Abstract
To determine whether Saccharomyces cerevisiae can serve as a host for efficient carotenoid and especially β-carotene production, carotenogenic genes from the carotenoid-producing yeast Xanthophyllomyces dendrorhous were introduced and overexpressed in S. cerevisiae. Because overexpression of these genes from an episomal expression vector resulted in unstable strains, the genes were integrated into genomic DNA to yield stable, carotenoid-producing S. cerevisiae cells. Furthermore, carotenoid production levels were higher in strains containing integrated carotenogenic genes. Overexpression of crtYB (which encodes a bifunctional phytoene synthase and lycopene cyclase) and crtI (phytoene desaturase) from X. dendrorhous was sufficient to enable carotenoid production. Carotenoid production levels were increased by additional overexpression of a homologous geranylgeranyl diphosphate (GGPP) synthase from S. cerevisiae that is encoded by BTS1. Combined overexpression of crtE (heterologous GGPP synthase) from X. dendrorhous with crtYB and crtI and introduction of an additional copy of a truncated 3-hydroxy-3-methylglutaryl-coenzyme A reductase gene (tHMG1) into carotenoid-producing cells resulted in a successive increase in carotenoid production levels. The strains mentioned produced high levels of intermediates of the carotenogenic pathway and comparable low levels of the preferred end product β-carotene, as determined by high-performance liquid chromatography. We finally succeeded in constructing an S. cerevisiae strain capable of producing high levels of β-carotene, up to 5.9 mg/g (dry weight), which was accomplished by the introduction of an additional copy of crtI and tHMG1 into carotenoid-producing yeast cells. This transformant is promising for further development toward the biotechnological production of β-carotene by S. cerevisiae.
Carotenoids are a class of pigments of commercial interest that have important biological functions. In humans, β-carotene is the precursor of vitamin A; it may function as an antioxidant, has properties protective against cancer, and stimulates the immune system (14, 22, 32). Because carotenoids are colored compounds, they are being used as pigments in the food and feed industries (20). Many carotenoids are being produced by chemical synthesis, which yields products that are pure and cheap (10). Several microorganisms, including fungi, bacteria, and algae, are able to produce carotenoids naturally (12). Biotechnological synthesis of certain carotenoids, by either homologous or heterologous production, may become more and more attractive (15, 17). One example of a carotenoid-producing yeast is the red yeast Xanthophyllomyces dendrorhous, which was formerly known as Phaffia rhodozyma (9). This yeast mainly produces the carotenoid astaxanthin but also accumulates some β-carotene as an intermediate of the astaxanthin biosynthesis pathway (1, 7, 35). The genes involved in β-carotene production in X. dendrorhous have been cloned previously (33, 34). In X. dendrorhous, the ergosterol and carotenoid biosynthetic pathways are connected by their utilization of prenyl diphosphates (Fig. 1). Farnesyl diphosphate (FPP) is converted into geranylgeranyl diphosphate (GGPP) by GGPP synthase, which is encoded by crtE. Next, the phytoene synthase activity of the bifunctional enzyme CrtYB results in the synthesis of phytoene from two GGPP molecules. Phytoene is subsequently converted into lycopene by four desaturation reactions catalyzed by the enzyme CrtI. Subsequently, two cyclization reactions catalyzed by CrtYB result in the conversion of lycopene into γ-carotene and finally into β-carotene. X. dendrorhous mutants with higher carotenoid production capacities have been obtained by chemical mutagenesis (1, 7) or by recombinant DNA technology (35, 39).
FIG. 1.
Overview of the ergosterol biosynthetic pathway in S. cerevisiae and the carotenogenic pathway in X. dendrorhous. The carotenogenic pathway in X. dendrorhous consists of GGPP synthase encoded by crtE, the bifunctional enzyme phytoene synthase and lycopene cyclase encoded by crtYB, and phytoene desaturase encoded by crtI. S. cerevisiae contains a GGPP synthase, encoded by BTS1, which is able to convert FPP into GGPP. HMG1 encodes HMG-CoA reductase, which is the main regulatory point in the ergosterol biosynthetic pathway in many organisms. IPP, isopentenyl diphosphate; DMAP, dimethylallyl diphosphate; GPP, geranyl diphosphate.
The food yeast Saccharomyces cerevisiae is widely used in the brewing and fermentation industries; it is generally recognized as safe and can be used for the production of biomass rich in high-quality proteins and metabolites. Furthermore, it has the advantage of easy genetic manipulation with established host-vector systems (2, 21). In order to transform S. cerevisiae into a β-carotene-producing organism, precursors of carotenoids should be present. Like X. dendrorhous, S. cerevisiae is able to produce FPP and converts it into GGPP, the basic building block of carotenoids. Conversion of FPP into GGPP is catalyzed by GGPP synthase encoded by BTS1 in S. cerevisiae (11; Fig. 1). Therefore, overexpression of only crtYB and crtI from X. dendrorhous in S. cerevisiae should generally be sufficient to transform S. cerevisiae into a β-carotene-producing organism. Additional overexpression of crtE from X. dendrorhous or BTS1 from S. cerevisiae might increase GGPP levels and thereby enhance β-carotene production. In an initial effort to produce β-carotene heterologously in S. cerevisiae, the carotenogenic genes crtE, crtB, crtI, and crtY from the bacterium Erwinia uredovora were introduced and overexpressed from episomal vectors with different yeast-derived promoters and terminators for each gene. This resulted in quite low β-carotene production levels of 103 μg/g (dry weight [dw]) (41). Overexpression of the same genes in the food yeast Candida utilis resulted in higher β-carotene production levels of 400 μg/g (dw) (18). Additional overexpression of the catalytic domain of 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase from C. utilis, which is considered to be a key regulatory step in ergosterol biosynthesis in yeasts (4), resulted in a 7.1-fold increase in lycopene content. The effect of HMG-CoA reductase on β-carotene production was not examined (29).
Attempts to produce carotenoids heterologously in Escherichia coli by using genes from X. dendrorhous resulted in poor enzyme expression and carotenoid production (G. Sandmann, unpublished results). Although heterologous carotenoid production in S. cerevisiae has already been studied by using bacterial genes from E. uredovora, we expect that β-carotene production levels will be much higher with genes from another yeast species. Therefore, we have constructed a series of carotenoid-producing S. cerevisiae strains by successive introduction and overexpression of carotenogenic genes from X. dendrorhous.
MATERIALS AND METHODS
Vector construction and transformation.
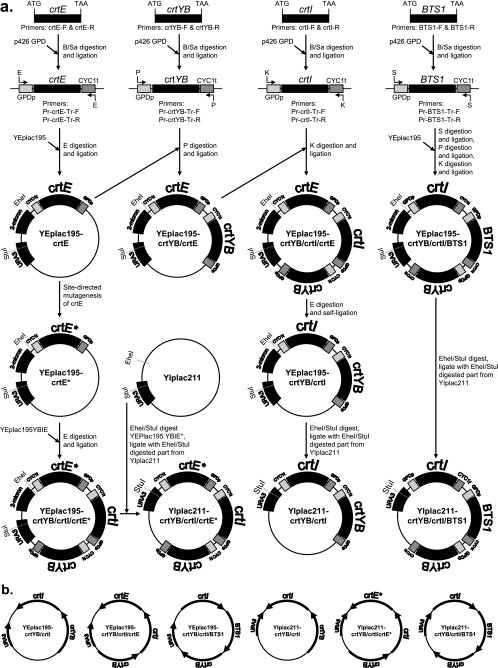
Episomal and integration vectors were constructed as depicted in Fig. 2a. The genes crtYB, crtI, and crtE were amplified with a cDNA library from X. dendrorhous as the template (36) and primers indicated in Table 1. The sequences of the carotenogenic genes can be accessed at NCBI (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Nucleotide&itool=toolbar). The BTS1 gene was amplified with genomic DNA prepared from S. cerevisiae strain CEN.PK 113-7D and primers indicated in Table 1. Standard PCR conditions and the proofreading enzyme super TAQ plus (SphaeroQ, Gorinchem, The Netherlands) were used in all PCRs, except for site-directed mutagenesis. The amplified PCR products were verified by sequencing. The crtYB, crtI, crtE, and BTS1 PCR products were BamHI/SalI ligated into vector p426GPD, containing 680 bp of the promoter region upstream of the start codon of the TDH3 gene and the first 250 bp of the CYC1 terminator from S. cerevisiae (19). Subsequently, TDH3 promoter-gene-CYC1 terminator products were amplified by PCR with the primers indicated in Table 1 (Isogen Life Science, IJsselstein, The Netherlands). To construct episomal expression vectors, TDH3p-crtE-CYC1t was EcoRI ligated into vector YEplac195 (6). Next, TDH3p-crtYB-CYC1t was PstI ligated and TDH3p-crtI-CYC1t was KpnI ligated into YEplac195E, resulting in vector YEplac195YB/I/E. To construct YEplac195YB/I/BTS1, in sequential order TDH3p-BTS1-CYC1t was SmaI ligated, TDH3p-crtYB-CYC1t was PstI ligated, and TDH3p-crtI-CYC1t was KpnI ligated into YEplac195. To construct YEplac195YB/I, YEplac195YB/I/E was restricted with EcoRI and the remaining vector was self-ligated. For targeted integration into the ura3-52 locus, integrative vectors were constructed (6). Targeted integration into the ura3-52 locus requires linearization of vector YIplac211 with StuI. The StuI restriction site present in the coding sequence of crtE was eliminated by site-directed mutagenesis. A at position 551 was mutated to G with vector YEplac195E as the template and the primers indicated in Table 1 (for the procedure used, see reference 37). THD3p-crtE*-CYC1t (* denotes the A-to-G mutation) was EcoRI ligated into YEplac195YB/I/E restricted with EcoRI. The 2μm part of YEplac195 was eliminated by ligating a 694-bp EheI/StuI fragment from vector YIplac211 into YEplac195YB/I/E*, YEplac195YB/I/BTS1, and YEplac195YB/I, which were restricted by EheI/StuI (6). In this manner, tandem head-to-tail orientation of the genes introduced was maintained. All promoter-gene-terminator fusions were indeed present in tandem head-to-tail orientation on the YEplac and YIplac expression vectors, as determined by restriction analysis and shown schematically in Fig. 2b. The YIplac211 vectors containing carotenogenic genes were linearized with StuI and transformed into strain CEN.PK 113-5D to create strains YB/I/BTS1 and YB/I/E. Single-copy integration of the constructs was confirmed by Southern blotting by standard laboratory procedures (27). To create vector YIplac128 crtI, vector YEplac195YB/I was restricted with KpnI and the TDH3p-crtI-CYC1t fragment was ligated into YIplac128 (6). For overexpression of the catalytic domain of HMG-CoA reductase (tHMG1 for truncated HMG1), the 1,575-bp C-terminal part of HMG1 was amplified with primers tHMG1-F and tHMG1-R and S. cerevisiae genomic DNA as the template. In the forward primer, a start codon (ATG) was included. Amplified tHMG1 was SpeI/XhoI ligated into vector p426 GPD (19). Subsequently, TDH3p-tHGM1-CYC1t was amplified with primers Pr-tHMG1-Tr-F and Pr-tHMG1-Tr-F and PstI ligated into YIplac204 (6) to create YIplac204 tHMG1. To create strain YB/I/E + extra I, YIplac128 crtI was linearized with ClaI for targeted integration into the leu2,3-112 locus. To create strain YB/I/E + tHMG1, YIplac204 tHMG1 was linearized with EcoRV to target integration into the trp1-289 locus. The linearized vectors were transformed into S. cerevisiae strain CEN.PK 113-6B previously transformed with YIplac211 YB/I/E*. To create strain YB/I/E + tHMG1 + extra I, strain YB/I/E + tHMG1 was transformed with ClaI-linearized YIPlac128 crtI for targeted integration into the leu2,3-112 locus. All constructs were transformed into yeast by electroporation (3). The constructs created in this study are indicated in Table 2.
FIG. 2.
Construction of expression vectors used in this study. (a) Scheme representing the construction of the different vectors used in this study. GPDp is the 680-bp sequence of the TDH3 promoter, and CYC1t is the 250-bp sequence of the CYC1 terminator. Restriction enzyme abbreviations: B, BamHI; Sa, SalI; E, EcoRI; P, PstI; K, KpnI; S, SmaI. Sequences used for primers are indicated in Table 1. Integration into the ura3-52 locus requires linearization of the YIplac211 constructs with StuI. Because crtE contains a StuI restriction site, A at position 551 was changed into G by site-directed mutagenesis (indicated by an asterisk). See Materials and Methods for details concerning the cloning strategy used. (b) Orientation of the carotenogenic genes in the vectors used in this study, as determined by restriction analysis. All genes are present in tandem head-to-tail orientation on the expression vectors.
TABLE 1.
| Primer | Sequence (5′ to 3′) |
|---|---|
| For PCR | |
| crtYB-F | CGCGGATCCATGACGGCTCTCGCATATTAC |
| crtYB-R | TGCGGTCGACTTACTGCCCTTCCCATCCGC |
| crtI-F | GCGGGATCCATGGGAAAAGAACAAGATCAGG |
| crtI-R | TGCGGTCGACTCAGAAAGCAAGAACACCAACG |
| crtE-F | CGCGGATCCATGGATTACGCGAACATCCTC |
| crtE-R | TGCGGTCGACTCACAGAGGGATATCGGCTAG |
| BTS1-F | CGCGGATCCATGGAGGCCAAGATAGATGAG |
| BTS1-R | TGCGGTCGACTCACAATTCGGATAAGTGGTCT |
| Pr-crtYB-Tr-F | AAAACTGCAGCAGTTCGAGTTTATCATTATCAAT |
| Pr-crtYB-Tr-R | TTTTCTGCAGGCGGCCGCGCAAATTAAAGCCTTCGAGCG |
| Pr-crtI-Tr-F | CGGGGTACCCAGTTCGAGTTTATCATTATCAAT |
| Pr-crtI-Tr-R | GCCGGTACCGGCCGGCCGCAAATTAAAGCCTTCGAGCG |
| Pr-crtE-Tr-F | GGAATTCCAGTTCGAGTTTATCATTATCAAT |
| Pr-crtE-Tr-R | CGAATTCGGCGCGCCGCAAATTAAAGCCTTCGAGCG |
| Pr-BTS1-Tr-F | TCCCCCGGGCAGTTCGAGTTTATCATTATCAAT |
| Pr-BTS1-Tr-R | AGGCCCGGGGGCGCGCCGCAAATTAAAGCCTTCGAGCG |
| crtE (551 A-to-G mutation)-F | CCATAGAGGGCAGGGCCTGGAGCTATTC |
| crtE (551 A-to-G mutation)-R | GAATAGCTCCAGGCCCTGCCCTCTATGG |
| tHMG1-F | GGACTAGTATGGACCAATTGGTGAAAACTGAAG |
| tHMG1-R | CCGCTCGAGTTAGGATTTAATGCAGGTGACG |
| Pr-tHMG1-Tr-F | AAAACTGCAGCAGTTCGAGTTTATCATTATCAAT |
| Pr-tHMG1-Tr-R | TTTTCTGCAGGCGGCCGCGCAAATTAAAGCCTTCGAGCG |
| For qPCR | |
| ACT1-F | GCCTTGGACTTCGAACAAGA |
| ACT1-R | CCAAACCCAAAACAGAAGGA |
| crtYB-F | TAGTCGCCTACGCAGAGGAT |
| crtYB-R | TCTCTCCGACGTCTCCTTTC |
| crtI-F | GGGCTGACTTAGTTGGTGGA |
| crtI-R | TTGTGTCGAATGATCCCTTG |
| crtE-F | CACAGAAACCCACTCATTCG |
| crtE-R | GTTCTTCCCCGTTCTTCCTT |
| BTS1-F | CGAAAGGTCAAACTGAGCAAC |
| BTS1-R | GCGAAGCCAAATCAGGTAAA |
Underlined letters indicate restriction enzyme cleavage sites corresponding to the primer description. Bold letters indicate the A-to-G mutation in the crtE gene. Bold and underlined letters indicate the start codon created for tHMG1.
TABLE 2.
Yeast strains and plasmids used in this studya
| Yeast strain or plasmid | Relevant features |
|---|---|
| Yeast strains | |
| CEN.PK 113-7Db | MATaSUC2 MAL2-8c |
| CEN.PK 113-5Db | MATaSUC2 MAL2-8cura3-52 |
| CEN.PK 113-6Bb | MATaSUC2 MAL2-8cura3-52 leu2,3-112 trp1-289 |
| Episomal vector transformants | |
| YB/I | CEN.PK 113-5D + YEplac195 YB/I |
| YB/I/BTS1 | CEN.PK 113-5D + YEplac195 YB/I/BTS1 |
| YB/I/E | CEN.PK 113-5D + YEplac195 YB/I/E |
| YB/I/E* | CEN.PK 113-5D + YEplac195 YB/I/E* |
| Integrative vector transformants | |
| YB/I/BTS1 | CEN.PK 113-5D + YIplac211 YB/I/BTS1 |
| YB/I/E | CEN.PK 113-5D + YIplac211 YB/I/E* |
| YB/I/E + extra I | CEN.PK 113-6B + YIplac211 YB/I/E* + YIplac128 crtI |
| YB/I/E + tHMG1 | CEN.PK 113-6B + YIplac211 YB/I/E* + YIplac204 tHMG1 |
| YB/I/E + tHMG1 + extra I | CEN.PK 113-6B + YIplac211 YB/I/E* + YIplac204 tHMG1 + YIplac128 crtI |
| Plasmids | |
| YEplac195 YB/I | YEplac195 TDH3p-crtYB-CYC1t; TDH3p-crtI-CYC1t |
| YEplac195 YB/I/BTS1 | YEplac195 TDH3p-crtYB-CYC1t; TDH3p-crtI-CYC1t; TDH3p-BTS1-CYC1t |
| YEplac195 YB/I/E | YEplac195 TDH3p-crtYB-CYC1t; TDH3p-crtI-CYC1t; TDH3p-crtE-CYC1t |
| YEplac195 YB/I/E* | YEplac195 TDH3p-crtYB-CYC1t; TDH3p-crtI-CYC1t; TDH3p-crtE*-CYC1t |
| YIplac211 YB/I/BTS1 | YIplac211 TDH3p-crtYB-CYC1t; TDH3p-crtI-CYC1t; TDH3p-BTS1-CYC1t |
| YIplac211 YB/I/E* | YIplac211 TDH3p-crtYB-CYC1t; TDH3p-crtI-CYC1t; TDH3p-crtE*-CYC1t |
| YIplac128 crtI | YIplac128 TDH3p-crtI-CYC1t |
| YIplac204 tHMG1 | YIplac204 TDH3p-tHMG1-CYC1t |
The asterisk indicates the 551A-to-G mutation in the crtE coding sequence. Functional genes are in uppercase italics; nonfunctional ones are in lowercase italics. Promoters and terminators are represented as follows: TDH3p, TDH3 promoter; CYC1t, CYC1 terminator. For targeted integration, integrative (YIplac) plasmids were transformed as described in Materials and Methods.
Institut für Mikrobiologie, J. W. Goethe Universität, Frankfurt am Main, Germany.
Strains and media.
The S. cerevisiae strains used in this study are indicated in Table 2. During batch culture, experiments yeast cells were grown on yeast nitrogen base (YNB) without amino acids (Difco, Boom, The Netherlands), supplemented with the appropriate amino acids when required and 2% (wt/vol) d-glucose. Cells were grown at 225 rpm and 30°C in a shaking incubator.
Carotenoid analysis.
Cells were resuspended in 1 ml sterile water, 1 g 0.50- to 0.75-mm glass beads was added, and cells were broken by vortexing for 3 min. A 2.5-ml volume of 0.2% (wt/vol) pyrogallol dissolved in methanol was added, and cells were vortexed for 10 s. After adding 1.25 ml 60% (wt/vol) KOH and vortexing for 10 s, the cells were incubated for 1 h at 75°C with vortexing every 15 min for saponification. Next, an appropriate amount of hexane was added to extract the carotenoid fraction. The tubes were centrifuged for 5 min at 2,800 rpm, and 1 ml of the hexane was pipetted into a cuvette. Absorption between 550 and 400 nm was monitored on a Shimadzu UV-2501 PC spectrophotometer (Shimadzu, Duisburg, Germany) to measure the amounts of colored carotenoids. A standard curve was determined by measuring known β-carotene concentrations. The total colored carotenoid concentration was calculated by the following formula: total carotenoids (in μg g−1 [dw]) = (A449 × ml hexane)/(0.2072 × g [dw]). An aliquot of the total carotenoid extractions was evaporated under nitrogen in the dark for subsequent analysis of the carotenoid composition by high-performance liquid chromatography (HPLC). HPLC separation and quantization were performed on a Nucleosil C18 3-μm column eluted isocratically with acetonitrile-methanol-2-propanol (50:40:10, vol/vol) at a flow rate of 1 ml min−1 (30). The separated carotenoids were detected with a Kontron 440 diode array detector and spectra were directly recorded online. Reference compounds for identification and quantitation were generated in transgenic E. coli cells (28). The dw of a sample was measured by taking the same culture volume as used for the carotenoid extraction, drying it overnight at 80°C, and measuring the weight of the dried yeast cells on an analytical balance.
qPCR studies.
Total RNA was isolated as previously described (38). To ensure similar amounts of starting materials, total RNA concentrations were carefully measured with a Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies Inc.). Next, chromosomal DNA was degraded by treatment with DNase I (amplification grade; Invitrogen) according to the manufacturer's instructions. Subsequently, cDNA was amplified with an Omniscript RT kit (QIAGEN, Venlo, The Netherlands) with the following modifications. A 10 mM deoxynucleoside triphosphate stock (Promega) and an Oligo-dT18 primer (Isogen Life Science, IJsselstein, The Netherlands) were used in the reaction mixtures. Pipetting was performed by a pipetting robot (Corbett Robotics), with the amplified cDNA, the quantitative PCR (qPCR) primers as indicated in Table 1, and ABsolute QPCR SYBR green mix (ABgene). These qPCR primers were designed with Primer3 (26). The qPCRs were performed in a Rotor Gene RG-3000 real-time PCR machine (Corbett Research). To correct for differences in the amounts of starting materials, ACT1 was chosen as a reference housekeeping gene. The results are presented as ratios of gene expression between the target gene (gene of interest) and the reference gene, ACT1 (23).
RESULTS AND DISCUSSION
Transformation with episomal vectors.
To determine whether S. cerevisiae is able to produce carotenoids, carotenogenic genes from X. dendrorhous were initially overexpressed from episomal vectors. Expression of each of the genes introduced was controlled by the constitutive TDH3 promoter and the CYC1 terminator (19). Overexpression of crtYB and crtI with YEplac195 resulted in faintly yellow transformants. Additional overexpression of the GGPP synthase BTS1 from S. cerevisiae or crtE from X. dendrorhous resulted in orange cells. When grown in YNB 2% glucose medium, all transformants reached optimal densities similar to the optical density of wild-type cells, indicating that carotenoid production in S. cerevisiae does not influence growth (data not shown). After centrifugation, cells overexpressing crtYB and crtI were faintly yellow, cells overexpressing crtYB, crtI, and BTS1 were yellow, and cells that overexpressed crtYB, crtI, and crtE were orange. HPLC studies confirmed that carotenoids were produced in cells overexpressing crtYB and crtI (3 μg total carotenoids/g [dw]) and that additional overexpression of BTS1 resulted in a 28-fold increase in carotenoid production (83 μg/g [dw]). Overexpression of crtYB, crtI, and crtE resulted in 162-fold higher total carotenoid levels compared to cells overexpressing crtYB and crtI (487 μg/g [dw]; Table 3). It was expected that overexpression of carotenogenic genes from X. dendrorhous would yield S. cerevisiae cells producing high levels of pure β-carotene. However, HPLC studies showed that besides β-carotene, cells overexpressing crtYB and crtI also produced phytoene and cells overexpressing crtYB, crtI, and BTS1 additionally produced phytoene and dihydro-β-carotene (a cyclization product of neurosporene). Phytoene, neurosporene, and lycopene were also accumulated in cells overexpressing crtYB, crtI, and crtE (Table 3).
TABLE 3.
Carotenoid compositions of S. cerevisiae strain transformed with the episomal vector YEplac195 containing different carotenogenic genes
| Carotenoid | Specific amt (μg/g [dw]) (% distribution) of carotenoidsa
|
||
|---|---|---|---|
| YB/I | YB/I/BTS1 | YB/I/E | |
| Phytoene | 1 (33) | 61 (73) | 310 (64) |
| Neurosporene | —b | — | 13 (3) |
| Lycopene | — | — | 79 (16) |
| β-Carotene | 2 (67) | 13 (16) | 85 (17) |
| 7,8-Dihydro-β-carotene | — | 9 (11) | — |
| Total | 3 | 83 (28) | 487 (162) |
The data are averages of two independent cultures. The n-fold increases in total carotenoid production levels compared to that of strain YB/I are in parentheses in the bottom row (the value for strain YB/I is set at 1).
—, not detected.
It was observed that about 10% of the cells transformed with episomal vectors lost their color after growth for 3 days in YNB medium and subsequent growth for 3 days on agar plates (data not shown). This result suggested instability of the strain or instability of the expression vector. Episomal expression vectors tend to be structurally unstable, especially when they contain large inserts (42). This was indeed confirmed by extraction of the expression vectors containing carotenogenic genes from white yeast cells and subsequent restriction analysis to determine the presence of the introduced genes on the vector (data not shown). To avoid the problem of instability, integrative vectors were used to generate a new series of transformants.
Transformation with integrative vectors.
To create genetically stable carotenoid-producing S. cerevisiae cells, integrative vectors were constructed as indicated in Fig. 2. Because integration into the ura3-52 locus required linearization of the vectors with StuI, which is present within the coding sequence of crtE, the StuI restriction site within the crtE gene was changed by site-directed mutagenesis. This mutation did not influence carotenoid accumulation (data not shown). Constructs containing different combinations of carotenogenic genes were integrated into genomic DNA. Integration of crtYB and crtI resulted in faintly yellow colonies, additional integration of crtE resulted in orange cells and integration of crtYB, crtI, and BTS1 resulted in yellow cells. Transformants were grown overnight in YNB medium and subsequently streaked onto nonselective agar plates. Less than 0.5% of the cells lost their color after 3 days of incubation, indicating that the stability of carotenoid-producing S. cerevisiae cells is greatly increased by genomic integration of carotenogenic genes. Because YB/I cells produced very low levels of carotenoids, this strain was excluded from further studies. To determine the exact composition of the accumulated carotenoids, cells were grown for 72 h in liquid cultures and HPLC studies were performed (Table 4). The growth properties of YB/I/E and YB/I/BTS1 cells were similar compared to those of wild-type cells (data not shown). Carotenoid production levels were higher in cells containing integrated carotenogenic genes compared to expression from episomal vectors (Tables 3 and 4). Copy numbers of episomal (YEplac) vectors are, in general, higher compared to those of integrative (YIplac) vectors (25), suggesting that higher protein levels and hence higher carotenoid production levels should be obtained. Results similar to ours were obtained with E. coli, where higher β-carotene production was obtained with a low-copy-number vector compared to a high-copy-number vector (13). The use of high-copy-number plasmids increases the demand for nucleotides during plasmid replication and might result in metabolic burden issues (8), resulting in decreased carotenoid production levels. Furthermore, it was observed that total carotenoid levels were higher in YB/I/E cells compared to YB/I/BTS1 cells (Tables 3 and 4). This might be caused by differences in substrate specificity for CrtE and Bts1. It has been reported that CrtE from E. uredovora uses both FPP and GPP as substrates (40), which might be similar for CrtE from X. dendrorhous. Only FPP can serve as a substrate for Bts1 (11). The ability to use both FPP and GPP as precursors might lead to higher carotenoid production levels. In both strains, about 90% of the produced carotenoids consisted of phytoene, whereas small amounts of neurosporene, β-zeacarotene, and 7,8-dihydro-β-carotene were also present (Table 4). It has been shown that overexpression of lycopene cyclase (crtY) from E. uredovora or Capsicum annuum in carotenoid-producing E. coli cells resulted in production of the bicyclic carotenoid 7,8-dihydro-β-carotene via monocyclic β-zeacarotene and neurosporene (31). Formation of these carotenoids in our carotenoid-producing S. cerevisiae strains is probably the result of the high cyclase activity of CrtYB (Fig. 1).
TABLE 4.
Carotenoid compositions of S. cerevisiae strains containing integrated carotenogenic gene overexpression cassettes and an additional crtI overexpression cassette, an additional tHMG1 overexpression cassette, or both
| Carotenoid | Specific amt (μg/g [dw]) (% distribution) of carotenoidsa
|
||||
|---|---|---|---|---|---|
| YB/I/BTS1 | YB/I/E | YB/I/E+tHMG1 | YB/I/E+I | YB/I/E+tHMG1+I | |
| Phytoene | 476 (94) | 1,323 (86) | 10,302 (92) | 682 (29) | 5,380 (48) |
| Neurosporene | 3 (0.5) | 5 (0.5) | 48 (0.5) | 75 (3) | —b |
| β-Carotene | 15 (3) | 141 (9) | 501 (4.5) | 1,627 (68) | 5,918 (52) |
| β-Zeacarotene | 4 (0.5) | 16 (1) | 109 (1) | — | — |
| 7,8-Dihydro-β-carotene | 10 (2) | 60 (3.5) | 262 (2) | — | — |
| Total | 508 (33) | 1,545 (100) | 11,222 (726) | 2,384 (154) | 11,298 (731) |
The data are averages of two independent cultures, except for YB/I/E+tHMG1+I, which are triplicates. The relative total carotenoid production levels compared to that of the YB/I/E strain are in parentheses in the bottom row (the value for strain YB/I/E is set at 100%).
—, not detected.
Accumulation of intermediates indicated that the flux through the carotenogenic pathway was not fully efficient. The high levels of phytoene and other intermediates with a degree of desaturation lower than that of β-carotene in YB/I/BTS1 and YB/I/E cells suggest that the phytoene desaturation reaction might be the bottleneck in heterologous β-carotene production by S. cerevisiae. This could be caused by poor transcription or translation efficiency of crtI or poor activity of the phytoene desaturase protein in S. cerevisiae. Accumulation of intermediates was not observed in the production of carotenoids in E. coli with the carotenogenic genes from E. uredovora (16). However, in S. cerevisiae cells overexpressing carotenogenic genes from E. uredovora on episomal vectors, intermediates also accumulated; 78% of the total carotenoids consisted of β-carotene, 11% accumulated as phytoene, and 11% accumulated as lycopene (41). Apparently, phytoene desaturation becomes a rate-limiting step in heterologous β-carotene production by S. cerevisiae when using carotenogenic genes from X. dendrorhous or E. uredovora. Accumulation of phytoene was also observed in a lycopene-producing C. utilis strain transformed with carotenogenic genes from E. uredovora (18). It was suggested that yeast membrane environments in which the conversion of phytoene into lycopene is likely to occur are not suitable for an efficient desaturation reaction. Suitable electron carriers, required for the dehydrogenation reaction, might be absent. It was presumed that active proteins would be present because of the presence of carotenogenic gene transcripts (18).
To determine whether the integrated carotenogenic genes in our strains were correctly expressed, qPCR studies were performed (Fig. 3). In wild-type cells, no signal was obtained with primers to detect the expression of crtYB, crtI, and crtE, whereas the GGPP synthase BTS1 was expressed at a low level. In YB/I/E cells, quite high expression of crtYB, crtI, and crtE and low expression of BTS1 were detected. In YB/I/BTS1 cells, no expression of crtE was detected, whereas crtYB, crtI, and BTS1 were expressed at high levels compared to the housekeeping gene ACT1, which encodes actin. In both YB/I/E and YB/I/BTS1 cells, the relative expression levels of crtI were the highest of the carotenogenic genes introduced. The results from the qPCR studies indicated that the high phytoene levels were presumably not caused by poor crtI transcription efficiency.
FIG. 3.
qPCR studies to determine overexpression of carotenogenic genes in carotenoid-producing yeast strains. Three cultures of each strain, the wild type, YB/I/E, and YB/I/BTS1, were inoculated in YNB with 2% glucose at the same optical density. Six hours after inoculation, samples were taken and further processed for cDNA synthesis. qPCR experiments were performed to determine the relative abundances of crtYB, crtI, crtE, and BTS1 in each strain with respect to that of ACT1, which encodes actin and served as an internal control. The data represent the average and standard deviation of three independently grown cultures.
Additional overexpression of crtI in strain YB/I/E.
Although high accumulation of phytoene was not caused by low crtI transcription levels, we tested whether additional overexpression of the crtI gene and integration at another locus in YB/I/E cells would have an effect on phytoene and β-carotene levels. The TDH3p-crtI-CYC1t cassette was integrated into the leu2,3-112 locus in YB/I/E cells. An increased copy number might result in higher levels of the phytoene desaturase protein and could lead to more efficient conversion of phytoene into downstream products of the carotenogenic pathway. The transformants displayed a more orange color compared to that of YB/I/E cells (Fig. 4a and b). Subsequent HPLC studies revealed that overexpression of crtI in strain YB/I/E resulted in a 1.5-fold increase in the total carotenoid levels compared to those of strain YB/I/E (Table 4). The major difference was a decrease in phytoene accumulation (86% to 29% of the total carotenoid levels) and an increase in β-carotene accumulation (9% to 68% of the total carotenoid levels), yielding β-carotene levels of around 1.5 mg/g (dw). Apparently, additional overexpression of crtI in a strain overexpressing crtYB, crtI, and crtE resulted in increased desaturation of phytoene and greatly improved the flux toward β-carotene. Possibly, the amount of CrtI protein should reach a certain level in order to efficiently convert phytoene into lycopene in the membrane environment of S. cerevisiae. This level might be reached after additional introduction and overexpression of the crtI gene.
FIG. 4.
Colors of different carotenoid-producing S. cerevisiae mutants. All strains were initially transformed with YIplac211 crtYB/crtI/crtE* to obtain carotenoid-producing cells and subsequently with the integrated vectors containing the indicated genes. (a) Strain YB/I/E, CEN.PK 113-5D; (b) strain YB/I/E+extra I, CEN.PK 113-6B transformed with YIplac204 crtI; (c) strain YB/I/E+tHMG1, CEN.PK 113-6B transformed with YIplac128 tHMG1; (d) strain YB/I/E+extra I+tHMG1, CEN.PK 113-6B transformed with YIplac128 crtI and YIplac204 tHMG1. Individual transformants were grown for 2 days on YNB-2% glucose medium supplemented with amino acids where required and streaked onto yeast extract-peptone-2% glucose plates. After 3 days of incubation at 30°C, the plates were photographed.
Overexpression of the catalytic domain of HMG1 in strain YB/I/E.
It has been shown that the flux through the ergosterol biosynthetic pathway, which is related to the carotenoid pathway by prenyl diphosphate utilization, can be increased by overexpression of the catalytic domain of HMG-CoA reductase (tHMG1) in S. cerevisiae (5, 24). Furthermore, overexpression of tHMG1 from C. utilis in C. utilis cells heterologously producing lycopene resulted in increased lycopene production, probably by increasing the supply of precursors (29). Therefore, it was determined whether overexpression of tHMG1 could increase carotenoid production in carotenoid-producing S. cerevisiae cells. For this purpose, the TDH3p-tHMG1-CYC1t cassette was integrated into the trp1-289 locus in YB/I/E cells. Transformation of tHMG1 resulted in a clear color difference; YB/I/E cells were orange, and YB/I/E+tHMG1 cells were yellow (Fig. 4a and c). Overexpression of the catalytic domain of Hmg1 results in a sevenfold increase in total carotenoid levels compared to those of YB/I/E cells (Table 4). This increase in total carotenoid accumulation is largely caused by a massive increase in phytoene levels, up to 10 mg/g (dw), which suggested that desaturation of phytoene is limiting without additional crtI overexpression.
Overexpression of tHMG1 and additional overexpression of crtI in strain YB/I/E.
The results obtained so far suggest that combined overexpression of crtI and tHMG1 in carotenoid-producing S. cerevisiae cells results in a strain that efficiently produces high β-carotene levels. Additional overexpression of crtI in strain YB/I/E+tHMG1 indeed increased the flux through the carotenogenic pathway and improved β-carotene production levels (Table 4). Phytoene levels decreased from 10.3 mg/g (dw) in strain YB/I/E+tHMG1 to 5.4 mg/g (dw) in strain YB/I/E+tHMG1+I, whereas β-carotene levels increased from 0.5 mg/g (dw) to 5.9 mg/g (dw). Total carotenoid accumulation levels were similar in strain YB/I/E+tHMG1 and strain YB/I/E+tHMG1+I (11 mg/g [dw]). The transformants were more orange than YB/I/E+tHMG1 cells (Fig. 4c and d). The absence of intermediates with a lower degree of desaturation than β-carotene (neurosporene, β-zeacarotene, and 7,8-dihydro-β-carotene) in strain YB/I/E+tHMG1+I indicated that the flux through the carotenogenic pathway was more efficient compared to that in strain YB/I/E+tHMG1. Although strains producing high levels of β-carotene were created by overexpression of tHMG1 and additional overexpression of crtI in strain YB/I/E, still relatively high phytoene levels accumulated, suggesting that a limitation at the level of phytoene desaturation still exists. Possibly, fine-tuning of tHMG1 expression levels to more efficiently control phytoene production might further increase β-carotene production levels.
Concluding remarks.
In summary, we have been able to construct S. cerevisiae strains that produce various amounts of carotenoids by integration and overexpression of carotenogenic genes from X. dendrorhous. We succeeded in the construction of a strain producing 5.9 mg β-carotene/g (dw), which is 57-fold more than previously reported for heterologous β-carotene production in S. cerevisiae (41). This was achieved by overexpression of the catalytic domain of HMG1 from S. cerevisiae and additional overexpression of the crtI gene from X. dendrorhous in carotenoid-producing S. cerevisiae cells transformed with carotenogenic genes from X. dendrorhous. Optimizing the culturing conditions, for instance, by growing the strains in large volumes under controlled fermentor conditions, might further increase β-carotene yields. This approach was successful for engineered β-carotene-producing E. coli strains (13). Possibly, β-carotene production levels can be further increased by chemical mutagenesis. This strategy has resulted in X. dendrorhous strains with increased astaxanthin production levels (1). The strains producing high β-carotene levels are promising for further development toward the biotechnological production of β-carotene by S. cerevisiae.
Acknowledgments
This work was funded by the Kluyver Center for Genomics of Industrial Fermentation, which is supported by The Netherlands Genomics Initiative.
Footnotes
Published ahead of print on 11 May 2007.
REFERENCES
- 1.An, G. H., D. B. Schuman, and E. A. Johnson. 1989. Isolation of Phaffia rhodozyma mutants with increased astaxanthin content. Appl. Environ. Microbiol. 55:116-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bui, K., and P. Galzy. 1990. Food yeast. Springer, Berlin, Germany.
- 3.Delorme, E. 1989. Transformation of Saccharomyces cerevisiae by electroporation. Appl. Environ. Microbiol. 55:2242-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dimster-Denk, D., M. K. Thorsness, and J. Rine. 1994. Feedback regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase in Saccharomyces cerevisiae. Mol. Biol. Cell 5:655-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donald, K. A., R. Y. Hampton, and I. B. Fritz. 1997. Effects of overproduction of the catalytic domain of 3-hydroxy-3-methylglutaryl coenzyme A reductase on squalene synthesis in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 63:3341-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 7.Girard, P., B. Falconnier, J. Bricout, and B. Vladescu. 1994. Beta-carotene producing mutants of Phaffia rhodozyma. Appl. Microbiol. Biotechnol. 41:183-191. [Google Scholar]
- 8.Glick, B. R. 1995. Metabolic load and heterologous gene expression. Biotechnol. Adv. 13:247-261. [DOI] [PubMed] [Google Scholar]
- 9.Golubev, W. I. 1995. Perfect state of Rhodomyces dendrorhous (Phaffia rhodozyma). Yeast 11:101-110. [DOI] [PubMed] [Google Scholar]
- 10.Gordon, H. T., and J. C. Bauernfeind. 1982. Carotenoids as food colorants. Crit. Rev. Food Sci. Nutr. 18:59-97. [DOI] [PubMed] [Google Scholar]
- 11.Jiang, Y., P. Proteau, D. Poulter, and S. Ferro-Novick. 1995. BTS1 encodes a geranylgeranyl diphosphate synthase in Saccharomyces cerevisiae. J. Biol. Chem. 270:21793-21799. [DOI] [PubMed] [Google Scholar]
- 12.Johnson, E. A., and W. A. Schroeder. 1996. Microbial carotenoids. Adv. Biochem. Eng. Biotechnol. 53:119-178. [DOI] [PubMed] [Google Scholar]
- 13.Kim, S. W., J. B. Kim, W. H. Jung, J. H. Kim, and J. K. Jung. 2006. Over-production of beta-carotene from metabolically engineered Escherichia coli. Biotechnol. Lett. 28:897-904. [DOI] [PubMed] [Google Scholar]
- 14.Machlin, L. J. 1995. Critical assessment of the epidemiological data concerning the impact of antioxidant nutrients on cancer and cardiovascular disease. Crit. Rev. Food Sci. Nutr. 35:41-50. [DOI] [PubMed] [Google Scholar]
- 15.Mijts, B. N., C. Schmidt-Dannert, B. E. Jackson, E. A. Hart-Wells, and S. P. Matsuda. 2003. Engineering of secondary metabolite pathways. Curr. Opin. Biotechnol. 14:597-602. [DOI] [PubMed] [Google Scholar]
- 16.Misawa, N., M. Nakagawa, K. Kobayashi, S. Yamano, Y. Izawa, K. Nakamura, and K. Harashima. 1990. Elucidation of the Erwinia uredovora carotenoid biosynthetic pathway by functional analysis of gene products expressed in Escherichia coli. J. Bacteriol. 172:6704-6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Misawa, N., and H. Shimada. 1997. Metabolic engineering for the production of carotenoids in non-carotenogenic bacteria and yeasts. J. Biotechnol. 59:169-181. [DOI] [PubMed] [Google Scholar]
- 18.Miura, Y., K. Kondo, T. Saito, H. Shimada, P. D. Fraser, and N. Misawa. 1998. Production of the carotenoids lycopene, beta-carotene, and astaxanthin in the food yeast Candida utilis. Appl. Environ. Microbiol. 64:1226-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mumberg, D., R. Muller, M. Funk, R. Niedenthal, K. Brinkmann, V. Ronicke, T. Henkel, A. M. Myers, A. Tzagoloff, D. M. Kinney, and C. J. Lusty. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156:119-122. [DOI] [PubMed] [Google Scholar]
- 20.Nelis, H. J., and A. P. De Leenheer. 1991. Microbial sources of carotenoid pigments used in foods and feeds. J. Appl. Bacteriol. 70:181-191. [Google Scholar]
- 21.Ostergaard, S., L. Olsson, and J. Nielsen. 2000. Metabolic engineering of Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 64:34-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palozza, P., and N. I. Krinsky. 1992. Antioxidant effects of carotenoids in vivo and in vitro: an overview. Methods Enzymol. 213:403-420. [DOI] [PubMed] [Google Scholar]
- 23.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Polakowski, T., U. Stahl, and C. Lang. 1998. Overexpression of a cytosolic hydroxymethylglutaryl-CoA reductase leads to squalene accumulation in yeast. Appl. Microbiol. Biotechnol. 49:66-71. [DOI] [PubMed] [Google Scholar]
- 25.Romanos, M. A., C. A. Scorer, and J. J. Clare. 1992. Foreign gene expression in yeast: a review. Yeast 8:423-488. [DOI] [PubMed] [Google Scholar]
- 26.Rozen, S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365-386. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 28.Sandmann, G. 2002. Combinatorial biosynthesis of carotenoids in a heterologous host: a powerful approach for the biosynthesis of novel structures. ChemBioChem 3:629-635. [DOI] [PubMed] [Google Scholar]
- 29.Shimada, H., K. Kondo, P. D. Fraser, Y. Miura, T. Saito, and N. Misawa. 1998. Increased carotenoid production by the food yeast Candida utilis through metabolic engineering of the isoprenoid pathway. Appl. Environ. Microbiol. 64:2676-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stickforth, P., S. Steiger, W. R. Hess, and G. Sandmann. 2003. A novel type of lycopene epsilon-cyclase in the marine cyanobacterium Prochlorococcus marinus MED4. Arch. Microbiol. 179:409-415. [DOI] [PubMed] [Google Scholar]
- 31.Takaichi, S., G. Sandmann, G. Schnurr, Y. Satomi, A. Suzuki, and N. Misawa. 1996. The carotenoid 7,8-dihydro-psi end group can be cyclized by the lycopene cyclases from the bacterium Erwinia uredovora and the higher plant Capsicum annuum. Eur. J. Biochem. 241:291-296. [DOI] [PubMed] [Google Scholar]
- 32.van Poppel, G., and R. A. Goldbohm. 1995. Epidemiologic evidence for beta-carotene and cancer prevention. Am. J. Clin. Nutr. 62:1393S-1402S. [DOI] [PubMed] [Google Scholar]
- 33.Verdoes, J. C., K. P. Krubasik, G. Sandmann, A. J. J. van Ooyen, C. Echavarri-Erasun, and E. A. Johnson. 1999. Isolation and functional characterisation of a novel type of carotenoid biosynthetic gene from Xanthophyllomyces dendrorhous. Mol. Gen. Genet. 262:453-461. [DOI] [PubMed] [Google Scholar]
- 34.Verdoes, J. C., N. Misawa, and A. J. J. van Ooyen. 1999. Cloning and characterization of the astaxanthin biosynthetic gene encoding phytoene desaturase of Xanthophyllomyces dendrorhous. Biotechnol. Bioeng. 63:750-755. [DOI] [PubMed] [Google Scholar]
- 35.Verdoes, J. C., G. Sandmann, H. Visser, M. Diaz, M. van Mossel, and A. J. J. van Ooyen. 2003. Metabolic engineering of the carotenoid biosynthetic pathway in the yeast Xanthophyllomyces dendrorhous (Phaffia rhodozyma). Appl. Environ. Microbiol. 69:3728-3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verdoes, J. C., J. Wery, T. Boekhout, and A. J. J. Van Ooyen. 1997. Molecular characterization of the glyceraldehyde-3-phosphate dehydrogenase gene of Phaffia rhodozyma. Yeast 13:1231-1242. [DOI] [PubMed] [Google Scholar]
- 37.Verwaal, R., M. Arako, R. Kapur, A. J. Verkleij, C. T. Verrips, and J. Boonstra. 2004. HXT5 expression is under control of STRE and HAP elements in the HXT5 promoter. Yeast 21:747-757. [DOI] [PubMed] [Google Scholar]
- 38.Verwaal, R., J. W. Paalman, A. Hogenkamp, A. J. Verkleij, C. T. Verrips, and J. Boonstra. 2002. HXT5 expression is determined by growth rates in Saccharomyces cerevisiae. Yeast 19:1029-1038. [DOI] [PubMed] [Google Scholar]
- 39.Visser, H., A. J. J. van Ooyen, and J. C. Verdoes. 2003. Metabolic engineering of the astaxanthin-biosynthetic pathway of Xanthophyllomyces dendrorhous. FEMS Yeast Res. 4:221-231. [DOI] [PubMed] [Google Scholar]
- 40.Wiedemann, M., N. Misawa, and G. Sandmann. 1993. Purification and enzymatic characterization of the geranylgeranyl pyrophosphate synthase from Erwinia uredovora after expression in Escherichia coli. Arch. Biochem. Biophys. 306:152-157. [DOI] [PubMed] [Google Scholar]
- 41.Yamano, S., T. Ishii, M. Nakagawa, H. Ikenaga, and N. Misawa. 1994. Metabolic engineering for production of beta-carotene and lycopene in Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 58:1112-1114. [DOI] [PubMed] [Google Scholar]
- 42.Zhang, Z., M. Moo-Young, and Y. Chisti. 1996. Plasmid stability in recombinant Saccharomyces cerevisiae. Biotechnol. Adv. 14:401-435. [DOI] [PubMed] [Google Scholar]