliosides, GM2 (13) or GM3 synthase (14), displayed no apparent defect in iNKT cell development (12). Upon testing a GSL from each of the lacto, globo, and isoglobo series [rabbit B-active GSL, globotrihexosylceramide (Gb3) and iGb3, respectively], only iGb3 had iNKT cell stimulatory activity. It was hypothesized that in SH disease, the storage of isoglobotetrahexosylceramide (iGb4, Fig. 1 a and b, not an iNKT cell stimulatory molecule) prevented the formation of iGb3 (the natural ligand), thus accounting for the iNKT cell deficiency of the hexb−/− mouse. It has also been suggested that iGb3 plays a role in activation of iNKT cells in the periphery (3). In this instance, lipopolysaccharide-activated DCs were suggested to present iGb3 in response to exposure to Salmonella typhimurium (3).
Fig. 1.
GSL biosynthetic and catabolic pathways and Galα1–3 biosynthetic enzymatic pathways. (a) Synthetic pathway of GSLs from ceramide with the full structures of the globo- and isoglobo-series GSLs. (b) Catabolic pathways of the globo- and isoglobo-series GSLs with the catabolic enzymes and the relevant lysosomal storage disorders highlighted. (c) The three different enzymes responsible for the addition of galactose in an α1–3 linkage are shown. R, glycoprotein or GSL.
However, there are currently no biochemical data on whether isoglobo-series GSLs are expressed in mouse or human thymus. Furthermore, the iGb3 synthase (iGb3S) gene in the human genome appears to be a pseudogene (15, 16). It therefore remains to be established whether iGb3 is a physiologically relevant selecting ligand in vivo or a molecule serendipitously capable of stimulating iNKT cells in vitro.
The isoglobo series of GSLs are derived from a relatively minor GSL biosynthetic pathway catalyzed by iGb3S (Fig. 1a). To date, there is only biochemical evidence for iGb3 in rat (17), dog (18) and cat intestine (19), rat lens (20), and rat spleen (21) [supporting information (SI) Table 1]. The biosynthetic derivative of iGb3, iGb4 (Fig. 1a), has been identified in multiple tissues in the rat, including intestine (17), stomach (22), kidney (23), spleen, glomerular mesangial cells, lymphosarcoma, granuloma (24), and a rat-derived metastatic cell line (25). It has been reported that mouse and human kidney lack iGb4 (23) (SI Table 1).
iGb3S was first identified in the rat (26), and a functional homolog is present in the mouse (12, 15). iGb3S is solely responsible for initiating the synthesis of isoglobo-series GSLs by the addition of galactose in an α1–3 linkage (Galα1–3) to lactosylceramide (LacCer) and to iGb3 itself to form polygalactose structures (15). Additional enzymes capable of synthesizing Galα1–3 are α1–3 galactosyltransferase (Ggta1) and the blood group B gene product (Fig. 1c). Ggta1 acts on N-acetyllactosamine (Galβ1–4GlcNAc-R), terminating glycoproteins or GSLs (15) (Fig. 1c). Two Ggta1 genes have been identified on chromosomes 9 and 12 in humans and shown to be inactive pseudogenes (27, 28), suggesting this enzyme activity is lacking in humans. This is consistent with the presence of a high natural antibody titer specific for Galα1–3 in humans and Old World primates (29, 30). The blood group B gene product acts exclusively on Fucose α1–2Galβ1-R terminating glycoproteins or GSLs (Fig. 1c) but does not produce an epitope recognized by the anti-Galα1–3 antibodies (19, 30).
We report here that, using a highly sensitive HPLC assay, we cannot detect iGb3 or iGb4 in mouse thymus or DCs. In a survey of mouse organs, iGb3 or iGb4 could be detected solely in the dorsal root ganglion (DRG) in wild-type mice. As would be predicted, these GSLs were stored in the DRG of α-galactosidase A-deficient (α-Gal A−/−, iGb3, Fig. 1b) and hexb−/− (iGb4) mice, respectively. However, when human thymus and DCs were analyzed, iGb3 and iGb4 were not detected. These data suggest that iGb3 is unlikely to be a natural iNKT cell-selecting ligand in the thymus of mouse or humans. Likewise, iGb3 is unlikely to be a peripheral iNKT cell-activating ligand in mouse or human DCs, despite its modest ability in vitro to stimulate iNKT cells (31).
Results
Generation of Standards and Validation of iGb3 and iGb4 Detection.
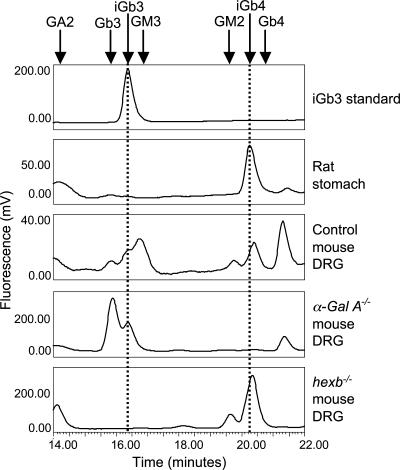
The identification of tissue- and cell-derived GSL oligosaccharides was ascertained by comparison of the retention times of detected peaks with those of authentic standards. A commercially available iGb3 trisaccharide and intact iGb3 GSL were both analyzed. The intact lipid was fully sensitive to ceramide glycanase digestion, consistent with previous studies for other GSL standards (32) (data not shown). The iGb3 trisaccharide and intact GSL-derived oligosaccharide gave the same retention times (data not shown). Using normal-phase HPLC (NP-HPLC), it was possible to separate iGb3 and Gb3 into discrete peaks (Fig. 2). For iGb4, no commercial standard was available. However, the major species in a neutral lipid preparation from rat stomach has been reported to be iGb4 (22). Therefore, neutral GSL-derived oligosaccharides from rat stomach were prepared and anthranilic acid (2AA) labeled. NP-HPLC revealed one major peak (Fig. 2), which was confirmed to be iGb4 by enzyme digests (data not shown). In contrast, the neutral GSL preparation from C57BL/6J mouse stomach did not reveal a peak for iGb4 (SI Table 1), and only Gb4 could be identified in this tissue.
Fig. 2.
NP-HPLC profile of mouse, rat, and commercial GSL-derived oligosaccharides. The iGb3 and iGb4 elution region of the chromatogram, for GSL-derived oligosaccharides from mouse DRG and rat stomach neutral GSLs, is shown. Dotted lines indicate the elution times of iGb3 and iGb4. α-Gal A−/− mice were aged 40 wk, and hexb−/− and controls were aged 11 wk.
Mouse DRG.
In normal mouse tissues and organs [brain, liver, kidney, spleen, thymus, testis, lung, stomach, intestine, eye (lens and retina), spinal cord, and plasma], isoglobo-series GSLs were undetectable (data not shown; summarized in SI Table 1). iGb3 was detected only in mouse DRG (≈40 fmol/μg protein) (Fig. 2). Furthermore, in the mouse model of Fabry disease (α-Gal A−/−), there was storage of iGb3 250-fold relative to wild type (10 pmol/μg protein; Fig. 2), as would be predicted as iGb3 is a substrate for α-galactosidase A (Fig. 1b). This was the only tissue in the α-Gal A−/− mouse where iGb3 was detectable (eye, stomach, intestine, and spleen were also analyzed, and all were negative).
As predicted (Fig. 1b), in the DRG from hexb−/− mice, there was storage of iGb4 (Fig. 2). iGb4 was undetectable in eye, spleen, stomach, and intestine of the hexb−/− mouse (data not shown). However, in contrast to iGb3 and Gb3, which run as discrete peaks, iGb4 and Gb4 run as one broad peak. Their identities, therefore, were confirmed after β-hex digestion that resulted in the generation of iGb3/Gb3 that were readily separated (data not shown).
Mouse Thymus.
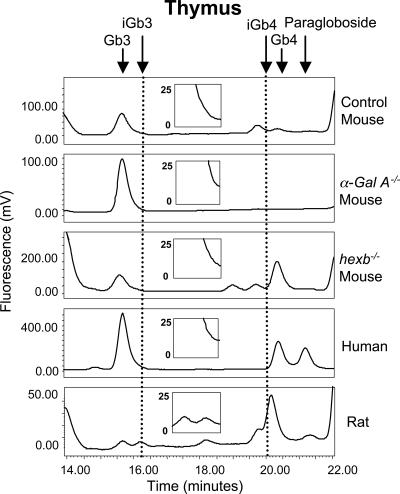
The thymus expresses multiple GSL species, including those of the globo series (Gb3 and Gb4). For example, Gb3 is present at similar levels in control and hexb−/− mice, which indicates the synthesis of this species in mouse thymus. Additionally, we established that the GSL profile of thymus was not developmentally regulated, because there was no detectable difference in the GSL profile from 10-day- and 4-, 9-, and 40-week-old control mouse thymi (data not shown). However, iGb3 or iGb4 were undetectable in control mouse thymus (Fig. 3). However, storage of iGb3 in α-Gal A−/− and iGb4 in hexb−/− thymus would be expected if isoglobo-series GSLs were present in this tissue, as shown for the DRG (Fig. 2). Because iGb3 and iGb4 could not be detected in the thymus of either of these mutant mice (Fig. 3), isoglobo series appear not to be expressed in mouse thymus.
Fig. 3.
NP-HPLC profile of mouse, human, and rat thymus neutral GSL-derived oligosaccharides. Mouse and human profiles are representative of three independent experiments, and the rat profile is representative of four independent experiments. Dotted lines indicate the elution times of iGb3 and iGb4 standards. α-Gal A−/− mice were aged 38 wk, and hexb−/− and control mice were 9 wk. (Insets) Profiles scaled to that of the rat thymus where iGb3 is detectable.
Detection Limit.
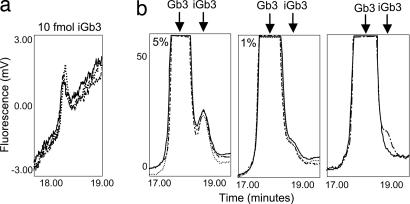
Using an iGb3 GSL-derived oligosaccharide standard, it was possible to detect a peak of 10 fmol (Fig. 4a) in the HPLC assay. The sensitivity of the assay is limited not by quantity of material but by the ratio of Gb3 to iGb3, because there is a limit to the degree to which these two molecules, which differ only in their linkages (Galα1–3 vs. Galα1–4) can be separated by HPLC. Using mixtures of Gb3 and iGb3 oligosaccharides, it was possible to detect iGb3 at 1% the level of Gb3 (Fig. 4b). In mouse thymocytes, Gb3 is present at ≈20,000 copies per cell, therefore 200 copies of iGb3 per cell would be detectable.
Fig. 4.
NP-HPLC profile of Gb3 and iGb3 standard oligosaccharide mixtures. (a) The profile shows the detection of 10 fmol of iGb3 standard oligosaccharide. (b) Gb3 was coinjected with different quantities of iGb3 (5% and 1%), as shown. The final image is the overlay of Gb3 alone (solid line) and Gb3 with 1% iGb3 (dashed line). This demonstrates that the presence of iGb3 at 1% the level of Gb3 is readily detected. The profiles are the overlay of three independent injections of standards.
Human Thymus and Other Tissues.
Four independent pediatric thymus samples were analyzed. The neutral GSL-derived oligosaccharide profile revealed that the major neutral species present were Gb3 and Gb4, with no evidence for the presence of iGb3 or iGb4 (Fig. 3). Additionally, human DRG, spinal cord, cerebrospinal fluid, and blood were processed and analyzed. All were found to express Gb3 and Gb4, but iGb3 and iGb4 were undetectable (data not shown; summarized in SI Table 1).
Distribution of Isoglobo-Series GSLs in Rat Tissues.
iGb4 (and to a lesser extent iGb3) were readily detected in rat thymus (Fig. 3). In rat thymus, iGb3 was present at ≈5,000 copies/cell and Gb3 at 6,000 copies per cell.
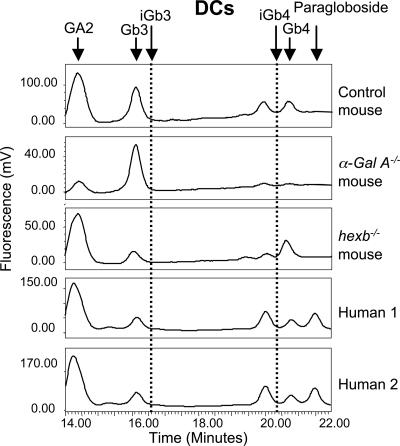
Mouse and Human DCs.
iGb3 and iGb4 were undetectable in the control mouse bone marrow-derived DC neutral GSL fraction (Fig. 5). This was further supported by the absence of iGb4 storage in hexb−/− DC and the absence of iGb3 storage in α-Gal A−/− DC (Fig. 5). Similarly, we failed to detect iGb3 or iGb4 in human DCs (Fig. 5). Additionally, neither human nor mouse DCs, after stimulation with Toll-like receptor agonists, had detectable iGb3 (data not shown).
Fig. 5.
NP-HPLC profile of mouse and human DC neutral GSL-derived oligosaccharides. Mouse profiles are representative of three independent experiments. Dotted lines indicate the elution times of iGb3 and iGb4 standards. α-Gal A−/− and control mice were aged 16 wk, and hexb−/− mice were aged 7 wk.
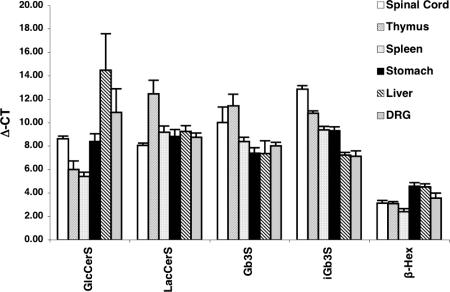
Quantitative PCR (Q-PCR) Analysis of Biosynthetic and Catabolic Gene Expression.
The expression level of transcripts for the catabolic enzyme, β-hex, was higher than that of the biosynthetic enzymes, irrespective of the tissue analyzed (Fig. 6). Expression level of ceramide glucosyltransferase varied among different organs, with higher mRNA expression in the spleen and thymus and lower expression in the liver. LacCer synthase mRNA expression appeared to be uniform among the tissues (with the exception of the thymus where its expression was lower). The expression level of iGb3S varied among the different organs tested, spinal cord having the lowest expression and DRG and liver the highest. DRG does have slightly higher expression levels of iGb3S than thymus, but the level was comparable to liver, where iGb3 was undetectable by NP-HPLC (data not shown). mRNA expression of Gb3S was also highest in the DRG and liver and lowest in thymus.
Fig. 6.
Q-PCR analysis of GSL synthetic and degradative enzyme expression in different organs. Δ-Ct values (inversely proportional to transcript levels) were calculated as described in Materials and Methods. The average value with the standard error of the mean is displayed from three experiments.
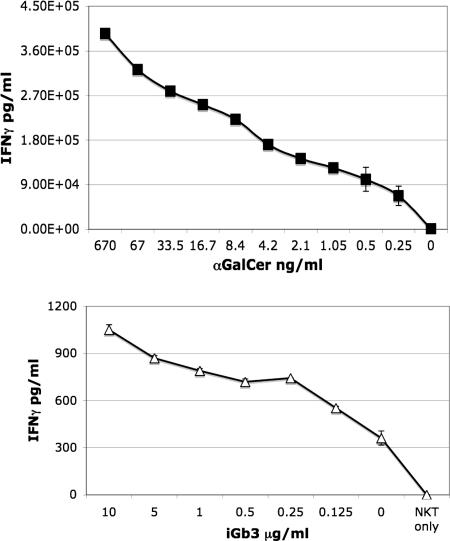
iGb3 and α-Galactosylceramide (αGalCer) Titration.
Using commercially available iGb3 GSL we pulsed C1R-CD1d cells with iGb3 and αGalCer to determine their relative potencies. Although αGalCer could sensitize secretion of IFN-γ from human iNKT cells at doses <0.25 ng/ml (Fig. 7 Upper), 1,000-fold higher doses of iGb3 (μg/ml range) were required to sensitize human iNKT cells to secrete detectable IFN-γ, and still the response was 100-fold lower (the difference therefore being five orders of magnitude; Fig. 7 Lower). These results are in agreement with a previous report (31). The μg/ml levels of iGb3 required to get detectable biological activity would be readily detectable by NP-HPLC under the experimental conditions used for the analysis of mouse or human thymus and DCs.
Fig. 7.
Stimulation of a human iNKT cell line with C1R-CD1d cells pulsed with different concentrations of iGb3 or αGalCer. IFN-γ release from iNKT cells in picograms per milliliter is plotted against the αGalCer (nanograms per milliliter in vehicle) or iGb3 (micrograms per milliliter in methanol) concentrations pulsed onto C1R-CD1d cells. No toxicity was observed at any concentration of either compound. NKT denotes iNKT cells alone with no C1R-CD1d cells, and 0 is the vehicle (no added αGalCer/iGb3) control showing background recognition of endogenous ligands. The experiment was performed twice in duplicate, and the average with the standard error of the mean is displayed.
Discussion
We have used a highly sensitive NP-HPLC assay to study the distribution of iGb3 in tissues from mouse, rat and human. iGb3 was found only in the DRG of control (C57BL/6J) mice and was stored in α-Gal A−/− mouse DRG. Similarly, iGb4 was detectable only in DRG of control mice and was stored in hexb−/− mice DRG (Fig. 2). We reasoned that if the isoglobo-series biosynthetic pathway were active in the mouse thymus or DCs, albeit at low levels, we should detect iGb3 in α-Gal A−/− and iGb4 in hexb−/− thymus or DCs because of the high levels of storage of GSLs with terminal Gal or GalNAc that occur in these engineered mice. However, when we analyzed neutral GSLs from the thymus and DCs of α-Gal A−/− and hexb−/− mice, we still failed to detect iGb3 and iGb4 (Figs. 3 and 5). When four human thymi and two human monocyte-derived DC samples were analyzed, they also lacked detectable isoglobo-series GSLs (Figs. 3 and 5). Using iGb3 and Gb3 of known concentration, it was possible by NP-HPLC to detect 10 fmol of iGb3 standard and to detect iGb3 when present at 1% the level of Gb3 (Fig. 4). Taken together, these data suggest that isoglobo-series GSLs are not expressed in mouse or human thymus or DCs.
Q-PCR analysis of biosynthetic and degradative enzyme expression indicates that measurement of mRNA expression does not relate to the amount of detected GSL. DRG (where isoglobo-series GSLs are detectable; Fig. 2) had higher iGb3S expression compared with thymus, but at a level comparable to liver where iGb3 was undetectable. This is not surprising, because the complement of GSLs found in a given tissue is the net result of differential flux through a number of competing pathways, all of which utilize LacCer as precursor substrate (Fig. 1a). It is therefore the relative flux through each competing pathway, catalyzed by specific glycosyltransferases, that determines the GSL repertoire detected in that tissue. Measuring transcript levels and enzyme activity does not, therefore, accurately predict the GSL complement found in any particular cell line or tissue. This is further compounded by the fact that different GSL species will also differ in their half-lives because of differences in their rates of turnover. There is, therefore, no substitute for measuring GSLs quantitatively in tissues to definitively determine the repertoire of GSLs present.
When the tissue and species pattern of expression of isoglobo-series GSLs was examined (see SI Table 1), it was clear that the rat utilizes this biosynthetic pathway in several tissues (17, 20–25). In mouse, iGb3 and iGb4 were detected only in DRG, among the tissues screened, including the CNS. This is in agreement with a previous report, where kidney from rat, but not from mouse, human, or pig, had the Galα1–3 linkage in ceramide tetrasaccharides (23). The reason for this differential pattern of GSL expression in rat and mouse is unknown. These data suggest that the balance of the competing transferases is both species- and tissue-specific.
The original suggestion that iGb3 was the iNKT cell-selecting ligand in the thymus arose because of the observed iNKT cell deficiency of hexb−/− mice (12). However, the α-Gal A−/− mouse has also been reported to have reduced numbers of iNKT cells (33, 34). If iGb3 were the natural ligand, these results would be hard to explain, because the amount of iGb3 would likely be increased (as a storage product) and would not, therefore, lead to a reduction in the number of iNKT cells. Reduced iNKT cell frequencies have also been reported in the NPC1 mouse (a model of Niemann–Pick disease type C 1, a lysosomal storage disorder) (34, 35) and the GM1 gangliosidosis mouse (β-galactosidase-deficient) (34). The iNKT cell deficiency in these mouse models cannot be explained by impaired generation of iGb3. For example, β-galactosidase deficiency in GM1 gangliosidosis plays no part in isoglobo-series GSL catabolism. As is clear from this study, iGb3 does not appear to be expressed in mouse thymus (Fig. 3) and therefore loss of iNKT cells in hexb−/− mice cannot be attributed to the involvement of iGb3.
In studies proposing a role for iGb3, there was no direct biochemical evidence for the presence of iGb3 (12). Instead, a lectin was used to block recognition of self ligand/CD1d complex by iNKT cells [IB4 from Griffonia (Bandeiraea) simplicifolia) (3, 12)]. However, multiple studies have demonstrated that this lectin is not specific for Galα1–3 but instead recognizes Galα1, -2, -3, -4, and -6 linkages (36–38).
There are currently only three known enzyme-catalyzed pathways by which galactose can be added in an α1–3 linkage (Fig. 1c): first, by iGb3S, exclusively to LacCer to form iGb3 and also to iGb3 to form poly Galα1–3 structures; second, Ggta1 adds exclusively to Galβ1–4GlcNAc structures present on glycoproteins or GSLs (e.g., paragloboside). Finally, the product of the human blood group B gene transfers galactose exclusively to Fucose α1–2Galβ1-R (where R is a GSL or a glycoprotein). Galα1–3 is an antigenic epitope in humans and Old World monkeys (29, 30). The Galα1–3 epitopes generated by Ggta1 are recognized by anti-Galα1–3 antibodies (19, 30, 39), which is not the case for the blood group B epitope because of the presence of the branched Fucose structure (19, 30). However, it is not clear whether iGb3 is recognized by the natural anti-Galα1–3 antibody repertoire present in humans. To date, this has been studied only by using a limited number of monoclonal antibodies (12, 16, 19, 39).
The presence of the anti-Galα1–3 antibodies correlates with the silencing of Ggta1 genes in Old World primates and humans (40). However, it is not clear whether iGb3S is silenced in Old World primates and humans. Monoclonal antibodies failed to bind to human tissue samples, and mRNA was not detected by Northern blot or RT-PCR from a range of human tissues (16, 41). Humans, therefore, are apparently unable to transcribe a functional iGb3S gene, which would be consistent with our inability to detect iGb3 in any of the human tissue samples analyzed in this study.
The present findings do not question the ability of iGb3 to serve as an iNKT cell ligand in vitro. However, there are inconsistent findings on the relative potency of iGb3 compared with αGalCer. They were reported to be equivalent (12), whereas in this study (Fig. 7) and that of Xia et al. (31), iGb3 was at least 5 orders of magnitude less potent than αGalCer. The reasons for these discrepancies are currently unclear and could relate to technical differences in the assays used. Despite the in vitro activity of iGb3 to stimulate iNKT cells, the findings in this study suggest it may not be a physiologically relevant iNKT cell-selecting ligand in vivo in the mouse and human thymus.
In view of the conservation of the CD1d/iNKT system, it can be speculated that, if there is a conserved endogenous GSL ligand(s) in the thymus of mammals, the analysis carried out in this study (Fig. 3) may be a first step toward its/their identification. There are numerous known and unknown GSLs species observed and potentially conserved between mouse, rat and human thymus (Fig. 3 and data not shown), and mouse and human DCs (Fig. 5 and data not shown).
In conclusion, this study does not support a role for iGb3 as the selecting ligand for iNKT cells in the thymus of mouse or human and suggests that other GSLs, or indeed other lipid species, need to be investigated. The analytical approach used in this study may help identify conserved GSL species that are candidates for playing a role in iNKT cell selection in the thymus.
Materials and Methods
Reagents.
Chemicals were purchased from Sigma-Aldrich (Poole, U.K.) and/or VWR (Lutterworth, U.K.), unless otherwise stated. All solvents were of analytical grade or higher (VWR, unless otherwise stated). HPLC standards were from Dextra Laboratories (Reading, U.K., iGb3 trisaccharide) or Alexis (Axxora U.K., Nottingham, U.K.; iGb3 lipid), prepared in-house (iGb4 (22), or kindly provided by Eric Samain (Centre de Recherches sur les Macromolécules Végétales–Centre Nationale de la Recherche Scientifique, Grenoble, France; Gb3). Tissue culture media were from GIBCO (Paisley, U.K.), unless otherwise stated. Water was Milli Q grade.
Animals.
Animals were housed and euthanized in accordance with the U.K. Home Office Animals (Scientific Procedures) Act, 1986. Mouse models of GSL storage disease were maintained and genotyped according to published methods [Fabry (α-Gal A−/−) (42) and SH (hexb−/−) (43) (both on C57BL/6J background)].
Cell Culture, GSL Extraction, Ceramide Glycanase Digestion, and 2AA Labeling.
Mouse DCs were prepared according to published methods (3). Blood was purchased from the United Kingdom National Blood Service, and human DCs were generated as described (44). An intact lobe of human thymus (containing cortex and medulla) was obtained after corrective congenital heart defect surgery. Human DRG and spinal cord were obtained from R. Liguori (University of Bologna, Bologna, Italy). GSL extraction, ceramide glycanase digestion, and 2AA labeling were performed as described (32).
Ion Exchange Chromatography.
QAE-Sephadex (Amersham Biosciences, Chalfont St. Giles, U.K.) in acetate form was placed in a 1.5-ml solid-phase extraction column (Alltech Associates, Carnforth, U.K.). The 2AA-labeled sample was applied and washed with 3 × 1 ml of water and neutral sugars eluted with 2 × 1 ml of 0.2 M acetic acid. Charged sugars were eluted with 2 × 1 ml 0.5 M ammonium acetate. Free 2AA label was removed by the addition of 3 volumes of ethyl acetate to 2 volumes of sample with mixing, spun (6,000 × g, 30 s), the upper phase was discarded, and the lower phase was extracted twice with ethyl acetate. Lower phases were dried (SpeedVac) and resuspended in water.
NP- HPLC.
NP-HPLC was performed as published (32). The sample was injected in water:acetonitrile 3:7 (vol/vol). Known (microgram) amounts of synthesized GSL oligosaccharides (as above) were 2AA-labeled and purified, and equimolar (1 pmol) amounts were analyzed by using NP-HPLC. Peaks were identified by using integration software (Waters Empower, Milford, MA). Peak areas were measured, and no significant differences in molar response factors were observed. Therefore, the peak area per picomole of GSL-derived oligosaccharide present can be determined and used to quantify the concentration of GSL in tissue and cell samples. Injections of different cell number equivalents of mouse and rat thymocytes allowed the calculation of picomolar GSL per thymocyte. Using Avogadro's number (6.03 × 1023), this was then converted to molecules per cell.
Enzyme Digests.
Coffee-bean α-galactosidase (Boerhinger, Mannheim, Germany), α1–3,6 galactosidase (Sigma, St. Louis, MO), and Jack Bean β-hex [purified in-house (45)] were used. GSL-derived 2AA-labeled oligosaccharides were dried in a SpeedVac and incubated overnight at 37°C with enzymes according to the manufacturer's instructions. Samples were made up to 30 μl with water, and 30 μl of acetonitrile was added, spun (4,000 × g, 45 min) through 10,000 Mr spin filters (Millipore, Watford, U.K.), and prewashed with 150 μl of water (4,000 × g, 45 min) to remove protein before NP-HPLC.
Analysis of Gene Expression by Q-PCR Analysis.
Total RNA was extracted by using guanidium isothiocyanate and isolated by ultracentrifugation over a cesium chloride cushion. cDNA was synthesized from 1 μg of total RNA with random hexamer primers and SuperScript reverse transcriptase (Promega, Madison, WI) by using standard procedures. cDNA (50 ng) were used as templates for PCR amplification by using the SYBR Green Master Mix and the ABI PRISM 7000 sequence detection system (Applied Biosystems, Foster City, CA). Primers (SI Table 2) specific for ceramide glucosyltransferase, LacCer synthase, Gb3S synthase, iGb3S, and β-hex were designed by the Primer Express Program (Applied Biosystems) and used for amplification in triplicate assays. For graphical representation of Q-PCR data, Δ-Ct values were obtained by deducting the raw cycle threshold (Ct values) obtained for GAPDH mRNA, the internal standard, from the Ct values obtained for investigated genes.
iGb3 and αGalCer Titration.
C1R-CD1d cells (5 × 104) were incubated with 3 × 104 NKT cells in a 96-well plate, in the presence or absence of different concentrations of αGalCer (Kirin Ltd., Gunma, Japan) or iGb3 (Axxora). The responder iNKT cell line was derived from healthy donors (46). The cell line is 98% CD1d αGalCer tetramer+ Vα24+ Vβ11+ (65% CD4+ and 35% CD4/8−). αGalCer (200 μg/ml in 150 mM NaCl and 0.5% Tween) was sonicated and diluted in complete medium. iGb3 was resuspended at 1 mg/ml in methanol, sonicated, and diluted in completed medium. Unpulsed control cells contained amounts of vehicle or methanol similar to those for the cells pulsed with the highest concentration of lipid. Supernatants (36 h) were analyzed by ELISA (IFN-γ; PharMingen, San Diego, CA).
Supplementary Material
Acknowledgments
We thank C. Vendeville for technical assistance and Kirin, Ltd., for the αGalCer. This work was funded by Cancer Research UK (Grant C399-A2291), the European Community (Grant DC-VACC LSHB-CT-503037), the Medical Research Council (V.C. and M.S.), Institut National de la Santé et de la Recherche Médicale (J.F. and F.T.), the Wellcome Trust (N.P.), and the National Institutes of Health Grant DK066917 (to M.A.E.). A.O.S., F.M.P., T.D.B., D.A.P., D.C.A.N., and T.H. were supported by the Glycobiology Institute endowment.
Abbreviations
- NKT
natural killer T
- iNKT
invariant NKT
- GSL
glycosphingolipid
- iGb3
isoglobotrihexosylceramide
- DC
dendritic cell
- DRG
dorsal root ganglion
- SH
Sandhoff disease
- hexb−/−
β-hexosaminidase A and B deficient
- β-hex
β-hexosaminidase
- Gb3
globotrihexosylceramide
- Gb4
globotetrahexosylceramide
- iGb4
isoglobotetrahexosylceramide
- iGb3S
iGb3 synthase
- Gal
galactose
- Galα1–3
α1–3-linked Gal
- LacCer
lactosylceramide
- Ggta1
α1–3 galactosyltransferase
- α-Gal A−/−
deficient in α-galactosidase A
- NP-HPLC
normal-phase HPLC
- 2AA
anthranilic acid
- Q-PCR
quantitative PCR
- αGalCer
α-galactosylceramide.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
See Commentary on page 5713.
GSL abbreviations are according to the International Union of Pure and Applied Chemistry–International Union of Biochemistry and Molecular Biology Commission (1977).
This article contains supporting information online at www.pnas.org/cgi/content/full/0607285104/DC1.
References
- 1.Brigl M, Brenner MB. Annu Rev Immunol. 2004;22:817–890. doi: 10.1146/annurev.immunol.22.012703.104608. [DOI] [PubMed] [Google Scholar]
- 2.Kinjo Y, Wu D, Kim G, Xing GW, Poles MA, Ho DD, Tsuji M, Kawahara K, Wong CH, Kronenberg M. Nature. 2005;434:520–525. doi: 10.1038/nature03407. [DOI] [PubMed] [Google Scholar]
- 3.Mattner J, Debord KL, Ismail N, Goff RD, Cantu C, III, Zhou D, Saint-Mezard P, Wang V, Gao Y, Yin N, et al. Nature. 2005;434:525–529. doi: 10.1038/nature03408. [DOI] [PubMed] [Google Scholar]
- 4.Wu D, Xing GW, Poles MA, Horowitz A, Kinjo Y, Sullivan B, Bodmer-Narkevitch V, Plettenburg O, Kronenberg M, Tsuji M, et al. Proc Natl Acad Sci USA. 2005;102:1351–1356. doi: 10.1073/pnas.0408696102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Godfrey DI, MacDonald HR, Kronenberg M, Smyth MJ, Van Kaer L. Nat Rev Immunol. 2004;4:231–237. doi: 10.1038/nri1309. [DOI] [PubMed] [Google Scholar]
- 6.Kronenberg M, Gapin L. Nat Rev Immunol. 2002;2:557–568. doi: 10.1038/nri854. [DOI] [PubMed] [Google Scholar]
- 7.Stanic AK, De Silva AD, Park JJ, Sriram V, Ichikawa S, Hirabyashi Y, Hayakawa K, Van Kaer L, Brutkiewicz RR, Joyce S. Proc Natl Acad Sci USA. 2003;100:1849–1854. doi: 10.1073/pnas.0430327100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiu YH, Park SH, Benlagha K, Forestier C, Jayawardena-Wolf J, Savage PB, Teyton L, Bendelac A. Nat Immunol. 2002;3:55–60. doi: 10.1038/ni740. [DOI] [PubMed] [Google Scholar]
- 9.Honey K, Benlagha K, Beers C, Forbush K, Teyton L, Kleijmeer MJ, Rudensky AY, Bendelac A. Nat Immunol. 2002;3:1069–1074. doi: 10.1038/ni844. [DOI] [PubMed] [Google Scholar]
- 10.Zhou D, Cantu C, III, Sagiv Y, Schrantz N, Kulkarni AB, Qi X, Mahuran DJ, Morales CR, Grabowski GA, Benlagha K, et al. Science. 2004;303:523–527. doi: 10.1126/science.1092009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang SJ, Cresswell P. Nat Immunol. 2004;5:175–181. doi: 10.1038/ni1034. [DOI] [PubMed] [Google Scholar]
- 12.Zhou D, Mattner J, Cantu C, III, Schrantz N, Yin N, Gao Y, Sagiv Y, Hudspeth K, Wu Y, Yamashita T, et al. Science. 2004;306:1786–1789. doi: 10.1126/science.1103440. [DOI] [PubMed] [Google Scholar]
- 13.Sheikh KA, Sun J, Liu Y, Kawai H, Crawford TO, Proia RL, Griffin JW, Schnaar RL. Proc Natl Acad Sci USA. 1999;96:7532–7537. doi: 10.1073/pnas.96.13.7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamashita T, Hashiramoto A, Haluzik M, Mizukami H, Beck S, Norton A, Kono M, Tsuji S, Daniotti JL, Werth N, et al. Proc Natl Acad Sci USA. 2003;100:3445–3449. doi: 10.1073/pnas.0635898100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taylor SG, McKenzie IF, Sandrin MS. Glycobiology. 2003;13:327–337. doi: 10.1093/glycob/cwg030. [DOI] [PubMed] [Google Scholar]
- 16.Milland J, Christiansen D, Sandrin MS. Immunol Cell Biol. 2005;83:687–693. doi: 10.1111/j.1440-1711.2005.01398.x. [DOI] [PubMed] [Google Scholar]
- 17.Breimer ME, Hansson GC, Karlsson KA, Leffler H. J Biol Chem. 1982;257:557–568. [PubMed] [Google Scholar]
- 18.Sung SS, Sweeley CC. Biochim Biophys Acta. 1979;575:295–298. doi: 10.1016/0005-2760(79)90031-6. [DOI] [PubMed] [Google Scholar]
- 19.Teneberg S, Lonnroth I, Torres Lopez JF, Galili U, Halvarsson MO, Angstrom J, Karlsson KA. Glycobiology. 1996;6:599–609. doi: 10.1093/glycob/6.6.599. [DOI] [PubMed] [Google Scholar]
- 20.Ogiso M, Ohta M, Irie A, Hoshi M, Komoto M. Exp Eye Res. 1995;60:193–198. doi: 10.1016/s0014-4835(95)80010-7. [DOI] [PubMed] [Google Scholar]
- 21.Arita H, Kawanami J. J Biochem (Tokyo) 1977;81:1661–1664. doi: 10.1093/oxfordjournals.jbchem.a131625. [DOI] [PubMed] [Google Scholar]
- 22.Bouhours D, Bouhours JF. J Biol Chem. 1985;260:2172–2177. [PubMed] [Google Scholar]
- 23.Siddiqui B, Kawanami J, Li YT, Hakomori S. J Lipid Res. 1972;13:657–662. [PubMed] [Google Scholar]
- 24.Stults CL, Sweeley CC, Macher BA. Methods Enzymol. 1989;179:167–214. doi: 10.1016/0076-6879(89)79122-9. [DOI] [PubMed] [Google Scholar]
- 25.Dumonceaux T, Carlsen SA. Arch Biochem Biophys. 2001;389:187–194. doi: 10.1006/abbi.2001.2320. [DOI] [PubMed] [Google Scholar]
- 26.Keusch JJ, Manzella SM, Nyame KA, Cummings RD, Baenziger JU. J Biol Chem. 2000;275:25308–25314. doi: 10.1074/jbc.M002629200. [DOI] [PubMed] [Google Scholar]
- 27.Joziasse DH, Shaper JH, Jabs EW, Shaper NL. J Biol Chem. 1991;266:6991–6998. [PubMed] [Google Scholar]
- 28.Larsen RD, Rivera-Marrero CA, Ernst LK, Cummings RD, Lowe JB. J Biol Chem. 1990;265:7055–7061. [PubMed] [Google Scholar]
- 29.Lee J, Cairns T, McKane W, Rashid M, George AJ, Taube D. Transplantation. 1998;66:1117–1119. doi: 10.1097/00007890-199810270-00028. [DOI] [PubMed] [Google Scholar]
- 30.Galili U, Macher BA, Buehler J, Shohet SB. J Exp Med. 1985;162:573–582. doi: 10.1084/jem.162.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xia C, Yao Q, Schumann J, Rossy E, Chen W, Zhu L, Zhang W, De Libero G, Wang PG. Bioorg Med Chem Lett. 2006;16:2195–2199. doi: 10.1016/j.bmcl.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 32.Neville DC, Coquard V, Priestman DA, te Vruchte DJ, Sillence DJ, Dwek RA, Platt FM, Butters TD. Anal Biochem. 2004;331:275–282. doi: 10.1016/j.ab.2004.03.051. [DOI] [PubMed] [Google Scholar]
- 33.Prigozy TI, Naidenko O, Qasba P, Elewaut D, Brossay L, Khurana A, Natori T, Koezuka Y, Kulkarni A, Kronenberg M. Science. 2001;291:664–667. doi: 10.1126/science.291.5504.664. [DOI] [PubMed] [Google Scholar]
- 34.Gadola SD, Silk JD, Jeans A, Illarionov PA, Salio M, Besra GS, Dwek R, Butters TD, Platt FM, Cerundolo V. J Exp Med. 2006;203:2293–2303. doi: 10.1084/jem.20060921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sagiv Y, Hudspeth K, Mattner J, Schrantz N, Stern RK, Zhou D, Savage PB, Teyton L, Bendelac A. J Immunol. 2006;177:26–30. doi: 10.4049/jimmunol.177.1.26. [DOI] [PubMed] [Google Scholar]
- 36.Kirkeby S, Moe D. Immunol Cell Biol. 2001;79:121–127. doi: 10.1046/j.1440-1711.2001.00992.x. [DOI] [PubMed] [Google Scholar]
- 37.Kirkeby S, Winter HC, Goldstein IJ. Xenotransplantation. 2004;11:254–261. doi: 10.1111/j.1399-3089.2004.00108.x. [DOI] [PubMed] [Google Scholar]
- 38.Wu AM, Song SC, Wu JH, Kabat EA. Biochem Biophys Res Commun. 1995;216:814–820. doi: 10.1006/bbrc.1995.2694. [DOI] [PubMed] [Google Scholar]
- 39.Galili U, Basbaum CB, Shohet SB, Buehler J, Macher BA. J Biol Chem. 1987;262:4683–4688. [PubMed] [Google Scholar]
- 40.Galili U, Clark MR, Shohet SB, Buehler J, Macher BA. Proc Natl Acad Sci USA. 1987;84:1369–1373. doi: 10.1073/pnas.84.5.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Milland J, Christiansen D, Lazarus BD, Taylor SG, Xing PX, Sandrin MS. J Immunol. 2006;176:2448–2454. doi: 10.4049/jimmunol.176.4.2448. [DOI] [PubMed] [Google Scholar]
- 42.Ohshima T, Murray GJ, Swaim WD, Longenecker G, Quirk JM, Cardarelli CO, Sugimoto Y, Pastan I, Gottesman MM, Brady RO, et al. Proc Natl Acad Sci USA. 1997;94:2540–2544. doi: 10.1073/pnas.94.6.2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sango K, McDonald MP, Crawley JN, Mack ML, Tifft CJ, Skop E, Starr CM, Hoffmann A, Sandhoff K, Suzuki K, et al. Nat Genet. 1996;14:348–352. doi: 10.1038/ng1196-348. [DOI] [PubMed] [Google Scholar]
- 44.Salio M, Shepherd D, Dunbar PR, Palmowski M, Murphy K, Wu L, Cerundolo V. J Immunol. 2001;167:1188–1197. doi: 10.4049/jimmunol.167.3.1188. [DOI] [PubMed] [Google Scholar]
- 45.Li SC, Li YT. J Biol Chem. 1970;245:5153–5160. [PubMed] [Google Scholar]
- 46.Gadola SD, Dulphy N, Salio M, Cerundolo V. J Immunol. 2002;168:5514–5520. doi: 10.4049/jimmunol.168.11.5514. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.