Abstract
The Ll.LtrB intron from the Gram-positive bacterium Lactococcus lactis is one of the most studied bacterial group II introns. Ll.LtrB interrupts the relaxase gene of three L. lactis conjugative elements. The relaxase enzyme recognizes the origin of transfer (oriT ) and initiates the intercellular transfer of its conjugative element. The splicing efficiency of Ll.LtrB from the relaxase transcript thus controls the conjugation level of its host element. Here, we used the level of sex factor conjugation as a read-out for Ll.LtrB splicing efficiency. Using this highly sensitive splicing/conjugation assay (107-fold detection range), we demonstrate that Ll.LtrB can trans-splice in L. lactis when fragmented at various positions such as: three different locations within domain IV, within domain I and within domain III. We also demonstrate that the intron-encoded protein, LtrA, is absolutely required for Ll.LtrB trans-splicing. Characteristic Y-branched trans-spliced introns and ligated exons are detected by RT-PCR from total RNA extracts of cells harbouring fragmented Ll.LtrB. The splicing/conjugation assay we developed constitutes the first model system to study group II intron trans-splicing in vivo. Although only previously observed in bacterial-derived organelles, we demonstrate that assembly and trans-splicing of a fragmented group II intron can take place efficiently in bacterial cells.
INTRODUCTION
Group II introns are large ribozymes that catalyse their own excision from RNA transcripts through a process called splicing (1–3). Some group II introns that harbour an open reading frame (ORF) are also mobile retroelements, which are capable of invading new DNA sites using an RNA intermediate (2). Group II introns are found in bacteria, archaea and bacterial-derived organelles of lower eukaryotes and higher plants (1).
Splicing of group II introns is achieved by two consecutive transesterification reactions (A) (2,3). The 2′-OH of a bulged adenosine (A) found near the 3′ end of the intron initiates the first nucleophilic attack on the exon 1–intron junction (Figure 1A, step 1). This results in a 2′–5′ branching of the intron on the bulged adenosine and the release of exon 1. The liberated 3′-OH of exon 1 initiates the second nucleophilic attack on the intron–exon 2 junction (Figure 1A, step 2), thereby releasing the intron in the form of a lariat and ligating the two exons. This splicing mechanism is identical to the removal of nuclear eukaryotic introns by the spliceosome, therefore suggesting a common origin for these two classes of introns (1,2,4).
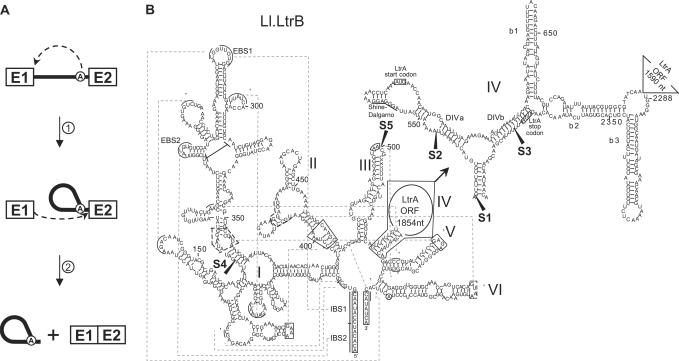
Figure 1.
Group II intron splicing pathway and Ll.LtrB secondary structure. (A) Splicing pathway of group II introns. The 2′OH of the bulged adenosine present in domain VI of the intron (circled A) performs the initial nucleophilic attack on the exon 1–intron junction (step 1), generating a 2′–5′ linkage, also known as a branch point. Then, the 3′-OH of the released exon 1 performs the second nucleophilic attack on the intron–exon 2 junction (step 2), releasing the intron lariat and ligating the flanking exons. Group II intron, black line; exon 1 and 2, E1 and E2; branch point, circled A. (B) Schematic of the Ll.LtrB secondary structure. The six domains of Ll.LtrB are indicated (I–VI) and the detailed secondary structure of a portion of domain IV is shown (top right). The LtrA start and stop codons are boxed and the Shine–Dalgarno sequence is underlined. Exon 1 and 2 on both sides of the intron are also boxed. Pairs of greek letters, linked by dashed lines, correspond to long-range interactions between different portions of the intron. The five chosen Ll.LtrB fragmentation sites are mapped with black arrowheads (S1–S5). EBS1 and EBS2, exon-binding site 1 and 2; IBS1 and IBS2, intron-binding site 1 and 2; branch point, circled A.
Group II introns share very little sequence similarity. However, their secondary structure is universally conserved and is composed of six domains radiating from a central hub (e.g. Figure 1B) (1–3). Domain I is transcribed first, and when folded, it provides the scaffold to dock the other folded intron domains (5). Domain IV plays little if any role in catalysis (2,3) and was shown to be dispensable for splicing in vitro (6). In many cases, the loop region of domain IV contains an ORF expressing an intron-encoded protein (IEP), which provides essential functions for both intron splicing and mobility (1–3,7). Domain V is the catalytic domain of these ribozymes and its sequence is the most conserved among group II introns (1–3,8). Domain VI carries the branch point nucleotide, which is most often an adenosine. The structure of this domain is believed to place the branch point nucleotide near the 5′ splice site in a position that allows initiation of the first nucleophilic attack (Figure 1A, step 1) (8).
In addition to local base pairing, a series of long-range interactions between the various folded domains were characterized (e.g. Figure 1B, greek symbols) (1–3,8). For instance, recognition of the exon–intron boundaries is achieved via long-range interactions between domain I and both exons. In general, domain I harbours the exon-binding sites 1 and 2 (e.g. Figure 1B, EBS1 and EBS2), which are involved in 5′ splice site recognition through base pairing interactions with exon 1. The sequences of these exon-binding sites are complementary to contiguous intron-binding sites 1 and 2 located at the 3′ end of exon 1 (e.g. Figure 1B, IBS1 and IBS2) (3). Similarly, the intron–exon 2 splice junction is recognized by either the δ–δ′ or EBS3–IBS3 interaction between domain I and exon 2 (1,3). Another important long-range interaction, ζ–ζ′, links the GNRA tetraloop of domain V to a phylogenetically conserved stem-loop structure within domain I (e.g. Figure 1B). This tertiary interaction is thought to form the intron catalytic core (8,9). It was previously demonstrated that tertiary interactions allow the assembly and splicing of an intron fragmented within domain IV in vitro, even if no base pairing can take place between the two intron fragments (6).
Remarkably, the fragments of some group II introns that became fragmented by genome rearrangements in organelles have the capacity to re-assemble and splice accurately in vivo (1,9). This phenomenon, known as trans-splicing, ligates exons expressed on two separate transcripts. Most fragmented group II introns are found interrupted within domain IV (1,9). When these introns harbour an IEP within domain IV, the fragmentation site is located either upstream or downstream from the ORF. There are two examples of group II introns fragmented within domain III; they are found in the chloroplastic genomes of the liverwort Marchantia polymorpha and the tobacco plant Nicotiana tabacum (10). In addition, two tripartite group II introns, i.e. introns fragmented into three pieces, have been reported to functionally trans-splice in their respective hosts. The first example ligates exons 1 and 2 of the psaA chloroplast gene of the green alga Chlamydomonas reinhardtii (11). The second example joins exons c and d of the nad5 subunit gene in mitochondria of the angiosperm plant Oenothera berteriana (12). These tripartite introns are both fragmented in domain I upstream of the EBS regions and in domain IV, which, in both cases, do not appear to contain an ORF.
The discovery of group II introns in bacteria greatly facilitated the study of their splicing and mobility pathways. One of the best characterized bacterial group II introns is Ll.LtrB, a 2.5-kb intron found in the low G + C Gram-positive bacterium Lactococcus lactis. It was the first bacterial group II intron shown to splice (13) and mobilize (14) in vivo. The splicing (15–18) and mobility pathways (19–22) of this intron have been extensively studied in vivo.
Ll.LtrB harbours an ORF in domain IV coding for a 599 amino acid protein, LtrA, with three enzymatic activities: reverse transcriptase, maturase and endonuclease (23). The maturase activity of LtrA is essential to promote intron splicing by assisting the folding of this large ribozyme into its catalytically active conformation (19,23). LtrA binds the intron RNA primarily at a distinctive structure found in domain IVa (Figure 1B, DIVa) (16). By binding to this structural motif, the IEP occludes its own Shine–Dalgarno sequence and autoregulates its translation (24).
The Ll.LtrB group II intron is found in three conjugative elements from L. lactis: the pRS01 (13) and pAH90 (25) plasmids, and the L. lactis 712 sex factor, an integrative and conjugative element closely related to pRS01 (26). Interestingly, in all three cases, the intron interrupts the gene coding for a putative relaxase at the same position. The relaxase enzyme is essential for conjugation; it initiates DNA transfer by inflicting a site and strand-specific nick at the origin of transfer (oriT ) of the conjugative element. Then, the enzyme carries one DNA strand to the recipient cell by rolling-circle replication via the mating pore, and re-ligates the ends of the conjugated element (27).
Since the relaxase enzyme is essential for conjugation, splicing of Ll.LtrB is a prerequisite for the intercellular transfer of its host elements (13,26). Klein and colleagues showed that the intimate relationship between splicing and conjugation can be exploited to monitor splicing of Ll.LtrB. They confirmed by real-time RT-PCR analyses that the conjugation efficiency of a mobilizable plasmid is directly proportional to splicing of Ll.LtrB from the relaxase transcript (17).
In this study, we use the Ll.LtrB intron from L. lactis as a model system to study group II intron trans-splicing in bacteria. We developed a splicing/conjugation assay that measures Ll.LtrB splicing efficiency by monitoring sex factor conjugation levels. We used this splicing/conjugation assay to investigate trans-splicing of the lactococcal intron. When fragmented at analogous locations to various naturally occurring fragmentation sites, Ll.LtrB consistently shows the ability to splice in trans, sometimes at very high efficiencies. Furthermore, we demonstrate that the LtrA protein is essential for trans-splicing. These findings constitute the first demonstration and characterization of trans-splicing of a group II intron in bacteria.
EXPERIMENTAL PROCEDURES
Strains and plasmids
Lactococcus lactis strains NZ9800ΔltrB::tet (21) and LM0231 (28) were grown at 30°C without shaking in M17 media supplemented with 0.5% glucose (GM17). Escherichia coli strain DH10β was used for cloning and was grown at 37°C by shaking in LB broth. Antibiotics were added at the following concentrations: 3 µg/ml for tetracycline (Tet), 300 µg/ml for spectinomycin (Spc), 25 µg/ml for fusidic acid (Fus) and 10 µg/ml for chloramphenicol (Cam).
Two 43-bp-long P23 constitutive promoters (P23 right and P23 left) (29) were obtained by annealing two complementary oligonucleotides (Supplementary Table S1, P23 right top/bottom, P23 left top/bottom). The pDL-P232 plasmid was constructed by cloning, in opposite directions, the two P23 constitutive promoters within the pDL278 shuttle plasmid (30). The P23 left promoter is oriented towards the rrnB terminator (PvuI) while the P23 right promoter is directed towards the λ terminator (BamHI). The two promoters are 135 bp apart and initiate transcription in opposite orientations (Figures 2A and 3B). When the oligonucleotides for P23 left were designed, a BssHII cloning site was introduced 6 nucleotides downstream of the transcription initiation site (Supplementary Table S1, P23 left top/bottom). Similarly, a NotI cloning site was introduced at the +6 position of P23 right (Supplementary Table S1, P23 right top/bottom). These two unique restriction sites were subsequently used to clone the various relaxase fragments.
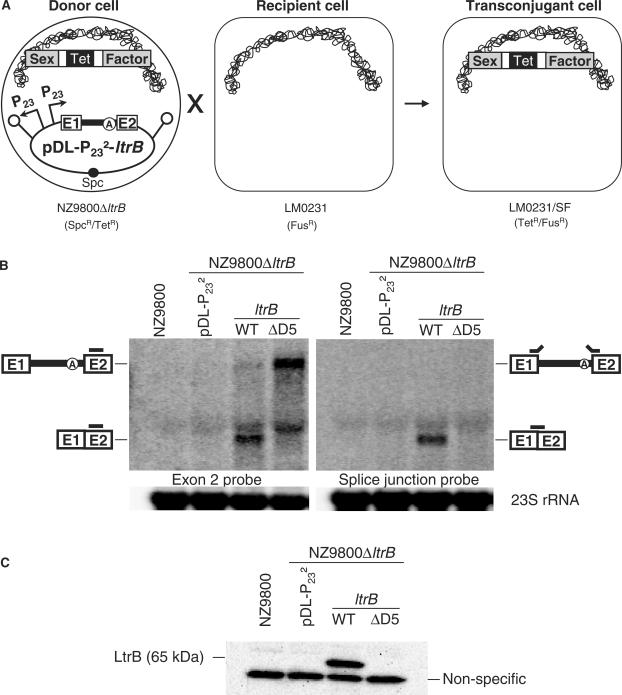
Figure 2.
Ll.LtrB splicing/conjugation assay and relaxase expression levels. (A) Schematic of the Ll.LtrB splicing/conjugation assay. The L. lactis strain used as the donor strain harbours a sex factor that has a defective relaxase (NZ9800▵ltrB::tet) while the recipient strain lacks the sex factor (LM0231). The Ll.LtrB intron, along with portions of its exons, was replaced within the chromosomal sex factor by a tetracycline resistance marker, which prevents relaxase expression. The ltrB relaxase is constitutively produced from the pDL-P232-ltrB complementation plasmid. When ltrB is interrupted by a splicing proficient variant of Ll.LtrB, relaxase is produced and mediates transfer of the sex factor from the donor to the recipient cell. The level of sex factor conjugation observed is proportional to the Ll.LtrB splicing efficiency. (B) Relaxase expression and Ll.LtrB splicing. Northern blots were performed using total RNA from NZ9800 and NZ9800ΔltrB harbouring different pDL-P232-based constructs. The amounts of total relaxase RNA (left) and mature relaxase transcript (right) produced were assessed (two independent blots, RNA was loaded in duplicate on the same gel). The exon 2 probe (left panel) (Supplementary Table S1, RT E2) and the splice-junction probe (right panel) (Supplementary Table S1, SJ) are depicted as grey bars. As an RNA loading control, the two membranes were stripped and probed with a 23S rRNA specific probe (Supplementary Table S1, 23S rRNA). (C) Western blot on total protein extracts using LtrB-specific antibodies. Ll.LtrB group II intron, black line; exon 1 and 2, E1 and E2; branch point, circled A; sex factor, grey; tetracycline resistance marker, Tet; spectinomycin resistance marker, Spc; P23 promoter, P23; transcription terminator, schematic stem-loop; L. lactis chromosome, scribble.
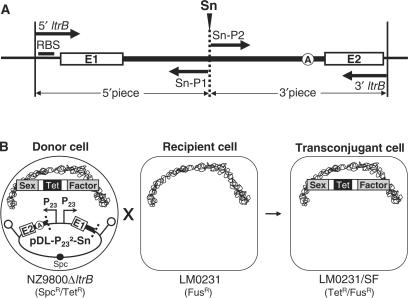
Figure 3.
Ll.LtrB trans-splicing/conjugation assay. (A) Strategy to generate a variant of Ll.LtrB fragmented at the Sn site (S1–S5). The relaxase gene is amplified as two non-overlapping fragments using the indicated primers (thick arrows, Supplementary Table S1). Each fragment is cloned under the control of a P23 promoter on pDL-P232. (B) Ll.LtrB trans-splicing/conjugation assay. The pDL-P232-Sn plasmid is transformed in NZ9800ΔltrB. If the two fragments of the intron can correctly fold, assemble and trans-splice, the ltrB exons are ligated allowing the production of relaxase, which is required for sex factor transfer. Ll.LtrB splicing efficiency is monitored by the level of sex factor conjugation from the donor (NZ9800ΔltrB) to the recipient L. lactis strain (LM0231). Ll.LtrB group II intron, thick black line; exon 1 and 2, E1 and E2; branch point, circled A; sex factor, grey; tetracycline resistance marker, Tet; spectinomycin resistance marker, Spc; P23 promoters, P23; transcription terminators, schematic stem-loops; L. lactis chromosome, scribble.
The pDL-P232-ltrB plasmid was obtained by cloning the promoter-less ltrB gene in pDL-P232 under the control of P23 right. The ltrB gene was PCR-amplified from genomic DNA extracted from the sex factor-containing L. lactis strain NZ9800 (31) using the 5′ltrB and 3′ltrB primers (Supplementary Table S1). To generate the intron-mutated versions of this plasmid, the BsrGI/HpaI fragment spanning most of the Ll.LtrB intron and a portion of the ltrB exon 2 was exchanged for the corresponding fragment from pLE12ΔD5, pLE12ΔORF and pLE12Mat−, respectively (23,31).
The pLE-P232 plasmid was generated by cloning the 1.8-kb SapI fragment from pDL-P232 containing the divergent P23 transcriptional units, into the unique SmaI site of the pLE1 plasmid (28). The resulting plasmid was used for expression of two of the three intron pieces when assessing tripartite variants of Ll.LtrB, and for overexpression of LtrA in ORF-complementation assays. For the latter experiments, the ltrA gene was PCR-amplified using primers 5′ltrA and 3′ltrA (Supplementary Table S1), and cloned in the NotI restriction site of pLE-P232.
To generate fragmented variants of Ll.LtrB, the sites of fragmentation, Sn (Figure 1B, S1–S5), were chosen according to the occurrence of fragmented group II introns in nature. Two primers were designed for each Sn fragmentation site. The Sn-P1 primer was always used in conjunction with the 5′ltrB primer to generate the 5′ fragment of the relaxase by PCR amplification (Figure 3A). Similarly, the Sn-P2 primer was always used with the 3′ltrB primer to generate the 3′ fragment of the relaxase (Figure 3A). The 5′ and 3′ fragments were cloned downstream from the P23 right and P23 left promoters, respectively. All relaxase fragments were PCR-amplified from genomic DNA of the sex factor-containing L. lactis strain NZ9800 (31). To generate the maturase-mutated and ΔORF variants of fragmented Ll.LtrB constructs, the ORF-containing intron fragment was PCR-amplified from the pLE12Mat− and pLE12ΔORF plasmids, respectively. The sequence integrity of all relaxase fragments generated by PCR amplification was confirmed by sequence analyses.
Conjugation assays
Mating was performed on milk plates made of 5% non-fat dried milk (Carnation™), 1% glucose and 1.5% agar (31). Donor and recipient L. lactis strains were inoculated from saturated overnight cultures (0.4 ml into 10 ml), and grown for 7 h. Cells were then collected by centrifugation, mixed and spread on milk plates. After a 16-h incubation at 30°C, cells were recovered in 1 ml 1X PBS, and appropriate dilutions were plated on selective medium for donors, recipients and transconjugants. Conjugation efficiencies were calculated as the ratio of transconjugant cells (FusR/TetR) to donor cells (SpcR), for three independent assays. The L. lactis strains NZ9800ΔltrB::tet and LM0231 (FusR) were used as the donor and recipient strains, respectively.
RNA isolation, northern blot hybridizations and RT-PCR
Total RNA was isolated from L. lactis cells inoculated from saturated overnight cultures (0.4 ml into 10 ml), and grown for 7 h. Cell pellets were mixed with 500 ml TRIzol (Invitrogen life technologies) and 250 µg of acid-washed glass beads (Sigma). The mixture was vortexed for 3 min and incubated at 55°C for 5 min; this treatment was repeated three times. The rest of the extraction was performed according to the TRIzol manufacturer's protocol.
For northern blot hybridization, 10 μg of total RNA was resolved on 1% agarose gel containing 5% formaldehyde and transferred by capillarity to nylon membrane (Hybond-N; Amersham Biosciences). The membranes were hybridized with the appropriate 5′-32P labelled oligonucleotide probe (see figure legends and Supplementary Table S1). Radiolabelling of the probes was performed in a final volume of 10 μl containing 10 pmol of the oligonucleotide, 6.4 pmol of [γ-32P]ATP (6000 Ci/mmol; Amersham Biosciences) and 5 U of T4 polynucleotide kinase (New England Biolabs) at 37°C for 1 h. The labelling mix was then purified on Sephadex G-50 columns. The membranes were hybridized for 2 h at 42°C, washed and exposed on a phosphor screen, which was visualized with the Molecular Imager FX software from Bio-Rad. As an RNA loading control, the membranes were stripped and re-probed with an oligonucleotide specific for the 23S rRNA of L. lactis (Figure 2C).
For reverse transcriptase (RT)-PCR analyses, cDNAs were synthesized using the SuperScript II cDNA synthesis kit from Invitrogen according to the manufacturer's instructions, using 5 µg of total RNA and 2 pmol of the appropriate primer. The BP cDNA primer was used to synthesize cDNA across the Ll.LtrB branch point, while the E2 cDNA primer was used to synthesize cDNA of the ltrB ligated exons (Supplementary Table S1). PCR amplification was performed using 10% of the cDNA obtained and 2 U of Taq DNA polymerase (New England Biolabs).
Protein isolation and western blot
Total proteins were extracted from L. lactis cells grown to mid-log phase. Cells were pelleted and resuspended in 100 µl TES buffer (10 mM Tris-HCl pH 8.0, 1 mM EDTA, 25% sucrose) containing 1 mg/ml lysozyme (Roche), 50 U/ml mutanolysin (Sigma®) and 0.1 mg/ml RNaseA (USB). This mixture was incubated for 1 h at 37°C. Cells were then lysed by mixing with 100 µl 1X TE buffer containing 4% SDS and boiling for 5 min. Samples were loaded on SDS-PAGE (10%) by mixing 10 µl of the total protein extract with 10 µl of SDS loading buffer containing 5% β-mercaptoethanol.
For western blot analysis, the gel was transferred electrophoretically onto PVDF membrane (Immobilon-P, Millipore). The membrane was blocked O/N in 1X PBS—0.1% Tween 20 (Fisher) (PBS-T) containing 10% non-fat dried milk (Carnation™). Anti-LtrB antibodies (18) were subjected to a membrane containing protein extracts from L. lactis cells not expressing LtrB to reduce the amounts of antibodies reactive to non-LtrB proteins, like previously described (15). Following blocking, the membrane containing the samples for analysis was incubated 1 h in PBS-T + 5% milk containing 1:5000 depleted anti-LtrB antibodies. After three 5-min washes in PBS-T, the membrane was incubated 1 h in PBS-T containing horseradish peroxidase (HRP)-conjugated anti-rabbit antibody (1:30 000; Bio-Rad), then washed four times for 5 min in PBS-T. The membrane was detected with Immobilon™ Western Chemiluminescent HRP Substrate (Millipore) and visualized with the Molecular Imager VersaDoc from Bio-Rad and on X-ray film. The software Quantity One® (Bio-Rad) was used for quantitative analysis of LtrB amounts.
RESULTS
A conjugation-based system to monitor Ll.LtrB splicing
We developed a genetic assay to monitor Ll.LtrB splicing by measuring the conjugation efficiency of the chromosomal sex factor between L. lactis strains (Figure 2A). This splicing/conjugation assay exploits the dependence of conjugative transfer of the sex factor on Ll.LtrB splicing from the relaxase transcript (17,26). The sex factor donor strain used in this assay is NZ9800ΔltrB::tet, in which the sex factor carries a deletion of the Ll.LtrB intron and portions of its flanking exons destroying the relaxase gene (21). In the absence of relaxase, the sex factor transfer efficiency drops from 7.33 × 10−3 (32) to 1.88 × 10−9 (Table 1, cf. SF wild-type and SFΔltrB/pDL-P232).
Table 1.
Sex factor conjugation efficiencies observed with different relaxase variants
| Donor strain | SF variant/complementation plasmid | SF conjugation efficiency |
|---|---|---|
| NZ9800 | SF wild-type | a(7.33 ± 0.58) × 10−3 |
| NZ9800ΔltrB | SFΔltrB | (1.88 ± 1.96) × 10−9 |
| NZ9800ΔltrB | SFΔltrB/pDL-P232-ltrB | (5.80 ± 2.89) × 10−2 |
| NZ9800ΔltrB | SFΔltrB/pDL-P232-ltrB ΔD5 | (3.79 ± 1.75) × 10−9 |
| NZ9800ΔltrB | SFΔltrB/pDL-P232-ltrB ΔORF | (1.93 ± 0.96) × 10−8 |
| NZ9800ΔltrB | SFΔltrB/pDL-P232-ltrB Mat− | (2.86 ± 0.86) × 10−4 |
SF; sex factor.
aBelhocine et al., 2005.
A complementation system was built where the intron-interrupted ltrB gene was cloned in the pDL-P232 plasmid and expressed under the control of the P23 constitutive promoter (Figure 2A, pDL-P232-ltrB). When the relaxase-deleted sex factor is thereby complemented, transfer efficiency is restored from 1.88 × 10−9 to 5.80 × 10−2 (Table 1, SFΔltrB/pDL-P232-ltrB). The difference in transfer efficiency between the complemented (5.80 × 10−2) and the wild-type sex factor (7.33 × 10−3) (∼8-fold) is likely due to overexpression of the relaxase gene from a constitutive promoter on a multiple copy plasmid. Notably, the relaxase transcript produced from the wild-type chromosomal sex factor is undetectable by northern blot. On the other hand, expression from the P23 promoter produces high levels of both the full-length relaxase transcript and mature mRNA (Figure 2B, compare lanes NZ9800 and ltrB-WT in both blots). Accordingly, significantly less ligated exons and no intron lariats are detected by RT-PCR in NZ9800, where the chromosomal sex factor expresses a functional relaxase, compared to expression of the relaxase gene from the P23 promoter (see below, Figure 4B and C, cf. NZ9800 and ltrB-WT). Western blot analysis confirms the significant increase in LtrB production from the plasmid when compared to its expression from the chromosomal sex factor (Figure 2C, cf. NZ9800 and ltrB-WT). Since no signal for LtrB is detected from NZ9800, the increase in LtrB expression from the P23 promoter was estimated to be of at least 112-fold. Therefore, the slight increase in conjugation efficiency between the wild-type and complemented sex factor (∼8-fold) is not comparable to the great difference in levels of relaxase expression observed (at least 112-fold). The complementation system is thus most likely saturated in relaxase enzyme, and the maximal rate of sex factor conjugation efficiency is probably reached (10−2 range). This suggests that the conjugation efficiency observed for the complemented sex factor underestimates the real splicing efficiency of Ll.LtrB. Therefore, the conjugation efficiencies observed throughout this study will be interpreted as increases from the background level rather than decreases from the saturated maximum level.
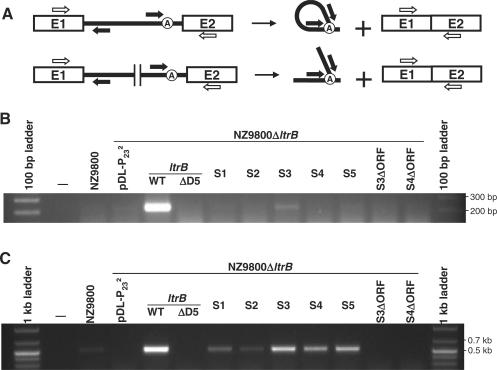
Figure 4.
RT-PCR analyses of excised Ll.LtrB introns and ligated exons. (A) Schematic of Ll.LtrB splicing (top) and trans-splicing (bottom). Primers used for RT-PCR amplifications of released intron lariats or Y-branched molecules are shown as black arrows (BP1 and 2). Ligated exons were amplified by RT-PCR with the RTE1 and 2 primers (open arrows). Agarose gels showing the RT-PCR amplification of Ll.LtrB across the branch point (B) and across the ltrB ligated exons (C) are presented. RT-PCR analyses were performed on total RNA extracts from NZ9800 or NZ9800ΔltrB harbouring different pDL-P232 plasmids expressing ltrB in one (WT, ΔD5) or two pieces (S1–S5). S3ΔORF and S4ΔORF represent the ΔORF variant of the S3 and S4 constructs without LtrA complementation. Ll.LtrB group II intron, black line; exon 1 and 2, E1 and E2; branch point, circled A.
To verify the dependence of sex factor conjugation on Ll.LtrB splicing, we tested an Ll.LtrB variant where the catalytic domain, i.e. domain V, was deleted (pDL-P232-ltrB-ΔD5) (23). Complementation with a relaxase gene harbouring the catalytically inactive intron failed to sustain sex factor transfer; the conjugation efficiency observed was only 3.79 × 10−9 (Table 1, SFΔltrB/pDL-P232-ltrB-ΔD5). This conjugation efficiency is similar to the background level observed for the ltrB-deficient sex factor (Table 1, SFΔltrB/pDL-P232). Northern blot analyses of total RNA extracted from donor cells harbouring the pDL-P232-ltrB-ΔD5 plasmid showed accumulation of the pre-mRNA precursor while no traces of ligated exons was detected (Figure 2B, lane ltrB-ΔD5 in both blots). Accordingly, neither ligated exons nor excised intron lariats were detected by RT-PCR analyses (see below, Figure 4B and C respectively). Taken together, these results demonstrate that transfer of the L. lactis sex factor is dependent on Ll.LtrB splicing from the relaxase transcript. We have therefore built a highly sensitive splicing/conjugation assay (107-fold detection range) to monitor Ll.LtrB splicing in L. lactis.
Trans-splicing of Ll.LtrB fragmented within domain IV
We used the above splicing/conjugation assay to assess if Ll.LtrB is capable of splicing when severed into two fragments at various positions. Several locations were chosen to mimic fragmentation sites found in naturally occurring trans-splicing group II introns (Figure 1B, S1–S5). To introduce a fragmentation site within Ll.LtrB (Sn, n varies from 1 to 5), the complete relaxase gene was PCR-amplified as two non-overlapping fragments (Figure 3A). To ensure equimolar production of each relaxase fragment, these were expressed from two constitutive P23 promoters located on the same plasmid, pDL- P232 (Figure 3B). The two promoters were cloned in opposite orientation to ensure that two independent RNA transcripts are generated. Proper expression of relaxase fragments was assessed by northern blot analyses and all the transcripts displayed the expected size. However, the fragments expressed from the P23-right promoter, consisting of exon 1 and the first portion of the Ll.LtrB intron, invariably showed slightly higher expression level than those expressed from the P23-left promoter, corresponding to the second portion of Ll.LtrB and exon 2 (data not shown). The lower expression of the 3′ relaxase fragments is likely due to their instability since transcription of these fragments is initiated at different positions within Ll.LtrB. This probably leads to a slight underestimation of the Ll.LtrB trans-splicing efficiency.
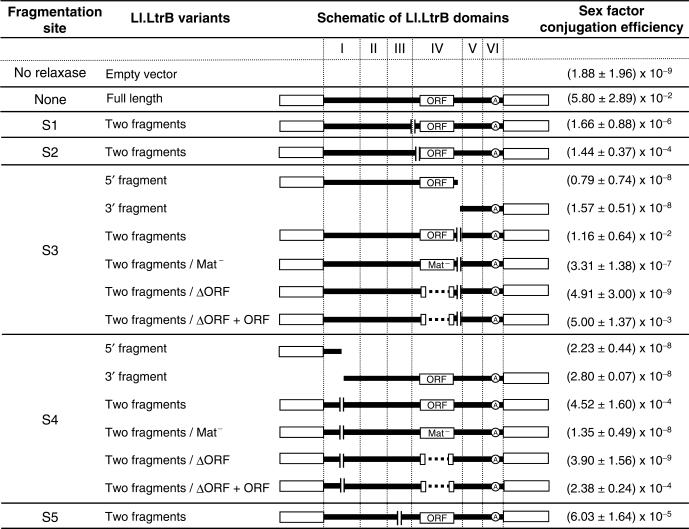
Most trans-splicing group II introns are found fragmented within domain IV. When these introns harbour an ORF, the fragmentation site can be found either upstream or downstream of the ORF. Therefore, we investigated the effect of fragmenting Ll.LtrB in domain IV, either upstream or downstream of the ltrA ORF. Two sites were chosen for fragmentation upstream of the ORF. The first site lies at the bottom of the DIV stem (position 524, Figure 1B, S1). Fragmentation of Ll.LtrB at this location does not allow base pairing interactions between the two intron RNA pieces, but leaves only a series of long-range tertiary interactions for intron fragment recognition and assembly (Figure 1B, greek letters). The second site lies within the DIVa stem at position 543 (Figure 1B, S2). According to the potential tertiary contacts between the two Ll.LtrB fragments, this fragmentation site is equivalent to S1. However, S2 also allows a potential 16 base pair interaction between the two intron pieces. When the intron is fragmented at the S1 site, the relaxase can promote sex factor conjugation to an efficiency of 1.66 × 10−6, ∼890-fold over the background (Figure 5, S1). This shows that tertiary interactions are sufficient to promote intron fragment assembly and splicing. On the other hand, when the relaxase is fragmented at the S2 site, sex factor conjugation is ∼87 times more efficient than S1, ∼76 000-fold over the background (1.44 × 10−4) (Figure 5, S2). These results suggest that base pairing interactions between intron fragments further stimulate their assembly and splicing in vivo.
Figure 5.
Sex factor conjugation efficiencies observed with different fragmented variants of Ll.LtrB. The fragmentation sites within Ll.LtrB correspond to those shown in Figure 1B (S1–S5). The Ll.LtrB variant column indicates which fragment(s) was/were overexpressed from the pDL-P232 plasmid, and which mutated version of the ORF was used when applicable. The ΔORF + ORF variants correspond to the assays where LtrA was overexpressed from a second plasmid to complement the deleted version within Ll.LtrB. The schematics show linear representations of Ll.LtrB and its six domains (I–VI), and the location of the fragmentation sites. Sex factor conjugation efficiency is the average of three independent assays and the error is the standard deviation of the three obtained values. Ll.LtrB group II intron, black line; exon 1 and 2, empty boxes; branch point, circled A.
Fragmentation of Ll.LtrB in domain IV downstream of the ORF was also tested. The intron was fragmented downstream from the ltrA stop codon at position 2375, which allows a potential 15 base pair interaction between the two intron fragments (Figure 1B, S3). Even though the potential tertiary interactions between the two intron pieces are the same as with S1 and S2, this Ll.LtrB trans-splicing variant harbours ltrA and the high affinity LtrA binding site (Figure 1B, DIVa) on the 5′ instead of the 3′ intron fragment. The relaxase gene fragmented at the S3 site promoted sex factor conjugation to very high levels (1.16 × 10−2) (Figure 5, S3, two fragments), ∼6.17 million-fold over the background (Table 1, SFΔltrB/pDL-P232). Moreover, when the two relaxase pieces were expressed independently, sex factor conjugation remained near basal levels (10−8), showing a slight increase compared to the background (Figure 5, S3, 5′ fragment and 3′ fragment). Therefore, expression of both relaxase fragments is absolutely required to promote sex factor conjugation. Taken together, these results demonstrate that when the Ll.LtrB intron is fragmented independently at three different positions within domain IV, a significant proportion of the two intron fragments can fold correctly, assemble and trans-splice to ligate their flanking exons.
Trans-splicing of Ll.LtrB fragmented at other naturally occurring group II intron fragmentation sites
Fragmentation sites outside domain IV can be found in domains I or III in some group II introns. The next Ll.LtrB trans-splicing variant that we analysed mimics the fragmentation site in domain I of the tripartite intron from the psaA chloroplast gene of the green algae C. reinhardtii (11). The fragmentation site was introduced at position 172 upstream of the EBS1 and 2 sequences. When Ll.LtrB is fragmented at this position, the possible tertiary interactions contributing to intron fragment assembly are different from those of the previous fragmentation sites (S1–S3). Moreover, a potential 19 base pair interaction can take place between the two fragments of the intron (Figure 1B, S4). When the two relaxase fragments are expressed independently, no significant increase in conjugation efficiency is observed (10−8 range) (Figure 5, S4, 5′ fragment and 3′ fragment). However, when both fragments are co-expressed, the rate of sex factor conjugation increases to 4.52 × 10−4 (Figure 5, S4, two fragments). The substantial increase observed (∼240 000-fold) in sex factor transfer efficiency from basal level when both relaxase fragments are produced demonstrates that Ll.LtrB can also trans-splice when fragmented in domain I at this position.
The last fragmentation site engineered in Ll.LtrB lies in the loop region of domain III at position 499 (Figure 1B, S5) and mimics the fragmentation site of the rps12 intron in M. polymorpha and N. tabacum chloroplasts (10). Fragmentation of Ll.LtrB at this location allows the same tertiary contacts between the two intron fragments as the S1, S2 and S3 sites. Moreover, 17 base pairs could be formed between the two intron fragments. When the ltrB gene is fragmented at this position, the amount of relaxase produced supports sex factor transfer to a level of 6.03 × 10−5 (Figure 5, S5), which represents an increase of approximately 32 000-fold over the background level.
These results demonstrate that when the intron is fragmented within domains I or III, a significant proportion of the two independently expressed Ll.LtrB fragments can fold correctly, assemble, splice in trans, and ligate their flanking exons.
Detection of trans-spliced intron molecules and ligated exons by RT-PCR
Excised group II intron lariats can be detected from total RNA extracts by RT-PCR across the branch point (Figure 4A, top, black arrows) (33). Using a similar approach, we designed an RT-PCR assay to detect the trans-spliced Y-shaped intron molecules (Figure 4A, bottom). This assay was designed to yield a 201-bp fragment when a lariat or a Y-shaped intron molecule is produced. The two RT-PCR primers lie outside of the region where the fragmentation sites were engineered, and could thus be used with all fragmented relaxase variants (Figure 4A, bottom, black arrows). The assay was performed on total RNA extracted from donor cells (NZ9800ΔltrB) containing various Ll.LtrB constructs. As shown in Figure 4B, donor cells harbouring the full-length relaxase gene yield a unique RT-PCR product of the expected size, representing the Ll.LtrB lariat (Figure 4B, ltrB-WT). On the other hand, Y-branched intron molecules are only detected with the most efficient trans-splicing construct, where the intron is fragmented downstream of the ORF (Figure 5, S3, two fragments). Failure to detect Y-branched introns from the other fragmented variants of Ll.LtrB is most likely due to the low level of these molecules as well as their instability. Indeed, Y-shaped molecules have three free ends and are more likely sensitive to degradation than intron lariats (Figure 4A, bottom).
The presence of ligated exons of the relaxase gene was also assessed by RT-PCR. This assay was designed to produce a 521-bp fragment when the exons are properly ligated (Figure 4A, open arrows). As can be seen in the agarose gel presented in Figure 4C, ligated exons are detected in total RNA extracted from all donor cells containing Ll.LtrB constructs that showed conjugation efficiencies above background. Specifically, ligated exons are detected in the case of the full-length ltrB gene but not from the catalytically inactive ltrB-ΔD5 variant (Figure 4C, ltrB-WT and ltrB-ΔD5). Ligated exons are also detected in all five cases of Ll.LtrB trans-splicing (Figure 4C, S1–S5). Sequence analyses of all RT-PCR products confirmed that the exons were ligated precisely at the intron splice junctions. These RT-PCR assays confirm that in all cases where sex factor conjugation increased over the background, properly ligated relaxase exons were readily detectable in donor cells. Furthermore, Y-branched intron molecules could be detected only for the fragmented relaxase construct that sustains the highest conjugation efficiency (S3).
The LtrA protein is essential for trans-splicing of Ll.LtrB
Splicing of group II introns in vivo requires the assistance of splicing co-factors that function as maturases to stabilize the catalytically active intron structure (2). Some group II introns, like Ll.LtrB, encode their own maturase. The role of IEPs in trans-splicing has never been investigated.
To assess the role of the LtrA protein in Ll.LtrB trans-splicing, we studied two different protein mutants. First, the Ll.LtrB-Mat− mutant carries point mutations that only affect the maturase activity of LtrA (23). Second, the Ll.LtrB-ΔORF intron carries a deletion of 88% of the ltrA coding sequence, which completely removes the maturase domain (23).
Cis-splicing of these Ll.LtrB mutants was first analysed in our splicing/conjugation assay. As expected, the Ll.LtrB-ΔORF mutant appears completely inactive. When the relaxase gene harbours this intron mutant, sex factor conjugation efficiency is only 10-fold higher than the background (Table 1, cf. SFΔltrB/pDL-P232-ltrB ΔORF and SFΔltrB/pDL-P232). Surprisingly, the maturase mutant efficiently induced sex factor transfer to a level ∼152 000-fold over the background (Table 1, cf. SFΔltrB/pDL-P232-ltrB Mat− and SFΔltrB/pDL-P232). Such a result suggests that this LtrA mutant can still bind and promote Ll.LtrB splicing.
Next, the two LtrA mutants were analysed in the context of two fragmented introns: the S4 variant fragmented within domain I, and the S3 variant fragmented downstream of the ORF within domain IV. These two introns were chosen because they trans-splice efficiently and are fragmented in different domains (DI versus DIV). The intron fragment that is bound by the LtrA protein also differs in these two variants. Indeed, when the intron is fragmented within domain I (S4), the LtrA primary-binding site (stem DIVa, Figure 1B) lies in the 3′ fragment of the intron. On the other hand, when the fragmentation site is located downstream of the ORF (S3), the LtrA primary binding site lies in the 5′ fragment of the intron (Figure 1B). As shown in Figure 5, the introduction of point mutations, that only abolish the maturase activity of LtrA to both fragmented introns, caused a consistent decrease of ∼35 000-fold in sex factor conjugation efficiency (Figure 5, S4 and S3, two fragments/Mat−). Moreover, removal of the ORF from Ll.LtrB completely abolishes trans-splicing as the sex factor conjugation efficiency drops to background levels with both variants tested (10−9) (Figure 5, S3 and S4, two fragments/ΔORF). Interestingly, if a second plasmid overexpressing LtrA (pLE-P232-ltrA) is introduced into cells harbouring the ΔORF variants of fragmented Ll.LtrB, conjugation efficiency is almost completely restored (Figure 5, S3 and S4, two fragments/ΔORF + ORF). This increase in sex factor conjugation efficiency is not a direct effect of LtrA on the sex factor since the LtrA-expressing plasmid alone does not sustain conjugation of the sex factor (1.31 ± 0.85 × 10−9). These results demonstrate that the LtrA protein, primarily through its maturase function, is an essential co-factor for Ll.LtrB trans-splicing in L. lactis.
DISCUSSION
In this study, we describe a highly sensitive conjugation-based system to monitor splicing of the Ll.LtrB group II intron in L. lactis. One of the important features of this experimental system lies in its extremely broad detection range. This simple genetic assay allows the detection and study of Ll.LtrB splicing quantitatively on a 107-fold detection range. As we demonstrate in this study, the Ll.LtrB splicing/conjugation assay will be particularly useful to study group II intron trans-splicing, an aspect of group II intron splicing that is not very well studied or understood.
Although this genetic assay was developed to study Ll.LtrB trans-splicing, some interesting facts regarding sex factor conjugation and Ll.LtrB cis-splicing were discovered. First, we observe that expressing higher amounts of the relaxase enzyme increased sex factor conjugation efficiency from 7.33 × 10−3 (32) to 5.80 × 10−2 (Table 1). Our data show that the sex factor conjugation machinery can sustain even higher transfer rates than what is observed with the wild-type element. This demonstrates that the presence of Ll.LtrB within the relaxase gene not only controls conjugation of the L. lactis sex factor, but also that the normal LtrB expression level in NZ9800 is sub-optimal for the sex factor conjugation machinery. This suggests that any mutation within Ll.LtrB that hinders splicing would also directly, and proportionally, reduce LtrB expression and sex factor conjugation efficiency.
The catalytic domain (DV) of Ll.LtrB was previously deleted to generate a splicing deficient mutant (23). When the relaxase gene contains this Ll.LtrB-ΔD5 variant, the sex factor conjugation efficiency is similar to the background observed when no relaxase is provided in trans (Table 1, 3.79 × 10−9 versus 1.88 × 10−9). Moreover, no RT-PCR signals for excised intron lariats or ligated exons were obtained for this Ll.LtrB mutant (Figure 4B and C). Taken together, these results confirm that deletion of domain V from Ll.LtrB prevents any residual splicing in L. lactis.
When the relaxase gene provided in trans is interrupted by the Ll.LtrB maturase-deficient mutant (Ll.LtrB-Mat−), sex factor conjugation efficiency increases by ∼152 000-fold over the background (Table 1, 2.86 × 10−4 versus 1.88 × 10−9). This shows that this LtrA maturase mutant can bind the intron and promote splicing to significant levels. When Ll.LtrB is missing most of the LtrA coding region (Ll.LtrB-▵ORF), we observe only a minor increase in sex factor conjugation efficiency (Table 1). In fact, this increase is comparable to the background conjugation rate obtained upon expression of individual fragments of the relaxase variants fragmented at position S3 and S4 (Figure 5, S3/5′ fragment and 3′ fragment, S4/5′ fragment and 3′ fragment). Conjugation efficiencies in the 10−8 range correspond to only a few transconjugant colonies per assay; very close to the limit of detection of our system.
Using our splicing/conjugation assay, we report the first experimental evidence that group II introns can trans-splice in bacteria. All the naturally occurring group II intron fragmentation sites were independently engineered within Ll.LtrB: three sites located in domain IV, two upstream and one downstream of the ORF, and one site located in both domains I and III. The trans-splicing efficiency of Ll.LtrB was found to vary greatly with the location of the fragmentation site (Figure 5). Considering the limited number of characterized trans-splicing introns and the lack of similarity between these introns and Ll.LtrB, it was surprising to detect trans-splicing with all fragmented variants of the lactococcal intron. The fragmentation site we engineered in domain I was actually never found as a unique fragmentation site. It was only observed in nature in conjunction with a second fragmentation within domain IV in two tripartite group II introns (11,12). Nevertheless, when fragmented at this location within domain I, Ll.LtrB trans-splices efficiently since the sex factor conjugation is ∼240 000-fold higher than the background.
In domain IV, two different intron fragmentation sites upstream of the ltrA coding region were studied. When the intron is fragmented at position S1 (Figure 1B), the trans-splicing efficiency is the lowest observed (1.66 × 10−6; Figure 5, S1). Due to the position of the fragmentation site, at the bottom of the DIV stem, the two intron fragments cannot interact by base pairing but only through long-range tertiary interactions. However, when the intron is fragmented at a position only 19 nt downstream (Figure 1B, S2), the two intron fragments potentially have an additional 16 nt base pair interaction. In this case, sex factor conjugation increases by ∼100-fold (1.44 × 10−4; Figure 5, S2). This finding suggests that base pairing between intron fragments plays an important role in intron assembly and splicing in vivo, contrary to what was previously proposed from in vitro data (6). Notably, the intron fragments of the vast majority of fragmented group II introns reported kept the potential to interact by a series of base pairs. Even though long range tertiary interactions are expected to be key in the assembly of folded group II intron fragments, it is conceivable that base pairing plays also a role in the assembly of fragmented group II introns at physiological conditions (1).
The trans-splicing efficiency of Ll.LtrB is greater when the fragmentation in domain IV occurs downstream rather than upstream from the ORF (Figure 5, cf. S3/two fragments to both S1 and S2). A reason for this difference may be that interrupting Ll.LtrB upstream of its ORF can disrupt the DIVa stem (Figure 1B), which is the primary binding site for LtrA (24,34). Since we showed that the LtrA protein is absolutely necessary for the trans-splicing reaction, it is likely that disruption of its primary interaction site within the intron RNA will decrease trans-splicing efficiency. Therefore, it would be difficult to generalize that group II introns trans-splice more efficiently when fragmented downstream rather than upstream from their carried ORFs within domain IV. Interestingly, a thorough comparative analysis by Qiu and Palmer showed that a mitochondrial group II intron in angiosperms, nad1i728, underwent fifteen independent incidences of fragmentation within domain IV throughout differentiation of these species (35). Among these cases, ten independent fragmentation events occurred upstream of the intron-encoded matR gene while only five occurred downstream. However, the authors did not conclude that fragmentation of a group II intron upstream rather than downstream of its carried ORF enabled better trans-splicing.
Since Ll.LtrB showed satisfactory trans-splicing efficiencies when fragmented in domain I and IV independently, we assessed the trans-splicing potential of two tripartite Ll.LtrB introns. We combined one fragmentation within domain IV, upstream or downstream of the ORF, with the fragmentation in domain I (Figure 1B, S2 and S4, S3 and S4). These two tripartite variants of Ll.LtrB failed to trans-splice since the sex factor conjugation efficiency observed was in the 10−8 range, which is comparable to overexpression of independent fragments of the relaxase fragmented at sites S3 and S4 (Figure 5). Notably, in chlamydomonas chloroplast, it was shown that at least 9 cellular factors are needed for trans-splicing of the psaA tripartite intron (36). One may imagine that following the fragmentation of a group II intron, the recruitment of splicing factors becomes important in maintaining reasonable splicing efficiency and gene expression.
Using the intron variants fragmented in domain I (S4) and domain IV downstream of the ORF (S3), we show that the LtrA protein is essential for Ll.LtrB trans-splicing in vivo (Figure 5). Indeed, trans-splicing of these introns is abolished in the absence of LtrA, and is almost completely restored when LtrA is produced in trans from a second plasmid. Interestingly, LtrA remains important for the assembly of Ll.LtrB fragments whether its primary binding site (DIVa) rests in the 5′ or the 3′ intron fragment. These results suggest that for both of these Ll.LtrB fragmented variants, as well as for the ORF-encoding fragmented introns (S1–S5), the LtrA binding structure is correctly folded in a significant proportion of the intron fragments to allow LtrA binding and Ll.LtrB splicing.
It is well established that the maturase activity of the LtrA protein is essential for Ll.LtrB splicing in vivo (19,23). In this study, we demonstrate that the maturase function of LtrA is also extremely important for Ll.LtrB trans-splicing. Using a maturase mutant for the introns fragmented at sites S3 and S4, we observe a significant and consistent drop of ∼35 000-fold in sex factor conjugation efficiency for both constructs (Figure 5, cf. S3/two fragments and S4/two fragments and their respective Mat− counterparts). In our system, it is impossible to use conjugation efficiencies to quantify the difference observed between the cis-splicing full-length intron and its maturase-deficient counterpart since conjugation is saturated when the full-length ltrB gene is overexpressed. Yet, one may expect that the assistance of the maturase function of LtrA is more important for bimolecular splicing reactions than for cis-splicing.
We demonstrated that a bacterial group II intron can trans-splice efficiently in vivo when fragmented at different positions. Although several splicing-competent fragmented introns were identified in organellar genomes, no trans-splicing group II intron was yet discovered in bacteria. Fragmentation of group II introns through genome rearrangements may occur more readily in organelles than in bacteria. Indeed, numerous trans-splicing group II introns were found in the mitochondrial genomes of higher plants, which are more plastic and subjected to high rearrangement rates (35,37). Nevertheless, this study suggests that trans-splicing group II introns may exist and eventually be uncovered in prokaryotes. Group II introns can splice in trans in bacterial cells and therefore could be maintained if ever generated.
Supplementary Material
ACKNOWLEDGEMENT
We thank Karen K. Yam and Greg T. Marczynski for providing comments on the manuscript, and Victoria Mandilaras for technical assistance. We are also thankful to Gary M. Dunny for kindly providing the anti-LtrB antibody and B. Franz Lang for helpful discussions on mitochondrial genome plasticity. This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada (NSERC) and the Canadian Institutes of Health Research (CIHR) to B.C. B.C. was a CIHR New Investigator Scholar and is currently an FRSQ Chercheur-Boursier Junior 2 and a McGill University William Dawson Scholar. K.B. holds a Postgraduate Studies Fellowship from NSERC. The Open Access publication charges were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Michel F, Ferat JL. Structure and activities of group II introns. Annu. Rev. Biochem. 1995;64:435–461. doi: 10.1146/annurev.bi.64.070195.002251. [DOI] [PubMed] [Google Scholar]
- 2.Lambowitz AM, Zimmerly S. Mobile group II introns. Annu. Rev. Genet. 2004;38:1–35. doi: 10.1146/annurev.genet.38.072902.091600. [DOI] [PubMed] [Google Scholar]
- 3.Lehmann K, Schmidt U. Group II introns: structure and catalytic versatility of large natural ribozymes. Crit Rev. Biochem. Mol. Biol. 2003;38:249–303. doi: 10.1080/713609236. [DOI] [PubMed] [Google Scholar]
- 4.Sharp PA. Five easy pieces. Science. 1991;254:663. doi: 10.1126/science.1948046. [DOI] [PubMed] [Google Scholar]
- 5.Su LJ, Waldsich C, Pyle AM. An obligate intermediate along the slow folding pathway of a group II intron ribozyme. Nucleic Acids Res. 2005;33:6674–6687. doi: 10.1093/nar/gki973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jarrell KA, Dietrich RC, Perlman PS. Group II intron domain 5 facilitates a trans-splicing reaction. Mol. Cell Biol. 1988;8:2361–2366. doi: 10.1128/mcb.8.6.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mohr G, Perlman PS, Lambowitz AM. Evolutionary relationships among group II intron-encoded proteins and identification of a conserved domain that may be related to maturase function. Nucleic Acids Res. 1993;21:4991–4997. doi: 10.1093/nar/21.22.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qin PZ, Pyle AM. The architectural organization and mechanistic function of group II intron structural elements. Curr. Opin. Struct. Biol. 1998;8:301–308. doi: 10.1016/s0959-440x(98)80062-6. [DOI] [PubMed] [Google Scholar]
- 9.Bonen L. Trans-splicing of pre-mRNA in plants, animals, and protists. FASEB J. 1993;7:40–46. doi: 10.1096/fasebj.7.1.8422973. [DOI] [PubMed] [Google Scholar]
- 10.Kohchi T, Umesono K, Ogura Y, Komine Y, Nakahigashi K, Komano T, Yamada Y, Ozeki H, Ohyama K. A nicked group II intron and trans-splicing in liverwort, Marchantia polymorpha, chloroplasts. Nucleic Acids Res. 1988;16:10025–10036. doi: 10.1093/nar/16.21.10025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldschmidt-Clermont M, Choquet Y, Girard-Bascou J, Michel F, Schirmer-Rahire M, Rochaix JD. A small chloroplast RNA may be required for trans-splicing in Chlamydomonas reinhardtii. Cell. 1991;65:135–143. doi: 10.1016/0092-8674(91)90415-u. [DOI] [PubMed] [Google Scholar]
- 12.Knoop V, Altwasser M, Brennicke A. A tripartite group II intron in mitochondria of an angiosperm plant. Mol. Gen. Genet. 1997;255:269–276. doi: 10.1007/s004380050497. [DOI] [PubMed] [Google Scholar]
- 13.Mills DA, McKay LL, Dunny GM. Splicing of a group II intron involved in the conjugative transfer of pRS01 in lactococci. J. Bacteriol. 1996;178:3531–3538. doi: 10.1128/jb.178.12.3531-3538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mills DA, Manias DA, McKay LL, Dunny GM. Homing of a group II intron from Lactococcus lactis subsp. lactis ML3. J. Bacteriol. 1997;179:6107–6111. doi: 10.1128/jb.179.19.6107-6111.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou L, Manias DA, Dunny GM. Regulation of intron function: efficient splicing in vivo of a bacterial group II intron requires a functional promoter within the intron. Mol. Microbiol. 2000;37:639–651. doi: 10.1046/j.1365-2958.2000.02033.x. [DOI] [PubMed] [Google Scholar]
- 16.Matsuura M, Noah JW, Lambowitz AM. Mechanism of maturase-promoted group II intron splicing. EMBO J. 2001;20:7259–7270. doi: 10.1093/emboj/20.24.7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klein JR, Chen Y, Manias DA, Zhuo J, Zhou L, Peebles CL, Dunny GM. A conjugation-based system for genetic analysis of group II intron splicing in Lactococcus lactis. J. Bacteriol. 2004;186:1991–1998. doi: 10.1128/JB.186.7.1991-1998.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Y, Klein JR, McKay LL, Dunny GM. Quantitative analysis of group II intron expression and splicing in Lactococcus lactis. Appl. Environ. Microbiol. 2005;71:2576–2586. doi: 10.1128/AEM.71.5.2576-2586.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cousineau B, Smith D, Lawrence-Cavanagh S, Mueller JE, Yang J, Mills D, Manias D, Dunny G, et al. Retrohoming of a bacterial group II intron: mobility via complete reverse splicing, independent of homologous DNA recombination. Cell. 1998;94:451–462. doi: 10.1016/s0092-8674(00)81586-x. [DOI] [PubMed] [Google Scholar]
- 20.Cousineau B, Lawrence S, Smith D, Belfort M. Retrotransposition of a bacterial group II intron. Nature. 2000;404:1018–1021. doi: 10.1038/35010029. Erratum in (2001) Nature, 414, 84. [DOI] [PubMed] [Google Scholar]
- 21.Ichiyanagi K, Beauregard A, Lawrence S, Smith D, Cousineau B, Belfort M. Retrotransposition of the Ll.LtrB group II intron proceeds predominantly via reverse splicing into DNA targets. Mol. Microbiol. 2002;46:1259–1272. doi: 10.1046/j.1365-2958.2002.03226.x. [DOI] [PubMed] [Google Scholar]
- 22.Smith D, Zhong J, Matsuura M, Lambowitz AM, Belfort M. Recruitment of host functions suggests a repair pathway for late steps in group II intron retrohoming. Genes Dev. 2005;19:2477–2487. doi: 10.1101/gad.1345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuura M, Saldanha R, Ma H, Wank H, Yang J, Mohr G, Cavanagh S, Dunny, G.M.Belfort M, et al. A bacterial group II intron encoding reverse transcriptase, maturase, and DNA endonuclease activities: biochemical demonstration of maturase activity and insertion of new genetic information within the intron. Genes Dev. 1997;11:2910–2924. doi: 10.1101/gad.11.21.2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh RN, Saldanha RJ, D'Souza LM, Lambowitz AM. Binding of a group II intron-encoded reverse transcriptase/maturase to its high affinity intron RNA binding site involves sequence-specific recognition and autoregulates translation. J. Mol. Biol. 2002;318:287–303. doi: 10.1016/S0022-2836(02)00054-2. [DOI] [PubMed] [Google Scholar]
- 25.O'Sullivan D, Ross RP, Twomey DP, Fitzgerald GF, Hill C, Coffey A. Naturally occurring lactococcal plasmid pAH90 links bacteriophage resistance and mobility functions to a food-grade selectable marker. Appl. Environ. Microbiol. 2001;67:929–937. doi: 10.1128/AEM.67.2.929-937.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shearman C, Godon JJ, Gasson M. Splicing of a group II intron in a functional transfer gene of Lactococcus lactis. Mol. Microbiol. 1996;21:45–53. doi: 10.1046/j.1365-2958.1996.00610.x. [DOI] [PubMed] [Google Scholar]
- 27.Byrd DR, Matson SW. Nicking by transesterification: the reaction catalysed by a relaxase. Mol. Microbiol. 1997;25:1011–1022. doi: 10.1046/j.1365-2958.1997.5241885.x. [DOI] [PubMed] [Google Scholar]
- 28.Mills DA, Choi CK, Dunny GM, McKay LL. Genetic analysis of regions of the Lactococcus lactis subsp. lactis plasmid pRS01 involved in conjugative transfer. Appl. Environ. Microbiol. 1994;60:4413–4420. doi: 10.1128/aem.60.12.4413-4420.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van der Vossen JM, van der Lelie D, Venema G. Isolation and characterization of Streptococcus cremoris Wg2-specific promoters. Appl. Environ. Microbiol. 1987;53:2452–2457. doi: 10.1128/aem.53.10.2452-2457.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LeBlanc DJ, Lee LN, bu-Al-Jaibat A. Molecular, genetic, and functional analysis of the basic replicon of pVA380-1, a plasmid of oral streptococcal origin. Plasmid. 1992;28:130–145. doi: 10.1016/0147-619x(92)90044-b. [DOI] [PubMed] [Google Scholar]
- 31.Belhocine K, Plante I, Cousineau B. Conjugation mediates transfer of the Ll.LtrB group II intron between different bacterial species. Mol. Microbiol. 2004;51:1459–1469. doi: 10.1111/j.1365-2958.2004.03923.x. [DOI] [PubMed] [Google Scholar]
- 32.Belhocine K, Yam KK, Cousineau B. Conjugative transfer of the Lactococcus lactis chromosomal sex factor promotes dissemination of the Ll.LtrB group II intron. J. Bacteriol. 2005;187:930–939. doi: 10.1128/JB.187.3.930-939.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li-Pook-Than J, Bonen L. Multiple physical forms of excised group II intron RNAs in wheat mitochondria. Nucleic Acids Res. 2006;34:2782–2790. doi: 10.1093/nar/gkl328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watanabe K, Lambowitz AM. High-affinity binding site for a group II intron-encoded reverse transcriptase/maturase within a stem-loop structure in the intron RNA. RNA. 2004;10:1433–1443. doi: 10.1261/rna.7730104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qiu YL, Palmer JD. Many independent origins of trans splicing of a plant mitochondrial group II intron. J. Mol. Evol. 2004;59:80–89. doi: 10.1007/s00239-004-2606-y. [DOI] [PubMed] [Google Scholar]
- 36.Perron K, Goldschmidt-Clermont M, Rochaix JD. A multiprotein complex involved in chloroplast group II intron splicing. RNA. 2004;10:704–711. doi: 10.1261/rna.5237804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fauron C, Casper M, Gao Y, Moore B. The maize mitochondrial genome: dynamic, yet functional. Trends Genet. 1995;11:228–235. doi: 10.1016/s0168-9525(00)89056-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.