Abstract
The auto-induction method of protein expression in E. coli is based on diauxic growth resulting from dynamic function of lac operon regulatory elements (lacO and LacI) in mixtures of glucose, glycerol and lactose. The results show that successful execution of auto-induction is strongly dependent on the plasmid promoter and repressor construction, on the oxygenation state of the culture, and on the composition of the auto-induction medium. Thus expression hosts expressing high levels of LacI during aerobic growth exhibit reduced ability to effectively complete the auto-induction process. Manipulation of the promoter to decrease the expression of LacI altered the preference for lactose consumption in a manner that led to increased protein expression and partially relieved the sensitivity of the auto-induction process to the oxygenation state of the culture. Factorial design methods were used to optimize the chemically defined growth medium used for expression of two model proteins, Photinus luciferase and enhanced green fluorescent protein, including variations for production of both unlabeled and selenomethionine-labeled samples. The optimization included studies of the expression from T7 and T7-lacI promoter plasmids and from T5 phage promoter plasmids expressing two levels of LacI. Upon the basis of the analysis of over 500 independent expression results, combinations of optimized expression media and expression plasmids that gave protein yields of greater than 1000 μg/mL of expression culture were identified.
Introduction
Reliable and reproducible methods for high throughput production of proteins are required for structural genomics (1–3), functional proteomics (4), drug discovery (5, 6) and other current protein biochemistry and enzymology initiatives.
As one approach to this problem, the auto-induction method has been used for production of recombinant proteins in E. coli (7–9). Auto-induction is based on the function of lac operon regulatory elements in mixtures of glucose, glycerol and lactose under diauxic growth conditions (10). Control of this metabolic circuit in wild type E. coli has been the subject of much research and is generally well understood (10–14). During the initial growth period, glucose is preferentially used as a carbon source and protein expression is low due to catabolite repression of alternative carbon utilization pathways (15–17) and binding interactions between lac repressors (LacI) and lac operators (lacO). As glucose is depleted, catabolite repression is relieved, which leads to a shift in cellular metabolism toward the import and consumption of lactose and glycerol. Lactose import results in the production of allolactose from lactose by a promiscuous reaction of β-galactosidase. Allolactose then acts as the physiological inducer of the lac operon.
An apparent advantage of the auto-induction method is that it places the transition from the un-induced to induced state under metabolic control of the expression host (7, 9). This minimizes the required handling of cultures from inoculation until cell harvest, which is a major advantage for high-throughput efforts. However, several factors complicate the use of auto-induction. Since multiple carbon sources are present in the auto-induction medium, their relative amounts and their patterns of usage are critically important contributors to the outcome of the auto-induction expression. Furthermore, for optimal utility, the auto-induction method should be easy to perform in both small-scale screening and large-scale production modes and should also provide correlation between the results obtained at the different scales of operation. This scaling requirement introduces variability arising from physical parameters such as the extent of aeration associated with different types of vessels used for cell culture. Indeed, it has been previously noted that the availability of O2 can affect the outcome of auto-induction experiments (7), but the origin of this effect was not clear.
For recombinant expression systems that operate under control of the lac operon, the appearance of allolactose during auto-induction also initiates the expression of heterologous proteins (7, 18). However, the construction of recombinant expression systems makes the circumstances of induction more complicated than in wild-type E. coli. For example, E. coli cells harboring a multi-copy expression plasmid may produce LacI at levels 200-fold higher than that present in wild-type cells. Currently, there is only limited experimental information on the diauxic behavior of cells expressing high concentrations of LacI (19).
Given the importance of bacterial protein expression studies (20–22), it is important to more fully understand the underlying metabolic and physical constraints to reproducibility and productivity of auto-induction approaches. This work addresses some fundamental questions of E. coli growth and heterologous expression in auto-induction medium. The results show that the level of LacI, the oxygenation state of the culture, and the medium composition have profound effects on the carbon consumption patterns required for successful execution of the auto-induction process. Arising from these findings, a factorial design approach was undertaken to optimize protein expression for three expression plasmid variants and two model proteins. In the discussed cases, the combination of fundamental and empirical studies led to an improvement in the volumetric productivity of protein expression from less than 100 μg/mL to more than 1000 μg/mL.
Materials and Methods
Chemicals
Unless otherwise stated, bacterial growth reagents, antibiotics, routine laboratory chemicals, and disposable labware were from Sigma-Aldrich (St. Louis, MO), Fisher (Pittsburgh PA), or other major distributors. L-SeMet was from Acros (Morris Plains, NJ). Preparations of standard laboratory reagents were as previously described (23). The 2-L polyethyleneterepthalate beverage bottles used for bacterial cell growth were from Ball Corporation (Chicago, IL).
Expression Strains
The methionine auxotroph Escherichia coli B834 [genotype F− ompT hsdSB(rB−mB−) gal dcm met, (24, 25)] was used for expression studies with T5 promoter plasmids, while E. coli B834(DE3) [genotype F− ompT hsdSB(rB− mB−) gal dcm met [DE3] was used for studies with T7 promoter plasmids (EMD Biosciences/Novagen, Madison, WI). Both expression hosts were transformed with pRARE2 (EMD Biosciences/Novagen) for rare codon adaptation. The pRARE2 plasmid conferred chloramphenicol resistance.
Expression Vectors
Table 1 summarizes relevant properties of the expression vectors evaluated in this work. pFN6K (Promega, Madison WI) and pET32 (EMD Biosciences/Novagen) are commercially available. The vectors pVP38K, pVP58K, pVP61K and pVP62K were created from pQE80 (Qiagen, Valencia CA) by removal of a non-functional chloramphenicol acetyltransferase coding region and by replacement of the beta-lactamase coding region with an aminoglycoside 3′-phosphotransferase coding region conferring kanamycin resistance. pVP38K and pVP61K contain the strong lacIq promoter from pQE80, 5′-GTGCAAAACCTTTCGCGGTATGGCATGAT-3′ [the point mutation responsible for the lacIq genotype is underlined (26)], while the wild-type lacI promoter was restored by PCR in pVP58K and pVP62K, 5′-GCGCAAAACCTTTCGCGGTATGGCATGAT-3′. pVP61K and pVP62K also incorporate the gene for tobacco vein mottling virus (TVMV) protease with low-level constitutive expression so that co-transformation with a separate plasmid encoding the protease is not needed to achieve in vivo proteolysis (27, 28).
Table 1.
Expression Vectors
| Expression Vectora | Target Gene Promoterb | Target Genec | Promoter for lacId | Relative LacI expressione | Fusion Tagf | Abbreviation in textg |
|---|---|---|---|---|---|---|
| pFN6K | T7 | Photinus luciferase | None | 1 | N-terminal-(HQ)3 | T7-Luc |
| pET32 | T7-lacO | Photinus luciferase | lacI | 20 | N-terminal-(HQ)3 | pET32-Luc or T7-lacI-Luc |
| pVP58K | T5-lacO1-lacO2 | Photinus luciferase | lacI | 20 | N-terminal-(HQ)3 | T5-lacI-Luc |
| pVP38K | T5-lacO1-lacO2 | Photinus luciferase | lacIq | 200 | N-terminal-(HQ)3 | T5-lacIq-Luc |
| pVP61K | T5-lacO1-lacO2 | Enhanced GFP | lacIq | 200 | MBP-TVMV-His8 -TEVh | T5-lacIq-eGFP |
| pVP62K | T5-lacO1-lacO2 | Enhanced GFP | lacI | 20 | MBP-TVMV-His8 -TEVh | T5-lacI-eGFP |
pFN6K is from Promega (Madison, WI). pET32 is from Novagen (Madison, WI). Other vectors were created as part of this work as described in Materials and Methods.
The promoter and operator construction used for expression of the target gene. In pET32, a single copy of lacO is located 3′ to the T7 promoter. In the T5 vectors, lacO1 is placed between the −35 and −10 regions of the promoter and lacO2 is located between the −10 region and the start codon of the expressed gene. lacO1 is truncated from the full length lacO2.
Target gene in the expression plasmid.
Promoter used for expression of LacI from the expression plasmid.
Relative level of LacI expression as compared to E. coli BL21 containing pFN6K, which includes contributions from copy number of the plasmid and relative strength of the lacI or lacIq promoters.
N-terminal fusion tag on the expressed target protein.
Abbreviation for the expression plasmid used in the text.
The fusion protein is cleaved in vivo by TVMV protease to release SerHis8GluAsnLeuTyrPheGln-AlaIleAle-eGFP.
Protein Targets
Structural genomics target At3g17820 is a putative glutamine synthase from Arabidopsis thaliana strain Columbia, target At1g65020 is an unknown Arabidopsis protein, and BC058837 is an unknown human protein.
pFN6K expresses Photinus luciferase as an N-terminal fusion to (HisGln)3 under control of the T7 promoter. Photinus luciferase was also expressed in the T7-lacI plasmid (pET32) and T5 promoter plasmids conferring both high (T5-lacIq, pVP38K) and medium (T5-lacI, pVP58K) levels of LacI. The luciferase gene was amplified by PCR from pFN6K and the appropriate restriction sites were incorporated into the 5′ and 3′ primers. Primers were from IDT (Coralville, IA). The NdeI and HindIII restriction sites were used for cloning into pET32; the NcoI and HindIII restriction sites were used for cloning into pVP38K and pVP58K. The luciferase expressed from each expression vector investigated had an identical primary sequence including an N-terminal (HisGln)3 tag.
The enhanced green fluorescent protein (eGFP) gene was assembled by standard molecular biology protocols (23) to incorporate the F64L, S65T, R81Q, F99S, M153T, and V163A mutations (29–31). The eGFP gene was subsequently amplified to add the SgfI and PmeI restriction sites required for Flexi-vector cloning (32) and transferred into pVP61K and pVP62K. eGFP was initially expressed from these vectors with an N-terminal maltose binding protein fusion that underwent in vivo proteolysis by tobacco vein mottling virus (TVMV) protease to liberate SerHis8AlaSerGluAsnLeuTyrPheGln-AlaIleAla-eGFP.
Media Formulations
The non-inducing and auto-induction media are derived from earlier reports on the development and use of auto-induction (7–9). All media contained 34 μg/mL of chloramphenicol and either 100 μg/mL of ampicillin or 50 μg/mL of kanamycin, depending on the selectable marker of the expression plasmid.
A 50× amino acids solution (1 L) was prepared from 10 g each of sodium glutamate, lysine-HCl, arginine-HCl, histidine-HCl, free aspartic acid, and zwitterionic forms of alanine, proline, glycine, threonine, serine, glutamine, asparagine, valine, leucine, isoleucine, phenylalanine and tryptophan.
A 5000× trace metals solution (100 mL) was prepared from 50 mL of 0.1 M FeCl3•6H2O dissolved in ~0.1 M HCl, 2 mL of 1 M CaCl2, 1 mL of 1 M MnCl2•4H2O, 1 mL of 1 M ZnSO4•7H2O, 1 mL of 0.2 M CoCl2•6H2O, 2 mL of 0.1 M CuCl2•2H2O, 1 mL of 0.2 M NiCl2•6H2O, 2 mL of 0.1 M Na2MoO4•5H2O, 2 mL of 0.1 M Na2SeO3•5H2O and 2 mL of 0.1 M H3BO3 and 36 mL of deionized water.
A 1000× vitamins solution (100 mL) for the non-inducing medium was prepared from 2 mL of 10 mM nicotinic acid, 2 mL of 10 mM pyridoxine-HCl, 2 mL of 10 mM thiamine-HCL, 2 mL of 10 mM p-aminobenzoic acid, 2 ml of 10 mM pantothenate, 5 mL of 100 μM folic acid, 5 mL of 100 μM riboflavin, 4 mL of 5 mM vitamin B12 solution and 76 mL of sterile water. A 1000× vitamins solution (100 mL) for the auto-induction medium was the same as above except that the volume of the vitamin B12 solution was replaced with sterile water.
A 20× source of nitrogen, sulfate, and phosphorous (1 L), was prepared using 68 g of KH2PO4, 71 g of Na2HPO4, 53.6 g of NH4Cl, and 14.2 g of Na2SO4 dissolved in sterile water.
A non-inducing medium for starting inocula (1 L) was prepared using 50 mL of 20× nitrogen, sulfate, and phosphorous mix, 0.5 g of MgSO4, 20 mL of the 50× amino acids solution, 0.2 mL of the 5000× trace metals solution, 1 mL of the 1000× vitamins solution for the non-inducing medium, appropriate antibiotics, and 0.8% (w/v) glucose with the balance sterile water.
The auto-induction medium contained the ingredients listed above for the non-inducing medium with the noted omission of B12 from the 1000× vitamins solution (9) and changes in the amino acids and carbon sources as described next. For expression of unlabeled proteins, the medium contained 0.2 mg/mL of methionine. For expression of selenomethionine labeled proteins, the medium contained 0.01 mg/mL of methionine and 0.125 mg/mL of selenomethionine. The concentrations of the carbon sources (glucose, glycerol, lactose) in the auto-induction medium were varied as part of a five level, three-parameter factorial design in the following range of carbohydrate concentrations (w/v): glucose, 0 to 0.1%; glycerol 0 to 1.2% and lactose from 0 to 0.6%. Succinate was maintained at 0.375% for all media formulations. The design points were based on two full three level cubic factorials, with one nested within the other (33). This gave a total of 53 independent medium compositions (the inner and outer factorial shared a common center point). In this design, the center points were replicated four times and the face-centered points along the lactose and glycerol axes were duplicated. These conditions were conveniently arranged into an 8 × 8 array within a 96-well growth block. The Supporting Information defines how the sugar concentrations were arranged in the growth block format.
Variations of the media containing either methionine alone or selenomethionine and methionine were tested separately. The composition of the media used for selenomethionine-labeling was tested in a factorial design space comprised of the inner factorial (32 data points per experiment including replicates) except in the case of work with the T7-lacI (pET32) expression vector where the full nested factorial was tested. This combination gave a total of 512 expression experiments.
Protein Expression
Starting inocula were grown to saturation overnight in the non-inducing medium using either 96-well growth blocks having a capacity of 2 mL per well (Qiagen) or in Erlenmeyer flasks. For the growth blocks, 400 μL of the medium was used per well. For the Erlenmeyer flasks, the volume of starting inoculum was less than 10% of the total flask volume in order to promote aerobic growth. All culture growth was done at 25 °C using either plate or platform shakers.
Small-scale expression trials were carried out in 96-well growth blocks. A 20-μL aliquot of the starting inoculum was transferred to 400 μL of the auto-induction medium and incubated for 24 h at 25 °C on a plate shaker. After the incubation period, an aliquot (100–200 μL) of each 400-μL culture was transferred into a 96-well PCR plate. These samples were directly frozen at −80°C without a preliminary cell pelleting centrifugation step. The plates were stored at −80°C until expression analysis. Large-scale expression was conducted in 2-L PET bottles containing 500 mL of culture medium (8, 9, 34). Samples for expression analysis were harvested and stored as for the small-scale expression trials.
Stirred Vessel Fermentations
A Sixfors parallel six fermenter system (Infors AG, Bottmingen, Switzerland) was used to investigate the influence of aeration on the auto-induction process. Two aeration states were developed to mimic the small- and large-scale cell culture environments. For the aerobic case, which best mimics the small-scale culture in the 96-well growth blocks, airflow and agitation rate were manually adjusted to maintain dissolved O2 above 10% of saturation. For the O2-limited condition, which best mimics the large-scale cell culture in shaken 2-L bottles, a fixed 12 volumes of air/h was added with low agitation. Samples were taken periodically to determine cell density, protein expression, and concentration of carbon sources remaining in the growth medium. The temperature was maintained at 25 °C and the pH was passively monitored during these experiments.
Carbon Source Analysis
An HPLC method was developed to measure the concentration of sugars and organic acids present in the expression medium. A 1-mL aliquot of the culture medium was centrifuged at 16,000g for 3 min to pellet the cells. A 900-μL aliquot of the clarified medium was added to 100 μL of a saturated Al2(SO4)3 solution to precipitate phosphate. This mixture was then heated to 90 °C for 5 min to inactivate any residual enzymatic activity. Samples were stable for at least 1 wk at 4 °C after this treatment. Prior to HPLC analysis, the samples were centrifuged briefly to remove aluminum phosphate precipitate. The clarified samples were analyzed using a Shimadzu 10A HPLC system (Shimadzu, Columbia, MD) with RID10A refractive index detector and Coregel 87H3 organic acid analysis column (Transgenomic, San Jose, CA). A 20-μL sample loop was used. An isocratic 0.08 N sulfuric acid mobile phase was used for elution. The elution times of the sugars, organic acids and phosphate were determined using the known compounds as standards.
Protein Expression Analysis
For analysis of protein expression, the PCR plates of frozen cell cultures were thawed and mixed with lysis buffer to obtain a final sample composition of 20 mM Tris-HCl, pH 7.5, 20 mM NaCl, 3 kU/mL of lysozyme (EMD Biosciences/Novagen), 0.7 U/mL of benzonase (EMD Biosciences/Novagen), 0.3 mM triscarboxyethylphosphine, and 1 mM MgSO4. The presence of culture media due to the lack of a centrifugation step prior to cell lysis did not interfere with the biological assays, SDS-PAGE, or capillary electrophoresis analysis. The samples were sonicated for 6–10 min on a plate sonicator (Misonix, Farmington, NY). Samples for total protein expression were prepared for analysis by LabChip90 capillary electrophoresis (Caliper Life Sciences, Hopkinton, MA) as recommended by the manufacturer and were prepared for SDS-PAGE analysis as previously reported(9). The soluble protein fraction used for the biological assays and LabChip90 analysis was obtained by centrifuging the sample plates for 30 min at 2200g. Expressed protein levels were determined by LabChip90 analysis (both eGFP and luciferase) and fluorescence (eGFP only).
Protein and Enzyme Assays
Assays for eGFP and luciferase were performed after dilution of the soluble lysate samples with buffer containing 10 mM Tris-HCl, pH 7.5, 20 mM NaCl, and 0.1 mg/mL of acetylated bovine serum albumin (Promega, Madison, WI). For eGFP, a 5-μL aliquot of the lysate sample was mixed with 75 μL of dilution buffer prior to measurement in the wells of a black Greiner 384 well plate (ISC Bioexpress, Kaysville, UT). Fluorescence measurements were conducted in duplicate using a Tecan Ultra 384 plate reader (Tecan Group LTD, Männedorf, Switzerland) with 485 nm (25 nm bandpass) excitation and 525 nM (20 nm bandpass) emission filters. Luciferase luminescence assays were performed using the Bright Glo luciferase assay system (Promega) after appropriate dilution of samples to bring the luciferase concentration into the linear assay measurement range. A serial dilution of purified recombinant luciferase (Promega) was assayed as a standard on every plate. Measurements were performed in duplicate with 80 μL total volume in black Greiner 384 well plates using the Tecan plate reader in luminescence mode.
Numerical analysis
Carbon source consumption patterns were analyzed using Microsoft Excel and the XLFit3 curve fitting add-in (ver. 3, ID Business Solutions Ltd., Guildford, UK). The changes in sugar and organic acid concentrations with respect to time and cell density were fitted to sigmoidal functions. The apparent carbon source consumption rate was determined by taking the first derivative of the sigmoidal curve fits. Results of factorial design experiments were analyzed with SAS version 9.1 (SAS Institute, Inc., Cary, NC). Where expression data was available for both eGFP and luciferase, the luciferase expression level was empirically found on average to be 1.58-fold higher than the eGFP expression level based on LabChip 90 quantitation of electrophherograms. This factor was used to normalize expression of luciferase and eGFP, and to simulate a response surface for eGFP from luciferase data as shown in Figure 3C. For model fitting purposes, the luciferase and eGFP expression data were merged into a single data set by normalizing the luciferase expression data to the eGFP expression data. This increased the number of observations available for model fitting. Expression levels were fit to either a first order model with two factor interactions (equation 1) or a second order model without factor interactions (equation 2), where sugar concentrations are expressed in % (w/v), EL is the expression level, RFn are the fitted response factors for the different media constituents and C is a fitting constant.
Figure 3.

Response surfaces arising from factorial design changes in the composition of auto-induction medium and changes in LacI dosing. A, expression from T5-lacI plasmids in media containing methionine (left side) or selenomethionine (right side). B, expression from T5-lacIq plasmids in media containing methionine (left side) or selenomethionine (right side). C, estimated expression of eGFP from T7-lacI plasmids in media containing methionine (left side) or selenomethionine (right side) using normalization factor for relationship of luciferase and eGFP expression levels described in Materials and Methods. The response models were not extended to zero lactose concentration due to highly non-linear response with these medium compositions.
| (eq 1) |
| (eq 2) |
Both models contained seven fitted parameters and the model with the higher R2 value was chosen for each data set. Data fits were significantly improved in some cases by excluding data at zero lactose concentration due to highly non-linear expression responses observed at low lactose concentrations. In order to simplify the graphical representation of the response surfaces, the effect of glucose was removed before generation of response surface plots by subtracting the fitted model estimate of the glucose contribution from the response at each data point. Response surface plots were generated using MathCAD version 13.0 (Mathsoft Engineering and Education, Inc.).
Results
Expression in Growth Blocks and 2-L Bottles
Our initial experiments with published auto-induction media (7–9) and T5-lacIq expression plasmids revealed substantial differences between small-scale expression trials run in 96-well blocks and large-scale expression trials run in 2-L bottles. Figure 1A shows three representative examples, which were typically characterized by low total expression in the small scale and more robust expression in the large scale. Surprisingly, higher cell densities were often obtained from the small-scale trials, which suggested more efficient use of the total carbon sources added. This poor correlation limited the predictive utility of the small-scale trials.
Figure 1.

SDS-PAGE demonstration of scale dependence during auto-induction. Total cell lysates are shown for three structural genomics target proteins (from left to right At3g17820, At1g65020, BC058837) expressed as MBP fusions from a T5-lacIq expression vector. A, expression in the original auto-induction medium formulation (7). The level of expression in growth blocks was typically much lower than obtained in 2-L bottles. B, expression of the same targets in a provisionally revised auto-induction medium. With the indicated modifications in carbon sources, the correlation between growth blocks (small-scale) and 2-L bottles (large-scale) was improved. This figure was assembled from pictures of different gels. No modifications were made to the images other than cutting, pasting, and resizing using Adobe Photoshop.
At first, we assumed that the large-scale trials had better aeration (34) than the small-scale and that O2-limitation led to lower protein expression in the smaller cultures. However, by comparing growth rates, pH profiles, and acetate production from the two growth methods, it became apparent that the opposite was true. In one representative experiment, the small-scale cultures reached saturation at OD600 of 22, did not produce acetate, and maintained a stable or increasing pH while cultures grown in 2-L bottles attained an OD600 of 8, produced significant amounts of acetate, and showed a drop in pH from 6.7 to 5.0 after 24 h of incubation. By undertaking a limited investigation of the medium composition, we were able to identify other formulations of glucose, glycerol and lactose that improved the correlation between small- and large-scale expression trials. Figure 1B shows this preliminary result for the three representative examples from Figure 1A. Although potentially useful, this finding did not yet clarify the origin of the differences in expression behavior dependent on culture scale.
Properties of Expression Plasmids Studied
Figure 2 shows maps of the four types of expression plasmids used in this study. Key elements of these plasmids related to the performance of auto-induction are the copy number of the plasmid, the promoter and regulator systems used to control inducible target expression, and the promoter used to control constitutive expression of LacI.
Figure 2.

Map of expression plasmids used in this study. pFN6K has a T7 promoter, pET32 has a T7-lacO promoter, and pVP38, pVP58K, pVP61K and pVP62K have a T5-lacO1-lacO2 promoter. pVP38K and pVP61K have the lacIq promoter controlling expression of LacI, while pVP58K and pVP62K contain the wild-type lacI promoter. Photinus luciferase was expressed from plasmids A, B, and C. Enhanced green fluorescent protein was expressed from pVP61K and pVP62K, shown in D. pVP61K and pVP62K also contain the coding region for tobacco vein mottling virus protease (TVMV) under control of the tet promoter. The expression strains used in this study do not overexpress the tet repressor, leading to low level, constitutive expression of TVMV. Due to the presence of a TVMV recognition site between the MBP and eGFP, the fusion protein is cleaved in vivo to liberate His7-eGFP.
All of the expression plasmids used in this work contain the pBR322 origin of replication and have similar copy numbers of ~15 to 20 per cell (35). Since only ~10 molecules of LacI are present in wild-type E. coli (26), several strategies have been developed to control basal expression from lac operator- repressed expression systems. pFN6K provides a simple T7 promoter for control of expression and no contributions from lacO or recombinant LacI to control basal expression. In contrast, pET32 provides a T7 promoter with an associated lacO sequence and constitutive expression of LacI from the plasmid (designated T7-lacI in the following). In this case, the copy number of the plasmid and the wild-type lacI promoter serve to supplement the level of LacI expression. Both pFN6K and pET32 plasmids require a lysogenic host containing T7 RNA polymerase under inducible control of the lacUV5 promoter such as E. coli B834(DE3) used here.
The pVP vectors used in this work have the T5 phage promoter (36–38) under control of two copies of the lac operator (lacO1 and lacO2 in Figure 2). The lacO1 sequence was truncated during the original construction of the T5 promoter vectors (37) to fit between the −35 and −10 regions of the promoter, so is distinct from lacO2, which retains the natural sequence from E. coli lac operon operator O1 (39). Biochemical data suggests that LacI is able to bind the truncated operator with an affinity similar to the full length operator (40). E. coli RNA polymerase recognizes the T5 promoter so many different E. coli expression strains can be used with this vector. pVP38K and pVP61K contain the strong lacIq promoter for overexpression of LacI [(26), originally present in pQE80], while pVP58K and pVP62K were mutated as part of this work to restore the wild type lacI promoter in order to attenuate expression of LacI.
Factorial Design of Medium Composition
Since the preliminary results of Figure 1 showed that increasing the amounts of glycerol, lactose and succinate and decreasing the amount of glucose could improve the correlation between small- and large-scale expression with the T5-lacIq expression system, we applied a factorial design approach to individually optimize the media for small-scale expression using the T5-lacI, T5-lacIq and T7-lacI plasmids. For this optimization, all media constituents were fixed except for glucose, glycerol and lactose, and these were independently varied in a factorial design approach (22, 33). A 3-dimensional representation of this design and the individual medium compositions are presented in the Supporting Information.
Figure 3 shows response surface models for expression results obtained in media containing methionine only (left side, including evaluation of 53 independent medium compositions) or selenomethionine (right side, including evaluation of 32 independent medium compositions for T5-lacI and T5-lacIq or 53 compositions for pET32). eGFP was used as an expression target for total soluble protein expression due to the ease of quantification through intrinsic fluorescence. Since eGFP requires O2 for fluorophore formation (41), only small-scale expression experiments where O2 was not limited were undertaken.
Figure 3A shows the response surfaces for expression using the T5-lacI-eGFP expression plasmid. The left response surface shows that variations of the carbon sources in a methionine medium can give a nearly 15-fold increase in soluble eGFP production based on the measured fluorescence, which corresponds to a range from ~100 μg/mL of eGFP in the poorest performing composition to ~1500 μg/mL of eGFP in the best performing composition. Figure 4A shows an SDS-PAGE analysis of the total eGFP expression obtained from one of the better performing media compositions (0.025% glucose, 0.9% glycerol, 0.45% lactose). In this case, stoichiometric proteolysis of the original fusion protein (70 kDa) to MBP (42 kDa) and the tagged-eGFP (29 kDa) was obtained from the constitutively expressed TVMV protease. The right side of Figure 3A shows the response surface for the same expression experiment in media containing selenomethionine. Overall, the response surfaces for T5-lacI-eGFP expression in the methionine and selenomethionine media tracked each other closely. Indeed, among the lesser number of compositions investigated for the selenomethionine medium, soluble eGFP expression was observed in excess of 1000 μg/mL (total recombinant protein expression exceeded 2000 μg/mL if MBP expression was also accounted for).
Figure 4.

SDS-PAGE analysis of eGFP expression from T5-lacI-eGFP. Lanes 1, 2 and 3 show total cell lysate, soluble fraction, and insoluble fraction obtained from expression in a methionine auto-induction medium containing 0.025% (w/v) glucose, 0.9% (w/v) glycerol, and 0.45% (w/v) lactose. Lanes 4, 5 and 6 show total cell lysate, soluble fraction and insoluble fraction obtained from expression in selenomethionine auto-induction medium with the same carbon source composition.
Table 2 shows the statistical factors for the model analysis of these two different media optimizations with the T5-lacI-eGFP expression vector. In both the methionine and selenomethionine media, a change in the glycerol concentration was most strongly correlated to a positive expression response, accounting for an estimated 38% or 36% of the modeled effect, respectively. In the methionine medium, increasing lactose concentration was also correlated with the expression response, accounting for 21% of the modeled effect. In the selenomethionine media, increasing lactose had less influence on the expression response, accounting for 13% of the modeled effect, while other higher order terms had a larger influence. Possible implications of these results are considered in the Discussion.
Table 2.
Response Surface Effect Estimates for Auto-Induction of eGFP Expression from the T5-lacI Expression Plasmid pVP62K
| Methionine medium | ||
|---|---|---|
| Model variablea | Scaled effect estimateb | p-valuec |
| Glucose | −0.15 | <0.001 |
| Glycerol | 0.38 | <0.001 |
| Lactose | 0.21 | <0.001 |
| Glucose2 | 0.1 | 0.003 |
| Glycerol2 | −0.14 | <0.001 |
| Lactose2 | −0.03 | 0.54 |
| Model R2 | 0.86 | |
| Selenomethionine medium | ||
| Model variable | Scaled effect estimate | p-value |
| Glucose | 0.13 | 0.17 |
| Glycerol | 0.36 | <0.001 |
| Lactose | 0.11 | 0.24 |
| Glucose2 | −0.18 | 0.049 |
| Glycerol2 | −0.21 | 0.026 |
| Lactose2 | 0.01 | 0.9 |
| Model R2 | 0.73 |
Variables from equation 2 used for response surface modeling based on concentrations of glucose, glycerol and lactose and measured expression results.
The estimated fractional contribution to the observed change in expression, with both positive and negative effects indicated.
p-values indicate the likelihood that the calculated fractional contribution contributes to the observed change; R2 value represents the overall predictive value of the models.
Figure 3B shows the response surfaces for expression from T5-lacIq-eGFP. This expression system gave lower total expression than T5-lacI-eGFP, with expression levels ranging from near zero at low lactose to ~600 μg/mL when glycerol and lactose were maximized. Figure 3C shows an estimated response surfaces for expression of eGFP from T7-lacI generated by normalization of scalar multiplication of the luciferase expression data with the factor 1.58 as described in Materials and Methods.
Basal Expression Studies Using Luciferase
Luciferase was used as an expression target due to the large linear range of the luminescence assay (5–6 orders of magnitude) and a low detection limit that was useful for quantifying basal expression. Table 3 compares the basal expression of luciferase from three of the plasmid types. The unregulated T7-Luc plasmid (pFN6K) gave the highest level of basal expression in non-inducing medium and a small increase in basal expression in auto-induction medium. This result arose through expression of T7 RNA polymerase from the poorly repressed genomic lacUV5 promoter and subsequent transcription from the plasmid T7 promoter upstream of the luciferase gene. In contrast, the highly regulated T5-lacIq-Luc plasmid (pVP38K) gave the lowest level of basal expression, around 1% of that from the T7-Luc plasmid, and no difference in basal expression was observed in either non-inducing or auto-induction media. The presence of two copies of lacO in the promoter region and overexpression of LacI from the plasmid contribute to this result. The T7-lacI plasmid (pET32-Luc) gave a 20-fold reduction in basal luciferase expression as compared to the T7 vector, but this level was still 5× higher than that observed with the T5-lacIq plasmids. Results (not shown) from the T5-lacI-Luc plasmid (pVP58K-Luc) suggested an expression level in the non-inducing medium similar to T7-lacI-Luc. Thus the higher basal expression observed for the T7-lacI and T5-lacI plasmids compared to T5-lacIq is likely a result of a decrease in cellular LacI and corresponding lower occupancy of the promoter lacO sites. Overall, the presence of lactose in the medium did not significantly increase basal expression of luciferase, indicating that the effects of catabolite repression and inducer exclusion (13, 16, 42) are sufficiently strong to prevent premature induction of the lac operon.
Table 3.
Basal Expression of Luciferase from Different Expression Plasmids in Auto-Induction Media
| ExpressionVector | −Lactosea | +Lactoseb |
|---|---|---|
| μg/mL | μg/mL | |
| T7-Luc | 2.7 ± 0.3 | 2.9 ± 0.4 |
| T7-lacI-Luc (pET32-Luc) | 0.19 ± 0.04 | 0.19 ± 0.04 |
| T5-lacIq-Luc | 0.03 ± 0.008 | 0.03 ± 0.004 |
Luciferase activity interpolated at a cell density of 2 (600 nm) based on measurements taken at lower and higher cell densities during exponential growth in a non-inducing medium containing 0.8% (w/v) glucose.
Luciferase activity interpolated at a cell density of 2 (600 nm) based on measurements taken at lower and higher cell densities during exponential growth in auto-inducing medium containing 0.8% (w/v) glucose and 0.1% (w/v) lactose.
When expressed at low levels, luciferase was found to be entirely soluble. However, as expression increased beyond 100 μg/mL of culture, an increasing fraction of the luciferase was insoluble. For this reason, total luciferase expression was determined using capillary electrophoresis (22). Figure 5 shows capillary electrophoresis elution profiles for luciferase expression in various plasmid constructs and auto-induction medium compositions.
Figure 5.

LabChip90 protein electropherograms showning luciferase expression from the indicated luciferase expression plasmids. Reported luciferase expression levels were 1820 μg/mL (T5-lacI, top), 500 μg/mL (T5-lacIq, middle), and 640 μg/mL (T7-lacI, bottom). Each protein expression was obtained from methionine auto-induction medium containing 0.025% (w/v) glucose, 0.45% (w/v) lactose and 0.9% (w/v) glycerol.
Fermentation Approach
An instrument-controlled fermenter was used to investigate the correlation between carbon source utilization, O2 saturation of the culture, and protein expression. In the fermenter, an aerobic growth condition was maintained during auto-induction by fixing O2 at greater than 10% of saturation during the entire cell growth. The aerobic growth condition in the fermenter best represents growth of small-scale cultures in 96-well growth blocks. For comparison, a microaerobic growth condition was maintained by completing the growth phase under O2-limitation (43–45). The microaerobic growth condition best represents growth of large-scale cultures in 2-L bottles. Figure 6 shows dissolved O2 and pH profiles for growth and auto-induction under these two conditions.
Figure 6.
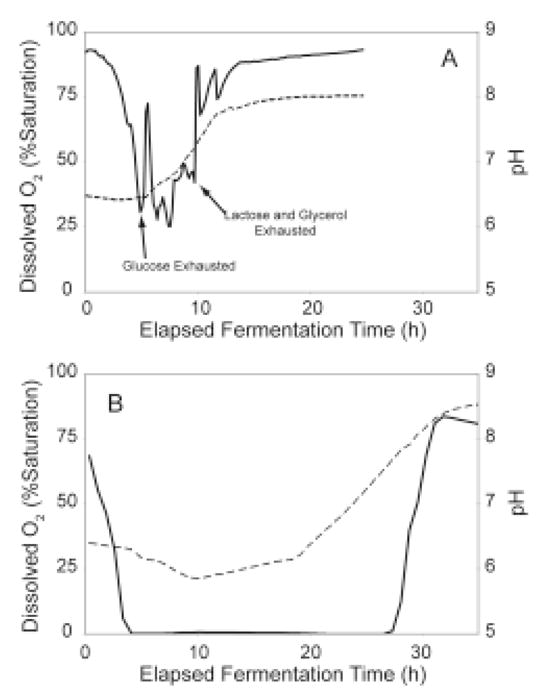
Dissolved O2 (solid lines) and pH (dashed lines) profiles for aerobic (top) and O2-limited (bottom) growth of E. coli B834(DE3) T7-Luc completed in a Sixfors instrumented fermenter. In both cases, the dissolved O2 initially dropped as increasing cell density raised the metabolic O2 demand. For the aerobic growth, the dissolved O2 was maintained above 10% of saturation during the course of the experiment. The dissolved O2 fluctuated in the aerobic fermentation during transitions from use of one carbon source to another and due to manual adjustments in agitation made to maintain aerobic conditions. The arrows indicate the times where glucose, lactose, and glycerol were exhausted. For the O2-limited growth, dissolved O2 was below measurable levels for much of the experiment because the metabolic demand exceeds the amount of O2 supplied. After ~10 h for the aerobic case and ~28 h for the O2-limited case, most of the carbon sources were consumed and the dissolved O2 increased rapidly as the metabolism ceased. For the aerobic growth, the pH was constant during glucose consumption and rose as succinate was consumed. In the O2-limited case, the pH dropped initially as acetate was produced by fermentation. The pH trend was reversed as succinate and eventually, acetate were consumed.
Carbon Source Consumption Patterns
Figure 7A shows representative HPLC traces obtained from the culture medium during the course of a growth of E. coli B834(DE3) with the simple T7-Luc plasmid in auto-induction medium. At t = 0, lactose, glucose, succinate and glycerol are present. At t = 8 h (cell density of ~5), the glucose was entirely consumed and lactose had become the preferred carbon source, so it was being depleted from the culture medium. It is notable that acetate accumulated early in the growth and auto-induction, but was later consumed. At t = 28 h (cell density of ~13), the growth was complete and the only identified carbon sources remaining were a residual small amount of acetate and a larger amount of galactose. Galactose accumulates in the culture medium when lactose is consumed as E. coli B834(DE3) is a galactose auxotroph (25).
Figure 7.

HPLC determination of carbon source levels and carbon consumption patterns during the time course of O2-limited auto-induction in E. coli B834 (DE3) transformed with T7-Luc. A, HPLC analysis of samples from different times during the fermentation. Peak identities are: a, lactose; b, glucose co-eluting with phosphate; c, galactose; d, unknown fermentation product; 5, succinate; 6, glycerol and 7, acetate. The sample from t = 0 was taken immediately after inoculation of the fermenter. The middle traces show accumulation of galactose and acetate during intermediate time points (t = 8, 19 h) and the top trace shows phosphate, galactose and acetate remained at the end of the fermentation (t = 28 h). Galactose cannot be metabolized by E. coli B834 and increased as a byproduct of lactose consumption, while acetate was a byproduct of anaerobic fermentation. B, sigmoidal curve fitting of the relationship between change in carbon source concentration and cell density. In all cases, glucose (circles) was consumed first, and followed successively by lactose (squares), glycerol (×), succinate (diamonds), and then acetate (triangles). Acetate was initially produced and later consumed as a carbon source. C, first derivative of the sigmoidal curve fits, defined to be the specific consumption for each carbon source. These series have the same markers as in B. The filled circles show luciferase expression from the T7-Luc expression plasmid as determined by luminescence assay.
Figure 7B shows the complete pattern of carbon source consumption during the aerobic growth of E. coli B834(DE3) transformed with T7-Luc (see Table 1) in the auto-induction medium. In these cells, LacI is only provided by low-level constitutive expression from the bacterial genome. The carbon source concentrations were fitted as sigmoidal functions (solid lines) for illustrative purposes, and Figure 7C shows the first derivative of these fits. In Figure 7B and 7C, the carbon consumption patterns are plotted relative to cell density (optical density at 600 nm), which provides a useful correlation between an easily measured experimental property and the status of the carbon sources during growth and auto-induction. This representation is independent of the time of the culture, but represents the progress toward consumption of carbon required to enter and complete the auto-induction program. For example, the transition from growth on glucose to growth on lactose occurs at a cell density of ~5, lactose consumption is complete at a cell density of ~7, and no consumable carbon sources are remaining when the cell density has reached ~13. The carbon consumption pattern of E. coli BL21 lacking an expression plasmid was indistinguishable (data not shown).
This pattern of carbon consumption is consistent with previous studies of E. coli diauxic growth (15). Thus glucose was preferentially consumed, followed by lactose, and finally glycerol. Furthermore, in these experiments, succinate was gradually consumed throughout the entire growth period and acetate was largely consumed by the end of the culture growth. In auto-induction, protein expression from the lac operon will be induced along with lactose consumption. For example, induction of T7 RNA polymerase expression under the control of a lacUV5 promoter in E. coli B834(DE3) would be expected to coincide with activation of the lac operon. Correspondingly, Figure 7C shows that luciferase activity was detected at a cell density of ~5 when lactose became the preferred carbon source, and continued to increase after lactose consumption was complete as glycerol and succinate were consumed.
Effect of LacI Dosing on Carbon Consumption Patterns
Figure 8 shows the effect of different levels of LacI on the carbon consumption patterns during auto-induction. The consumption patterns for glycerol and lactose for E. coli B834 expressing T5-lacI-Luc (pVP58K) by aerobic auto-induction are shown in Figure 8A. This construct provides expression of plasmid-encoded LacI from the weak lacI promoter. Increasing LacI shifts the order of preference from glucose/lactose/glycerol to glucose/glycerol/lactose in aerobic culture. Thus there is a dramatic shift in the pattern of carbon consumption relative to the T7-Luc data shown in Figure 7C.
Figure 8.

The timing of lactose consumption as a consequence of LacI dosing. A, glycerol is preferentially consumed before lactose in an aerobic growth with the T5-lacI expression plasmid. This result can be contrasted with Figure 7C, where lactose is preferentially consumed before glycerol. B, specific consumption of lactose during auto-induction growth with the indicated expression plasmids. The T7-Luc expression plasmid does not supplement LacI expression. The T7-lacI-Luc and T5-lacI-Luc plasmids contain a plasmid borne copy of the lac repressor gene with a wild-type promoter and give ~20-fold increase in the level of LacI relative to T7-Luc. The T5-lacIq-Luc plasmid also contains a plasmid borne copy of the lac repressor with a lacIq promoter that increases the level of LacI by ~10-fold higher than from T7-lacI-Luc and T5-lacI-Luc. With this latter plasmid, only a small amount of lactose was consumed and culture growth was halted at a cell density of 16 (OD600 units). In contrast, the other cultures were able to fully consume the lactose and achieved a cell density of ~21.
Figure 8B shows the consequences of LacI levels on lactose consumption for each of the luciferase expression plasmids from Table 1. T7-Luc, which provides no recombinant LacI, maximally consumed lactose at a cell density of ~10. In contrast, T7-lacI-Luc, with constitutive plasmid-encoded expression of LacI, shifted the maximal consumption of lactose to a cell density of ~15, while T5-lacI-Luc, also providing constitutive plasmid-encoded expression of LacI, behaved in a similar manner and shifted the maximal consumption of lactose to a cell density of ~18. Finally, T5-lacIq-Luc, giving overexpression of plasmid-encoded LacI from the strong lacIq promoter), the shift in carbon consumption pattern was so extreme that culture growth stopped in aerobic conditions before lactose could be substantially consumed (Figure 8B, × symbols).
Consequences of O2 Availability During Auto-Induction
Figure 9A shows the consequences of aerobic or O2-limited growth on the lactose consumption pattern for T5-lacI-Luc expression in E. coli B834. During aerobic growth, the maximal lactose consumption occurred at a cell density of ~18, as shown in Figure 8. The appearance of luciferase activity closely tracked this maximal consumption pattern, which is consistent with the relatively strong control of basal expression given by aerobic growth and the presence of recombinant LacI. For comparison, Figure 9A also shows that O2-limited growth during auto-induction shifted the maximal lactose consumption to a lower cell density. Thus changes in oxygenation state of the medium dramatically affected the preference for lactose consumption relative to other carbon sources. Furthermore, in the O2-limited growth, the appearance of luciferase activity no longer closely tracked the lactose consumption pattern, but was shifted to earlier in the overall growth period. These results are consistent with a weakening of catabolite repression and consequent increase in basal expression from both the genomic lac operon (generating allolactose) and from the recombinant expression system (generating luciferase).
Figure 9.

The effect of aeration on lactose consumption with the T5-lacI-Luc expression plasmid. A, lactose consumption (open triangles) and protein expression (filled triangles) occurred at an earlier stage of growth in O2-limited cultures as compared to the aerobic cultures (lactose consumption and expression measurements represented with either open or filled squares, respectively). B, effect of the T5-lacIq expression plasmid on lactose consumption. In O2-limited cultures, all lactose was consumed by 30 h after inoculation. In the aerobic cultures, the cell density stopped increasing at 20 h and lactose was only slowly consumed thereafter.
Figure 9B emphasizes the strong influence of oxygenation state on the consumption of lactose with the E. coli T5-lacIq expression vector. Under aerobic auto-induction conditions, lactose utilization was only weakly initiated and ~70% of the initial lactose remained after ~40 h. After the time when glucose, glycerol, and succinate were consumed (~15 h), little additional cell growth or protein expression were observed. For comparison, O2-limited auto-induction gave complete utilization of lactose between 10 and 30 h. During this time, continued cell growth and protein expression were both obtained.
Discussion
Auto-Induction Method
Auto-induction medium contains a mixture of carbon and energy sources. Glucose is the preferred source for E. coli and is utilized during the early stages of growth. Lactose and glycerol serve as carbon and energy sources during later stages of growth and recombinant protein production. Succinate (or other organic acids such as aspartate or glutamate) may be included to help maintain the culture pH and to act as additional sources of carbon and nitrogen. The consumption of these individual carbon sources by E. coli has been extensively studied (11–17, 46) and in some cases, in combination (as is the case for glucose-lactose diauxic growth). This work demonstrates the importance and possible advantages of considering the interactions between media composition, LacI expression and oxygenation state in the function of auto-induction systems for protein production in E. coli.
Comparison of Response Surfaces
Figure 10 shows a two-dimensional plane through the response surfaces of Figure 3 starting from zero glycerol and lactose and ending at 1.2% (w/v) glycerol and 0.6% (w/v) lactose, a trajectory that includes the highest response for all cases. This simplified representation offers a direct comparison of expression results achieved from the three expression systems. Expression from T5-lacI (solid line) was higher than from T7-lacI (pET32, dashed line) at all compositions except at the lowest lactose concentrations, where basal expression from T7-lacI was higher (Table 3). T5-lacIq (diamonds, methionine medium; circles, selenomethionine medium) exhibited the lowest expression levels. Surprisingly, the combination of T5-lacIq with selenomethionine medium gave a higher level of expression than the same plasmid with methionine medium, and selenomethionine-labeled protein was obtained with yield of ~500 μg/mL. This enhanced performance occurred because the presence of selenomethionine shifted the maximal lactose consumption to lower cell density (data not shown), allowing more complete execution of the auto-induction program. How this response is mediated at the cellular level is not known.
Figure 10.

Comparison of modeled expression levels for T5-lacI (solid line), T7-lacI (pET32, dashed line), T5-lacIq in methionine auto-induction medium (filled diamonds) and T5-lacIq in selenomethionine auto-induction medium (filled circles). This figure is a two-dimensional plane through the response surfaces of Figure 3A (T5-lacI), 3C (T5-lacIq, methionine medium), 3D (T5-lacIq, selenomethionine medium) and 3E (T7-lacI, pET32) starting from zero glycerol and lactose and ending at 1.2% (w/v) glycerol and 0.6% (w/v) lactose.
Figure 11 shows a two-dimensional surface plot that reveals additional features about the composition of the optimal medium for the T5-lacI plasmid. For this plot, the range of carbon source concentrations investigated was intentionally extended beyond that shown in Figure 3, and notably resulted in medium compositions that decreased the expression. These results indicate that with the present composition of non-carbon source components, maximum expression from the T5-lacI plasmid was obtained near the limits of the lower factorial (dotted line), specifically 0.6% lactose and 1.2% glycerol and that slightly lower glycerol or lactose concentrations had little effect on expression while higher concentrations of glycerol adversely affected expression (region bounded by the dashed line). This behavior is also summarized in Table 2, where glycerol had a positive first order scaled effect estimate but a negative second order scaled estimate effect (qualitatively similar behavior was obtained from both methionine and selenomethionine media). The region where highest expression occurred is a broad plateau, indicating overall tolerance to minor variations in medium composition without altering the expression outcome. Figure 11 shows that there are choices for medium composition that give gradual change between lower and higher expression levels. Knowledge of this may be useful to maximize the soluble production of some proteins like luciferase that apparently have an intrinsic solubility limit within cells. Other choices for change in medium composition give precipitous changes in the expression level. Knowledge of these is important to avoid experimental conditions that are likely to give poor or irreproducible results.
Figure 11.
A topographical map that includes expression data for higher carbon source concentrations. Lower expression is indicated with blue hues and higher expression with yellow hues. Experimental design points are shown as black circles. The design space explored in the first, lower concentration study is surrounded by dotted lines. For this experiment, a second factorial was completed at higher concentrations of glycerol and lactose for T5-lacI-Luc with methionine media. Dashed lines surround the second factorial, which covers higher concentrations of lactose and glycerol. The contour plot shown here represents a quadratic spline fit to the experimental data, as a single low order model could not adequately model the results due to multiple curvatures. Some fine features of the quadratic spline surface contain experimental uncertainty (such as the “valley” between the two highest expression regions) that were smoothed out in the response surface models.
Figure 11 also shows that additional increases in lactose and glycerol near the high end of the experimental range investigated did not increase expression, but in some circumstances actually decreased expression. The reason(s) for the decrease in expression are not known. In the present media, the cell density appeared to be limited to OD600 ~25 and was not affected by further increases in lactose or glycerol, suggesting that some non-carbon source component may have become limiting at this cell density. Systematic evaluation of the contribution of other media components to expression results in a manner similar to that used here for carbon sources may yield further increases in cell density and volumetric protein expression.
As indicated by Figure 7C and 8A, glucose was always the preferred carbon source. Thus changes in the level of glucose added to the medium control the cell density at which the auto-induction protocol will be initiated. This concept has been discussed previously in the context of lactose-induced high-density fed-batch fermentation (18) and use of this approach for isotopic labeling studies (47). However, increasing the level of glucose will increase the cell mass and biological demand for carbon sources, leading to more rapid consumption of lactose and glycerol during the auto-induction phase without compensating changes in the levels of lactose and glycerol. This would shorten the time of auto-induction. Depending on circumstances, this may be beneficial or not.
Influence of LacI on Auto-Induction
LacI acts in two ways to delay the onset of lactose consumption required for auto-induction. First, high intracellular concentrations of LacI increase the occupancy of the lacO sites located upstream of the lac operon structural genes. This occupancy strongly decreases the basal expression of β-galactosidase and lac permease, which in turn decreases the rate of allolactose production. Second, a larger absolute amount of allolactose is required in order to dissociate intracellular LacI from lacO sites so that induction of the lac operon and heterologous protein expression can begin. These combined effects are sufficiently dominant to completely change the order of carbon source consumption from glucose/lactose/glycerol to glucose/glycerol/lactose for E. coli growths with each of the plasmids tested that supplement LacI expression.
Maximal lactose consumption occurred at a higher cell density for the growths with the T5-lacI plasmid (Figure 8) as compared with the T7-lacI (pET32) plasmid. Since both plasmids have the pBR322 origin, the copy number should not differ greatly (48). Moreover, since both use the lacI promoter to express LacI from the plasmid, the level of LacI should be similar. Small differences in LacI expression due to positional effects in the plasmid may account for the difference in behavior. Thus a previous study has demonstrated that such positional effects can influence basal levels of heterologous protein expression (49, 50) and it is plausible that positional effects could influence constitutive expression of LacI in a similar way.
It is not clear why expression levels from the T5-lacI plasmid were ~70% higher than those determined for the pET32 plasmid (T7-lacI, compare Figure 3A and 3C). The T5 promoter uses E. coli RNA polymerase, while pET32 requires that T7 RNA polymerase must also be made. It seems unlikely that this difference alone accounts for the lower expression from the pET32 plasmid. T7 RNA polymerase is highly active (51) and might be expected to make more mRNA than E. coli polymerase, especially upon considering that the T7 polymerase is dedicated to the production of target gene transcripts (52) while the T5 promoter must compete with other host promoters. Thus it is possible that high transcription levels may excessively direct energy fluxes towards mRNA production and away from protein expression. Furthermore, transcript instability due to a decoupling of transcription and translation caused by the high transcription rate of T7 RNA polymerase may play a role (53, 54).
Influence of Oxygenation State on Auto-Induction
The consequence of oxygenation state in the auto-induction culture is apparent from Figure 9. In all cases investigated, lactose consumption and protein expression were shifted to a lower cell density by O2-limitation. For T5-lacIq, this effect was dramatic enough that the final expression levels were higher when O2 was limited.
E. coli is known to control glucose and lactose import as a response to O2-limitation through a variety of transcriptional and post-translational mechanisms. As one example, phosphorylated ArcA is a negative transcriptional regulator of the IICBGlc component of the bacterial phosphoenolpyruvate:sugar phosphotransferase (PTS) system (46, 55). Decreased expression of IICBGlc leads to an accumulation of the phosphorylated PTS enzyme component IIAGlc. Since dephosphorylated IIAGlc is a known inhibitor of lac permease (15–17), O2-limitation and accumulation of phosphorylated IIAGlc relieve the inhibition of lac permease, allowing higher lactose import rates.
The results elucidate the origin of the difference in expression behavior observed between small- and large-scale experiments. In the tests of Figure 1A, the elevated level of LacI postponed lactose utilization in the aerobic conditions of the small-scale, while the O2-limited conditions of the large-scale shifted lactose utilization to lower cell density and thus promoted protein expression. Reformulation of the carbon sources promoted growth to higher cell density by increases in glycerol and lactose, utilization of lactose at lower cell density through lowered glucose, and more complete utilization of the provided carbon sources, regardless of the culture oxygenation state (Figure 1B).
Role in High-throughput Protein Expression Studies
For high throughput studies, which provided the motivation for the development of auto-induction (7) and the experimental basis for this work (8, 9), it is often desirable to screen for suitable protein expression in small volumes using multi-well growth blocks. We found that this expression environment was aerobic, but surprisingly, led to significantly lower protein expression in the initially defined auto-induction medium. In contrast, O2-limitation was previously noted to increase the yield of recombinant protein (7) and we have found this limitation most closely corresponded to the actual conditions in 2-L bottles used for large-scale protein expression (34). It is noted that fully anaerobic conditions are not conducive to either rapid cell growth or high-yield production of biomass.
This work was further prompted by the need for improved correlation between small-scale protein expression screening and large-scale protein production at the University of Wisconsin Center for Eukaryotic Structural Genomics (8, 9). Through an initial set of media optimization experiments, the discrepancy between small and large scale culture results was addressed by increasing the concentration of carbon sources available, which on average gave a 2- to 3-fold increase in target protein expression in the small-scale cultures. However, protein expression in large-scale cultures was only marginally improved with this initial change in medium composition. Therefore, we were interested in whether other manipulations of the biochemical apparatus used for auto-induction might also improve protein expression yields. By decreasing the LacI expression level provided by the expression plasmid, lactose consumption and heterologous protein expression were shifted to an earlier phase of growth. Although culture oxygenation still contributed to the timing of induction, both small- and large-scale growths were able to produce enough allolactose to derepress the lac operon, fully consume the available lactose, and achieve high levels of protein expression.
These studies also give insight into the use of auto-induction for production of 13C- and 15N-labeled proteins for NMR structure determination. First, efficient consumption of succinate during the auto-induction process limits the utility of these media formulations for production of 13C-labeled samples for NMR structure determination, unless 13C-labeled succinate is used. The substitution of other amino acids (aspartate, glutamate) will not correct this problem and potentially introduce problematic dilution of 15N-labeling unless the more expensive 13C- and 15N-labeled analogs are used for these components. Furthermore, our previously described changes in medium composition for NMR studies (8) are now recognized to fall into a low productivity region of the T5-lacIq response surface shown in Figure 3B (0.5% glycerol, 0.2% lactose). In the previous study, unlabeled lactose (not cost-effective for use as a 13C-labeled compound) was intentionally minimized in order to avoid isotopic dilution of 13C-labeled glycerol. As an alternative, the response curve in Figure 3A suggests that this same mixture of glycerol and lactose may give considerably better expression results when coupled with a T5-lacI plasmid.
Other Possible Uses of Factorial Medium Design
Small-scale protein expression in 24- or 96-well blocks was originally intended to be a screening tool for numerous structural genomics targets, whose expression properties were not known. The array format for the various auto-induction media provides a simple way to test the performance of other plasmid vectors and host strains for conditions that maximize the expression of these unknown proteins. Furthermore, based on expression levels possibly exceeding 1000 μg/mL of culture fluid (actually exceeding 2000 μg/mL for the combination of in vivo proteolysis of MBP and eGFP from pVP62K, see Figure 4), it is reasonable to consider other applications for small-scale expression. For example, the amount of protein produced from a few mL of these cultures may be sufficient for automated protein purification (56), microfluidics-based crystallization screening (57, 58), initial nL-scale crystallization trials (59, 60), 15N HSQC NMR measurements (61, 62), or functional and enzymatic characterizations (63, 64).
Other Experience with Use of Optimized Auto-induction Conditions
The work presented here includes expression studies in 96-well growth blocks, 2-L shaken bottles and automated stirred-vessel fermenters. In each case, the combination of a designed auto-induction medium and matched expression plasmid gave strong expression results, demonstrating utility in several different formats used to grow bacterial cells. The results presented here derive from study of only two target proteins, eGFP and luciferase, that were chosen due to the attractiveness of their assays. Nevertheless, our preliminary experience with other proteins suggests that these modifications to auto-induction media composition and LacI dosing may have general utility in improving the level of recombinant protein expression. Thus combination of a T5-lacI expression plasmid with a terrific broth medium supplemented with an auto-induction mixture of 0.015% glucose, 0.8% glycerol, 0.5% lactose, 0.375% aspartic acid and 2 mM MgSO4 contributed to an ~5-fold increase in expression of soluble TEV protease (64) when compared to previous reports (66, 67). Moreover, preliminary expression studies with other proteins such as toluene 4-monooxygenase hydroxylase (68), stearoyl-ACP Δ9 desaturase (18), cytochrome b5 (Sobrado, P. and Fox, B.G., unpublished work) and various bacterial and plant FMN-containing oxidoreductases (Malone, T.E. and Fox, B.G., unpublished work), indicate the combination of a factorial designed auto-induction media with T5-lacI plasmids offers substantial promise for structural and functional work with known proteins.
Supplementary Material
Figure S1. Two representations of the factorial design experimental space. The design points limited to glycerol and lactose are shown in A while B is a three dimensional projection of all design points from the three-factor five-level factorial.
Figure S2. A picture taken of a plate containing diluted eGFP lysates obtained from expression in the different medias listed in Table S1. The clear 96 well plate shown here was illuminated with a 340 nm light source.
Table S1. Concentrations of glucose (top), lactose (middle), and glycerol (bottom) for each expression medium tested are shown in the 96-well plate format used for expression testing. Columns 1–8 contain methionine media while columns 9–12 contain selenomethionine labeling media.
Acknowledgments
We thank Dr. R. McClain (University of Wisconsin Chemistry Department) for the use of the HPLC refractive index detector and Dr. R.O. Frederick (University of Wisconsin Center for Eukaryotic Structural Genomics) for supplying the eGFP clone. The NIH Protein Structure Initiative (1 U54 GM 74901, J.L. Markley, Principal Investigator; G.N. Phillips, Jr. and B.G. Fox, Co-Investigators) and Promega Corporation (B.G. Fox, Principal Investigator) supported this work.
Footnotes
Supporting information available: Photograph of eGFP fluorescence in a 96-well plate format, composition of the auto-induction medium present in each well of the plate, and three dimensional representation of factorial design experimental space. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Zhang C, Kim SH. Overview of Structural Genomics: From Structure to Function. Curr Opin Chem Biol. 2003;7:28–32. doi: 10.1016/s1367-5931(02)00015-7. [DOI] [PubMed] [Google Scholar]
- 2.Terwilliger TC. Structural Genomics in North America. Nat Struct Mol Biol. 2000;7(Suppl):935–9. doi: 10.1038/80700. [DOI] [PubMed] [Google Scholar]
- 3.Chandonia JM, Brenner SE. The Impact of Structural Genomics: Expectations and Outcomes. Science. 2006;311:347–51. doi: 10.1126/science.1121018. [DOI] [PubMed] [Google Scholar]
- 4.Becker KF, Metzger V, Hipp S, Hofler H. Clinical Proteomics: New Trends for Protein Microarrays. Curr Med Chem. 2006;13:1831–7. doi: 10.2174/092986706777452506. [DOI] [PubMed] [Google Scholar]
- 5.Arcus VL, Lott JS, Johnston JM, Baker EN. The Potential Impact of Structural Genomics on Tuberculosis Drug Discovery. Drug Discovery Today. 2006;11:28–34. doi: 10.1016/S1359-6446(05)03667-6. [DOI] [PubMed] [Google Scholar]
- 6.Lundstrom K. Structural Genomics for Membrane Proteins. Cell Mol Life Sci. 2006;63:2597–607. doi: 10.1007/s00018-006-6252-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Studier FW. Protein Production by Auto-Induction in High Density Shaking Cultures. Protein Expression Purif. 2005;41:207–34. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 8.Tyler RC, Sreenath HK, Singh S, Aceti DJ, Bingman CA, Markley JL, Fox BG. Auto-Induction Medium for the Production of [U-15N]- and [U-13C, U-15N]-Labeled Proteins for NMR Screening and Structure Determination. Protein Expression Purif. 2005;40:268–78. doi: 10.1016/j.pep.2004.12.024. [DOI] [PubMed] [Google Scholar]
- 9.Sreenath HK, Bingman CA, Buchan BW, Seder KD, Burns BT, Geetha HV, Jeon WB, Vojtik FC, Aceti DJ, Frederick RO, Phillips GN, Jr, Fox BG. Protocols for Production of Selenomethionine-Labeled Proteins in 2-L Polyethylene Terephthalate Bottles Using Auto-Induction Medium. Protein Expression Purif. 2005;40:256–67. doi: 10.1016/j.pep.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 10.Jacob F, Monod J. Genetic Regulatory Mechanisms in the Synthesis of Proteins. J Mol Biol. 1961;3:318–56. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- 11.Holms H. Flux Analysis and Control of the Central Metabolic Pathways in Escherichia Coli. FEMS Microbiol Rev. 1996;19:85–116. doi: 10.1111/j.1574-6976.1996.tb00255.x. [DOI] [PubMed] [Google Scholar]
- 12.Mahadevan R, Edwards JS, Doyle FJ. 3rd, Dynamic Flux Balance Analysis of Diauxic Growth in Escherichia Coli. Biophys J. 2002;83:1331–40. doi: 10.1016/S0006-3495(02)73903-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong P, Gladney S, Keasling JD. Mathematical Model of the Lac Operon: Inducer Exclusion, Catabolite Repression, and Diauxic Growth on Glucose and Lactose. Biotechnol Prog. 1997;13:132–43. doi: 10.1021/bp970003o. [DOI] [PubMed] [Google Scholar]
- 14.Reznikoff WS. The Lactose Operon-Controlling Elements: A Complex Paradigm. Mol Microbiol. 1992;6:2419–22. doi: 10.1111/j.1365-2958.1992.tb01416.x. [DOI] [PubMed] [Google Scholar]
- 15.Inada T, Kimata K, Aiba H. Mechanism Responsible for Glucose-Lactose Diauxie in Escherichia Coli: Challenge to the cAMP Model. Genes Cells. 1996;1:293–301. doi: 10.1046/j.1365-2443.1996.24025.x. [DOI] [PubMed] [Google Scholar]
- 16.Hogema BM, Arents JC, Bader R, Eijkemans K, Inada T, Aiba H, Postma PW. Inducer Exclusion by Glucose 6-Phosphate in Escherichia Coli. Mol Microbiol. 1998;28:755–65. doi: 10.1046/j.1365-2958.1998.00833.x. [DOI] [PubMed] [Google Scholar]
- 17.Hogema BM, Arents JC, Bader R, Postma PW. Autoregulation of Lactose Uptake through the LacY Permease by Enzyme IIAglc of the PTS in Escherichia Coli K-12. Mol Microbiol. 1999;31:1825–33. doi: 10.1046/j.1365-2958.1999.01319.x. [DOI] [PubMed] [Google Scholar]
- 18.Hoffman BJ, Broadwater JA, Johnson P, Harper J, Fox BG, Kenealy WR. Lactose Fed-Batch Overexpression of Recombinant Metalloproteins in Escherichia Coli Bl21 (DE3): Process Control Yielding High Levels of Metal-Incorporated, Soluble Protein. Protein Expression Purif. 1995;6:646–54. doi: 10.1006/prep.1995.1085. [DOI] [PubMed] [Google Scholar]
- 19.Chen W, Lee SB, Bailey JE. Molecular Design of Expression Systems: Comparison of Different Repressor Control Configurations Using Molecular Mechanism Models. Biotechnol Bioeng. 1991;38:679–687. doi: 10.1002/bit.260380702. [DOI] [PubMed] [Google Scholar]
- 20.Hammarstrom M, Hellgren N, van Den Berg S, Berglund H, Hard T. Rapid Screening for Improved Solubility of Small Human Proteins Produced as Fusion Proteins in Escherichia Coli. Protein Sci. 2002;11:313–21. doi: 10.1110/ps.22102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Terpe K. Overview of Bacterial Expression Systems for Heterologous Protein Production: From Molecular and Biochemical Fundamentals to Commercial Systems. Appl Microbiol Biotechnol. 2006;72:211–22. doi: 10.1007/s00253-006-0465-8. [DOI] [PubMed] [Google Scholar]
- 22.Swalley SE, Fulghum JR, Chambers SP. Screening Factors Effecting a Response in Soluble Protein Expression: Formalized Approach Using Design of Experiments. Anal Biochem. 2006;351:122–7. doi: 10.1016/j.ab.2005.11.046. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J, Russell DW, editors. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- 24.Wood WB. Host Specificity of DNA Produced by Escherichia coli: Bacterial Mutations Affecting the Restriction and Modification of DNA. J Mol Biol. 1966;16:118–33. doi: 10.1016/s0022-2836(66)80267-x. [DOI] [PubMed] [Google Scholar]
- 25.Arber W. Host Specificity of DNA Produced by Escherichia coli. 3. Effects on Transduction Mediated by _dg. Virology. 1964;23:173–82. doi: 10.1016/0042-6822(64)90280-6. [DOI] [PubMed] [Google Scholar]
- 26.Calos MP. DNA Sequence for a Low-Level Promoter of the Lac Repressor Gene and an ‘up’ Promoter Mutation. Nature. 1978;274:762–5. doi: 10.1038/274762a0. [DOI] [PubMed] [Google Scholar]
- 27.Nallamsetty S, Kapust RB, Tozser J, Cherry S, Tropea JE, Copeland TD, Waugh DS. Efficient Site-Specific Processing of Fusion Proteins by Tobacco Vein Mottling Virus Protease in Vivo and in Vitro. Protein Expression Purif. 2004;38:108–15. doi: 10.1016/j.pep.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 28.Donnelly MI, Zhou M, Millard CS, Clancy S, Stols L, Eschenfeldt WH, Collart FR, Joachimiak A. An Expression Vector Tailored for Large-Scale, High-Throughput Purification of Recombinant Proteins. Protein Expression Purif. 2006;47:446–54. doi: 10.1016/j.pep.2005.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patterson GH, Knobel SM, Sharif WD, Kain SR, Piston DW. Use of the Green Fluorescent Protein and Its Mutants in Quantitative Fluorescence Microscopy. Biophys J. 1997;73:2782–90. doi: 10.1016/S0006-3495(97)78307-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang TT, Cheng L, Kain SR. Optimized Codon Usage and Chromophore Mutations Provide Enhanced Sensitivity with the Green Fluorescent Protein. Nucleic Acids Res. 1996;24:4592–3. doi: 10.1093/nar/24.22.4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crameri A, Whitehorn EA, Tate E, Stemmer WP. Improved Green Fluorescent Protein by Molecular Evolution Using DNA Shuffling. Nat Biotechnol. 1996;14:315–9. doi: 10.1038/nbt0396-315. [DOI] [PubMed] [Google Scholar]
- 32.Blommel PG, Martin PA, Wrobel RL, Steffen E, Fox BG. High Efficiency Single Step Production of Expression Plasmids from cDNA Clones Using the Flexi Vector Cloning System. Protein Expression Purif. 2006;47:562–70. doi: 10.1016/j.pep.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 33.Myers RH, Montgomery DC. Response Surface Methodology: Process and Product Optimization Using Designed Experiments. 2. J. Wiley; New York: 2002. p. 798. [Google Scholar]
- 34.Millard CS, Stols L, Quartey P, Kim Y, Dementieva I, Donnelly MI. A Less Laborious Approach to the High-Throughput Production of Recombinant Proteins in Escherichia coli Using 2-Liter Plastic Bottles. Protein Expression Purif. 2003;29:311–20. doi: 10.1016/s1046-5928(03)00063-9. [DOI] [PubMed] [Google Scholar]
- 35.Lee C, Kim J, Shin SG, Hwang S. Absolute and Relative qPCR Quantification of Plasmid Copy Number in Escherichia coli. J Biotechnol. 2006;123:273–80. doi: 10.1016/j.jbiotec.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 36.Brunner M, Bujard H. Promoter Recognition and Promoter Strength in the Escherichia coli System. EMBO J. 1987;6:3139–44. doi: 10.1002/j.1460-2075.1987.tb02624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bujard H, Gentz R, Lanzer M, Stueber D, Mueller M, Ibrahimi I, Haeuptle MT, Dobberstein B. A T5 Promoter-Based Transcription-Translation System for the Analysis of Proteins in Vitro and in Vivo. Methods Enzymol. 1987;155:416–33. doi: 10.1016/0076-6879(87)55028-5. [DOI] [PubMed] [Google Scholar]
- 38.Gentz R, Bujard H. Promoters Recognized by Escherichia coli RNA Polymerase Selected by Function: Highly Efficient Promoters from Bacteriophage T5. J Bacteriol. 1985;164:70–7. doi: 10.1128/jb.164.1.70-77.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oehler S, Eismann ER, Kramer H, Muller-Hill B. The Three Operators of the Lac Operon Cooperate in Repression. EMBO J. 1990;9:973–9. doi: 10.1002/j.1460-2075.1990.tb08199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bahl CP, Wu R, Stawinsky J, Narang SA. Minimal Length of the Lactose Operator Sequence for the Specific Recognition by the Lactose Repressor. Proc Natl Acad Sci U S A. 1977;74:966–70. doi: 10.1073/pnas.74.3.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang L, Patel HN, Lappe JW, Wachter RM. Reaction Progress of Chromophore Biogenesis in Green Fluorescent Protein. J Am Chem Soc. 2006;128:4766–72. doi: 10.1021/ja0580439. [DOI] [PubMed] [Google Scholar]
- 42.Saier MH., Jr Protein Phosphorylation and Allosteric Control of Inducer Exclusion and Catabolite Repression by the Bacterial Phosphoenolpyruvate: Sugar Phosphotransferase System. Microbiol Rev. 1989;53:109–20. doi: 10.1128/mr.53.1.109-120.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alexeeva S, de Kort B, Sawers G, Hellingwerf KJ, de Mattos MJ. Effects of Limited Aeration and of the ArcAB System on Intermediary Pyruvate Catabolism in Escherichia coli. J Bacteriol. 2000;182:4934–40. doi: 10.1128/jb.182.17.4934-4940.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alexeeva S, Hellingwerf KJ, Teixeira de Mattos MJ. Quantitative Assessment of Oxygen Availability: Perceived Aerobiosis and Its Effect on Flux Distribution in the Respiratory Chain of Escherichia coli. J Bacteriol. 2002;184:1402–6. doi: 10.1128/JB.184.5.1402-1406.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alexeeva S, Hellingwerf KJ, Teixeira de Mattos MJ. Requirement of ArcA for Redox Regulation in Escherichia coli under Microaerobic but Not Anaerobic or Aerobic Conditions. J Bacteriol. 2003;185:204–9. doi: 10.1128/JB.185.1.204-209.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jeong JY, Kim YJ, Cho N, Shin D, Nam TW, Ryu S, Seok YJ. Expression of ptsG Encoding the Major Glucose Transporter Is Regulated by ArcA in Escherichia coli. J Biol Chem. 2004;279:38513–8. doi: 10.1074/jbc.M406667200. [DOI] [PubMed] [Google Scholar]
- 47.Studts JM, Fox BG. Application of Fed-Batch Fermentation to the Preparation of Isotopically Labeled or Selenomethionyl-Labeled Proteins. Protein Expression Purif. 1999;16:109–19. doi: 10.1006/prep.1999.1067. [DOI] [PubMed] [Google Scholar]
- 48.Polisky B. ColE1 Replication Control Circuitry: Sense from Antisense. Cell. 1988;55:929–32. doi: 10.1016/0092-8674(88)90235-8. [DOI] [PubMed] [Google Scholar]
- 49.Dubendorff JW, Studier FW. Controlling Basal Expression in an Inducible T7 Expression System by Blocking the Target T7 Promoter with Lac Repressor. J Mol Biol. 1991;219:45–59. doi: 10.1016/0022-2836(91)90856-2. [DOI] [PubMed] [Google Scholar]
- 50.Saida F, Uzan M, Odaert B, Bontems F. Expression of Highly Toxic Genes in E. Coli: Special Strategies and Genetic Tools. Curr Protein Pept Sci. 2006;7:47–56. doi: 10.2174/138920306775474095. [DOI] [PubMed] [Google Scholar]
- 51.Makarova OV, Makarov EM, Sousa R, Dreyfus M. Transcribing of Escherichia coli Genes with Mutant T7 RNA Polymerases: Stability of lacZ mRNA Inversely Correlates with Polymerase Speed. Proc Natl Acad Sci U S A. 1995;92:12250–4. doi: 10.1073/pnas.92.26.12250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Studier FW, Moffatt BA. Use of Bacteriophage T7 RNA Polymerase to Direct Selective High-Level Expression of Cloned Genes. J Mol Biol. 1986;189:113–30. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 53.Iost I, Guillerez J, Dreyfus M. Bacteriophage T7 RNA Polymerase Travels Far Ahead of Ribosomes in Vivo. J Bacteriol. 1992;174:619–22. doi: 10.1128/jb.174.2.619-622.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lopez PJ, Iost I, Dreyfus M. The Use of a tRNA as a Transcriptional Reporter: The T7 Late Promoter Is Extremely Efficient in Escherichia coli but Its Transcripts Are Poorly Expressed. Nucleic Acids Res. 1994;22:2434. doi: 10.1093/nar/22.12.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Perrenoud A, Sauer U. Impact of Global Transcriptional Regulation by ArcA, ArcB, Cra, Crp, Cya, Fnr, and Mlc on Glucose Catabolism in Escherichia coli. J Bacteriol. 2005;187:3171–9. doi: 10.1128/JB.187.9.3171-3179.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murphy MB, Doyle SA. High-Throughput Purification of Hexahistidine-Tagged Proteins Expressed in E. Coli. Methods Mol Biol. 2005;310:123–30. doi: 10.1007/978-1-59259-948-6_9. [DOI] [PubMed] [Google Scholar]
- 57.van der Woerd M, Ferree D, Pusey M. The Promise of Macromolecular Crystallization in Microfluidic Chips. J Struct Biol. 2003;142:180–7. doi: 10.1016/s1047-8477(03)00049-2. [DOI] [PubMed] [Google Scholar]
- 58.Segelke B. Macromolecular Crystallization with Microfluidic Free-Interface Diffusion. Expert Rev Proteomics. 2005;2:165–72. doi: 10.1586/14789450.2.2.165. [DOI] [PubMed] [Google Scholar]
- 59.Zheng B, Gerdts CJ, Ismagilov RF. Using Nanoliter Plugs in Microfluidics to Facilitate and Understand Protein Crystallization. Curr Opin Struct Biol. 2005;15:548–55. doi: 10.1016/j.sbi.2005.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lion N, Reymond F, Girault HH, Rossier JS. Why the Move to Microfluidics for Protein Analysis? . Curr Opin Biotechnol. 2004;15:31–7. doi: 10.1016/j.copbio.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 61.Galvao-Botton LM, Katsuyama AM, Guzzo CR, Almeida FC, Farah CS, Valente AP. High-Throughput Screening of Structural Proteomics Targets Using NMR. FEBS Lett. 2003;552:207–13. doi: 10.1016/s0014-5793(03)00926-8. [DOI] [PubMed] [Google Scholar]
- 62.Kennedy MA, Montelione GT, Arrowsmith CH, Markley JL. Role for NMR in Structural Genomics. J Struct Funct Genomics. 2002;2:155–69. doi: 10.1023/a:1021261026670. [DOI] [PubMed] [Google Scholar]
- 63.Kumar RA, Clark DS. High-Throughput Screening of Biocatalytic Activity: Applications in Drug Discovery. Curr Opin Chem Biol. 2006;10:162–8. doi: 10.1016/j.cbpa.2006.02.033. [DOI] [PubMed] [Google Scholar]
- 64.Blommel PG, Fox BG. A combined approach to improving large-scale production of tobacco etch virus protease. Protein Express Purif. 2007 doi: 10.1016/j.pep.2007.04.013. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schmidt M, Bornscheuer UT. High-Throughput Assays for Lipases and Esterases. Biomol Eng. 2005;22:51–6. doi: 10.1016/j.bioeng.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 66.Fang L, Jia KZ, Tang YL, Ma DY, Yu M, Hua ZC. An Improved Strategy for High-Level Production of TEV Protease in Escherichia coli and Its Purification and Characterization. Protein Expression Purif. 2006 doi: 10.1016/j.pep.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 67.van den Berg S, Lofdahl PA, Hard T, Berglund H. Improved Solubility of TEV Protease by Directed Evolution. J Biotechnol. 2006;121:291–8. doi: 10.1016/j.jbiotec.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 68.Studts JM, Mitchell KH, Pikus JD, McClay K, Steffan RJ, Fox BG. Optimized Expression and Purification of Toluene 4-Monooxygenase Hydroxylase. Protein Expression Purif. 2000;20:58–65. doi: 10.1006/prep.2000.1281. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Two representations of the factorial design experimental space. The design points limited to glycerol and lactose are shown in A while B is a three dimensional projection of all design points from the three-factor five-level factorial.
Figure S2. A picture taken of a plate containing diluted eGFP lysates obtained from expression in the different medias listed in Table S1. The clear 96 well plate shown here was illuminated with a 340 nm light source.
Table S1. Concentrations of glucose (top), lactose (middle), and glycerol (bottom) for each expression medium tested are shown in the 96-well plate format used for expression testing. Columns 1–8 contain methionine media while columns 9–12 contain selenomethionine labeling media.



