Abstract
Five Arabidopsis thaliana genes that encode UDP-glucose 4-epimerase (UGE) and represent two ancient plant UGE clades might be involved in the regulation of cell wall carbohydrate biosynthesis. We tested this hypothesis in a genome-wide reverse genetic study. Despite significant contributions of each gene to total UGE activity, none was essential for normal growth on soil. uge2 uge4 displayed dramatic general growth defects, while other mutant combinations were partially aberrant. UGE2 together with UGE3 influenced pollen development. UGE2 and UGE4 synergistically influenced cell wall galactose content, which was correlated with shoot growth. UGE2 strongly and UGE1 and UGE5 lightly supported UGE4 in influencing root growth and cell wall galactose content by affecting galactan content. By contrast, only UGE4 influenced xyloglucan galactosylation in roots. Secondary hypocotyl thickening and arabinogalactan protein carbohydrate structure in xylem parenchyma depended on the combination of UGE2 and UGE4. As opposed to cell wall galactose content, tolerance to external galactose strictly paralleled total UGE activity. We suggest a gradual recruitment of individual UGE isoforms into specific roles. UGE2 and UGE4 influence growth and cell wall carbohydrate biosynthesis throughout the plant, UGE3 is specialized for pollen development, and UGE1 and UGE5 might act in stress situations.
INTRODUCTION
Higher plants make a multitude of complex carbohydrates, the most diverse and complex of which exist in the cell wall. Cell wall polysaccharide structures vary strongly between individual cell types and different developmental phases. Ultimately, there has to be a relation between cell wall structure and the functional properties of every differentiated cell (Somerville et al., 2004). It is believed that the structural–functional diversity of cell wall polymers is generated both at the level of their biosynthesis and by remodeling deposited cell wall polysaccharides. One of the recurring messages from the analysis of complete genomes is the large proportion of genes related to cell wall biosynthesis and remodeling (Arabidopsis Genome Initiative, 2000; International Rice Genome Sequencing Project, 2005; Tuskan et al., 2006). Complex carbohydrate biosynthesis requires the action of linkage-specific glycosyl transferases and a metabolic network of nucleotide sugar interconversion pathways. Most of the glycosyl transferases involved in cell wall carbohydrate biosynthesis remain to be identified (Scheible and Pauly, 2004). However, large strides have been made in elucidating the genetic identities of plant nucleotide sugar interconversion enzymes, all encoded by small gene families (Reiter and Vanzin, 2001; Seifert, 2004). Direct comparative analyses of genetically encoded isoforms of nucleotide sugar interconversion enzymes (Pattathil et al., 2005; Barber et al., 2006; Zhang et al., 2006) as well as analyses performed on individual isoforms in different laboratories (Gu and Bar-Peled, 2004; Molhoj et al., 2004; Usadel et al., 2004) highlight variations in their biochemical properties, transcriptional regulation, and subcellular localization—a reason for speculating that this genetic diversity is related to the regulation of cell wall carbohydrate complexity during development, growth, and stress (Bonin et al., 2003; Seifert, 2004; Pattathil et al., 2005). Although roles for isoforms of nucleotide sugar interconversion enzymes in cell wall biosynthesis have been demonstrated by forward genetics, the isolation of Arabidopsis thaliana mutants defective in cell wall structure or function (Bonin et al., 1997; Seifert et al., 2002; Burget et al., 2003; Diet et al., 2006), the biological meaning of genetic redundancy, and the molecular diversity of nucleotide sugar interconversion enzymes remain elusive.
UDP-glucose 4-epimerase (UGE) freely interconverts UDP-glucose and UDP-galactose, and a family of five UGE isoforms is encoded in the Arabidopsis genome. UGE1 to UGE5 show in vitro variations in substrate affinity, cofactor requirement, and metabolite inhibition profile (Barber et al., 2006). Their global expression pattern suggests that UGE1 and UGE3 are coregulated with carbohydrate catabolic enzymes, while UGE2, UGE4, and UGE5 are coregulated with carbohydrate biosynthetic enzymes. All isoforms can act in vivo in both directions, as indicated by overexpression experiments (Barber et al., 2006). Expression levels and experiments with UGE1 antisense lines both suggest that UGE1 might be the dominant isoform in green plant parts (Dörmann and Benning, 1998). Antisense interference of UGE1, however, neither induces a morphological phenotype nor alters cell wall polymers or any other galactose-containing carbohydrate. This suggests that other isoforms are required for the biosynthesis of glycoconjugates. Mutants in the UGE4 locus, which have been isolated in three independent screens for root hair development, root morphology, and susceptibility to nematodes (Schiefelbein and Somerville, 1990; Baskin et al., 1992; Baum et al., 2000), show that this gene is involved in the galactosylation of xyloglucan and arabinogalactan II in a cell type–dependent manner (Andeme-Onzighi et al., 2002; Seifert et al., 2002; Nguema-Ona et al., 2006). By contrast, other cell wall polymers such as pectic galactan and specific carbohydrate structures on arabinogalactan proteins (AGPs) are not affected (Andeme-Onzighi et al., 2002; Seifert et al., 2002, 2004; Nguema-Ona et al., 2006), prompting speculations that the biosynthesis of some carbohydrates might be regulated by metabolite channeling of UDP-galactose between cytosolic UGE4 and Golgi-localized glycosyl transferases (Seifert, 2004; Seifert et al., 2004; Nguema-Ona et al., 2006). Other carbohydrates might receive UDP-galactose from coexpressed UGE isoforms interacting with different glycosyl transferases (Seifert, 2004). However, the substrate-channeling model does not exclude the possibility that other mechanisms, such as transcriptional coregulation between UGE isoforms and diverse metabolically adjacent enzymes as well as kinetic diversification, might all contribute to functional specificity.
Here, we take a genome-wide reverse genetics approach to disrupt the function of every Arabidopsis UGE isoform and test the contribution of individual and combinations of UGE loci to cell wall biosynthesis and growth. We quantify cell wall galactose content, specifically in xyloglucan and pectin, qualitatively analyze AGP carbohydrates, and assay galactose tolerance in single mutants and most mutant combinations. We report that the isoforms UGE2 and UGE4 together play a major role in vegetative growth and cell wall carbohydrate biosynthesis. UGE3 and UGE2 cooperate in pollen development, and UGE1 and UGE5 contribute nonspecifically to UGE activity and growth under unstressed conditions but might be more specifically involved in stress situations. Our results suggest a gradual specialization of UGE isoforms throughout plant evolution that is likely to involve transcriptional as well as cellular mechanisms.
RESULTS
Two Clades of UGE Isoforms in Higher Plants
Five Arabidopsis genes encode functional UGE isoforms (Barber et al., 2006). In Populus trichocarpa, there are at least three, and in Oryza sativa, there are at least four putative UGE genes (Zhang et al., 2006). In Physcomitrella patens, there are at least five potential UGE genes. However, only a single gene is found in the unicellular green alga Chlamydomonas reinhardtii genome. A phylogenetic tree of the UGE peptide sequences rooted with the C. reinhardtii protein (Figure 1) shows three main clades. One clade contains all of the P. patens sequences. The other two clades contain all sequences of higher plants. One of the two higher plant clades contains Arabidopsis UGE1 and UGE3 as well as sequences from poplar and rice. The other higher plant clade contains Arabidopsis UGE2, UGE4, and UGE5, three rice sequences, and two poplar sequences. Within the two higher plant clades, the sequences derived from different plant species cluster into separate subclades. We conclude that UGE isoforms from higher plants occur in two separate sequence clades whose separation predates the separation of dicotyledons and monocotyledons. Other gene duplications are more recent events. Bryophytes appear to have obtained a family of UGE genes only after the evolution of higher plants.
Figure 1.
Phylogeny of Plant UGE Isoforms.
Phylogeny of UGE proteins from Chlamydomonas reinhardtii (Cr), Physcomitrella patens (Pp), Populus trichocarpa (Pt), Arabidopsis thaliana (At), and Oryza sativa (Os) excluding N- and C-terminal sequences. The tree was rooted with the Chlamydomonas sequence. Percentages are bootstrap values obtained from 100 bootstrap replicates.
Spatial Patterns of UGE Expression Overlap
With the exception of UGE5:GUS (for β-glucuronidase), UGE:GUS constructs are expressed throughout the shoot of 8-d-old seedlings with no apparent cell type–specific pattern (Figure 2A). In the root tip of transgenic plants, UGE1:GUS and UGE2:GUS are expressed in the root cap. In the elongation zone, mostly UGE4:GUS and UGE1:GUS are expressed. In the mature zone of the root, all five constructs are expressed. In dark-germinated 4-d-old seedlings, UGE5:GUS is expressed only in the distal part of the cotyledons, whereas the other constructs are expressed throughout the cotyledons and the upper third of the hypocotyl (Figure 2A). In 6-week-old hypocotyls undergoing secondary thickening, UGE1:GUS and UGE3:GUS are expressed throughout the cross section. The expression of UGE2:GUS, UGE4:GUS, and UGE5:GUS, on the other hand, is concentrated at the cambium and the differentiation zone of xylem and phloem (Figure 2B).
Figure 2.
Spatial Expression Patterns of UGE:GUS Constructs.
(A) Top, UGE1:GUS to UGE5:GUS expression in 8-d-old light-grown seedlings (left) and 4-d-old dark-grown seedlings (right). Bottom, details of expression in root tips of 5-d-old light-grown seedlings.
(B) UGE1:GUS to UGE5:GUS expression in sections of secondary thickened hypocotyls of 6-week-old plants.
Multiple UGE Genes Are Required for Normal Growth and Development
The uge4 root phenotype apart, all uge single mutants and most double mutant combinations show apparently normal shoot and root growth and fecundity. Some uge mutant combinations develop significantly smaller rosettes than wild-type plants (Figure 3A, Table 1). The uge1,2 double mutant has a rosette that is 50% smaller than the wild type, and this is combined with curly leaves with necrotic patches in the uge1,2,5 triple mutant (Figure 3A; see Supplemental Figure 1A online). The combination of the uge2 and uge4 mutant alleles leads to a dramatic reduction in rosette size to approximately one-third of the wild-type size. While only the young leaves of uge1,2 double and uge1,2,5 triple mutants show light chlorosis at their base, most leaves of the uge2,4 double mutant are strongly chlorotic. The strongly dwarfed uge1,2,4 triple mutant also is pale green, but not as dramatically as the uge2,4 double mutant (Figure 3A) or the uge2,4,5 triple mutant (see Supplemental Figure 1B online). uge1,2,4 displays a delay in development, notably in germination and seedling establishment but also later in the formation of rosette leaves (see Supplemental Table 1 online). The most severe phenotype is seen in the uge1,2,4,5 quadruple mutant, which is only rarely obtained from a homozygous uge1,2,5 population segregating for the uge4 allele. Homozygous uge1,2,4,5 mutants fail to germinate on galactose-free medium (tested population of 300 seeds), while on medium supplemented with 3 mM d-galactose, only a few uge1,2,4,5 mutants can be rescued (2 out of 100 seeds). Despite the presence of d-galactose, uge1,2,4,5 growth is extremely slow. Although root, hypocotyl, cotyledon, and leaf tissues do form, the tissue appearance is partially disorganized, resembling callus. Upon transfer to galactose-free medium, the uge1,2,4,5 mutant turns brown within 48 h and dies (see Supplemental Figure 1C online). Due to their galactose auxotrophy, uge1,2,4,5 mutants are not included in further aspects of this study. In 4-d-old dark-germinated seedlings, a reduction in hypocotyl length by 50 to 60% compared with the wild type occurs in uge1,3,4,5, uge2,4, and uge1,2,4, and a reduction by ∼20% occurs in uge2, uge1,2,5, and uge1,3,4 (Table 1). Secondary thickening of the hypocotyl proceeds normally in most of the uge mutants except for uge2,4 and uge1,2,4 combinations, in which the hypocotyl diameter is 62 and 50% of the wild-type value after 6 weeks of growth, respectively (Table 1).
Figure 3.
Rosette and Root Phenotypes of uge Mutants.
(A) Rosettes of 22-d-old wild-type Columbia (Col-0) and selected uge mutant plants.
(B) Top, roots of 4-d-old wild-type Col-0 and selected uge mutant plants. Bottom, root tips with division and elongation zones magnified 2.5 times. Bars = 1 mm (top) and 0.4 mm (bottom).
Table 1.
Summary of Qualitative and Quantitative Morphological Traits of UGE Mutant Combinations
| Rosette Diameter (mm) | Root Length (mm) | Hypocotyl
|
||||
|---|---|---|---|---|---|---|
| Abbreviation | Genotype | Length (mm) | Diameter (mm) | Phenotype | ||
| Col-0 | Columbia-0 wild type | 66.1 | 40.4 | 13.8 | 1.23 | Normal |
| uge1 | uge1-1 | 63.7 | 37.9 | 13.4 | 1.30 | Normal |
| uge2 | uge2-2 | 60.3 | 39.3 | 11.3a | 1.23 | Normal |
| uge3 | uge3-2 | 59.1 | 39.6 | 14.1 | ND | Normal |
| uge4 | uge4-4 | 65.1 | 17.7a | 13.3 | ND | Root epidermal bulging |
| uge5 | uge5-2 | 70.5 | 43.5 | 13.7 | ND | Normal |
| uge1,2 | uge1-1uge2-2 | 38.5a | 45.1 | 12.5 | 1.16 | Slightly pale |
| uge1,3 | uge1-1uge3-2 | 58.1b | 47.7 | 14.3 | ND | Normal |
| uge1,4 | uge1-1uge4-4 | 50.8a | 15.8a | 12.8 | 1.17 | Root epidermal bulging |
| uge2,3 | uge2-1uge3-2 | ND | ND | ND | ND | Male sterile |
| uge2,4 | uge2-2uge4-4 | 24.3a | 7.3a | 8.5a | 0.76a | Pale dwarf, root epidermal bulging |
| uge2,5 | uge2-1uge5-2 | 68.0 | 39.7 | 14.8 | ND | Normal |
| uge3,4 | uge3-2uge4-4 | 59.8 | 18.6a | 13.4 | ND | Root epidermal bulging |
| uge3,5 | uge3-2uge5-2 | 58.9 | 45.4 | 12.3b | ND | Normal |
| uge4,5 | uge4-1uge5-2 | 64.4 | 14.6a | 12.4 | ND | Root epidermal bulging |
| uge1,2,4 | uge1-1uge2-2uge4-1 | 9.1a | 2.7a | ND | 0.62a | Sterile dwarf, root epidermal bulging |
| uge1,2,5 | uge1-1uge2-1uge5-2 | 30.0a | 46.2 | 11.6a | ND | Small, necrosis |
| uge1,3,4 | uge1-1uge3-2uge4-4 | 63.1 | 14.8a | 11.6a | ND | Root epidermal bulging |
| uge1,4,5 | uge1-1uge4-4uge5-2 | 55.8a | 11.5a | 8.0a | 1.54 | Root epidermal bulging |
| uge2,3,5 | uge2-1uge3-2uge5-2 | 63.9 | 48.2 | ND | ND | Male sterile |
| uge2,4,5 | uge2-1uge4-4uge5-2 | ND | ND | ND | ND | Pale sterile dwarf, root epidermal bulging |
| uge3,4,5 | uge3-2uge4-4uge5-2 | 67.9 | 14.1a | 13.5 | ND | Root epidermal bulging |
| uge1,2,4,5 | uge1-1uge2-2uge4-4uge5-2 | ND | ND | ND | ND | Galactose auxotroph |
| uge1,3,4,5 | uge1-1uge3-2uge4-4uge5-2 | 57.8b | 10.9a | 6.8a | 1.19 | Root epidermal bulging |
Rosette diameter, primary root length, primary hypocotyl length, and secondary hypocotyl diameter were measured as described in Methods. The qualitative phenotypes are described in the text. ND, not determined.
Significantly different from the wild type at P < 0.01.
Significantly different from the wild type at P < 0.05.
All lines with the uge4 mutant allele produce roots of less than half the length of wild-type roots and display abnormal morphology (Figure 3B). Combination of the uge4 mutant allele with the uge1, uge2, or uge5 mutant allele leads to an additional reduction of root length (Table 1). Combination of the uge4 and uge2 mutant alleles displays exacerbated root epidermal bulging, and the radially swollen elongation zone is visibly shortened compared with the uge4 single mutant. The uge1,2,4 triple mutant develops only a very short root with heavily bulging cells, similar to the uge2,4 double mutant; however, radial swelling already occurs in the division zone (Figure 3B).
Consistent with their general growth defects, uge2,4 and uge1,2,5 display reduced fecundity, while uge1,2,4 and uge2,4,5 triple mutants are completely sterile. Despite its otherwise normal appearance, uge2,3 is almost completely sterile, and mature plants form short siliques that very rarely contain seeds. uge2,3 can be pollinated using wild-type pollen, but pollination trials using uge2,3 pollen have remained unsuccessful. Pollen of uge3 is fully fertile provided that UGE2 is present and vice versa. Flowers of uge1,2,5, uge2,3, and uge1,3,4,5 at a comparable developmental stage appear similar to wild-type flowers (Figure 4A). uge1,2,4 displays a severe general growth defect in flowers, and although all floral organs are formed in normal number and position, they are very small and abnormally shaped (Figure 4B). In uge2,4, general floral growth defects are also apparent, although more variable (Figure 4A). Mature wild-type stamens after dehiscence are heavily covered with uniformly sized, rugby ball–shaped pollen grains. This is also the case for other mutants except uge2,3, in which the pollen grains are collapsed (Figure 4C).
Figure 4.
Floral Phenotypes of uge Multiple Mutants.
(A) General growth phenotypes in uge2,4 and uge1,2,4. Bar = 500 μm.
(B) Floral organs are formed but expand abnormally in uge1,2,4. Bar = 200 μm.
(C) Pollen phenotype in uge2,3. Bar = 50 μm.
Together, our analysis of uge mutants suggests that no individual UGE isoform is sufficient for completely normal plant growth even under unstressed conditions and that combinations of UGE genes are necessary for several processes. UGE2 and UGE4 are sufficient for normal growth and development of the shoot and root, respectively. The galactose-dependent germination of uge1,2,4,5 suggests that UGE3 is at least partially sufficient for gametophyte and embryo development but not for postembryonic development. UGE2 and UGE4 play an important overlapping role in shoot growth. The essential function of UGE3 in pollen development overlaps with that of UGE2. The role of UGE3, which appears limited to pollen development, and the observation that uge1,2,4 and uge2,4,5 triple mutants form all organs in the correct position and number suggest that UGE5 and UGE1 are sufficient for overall plant pattern formation but insufficient for normal growth.
Differential Contribution of UGE Isoforms to Total UGE Activity, Cell Wall Carbohydrate Galactose Content, and Growth
In uge1 and uge2 single mutant shoots, wild-type UGE activity is decreased by 30 and 45%, respectively. Mutations in UGE3 and UGE5 cause reductions of 9 and 16%, respectively. The fact that the sum of the activity reductions in each uge single mutant is approximately equivalent to the total wild-type activity suggests no stimulatory or inhibitory interaction between most isoforms. However, for reasons that are unclear at present, uge4 mutants and its combinations uge1,4, uge2,4, and uge1,2,4 display an increase of UGE activity in shoots by ∼13% compared with the corresponding UGE4 background (see Supplemental Table 2 online). Roots of uge2 and uge5 mutant seedlings produce only two-thirds of the wild-type UGE activity, and mutations in UGE1, UGE3, and UGE4 lead to reductions of ∼15, 8, and 20%, respectively. There exists a weak trend of decreasing cell wall galactose content at lower UGE activities among all mutant combinations containing a wild-type UGE2 or UGE4 gene. However, the reduction of cell wall galactose by 20% in uge2,4 and uge1,2,4 mutants is considerably stronger than expected from this trend (Figure 5A). This indicates that the effect of the uge2 mutant allele on cell wall galactose content is strongly enhanced by the uge4 mutation, in contrast with the negligible contribution of UGE4 to total UGE activity in shoots. The reduction of cell wall galactose content by up to 20% in uge shoots correlates (r2 = 0.855) with the reduction of rosette diameter by up to 80% (Figure 5B), indirectly implying that defective cell wall biosynthesis might strongly affect growth. The relative abundance of galactose-containing xyloglucan oligosaccharides in the genotypes uge1,2, uge2,4, uge1,2,4, and uge1,2,5 is decreased compared with that in the wild type (Figure 5C; see Supplemental Table 4 online) and moderately correlates with cell wall galactose content (r2 = 0.627). The relative content of pectic galactan in uge mutant shoots also correlates with total cell wall galactose. The uge1,2-containing combinations have the lowest pectic galactan content, between 49 and 60% of that in wild-type plants. The uge2,4 double mutant, on the other hand, contains 86% of the wild-type pectic galactan content (Figure 5D; see Supplemental Table 5 online).
Figure 5.
Correlation of Biochemical and Growth Parameters of Shoots of uge Mutants.
The dot plots show the comparison between UGE activity and galactose content of cell walls (A), rosette diameter (B), and the amount of galactose-containing xyloglucan oligosaccharides (XGO) (C) and pectic galactan (D). All values are represented in percentages of the wild-type level. The plots also show linear regressions including all genotypes ([B] to [D]; r2 = 0.855, 0.627, and 0.583 respectively) or excluding uge2,4 and uge1,2,4 ([A]; r2 = 0.391). For clarity, only some data points are labeled. All numerical values can be found in Supplemental Tables 2 to 5 online.
Compared with shoots, the galactose content of root cell walls displays a larger variation between different uge mutant combinations (see Supplemental Table 3 online). The strongest reduction is 52% in uge1,2,4 (Figure 6A). Consistent with previous results (Seifert et al., 2002; Nguema-Ona et al., 2006), roots of the uge4 single mutant show a 20% decrease in cell wall galactose content compared with wild-type roots. Mutations in UGE1, UGE2, and UGE5 enhance the effect of the uge4 mutant allele on cell wall galactose content by 7, 17, and 9%, respectively. There is a low correlation between UGE activity and cell wall galactose content if all genotypes are included (r2 = 0.354) (Figure 6A). However the correlation improves for lines combining the uge4 mutant and the UGE2 wild-type allele (r2 = 0.674) and for combinations containing the UGE4 wild-type allele (r2 = 0.414). Combinations of the uge2 and uge4 mutant alleles, on the other hand, have a disproportionate effect on cell wall galactose over the reduction in UGE activity, indicating a synergistic role of these two isoforms influencing cell wall galactose content. The comparison between root length and cell wall galactose content reveals two clusters reflecting the strong reduction of root length caused by mutations in UGE4 (Figure 6B). Within the cluster of lines containing the uge4 mutant allele, root length correlates with cell wall galactose content. By contrast, the UGE4 wild-type allelic combinations show normal root length, although some lines, such as uge1,2,5, display reduced cell wall galactose content (Figure 6B). The large variation in UGE activity in UGE4 wild-type combinations also has no effect on root length (Figure 6C). On the other hand, the two variables show a strong correlation in genotypes combining the uge4 mutant and the UGE2 wild-type allele (r2 = 0.894). Again, the combinations between uge4 and uge2 deviate from this trend. uge2,4 and uge1,2,4 roots are shorter than expected, indicating a synergistic effect of these mutations on root elongation. In roots of all genotypes containing the uge4 mutant allele, the relative total abundance of galactose-containing xyloglucan oligosaccharides is consistently reduced compared with that in the wild type by ∼40% (Figure 6D; see Supplemental Table 4 online). Mutations in the other UGE genes exert no additional effect on xyloglucan galactosylation in roots. The content of pectic galactan varies strongly between different mutant combinations and the wild type (Figure 6E; see Supplemental Table 5 online). Pectic galactan content correlates with total cell wall galactose content; however, in contrast with root length and xyloglucan galactosylation, this parameter fails to separate UGE4 wild type and uge4 mutant combinations (Figure 6E). In summary, UGE2 and UGE4 together play a major role in determining cell wall galactose content in shoots, which correlates with rosette growth. Root cell wall galactose and pectic galactan content are influenced by various UGE isoforms, with UGE4 specifically affecting xyloglucan galactosylation and UGE4 and UGE2 synergistically providing UDP-galactose for a process that is crucial for normal root elongation.
Figure 6.
Correlation of Biochemical and Growth Parameters of Roots of uge Mutants.
The dot plots show the comparison between UGE activity and galactose content of cell walls (A), root length ([B] and [C]), and the amount of galactose-containing xyloglucan oligosaccharides (XGO) (D) and pectic galactan (E). All values are represented in percentages of the wild-type level. The plots also show linear regressions including all genotypes tested ([E]; r2 = 0.617), lines with the UGE4 wild-type background ([A], black line; r2 = 0.414), the uge4 mutant allele ([B]; r2 = 0.871), or with a UGE2 wild-type allele and a uge4 mutant allele ([A] and [C], gray lines; r2 = 0.674 and 0.894, respectively). For clarity, only some data points are labeled. All numerical values can be found in Supplemental Tables 2 to 5 online.
Defects in Secondary Hypocotyl Thickening of uge2,4 Mutants Coincide with Alterations of AGP Epitopes
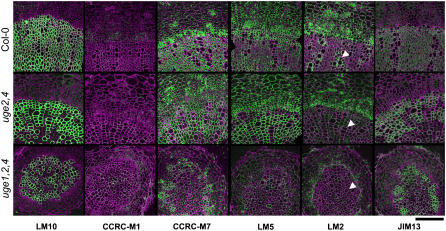
In 8-week-old hypocotyls undergoing secondary thickening, the monoclonal antibody LM10, which reacts with an epitope on xylan, a major matrix polymer of secondary walls in Arabidopsis (McCartney et al., 2005), strongly labels the secondary walls of xylem vessels in the wild type and in uge mutants without apparent variation between genotypes (Figure 7). This indicates the overall integrity of this tissue and that the structure of this xylan epitope is not dramatically affected by mutations in UGE genes. However, monoclonal antibodies recognizing d-galactose–containing epitopes on cell wall polymers differentially label wild-type, uge2,4, and uge1,2,4 hypocotyl sections. CCRC-M1, which binds to α-l-fucosyl (1→2)-β-d-galactosyl side chains of xyloglucan (Puhlmann et al., 1994), only weakly labels the phloem region of wild-type and mutant plants (Figure 7). By contrast, CCRC-M7, which recognizes an arabinosylated (1→6)-β-d-galactan epitope present on AGPs (Steffan et al., 1995), strongly labels cambium, xylem parenchyma, and phloem cells but is absent from vessel elements. The strong labeling of the cambium observed in the wild type is gradually reduced in uge2,4 and uge1,2,4 (Figure 7). LM5, which recognizes (1→4)-β-d-galactan side chains of pectic rhamnogalacturonan I (Jones et al., 1997), shows strong labeling of pith parenchyma, phloem, cambium, and xylem parenchyma but not of xylem vessels. This pattern is not altered in uge2,4. However, in uge1,2,4, LM5 labeling is reduced dramatically (Figure 7). LM2, which recognizes a β-d-glucuronosyl residue on AGP (Yates et al., 1996), labels cambium, phloem, and small cells in the pith region. Moreover, specific small xylem parenchyma cells adjacent to xylem vessel elements, presumably companion cells, are strongly labeled by LM2 (arrowheads in Figure 7). Labeling of the cambium, phloem, and pith region is comparable between uge2,4 and the wild type but generally weaker in uge1,2,4. In uge2,4 and uge1,2,4, the presumptive xylem companion cells are unlabeled (arrowheads in Figure 7). JIM13, which binds to AGPs (Yates et al., 1996), exclusively labels xylem parenchyma in all genotypes. In contrast with other AGP epitopes, JIM13 labeling intensity increases in uge2,4 and uge1,2,4 compared with the wild type (Figure 7). Sections of uge1, uge2, uge4, uge1,2, and uge1,4 JIM13 are labeled with CCRC-M7, LM5, LM2, or like wild-type sections (data not shown).
Figure 7.
Distribution of Cell Wall Epitopes in Secondary Thickened Hypocotyls of uge Mutants.
Transverse sections through secondary thickened hypocotyls of 8-week-old wild-type (top), uge2,4 (middle), and uge1,2,4 (bottom) plants. LM10 recognizes an unsubstituted xylan in the secondary walls of xylem (McCartney et al., 2005); CCRC-M1 binds to α-l-fucosyl (1→2)-β-d-galactosyl side chains of xyloglucan (Puhlmann et al., 1994); LM5 recognizes (1→4)-β-d-galactan side chains of pectic rhamnogalacturonan I (Jones et al., 1997); LM2 recognizes a β-d-glucuronosyl residue on AGP (Yates et al., 1996); CCRC-M7 recognizes an arabinosylated (1→6)-β-d-galactan of arabinogalactan II (Steffan et al., 1995); and JIM13 binds to AGPs (Yates et al., 1996). Arrowheads indicate xylem vessels. Antibody labeling is shown in green, and autofluorescence counterstain is shown in purple. Bar = 100 μm.
Galactose Tolerance Parallels Total UGE Activity
d-Galactose causes root hair and root elongation defects, root tip necrosis, leaf darkening or chlorosis, growth arrest, whole seedling necrosis, and inhibition of germination to various degrees in the wild type and uge mutants (Figure 8; see Supplemental Figure 2 online). Wild-type roots become increasingly stunted and root hairs are more variable in length (see Supplemental Figure 2 online), and shoot growth is inhibited at 72 mM d-galactose and higher (Figure 8). On the other end of the spectrum, uge1,2,5 root hair elongation is strongly inhibited at 9 mM d-galactose and root tips necrotize soon after germination in 18 mM d-galactose (see Supplemental Figure 2 online), and seedlings are white and die before true leaves are formed (Figure 8). The uge2,4 double mutant, which is strongly affected in growth and cell wall composition, is much less affected in its ability to cope with galactose toxicity (Figure 8). Galactose tolerance is ranked Col-0 > uge1,3 = uge1,4 > uge2,4 > uge1,3,4,5 > uge1,2 > uge2,5 > uge1,2,5 in shoots and Col-0 > uge1,3 = uge1,4 > uge2,4 > uge1,2 > uge2,5 > uge1,3,4,5 > uge1,2,5 in roots. Hence, galactose tolerance in shoots and roots parallels total UGE activity.
Figure 8.
Effects of d-Galactose on uge Mutants.
Seedlings were germinated on plates containing standard medium supplemented with different concentrations of d-galactose and documented 12 d after germination. Signs of galactose toxicity are reduced growth, anthocyanin production, and, in the most severe cases, chlorosis and necrosis. Toxicity is moderate in the wild type but most strongly enhanced in multiple uge mutants that display the strongest reduction of UGE activity. The complete lack of pigmentation in uge1,2,5 und uge2,5 seedlings caused by galactose is currently unexplained. However, it might indicate that one mode of action of galactose toxicity in plants is the inhibition of chlorophyll biosynthesis or chloroplast development.
DISCUSSION
Two Major UGE Clades in Higher Plants
At present, only genomes of higher plants, not of unicellular algae, are known to contain multiple genes encoding UGE. The retention of paralogs might be beneficial or neutral in selective terms. Arguing for selective benefit, all five Arabidopsis genes code for functional enzymes, subsets of isoforms are functionally nonredundant, and two major UGE clades are conserved between monocotyledons and dicotyledons. Although the precise number of UGE genes varies between higher plant species, selected gene expression experiments are suited for interspecific comparison of the two large UGE clades. For instance, the subgroup consisting of Arabidopsis UGE2, UGE4, and UGE5 is upregulated in the differentiation zone of the xylem and phloem during secondary hypocotyl thickening (Figure 2), and the subgroup consisting of UGE1:GUS and UGE3:GUS constructs are expressed in pollen tubes growing through the style (see Supplemental Figure 3 online). It remains to be determined whether similar expression patterns will be obtained for groups of orthologous UGE genes in other plant species.
UGE Genes Are Essential for Growth and Galactose Metabolism
The uge1,2,4,5 quadruple mutants only germinate in the presence of low levels of externally applied galactose, suggesting an essential role of UGE genes in the de novo biosynthesis of UDP-galactose. This is consistent with the inability of UDP-xylose and UDP-glucuronic acid 4-epimerases to interconvert UDP-glucose and UDP-galactose (Burget et al., 2003; Gu and Bar-Peled, 2004; Molhoj et al., 2004; Usadel et al., 2004). Galactose tolerance correlates with the level of bulk UGE activity in uge mutants (Figure 8; see Supplemental Figure 2 online), which suggests that all UGE isoforms can act in the conversion of UDP-galactose to UDP-glucose in vivo. As testing galactose tolerance requires artificially high galactose levels, it remains to be investigated whether individual isoforms specifically act in the salvage pathway under normal growth conditions.
Specific Roles for UGE2 and UGE4 in Cell Wall Carbohydrate Biosynthesis and Growth
UGE2 and UGE4 strongly interact in their influence on growth and the composition of cell wall carbohydrates. Among uge double mutants, only uge2,4 displays a reduction in rosette and root growth, hypocotyl elongation, and secondary hypocotyl thickening and shows chlorosis (Table 1). Despite a stronger reduction of UGE activity, uge1,2 displays a less severe reduction of rosette growth and uge1,2,5 roots are apparently normal. Only roots of uge4 single and multiple mutants are shorter than wild-type roots, and within this group the combinations containing uge2,4, are markedly shorter than the rest (Figure 6C). This means that UGE2 and UGE4 cooperate in a process required for normal plant growth.
The suppression of uge mutant growth defects by galactose (Figure 8; see Supplemental Figure 2 online) indicates the requirement of UGE in the provision of UDP-galactose for growth. As a general rule, the reduction of growth in the various uge mutants correlates with their total cell wall galactose content (Figures 5B and 6B). However, the correlations between total cell wall galactose, xyloglucan galactosylation, pectic galactan, and growth differ between shoots and roots.
Rosette diameter, xyloglucan galactosylation, and pectic galactan content correlate with cell wall galactose content (Figures 5B to 5D). The double mutant combinations mainly causing a reduction of cell wall galactose are uge2,4 and uge1,2. However, while the more dramatic growth phenotype of uge2,4 compared with uge1,2 coincides with a stronger reduction in total cell wall galactose (Figure 5B), it contrasts with the higher pectic galactan content in uge2,4 compared with uge1,2 (Figure 5D) and with a 9 to 11% reduction in xyloglucan galactosylation equally affecting uge1,2 and uge2,4 (Figure 5C). The lack of any correlation between growth and xyloglucan or pectin composition in uge2,4 and uge1,2 suggests that other carbohydrates might account for the stronger reduction of cell wall galactose content and growth in uge2,4 compared with uge1,2. Specific AGP-associated carbohydrates in specific cell types depend on the cooperative action of UGE2 and UGE4. This is shown by the loss of LM2 labeling but not of CCRC-M7 and JIM13 labeling in putative xylem companion cells of uge2,4 (Figure 7). The glycosylation of synthetic AGPs expressed in Arabidopsis underlies strong developmental control (Estevez et al., 2006). The synergism between UGE2 and UGE4 could be involved in AGP glycosylation. AGPs have previously been implicated in normal cell expansion (Willats and Knox, 1996; Lee et al., 2005), and it is possible that AGP composition might influence plant growth.
Root elongation is normal in all combinations with the UGE4 wild-type gene, although in some cases, such as uge1,2,5, cell wall galactose content is reduced to a similar extent as in uge4 (Figure 6B, Table 1; see Supplemental Table 3 online). The separate effects of uge4 and UGE4 combinations on the root length axis suggest that root elongation specifically depends on UGE4. However, the correlation between root length and cell wall galactose apparent in uge4 mutant combinations (Figure 6B) could mean that, in the absence of UGE4, other isoforms can partially provide UDP-galactose for a growth-related process and that their loss results in a proportional reduction of cell wall galactose and of growth capacity (Figure 6B). Xyloglucan galactosylation is either like that in the wild type in all allelic combinations containing the UGE4 wild-type allele or at a constant value of ∼60% of the wild-type value in all uge4 mutant allele combinations. By contrast, pectic galactan correlates with total cell wall galactose independently of the presence or absence of UGE4. This might mean that xyloglucan galactosylation in the root crucially depends on UGE4, while pectic galactan structure is determined in a less rigorous manner, accepting UDP-galactose from various UGE isoforms. Thus, the difference between normal roots and shortened roots resembles that between normal and reduced xyloglucan galactosylation (cf. Figures 6B and 6D), while the additional dependence of root length on cell wall galactose content matches the isoform-independent correlation between galactose content and pectic galactan content (cf. Figures 6B, bottom, and 6E). This could mean that reduction of xyloglucan galactosylation in the uge4 single mutant has a major influence on root elongation, while additional reduction of pectic galactan in uge4 mutant combinations, especially in uge2,4 and uge1,2,4, causes additional reduction of root growth.
In summary, UGE2 and UGE4 play an important role in growth that exceeds their individual contributions to total UGE activity and that might be explained by a function of these isoforms in the biosynthesis of specific cell wall polysaccharides, such as xyloglucan, pectic galactan, and AGPs.
The Synergistic Action of UGE2 and UGE4 Potentially Involves Coexpression and Colocalization
The regulation of cell wall biosynthesis is an extremely complex process, which to date has eluded in vitro reconstitution. Genetic whole organism analyses are inherently correlative, and mechanistic models have to be formulated with caution. There are several interesting parallels between the synergistic genetic role of UGE2 and UGE4 and recent biochemical, cytological, and systems biological observations of Arabidopsis UGE isoforms (Barber et al., 2006). The expression of UGE2 and UGE4 is coordinated with that of genes encoding cell wall biosynthetic genes, such as xyloglucan-specific galactosyl transferases (Madson et al., 2003; Li et al., 2004). It is possible that the precise coordination of the expression of individual isoforms with metabolically adjacent enzymes is required for efficient control. However, as UGE5 also is coregulated with UGE2 and UGE4 but does not display an apparent synergism in growth, other mechanisms might come into play as well. Considering the expression of UGE4 and UGE1 in the root elongation zone and of UGE2 in the lateral root cap and the mature zone (Seifert et al., 2004; Barber et al., 2006), it is surprising that uge2,4 exceeds uge1,4 in its effect on root elongation and cell wall composition (Figures 3B and 6, Table 1; see Supplemental Tables 2 to 5 online). There is genetic and cytological evidence of peripheral Golgi localization of UGE4 in the root elongation zone (Seifert et al., 2002, 2004; Barber et al., 2006; Nguema-Ona et al., 2006), and a specific role of uge4 in normal xyloglucan galactosylation in roots is compatible with a tight link between this isoform and Golgi-localized xyloglucan-specific galactosyl transferases. By analogy, the synergistic role of UGE2 and UGE4 could be explained by an accumulation of UGE2 at the Golgi membrane. UGE2 and UGE4 both depend on exogenously supplied NAD+ for maximal in vitro activity, while the cofactor is tightly bound in other isoforms in Arabidopsis and barley (Hordeum vulgare) (Barber et al., 2006; Zhang et al., 2006). Together with the genetic role of UGE2 and UGE4 in cell wall biosynthesis, this could imply that fluctuations of cofactor redox state might fine-tune the incorporation of galactose into cell wall carbohydrates (Seifert, 2004).
Specific Roles of UGE3 and UGE5 Might Be Defined by Their Transcriptional Pattern
The male sterility and the defective pollen of uge2,3 mutants (Figure 4C) suggest an overlapping role for UGE3 and UGE2 in pollen development. As homozygous uge2,3 double mutants segregate from parents homozygous for uge2 and heterozygous for uge3, the uge2,3 pollen per se must be viable, also suggesting a defect in pollen development rather than in pollen germination. The role of UGE3 in pollen development coincides with its high expression in flowers, particularly in stamens (Barber et al., 2006).
We failed to observe growth phenotypes in double mutants carrying the uge5 mutant allele. Nevertheless, uge5 enhances the uge1,2 and uge2,4 growth phenotypes and galactose sensitivity, demonstrating UGE5 functionality. Despite the large contribution of UGE5 to total UGE activity in roots, the uge5 mutation does not influence cell wall galactose content. However, as UGE5 transcription is elevated by abscisic acid and by salt and temperature stress (G.J. Seifert and J. Rösti, unpublished data), it might act in stress situations by supplying UDP-galactose for galactinol biosynthesis. Galactinol and raffinose were previously hypothesized to function as osmoprotectants in drought stress tolerance (Taji et al., 2002).
Conclusions and Perspectives
We have shown that individual UGE isoforms perform essential overlapping functions. UGE2 and UGE4 overlap in general growth, and UGE2 and UGE3 act jointly in pollen development. The synergistic role of UGE2 and UGE4 in plant growth might be related to an efficient provision of UDP-galactose for the biosynthesis of specific carbohydrates in specific cell types. Functional specialization parallels transcriptional coregulation within the carbohydrate biosynthetic network and might also involve metabolite channeling between UGE4 and Golgi-localized galactosyl transferases in roots. Our findings argue against precisely separated functions of UGE isoforms but for a more gradual diversification and partial functional overlap. The specific effects of different mutant combinations suggest that synergisms between sets of isoforms in different metabolic situations might provide the selective benefit that has led to the retention of multiple paralogs. Besides testing the contribution of transcriptional and other regulatory mechanisms to cell wall biosynthesis, it will be interesting in future work to identify stress situations revealing specific roles for UGE1 and UGE5, to analyze the influence on galactinol and galactolipid biosynthesis by UGE isoforms, and to test whether important metabolic pathways such as sucrose and starch metabolism interact with UGE.
METHODS
Database Search and Phylogenetic Analysis of UGE Genes
To compare UGE peptide sequences encoded by completely sequenced plant genomes, the following databases were searched for genes homologous with Arabidopsis thaliana UGEs: Populus trichocarpa and Chlamydomonas reinhardtii databases of the U.S. Department of Energy Joint Genome Institute version 1.1 (http://genome.jgi-psf.org/Poptr1_1/Poptr1_1.home.html) and version 3.0 (http://genome.jgi-psf.org/Chlre3/Chlre3.home.html), respectively, and the Physcomitrella patens database (PHYSCObase) of the National Institute for Basic Biology, Japan (http://moss.nibb.ac.jp). The annotated or whole genome shotgun sequence databases were queried with all five Arabidopsis UGE protein sequences using the TBLASTN algorithm. Resulting coding sequences or contigs were electronically translated and peptide sequences aligned with UGE, UXE, and GAE sequences of Arabidopsis. Sequences clustering with UGEs were truncated at the C and N termini to the first and last conserved residue, respectively, and used for the generation of a maximum likelihood phylogenetic tree with the program PROML from the Phylip version 3.6 software package. Bootstrap values were obtained from 100 bootstrap replicates. The tree was rooted with the C. reinhardtii sequence.
GUS Expression Analysis
For expression analysis of UGE 5′ and 3′ regulatory regions in the wild-type background, several independent transformed UGE promoter:GUS:UGE 3′ untranslated region (UGE:GUS) lines (Seifert et al., 2004) were outcrossed to Col-0, and homozygous UGE4 F3 and F4 populations were analyzed as described previously (Jefferson et al., 1987).
Plant Material
The following uge mutant lines obtained from the ABRC and the Torrey Mesa Research Institute were used: uge1-1 (SALK_019587; T-DNA insertion site at base A1746 in the seventh exon), uge2-1 (SALK_006761; T-DNA insertion site at base C949 in the third intron), uge2-2 (SALK_024044; T-DNA insertion site at base T1560 in the sixth exon), uge3-2 (Garlic_508_E09; T-DNA insertion site at base C1106 in the fifth exon), and uge5-2 (SALK_127988; T-DNA insertion site at base A2296 in the eighth intron). RNA was isolated from mutant and wild-type plants, and cDNA was generated using standard procedures. PCR did not detect specific products in the corresponding mutant lines indicating loss of function. The UGE4 loss-of-function alleles rhd1-1 and rhd1-4 (Seifert et al., 2002) are here named uge4-1 and uge4-4. Additional mutant alleles of UGE3 and UGE5 isolated earlier were not used in this study. Mutant combinations were obtained from repeated crosses and generally kept as homozygous lines. Homozygosity of the uge mutant alleles was determined using PCR of genomic DNA (for primer combinations and sequences, see Supplemental Tables 6 and 7 online). Lethal mutant combinations are kept as populations segregating for the phenotypically obvious uge4 mutant.
Plant Growth
Arabidopsis seeds were surface-sterilized in 5% cleaning bleach (Sanmex) for 30 min and washed twice in sterile water before sowing on standard growth medium (0.5% Phytagel, 4.3 g/L Murashige and Skoog basal salt mixture [Duchefa], and 1% sucrose, pH 5.8). The seeds were stratified at 4°C for 3 d in the dark before germinating and growing in a growth cabinet at 25°C with continuous light at 70 to 90 μmol·m−2·s−1. To assay galactose tolerance, medium was supplemented with 9 to 180 mM d-galactose. Control medium, supplemented with 180 mM d-glucose, led to moderate suppression of growth and leaf darkening. For growth analysis, seeds of each line were sown onto three square plates (100 × 100 mm). The growth progression of the seedlings was monitored each day according to Boyes et al. (2001); growth stages were as follows: 0.5, radicle emergence; 0.7, hypocotyl and cotyledon emergence; 1.0, cotyledons fully opened; 1.02, two rosette leaves > 1 mm; 1.04, four rosette leaves > 1 mm. The root length after 7 d was measured with a ruler, and roots of 5-d-old seedlings were imaged using a dissecting microscope (Wild M10; Leica) with a digital camera (Coolpix 4500; Nikon).
For the morphological analysis of plants grown on soil, seedlings were grown on plates as described above. After 9 d in the growth room, 12 plants of each line were transferred onto soil pots in a randomized arrangement and transferred into a controlled-environment cabinet with 16 h of light (90 μmol·m−2·s−1) and 8 h of dark at temperatures of 23 and 20°C and with 70 and 75% RH, respectively. The growth progression of the plants was monitored every 2 d according to Boyes et al. (2001); growth stages were as follows: 1.08, eight rosette leaves > 1 mm; 5.00, first flower bud visible; 6.00, first flower open. The rosette was measured with a ruler as the largest distance between two leaf tips and photographed at 22 d after germination. For measurement of secondary thickening of hypocotyls, seedlings were grown on standard growth medium for 2 weeks under standard conditions before transferring to soil in a long-day greenhouse for an additional 7 weeks. During this time, inflorescence stems were removed on a weekly basis to encourage vegetative growth. The hypocotyl diameter was measured in the middle section using calipers.
Scanning Electron Microscopy
Flowers of 5-week-old plants were mounted on an aluminum stub using Tissue Tek optimum cutting temperature compound (BDH Laboratory Supplies). The stub was immediately plunged into liquid nitrogen slush at ∼−210°C to cryopreserve the material. The sample was transferred onto the cryostage of an ALTO 2500 cryotransfer system (Gatan) attached to a Zeiss Supra 55 VP FEG scanning electron microscope. Sublimation of surface frost was performed at −95°C for 3 min before sputter-coating of the sample with platinum for 2 min at 10 mA at colder than −110°C. After sputter-coating, the sample was moved onto the cryostage in the main chamber of the microscope and held at ∼−130°C. The sample was imaged at 3 kV, and digital TIFF files were stored.
UGE Activity Assay
Seedlings of wild-type and uge mutant plants were grown on standard growth medium under standard conditions for 11 d. The following uge mutants were used: uge1-1, uge2-2, uge3-2, uge4-4, and uge5-2. For each line, a separate plate was used, and three replicates of all lines were grown at 1-d intervals. On each plate, 60 seedlings were grown on two rows, and all shoots of the lower row and the roots of both rows were harvested separately and homogenized immediately. Plant material was homogenized in a mortar with a pestle and acid-washed glass beads (425 to 600 μm; Sigma-Aldrich) in 10 volumes (v/w) of homogenization buffer (100 mM Tris-HCl, pH 8.7, 1 mM EDTA, 1 mM NAD+, 2 mM DTT, and 1% [v/v] protease inhibitor cocktail [Sigma-Aldrich]). The extract was cleared of cell debris by centrifugation before diluting in an equal volume of homogenization buffer. In order to remove endogenous metabolites, 0.5 mL of the extract was separated on a NAP 5 column (Sephadex G-25; Amersham) equilibrated with homogenization buffer according to the manufacturer's instructions and eluted in 1 mL of homogenization buffer on ice or at 4°C. Three 50-μL aliquots of the protein extract (final dilution, 40 times [v/w]) or homogenization buffer as blank were transferred onto a clear polystyrene, flat-bottom, 96-well microtiter plate (Nunc) and mixed with 200 μL of assay mix (100 mM Tris-HCl, pH 8.7, 2 mM NAD+, 2 mM DTT, 1% [v/v] Tween 20, 132 microunits/μL recombinant UDP-glucose dehydrogenase from Escherichia coli [Calbiochem], and 1 mM UDP-galactose [Calbiochem]) preincubated at ambient temperature for 10 min in order to convert any contaminating UDP-glucose. The increase in absorbance at 340 nm was read every 10 s for 45 min at 25°C on a SpectraMax Plus microplate spectrophotometer (Molecular Devices) controlled by SoftMax Pro version 3.1.1 software (Molecular Devices). Due to a short lag phase at the start of the assay and deviation from linear absorbance increase after 25 min, the slope between 5 and 15 min was used for activity calculations. Saturating conditions were verified by adding double amounts of substrates or coupling enzyme to an assay with twice the wild-type activity in the shoot. Assay linearity in the range used was confirmed with different dilutions of wild-type extract. Without UDP-galactose, no background activity was detected in the wild-type extract. The activity was related to total protein concentration, which was determined using a BSA standard (2 to 25 μg/mL) and the Bio-Rad protein assay dye reagent according to the manufacturer's instructions for cuvette-based or microtiter plate–based microassay procedure. The contribution of each isoform to total UGE activity was experimentally determined from the reduction of the UGE activity in single uge mutants. UGE activity for multiple mutants was estimated by subtracting the combined contributions of the uge mutant alleles from the wild-type value.
Cell Wall Extraction and Analysis
Cell wall extraction was modified from Barton et al. (2006). Ten days after germination, three samples of shoots and roots were transferred separately into tubes, freeze-dried, and ground in a ball mill (Retsch TissueLyser; Qiagen) for 3 min at 30 rpm using 3-mm tungsten carbide beads. The material was suspended in 65% (v/v) ethanol and mixed with the ball mill for 1 min at 30 rpm and incubated for 10 min at 65°C. The homogenate was cooled on ice and centrifuged at 3220g for 10 min. The alcohol-insoluble residue was washed once in 65% (v/v) ethanol and twice in 2:3 (v/v) methanol:chloroform, incubating for 2 to 3 h during the third washing step. The pellet was washed twice in 100% acetone and air-dried. For analysis of the monosaccharide composition of the alcohol-insoluble residue of total cell walls, 1 to 4 mg of dry material was mixed with 200 μL of water containing 50 μg of inositol as an internal standard. To the suspension, 200 μL of 4 M trifluoroacetic acid (final concentration, 2 M) was added before hydrolysis during 1 h at 121°C. The hydrolyzed sample was mixed with 300 μL of 2-propanol and dried at 40°C three times. The dry hydrolysate was resuspended in 400 μL of water and spun down before removing 100 μL of the supernatant for the subsequent alditol acetate derivatization and gas chromatography (York et al., 1986). The xyloglucan oligosaccharide profile was determined as described previously (Lerouxel et al., 2002). The resuspended alcohol-insoluble residue of total cell walls from young shoots and roots was digested overnight at 37°C with 660 milliunits of xyloglucan-specific endoglucanase (Pauly et al., 1999) in 50 mM ammonium formate buffer, pH 4.5. The solubilized xyloglucan oligosaccharides present in the supernatant after centrifugation at 1000g for 10 min were incubated for 10 min with high-performance Q-Sepharose beads (Amersham Biosciences) to remove ions interfering with the subsequent mass spectrometric analysis. The ion-exchanged solution was mixed 1:1 with 10 mg/mL 2,5-dihydroxybenzoic acid as matrix before obtaining the mass spectra using a Voyager DE-Pro matrix-assisted laser-desorption ionization time-of-flight instrument (Applied Biosystems). Enzymatic fingerprinting of pectic polysaccharides by carbohydrate gel electrophoresis and quantification of galactanase-accessible galactan and arabinanase-accessible arabinan were performed as described previously (Barton et al., 2006).
Immunoreflection Microscopy
To analyze xylem differentiation, plants were grown on soil and inflorescences were cut back weekly to stimulate secondary thickening. Hypocotyls were excised from 8-week-old plants. Fixation embedding and immunoreflection imaging were performed as described previously (Bush and McCann, 1999).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: At1g12780 (UGE1), At4g23920 (UGE2), At1g63180 (UGE3), At1g64440 (UGE4), and At4g10960 (UGE5).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Strong Morphological Phenotypes of Multiple uge Mutants.
Supplemental Figure 2. Effect of d-Galactose on Roots of uge Mutants.
Supplemental Figure 3. UGE Promoter Activity in Growing Pollen Tubes.
Supplemental Table 1. Progression of Growth Stages in uge Mutants.
Supplemental Table 2. UGE Activity in uge Single Mutants and Mutant Combinations.
Supplemental Table 3. Neutral Sugar Composition of Cell Walls in uge Mutant Shoots and Roots.
Supplemental Table 4. Xyloglucan Oligosaccharides of uge Mutants.
Supplemental Table 5. Contents of Galactanase- and Arabinanase-Accessible Pectin Side Chains in uge Mutants.
Supplemental Table 6. Primer Combinations to Genotype uge Mutant Alleles.
Supplemental Table 7. Oligonucleotide Sequences for Genotyping Primers.
Supplementary Material
Acknowledgments
We thank Nicolai Obel and David Rico Arcos for their kind help with cell wall analysis; Sandrine J. Rösti for help with bioinformatics; Kay Denyer and members of her laboratory for advice on the activity assay; Michael Hahn and Paul Knox for the provision of monoclonal antibodies; and the ABRC and Syngenta for mutant seeds. J.R. was financially supported by the John Innes Foundation, the United Kingdom Scholarship for International Research Students, and the Janggen-Pöhn Foundation (St. Gallen, Switzerland). G.J.S. was supported by Biotechnology and Biological Science Research Council (BBSRC) Grant 208/D10332, European Union Grant QLK5-CT-2001-00443 (EDEN), and the Austrian Research Fund (FWF Grant P19778-B12). K.F. was supported by the BBSRC. S.A. was supported by European Community Framework V Research Training Network Contract HPRN–CT–2002–00262 (Biointeractions). C.J.B. was supported by a BBSRC Ph.D. studentship.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Georg J. Seifert (georg.seifert@boku.ac.at).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Andeme-Onzighi, C., Sivaguru, M., Judy-March, J., Baskin, T.I., and Driouich, A. (2002). The reb1-1 mutation of Arabidopsis alters the morphology of trichoblasts, the expression of arabinogalactan-proteins and the organization of cortical microtubules. Planta 215 949–958. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815. [DOI] [PubMed] [Google Scholar]
- Barber, C., Rosti, J., Rawat, A., Findlay, K., Roberts, K., and Seifert, G.J. (2006). Distinct properties of the five UDP-D-glucose/UDP-D-galactose 4-epimerase isoforms of Arabidopsis thaliana. J. Biol. Chem. 281 17276–17285. [DOI] [PubMed] [Google Scholar]
- Barton, C.J., Tailford, L.E., Welchman, H., Zhang, Z., Gilbert, H.J., Dupree, P., and Goubet, F. (2006). Enzymatic fingerprinting of Arabidopsis pectic polysaccharides using polysaccharide analysis by carbohydrate gel electrophoresis (PACE). Planta 224 163–174. [DOI] [PubMed] [Google Scholar]
- Baskin, T.I., Betzner, A.S., Hoggart, R., Cork, A., and Williamson, R.E. (1992). Root morphology mutants in Arabidopsis thaliana. Aust. J. Plant Physiol. 19 427–437. [Google Scholar]
- Baum, T.J., Wubben, M.J., 2nd, Hardy, K.A., Su, H., and Rodermel, S.R. (2000). A screen for Arabidopsis thaliana mutants with altered susceptibility to Heterodera schachtii. J. Nematol. 32 166–173. [PMC free article] [PubMed] [Google Scholar]
- Bonin, C.P., Freshour, G., Hahn, M.G., Vanzin, G.F., and Reiter, W.D. (2003). The GMD1 and GMD2 genes of Arabidopsis encode isoforms of GDP-D-mannose 4,6-dehydratase with cell type-specific expression patterns. Plant Physiol. 132 883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonin, C.P., Potter, I., Vanzin, G.F., and Reiter, W.D. (1997). The MUR1 gene of Arabidopsis thaliana encodes an isoform of GDP-D-mannose-4,6-dehydratase, catalyzing the first step in the de novo synthesis of GDP-L-fucose. Proc. Natl. Acad. Sci. USA 94 2085–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes, D.C., Zayed, A.M., Ascenzi, R., McCaskill, A.J., Hoffman, N.E., Davis, K.R., and Görlach, J. (2001). Growth stage-based phenotypic analysis of Arabidopsis: A model for high throughput functional genomics in plants. Plant Cell 13 1499–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burget, E.G., Verma, R., Molhoj, M., and Reiter, W.D. (2003). The biosynthesis of L-arabinose in plants: Molecular cloning and characterization of a Golgi-localized UDP-D-xylose 4-epimerase encoded by the MUR4 gene of Arabidopsis. Plant Cell 15 523–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush, M.S., and McCann, M.C. (1999). Pectic epitopes are differentially distributed in the cell walls of potato (Solanum tuberosum) tubers. Physiol. Plant. 107 201–213. [Google Scholar]
- Diet, A., Link, B., Seifert, G.J., Schellenberg, B., Pauly, M., Reiter, W.D., and Ringli, C. (2006). The Arabidopsis root hair cell wall formation mutant lrx1 is suppressed by mutations in the RHM1 gene encoding a UDP-L-rhamnose synthase. Plant Cell 18 1630–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörmann, P., and Benning, C. (1998). The role of UDP-glucose epimerase in carbohydrate metabolism of Arabidopsis. Plant J. 13 641–652. [DOI] [PubMed] [Google Scholar]
- Estevez, J.M., Kieliszewski, M.J., Khitrov, N., and Somerville, C. (2006). Characterization of synthetic hydroxyproline-rich proteoglycans with arabinogalactan protein and extensin motifs in Arabidopsis. Plant Physiol. 142 458–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, X., and Bar-Peled, M. (2004). The biosynthesis of UDP-galacturonic acid in plants. Functional cloning and characterization of Arabidopsis UDP-D-glucuronic acid 4-epimerase. Plant Physiol. 136 4256–4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005). The map-based sequence of the rice genome. Nature 436 793–800. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, L., Seymour, G.B., and Knox, J.P. (1997). Localization of pectic galactan in tomato cell walls using a monoclonal antibody specific to (1->4)-β-D-galactan. Plant Physiol. 113 1405–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K.J.D., Sakata, Y., Mau, S.L., Pettolino, F., Bacic, A., Quatrano, R.S., Knight, C.D., and Knox, J.P. (2005). Arabinogalactan proteins are required for apical cell extension in the moss Physcomitrella patens. Plant Cell 17 3051–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerouxel, O., Choo, T.S., Seveno, M., Usadel, B., Faye, L., Lerouge, P., and Pauly, M. (2002). Rapid structural phenotyping of plant cell wall mutants by enzymatic oligosaccharide fingerprinting. Plant Physiol. 130 1754–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., Cordero, I., Caplan, J., Molhoj, M., and Reiter, W.D. (2004). Molecular analysis of 10 coding regions from Arabidopsis that are homologous to the MUR3 xyloglucan galactosyltransferase. Plant Physiol. 134 940–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madson, M., Dunand, C., Li, X., Verma, R., Vanzin, G.F., Caplan, J., Shoue, D.A., Carpita, N.C., and Reiter, W.D. (2003). The MUR3 gene of Arabidopsis encodes a xyloglucan galactosyltransferase that is evolutionarily related to animal exostosins. Plant Cell 15 1662–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney, L., Marcus, S.E., and Knox, J.P. (2005). Monoclonal antibodies to plant cell wall xylans and arabinoxylans. J. Histochem. Cytochem. 53 543–546. [DOI] [PubMed] [Google Scholar]
- Molhoj, M., Verma, R., and Reiter, W.D. (2004). The biosynthesis of D-galacturonate in plants. Functional cloning and characterization of a membrane-anchored UDP-D-glucuronate 4-epimerase from Arabidopsis. Plant Physiol. 135 1221–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguema-Ona, E., Andeme-Onzighi, C., Aboughe-Angone, S., Bardor, M., Ishii, T., Lerouge, P., and Driouich, A. (2006). The reb1-1 mutation of Arabidopsis: Effect on the structure and localization of galactose-containing cell wall polysaccharides. Plant Physiol. 140 1406–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattathil, S., Harper, A.D., and Bar-Peled, M. (2005). Biosynthesis of UDP-xylose: Characterization of membrane-bound AtUxs2. Planta 221 538–548. [DOI] [PubMed] [Google Scholar]
- Pauly, M., Andersen, L.N., Kauppinen, S., Kofod, L.V., York, W.S., Albersheim, P., and Darvill, A. (1999). A xyloglucan-specific endo-beta-1,4-glucanase from Aspergillus aculeatus: Expression cloning in yeast, purification and characterization of the recombinant enzyme. Glycobiology 9 93–100. [DOI] [PubMed] [Google Scholar]
- Puhlmann, J., Bucheli, E., Swain, M.J., Dunning, N., Albersheim, P., Darvill, A.G., and Hahn, M.G. (1994). Generation of monoclonal antibodies against plant cell-wall polysaccharides. I. Characterization of a monoclonal antibody to a terminal α-(1→2)-linked fucosyl-containing epitope. Plant Physiol. 104 699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter, W.D., and Vanzin, G.F. (2001). Molecular genetics of nucleotide sugar interconversion pathways in plants. Plant Mol. Biol. 47 95–113. [PubMed] [Google Scholar]
- Scheible, W.R., and Pauly, M. (2004). Glycosyltransferases and cell wall biosynthesis: Novel players and insights. Curr. Opin. Plant Biol. 7 285–295. [DOI] [PubMed] [Google Scholar]
- Schiefelbein, J.W., and Somerville, C. (1990). Genetic control of root hair development in Arabidopsis thaliana. Plant Cell 2 235–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert, G.J. (2004). Nucleotide sugar interconversions and cell wall biosynthesis: How to bring the inside to the outside. Curr. Opin. Plant Biol. 7 277–284. [DOI] [PubMed] [Google Scholar]
- Seifert, G.J., Barber, C., Wells, B., Dolan, L., and Roberts, K. (2002). Galactose biosynthesis in Arabidopsis: Genetic evidence for substrate channeling from UDP-D-galactose into cell wall polymers. Curr. Biol. 12 1840–1845. [DOI] [PubMed] [Google Scholar]
- Seifert, G.J., Barber, C., Wells, B., and Roberts, K. (2004). Growth regulators and the control of nucleotide sugar flux. Plant Cell 16 723–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville, C., Bauer, S., Brininstool, G., Facette, M., Hamann, T., Milne, J., Osborne, E., Paredez, A., Persson, S., Raab, T., Vorwerk, S., and Youngs, H. (2004). Toward a systems approach to understanding plant cell walls. Science 306 2206–2211. [DOI] [PubMed] [Google Scholar]
- Steffan, W., Kovac, P., Albersheim, P., Darvill, A.G., and Hahn, M.G. (1995). Characterization of a monoclonal antibody that recognizes an arabinosylated (1→6)-β-D-galactan epitope in plant complex carbohydrates. Carbohydr. Res. 275 295–307. [DOI] [PubMed] [Google Scholar]
- Taji, T., Ohsumi, C., Iuchi, S., Seki, M., Kasuga, M., Kobayashi, M., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2002). Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J. 29 417–426. [DOI] [PubMed] [Google Scholar]
- Tuskan, G.A., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313 1596–1604. [DOI] [PubMed] [Google Scholar]
- Usadel, B., Schluter, U., Molhoj, M., Gipmans, M., Verma, R., Kossmann, J., Reiter, W.D., and Pauly, M. (2004). Identification and characterization of a UDP-D-glucuronate 4-epimerase in Arabidopsis. FEBS Lett. 569 327–331. [DOI] [PubMed] [Google Scholar]
- Willats, W.G., and Knox, J.P. (1996). A role for arabinogalactan-proteins in plant cell expansion: Evidence from studies on the interaction of β-glucosyl Yariv reagent with seedlings of Arabidopsis thaliana. Plant J. 9 919–925. [DOI] [PubMed] [Google Scholar]
- Yates, E.A., Valdor, J.F., Haslam, S.M., Morris, H.R., Dell, A., Mackie, W., and Knox, J.P. (1996). Characterization of carbohydrate structural features recognized by anti-arabinogalactan-protein monoclonal antibodies. Glycobiology 6 131–139. [DOI] [PubMed] [Google Scholar]
- York, W.S., Darvill, A., McNell, M., Stevenson, T.T., and Albersheim, P. (1986). Isolation and characterization of plant cell walls and cell wall components. Methods Enzymol. 118 3–40. [Google Scholar]
- Zhang, Q., Hrmova, M., Shirley, N.J., Lahnstein, J., and Fincher, G.B. (2006). Gene expression patterns and catalytic properties of UDP-D-glucose 4-epimerases from barley (Hordeum vulgare L.). Biochem. J. 394 115–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.