Abstract
We previously identified a sperm-specific Na+/H+ exchanger (sNHE) principally localized to the flagellum. Disruption of the sNHE gene in mice resulted in absolute male infertility associated with a complete loss of sperm motility. Here, we show that the sNHE-null spermatozoa fail to develop the cAMP-dependent protein tyrosine phosphorylation that coincides with the functional maturation occurring upon incubation in capacitating conditions in vitro. Both the sperm motility defect and the lack of induced protein tyrosine phosphorylation are rescued by the addition of cell-permeable cAMP analogs, suggesting that cAMP metabolism is impaired in spermatozoa lacking sNHE. Our analyses of the bicarbonate-dependent soluble adenylyl cyclase (sAC) signaling pathway in sNHE-null sperm cells reveal that sNHE is required for the expression of full-length sAC, and that it is important for the bicarbonate stimulation of sAC activity in spermatozoa. Furthermore, both codependent expression and coimmunoprecipitation experiments indicate that sNHE and sAC associate with each other. Thus, these two proteins appear to be components of a signaling complex at the sperm flagellar plasma membrane. We propose that the formation of this complex efficiently modulates intracellular pH and bicarbonate levels through the rapid and effective control of sAC and sNHE activities to facilitate sperm motility regulation.
Keywords: cAMP, sperm motility
Intracellular pH has long been speculated to play an important role in both invertebrate and vertebrate sperm motility. In particular, Na+/H+ exchangers (NHEs) have been suggested to regulate intracellular pH of spermatozoa in various species (1–3). Both NHE1 and NHE5 are expressed in spermatozoa, but male mice lacking NHE1 are fertile, demonstrating that NHE1 is not critical for sperm function (4). The contribution of NHE5 to sperm motility or male fertility is yet to be assessed.
Sperm-specific NHE (sNHE) is a member of the mammalian NHE superfamily (5, 6). The sNHE protein is located on the principal piece of the sperm flagellum and is predicted to contain 14 membrane-spanning helices based on hydropathy analysis (7). The N terminus of sNHE contains a domain with 10 putative transmembrane segments that is conserved within the NHE superfamily. The C terminus of sNHE contains a long nonconserved region (≈700 aa residues) presumably located predominantly in the sperm cell cytoplasm. All of the eukaryotic NHEs that localize to the plasma membrane have long cytoplasmic carboxyl tails (≈300–700 aa residues), which are thought to regulate the exchanger activity through changes in phosphorylation and their interactions with various binding proteins (5). Quite distinct from other mammalian NHEs, sNHE contains a consensus sequence for a putative cyclic nucleotide-binding domain near the C terminus and four putative transmembrane helices analogous to the voltage-sensing domain of voltage-gated ion channels (7), implying that sNHE function could be regulated by cyclic nucleotide(s) and/or sperm membrane potential.
Numerous attempts to express sNHE in a functional state on the plasma membrane of various eukaryotic cell lines, including NHE-deficient fibroblast cell lines, such as PS200 and AP-1, have all failed, limiting exploration of sNHE function. To determine the physiological significance of the protein, we disrupted the corresponding gene in mice. Loss of the sNHE gene causes complete male-specific infertility due to sperm motility failure (7). The cell-permeable weak base NH3 (delivered as 20 mM NH4Cl) reverses the motility defect in ≈20% of sperm cells, suggesting that lower intracellular pH is at least partially responsible for the motility defect. Unexpectedly, the most effective means of reversing the motility failure phenotype is through the addition of cAMP analogs. Cell-permeable cAMP analogs restore both the percentage of motile spermatozoa and basal cellular velocities to levels comparable with those of wild-type animals (7).
cAMP has been suggested to mediate signaling for motility initiation (8). In mammalian spermatozoa, cAMP synthesis is predominantly controlled by a soluble isoform of the adenylyl cyclase (sAC) family that is not regulated by G proteins (9, 10). This sAC form is thought to be regulated by divalent cations, such as Mg2+ and Ca2+ (11–13), and physiological concentrations of bicarbonate (10, 14).
The sAC gene encodes a Mr ≈187,000 full-length protein with low specific activity and a Mr ≈ 53,000 form with higher specific activity in vitro (9, 15). Both the full-length and truncated forms of sAC contain two apparent catalytic domains (C1 and C2) that resemble the catalytic regions of adenylyl cyclases from cyanobacteria (10). In humans, additional alternatively spliced transcripts are known to exist (16). Little is known about the functions and regulation of the different sAC polypeptides in vivo. The disruption of the sAC gene causes male sterility and a profound loss of sperm motility (17). In addition, capacitation-associated changes in tyrosine phosphorylation are largely absent in sAC-null spermatozoa (18, 19). Similar to the sNHE-null spermatozoa, cell-permeable cAMP analogs rescue these defects.
The near complete rescue of sNHE-null spermatozoa motility by cAMP analogs suggests that cAMP metabolism is impaired in the sNHE-null spermatozoa. In this study, we strove to elucidate the role of sNHE in the cAMP signaling pathway of spermatozoa by analyzing the expression and function of sAC in the sNHE-null sperm cells. We report evidence that sNHE and sAC are associated in a signaling complex that is essential for the generation of the cAMP second messenger in spermatozoa. Furthermore, we successfully express sNHE on the plasma membrane of mammalian somatic cells, showing that it is a functional NHE.
Results
sNHE-Null Spermatozoa Do Not Show Capacitation-Induced Protein Tyrosine Phosphorylation.
Cauda epididymal mouse sperm undergo tyrosine phosphorylation on a distinct set of proteins in a time-dependent manner when incubated in capacitating medium containing calcium, bicarbonate, and albumin (20, 21). Sperm cells from wild-type animals showed typical patterns of protein tyrosine phosphorylation after incubation in capacitating medium reaching maximal levels by 60 min [supporting information (SI) Fig. 7A Left]. However, in sNHE-null spermatozoa, capacitation-induced tyrosine phosphorylation failed to occur even though tyrosine phosphorylation of hexokinase (22) was normal (SI Fig. 7A Right). The addition of a cAMP analog (Sp-cAMP) and a phosphodiesterase inhibitor (3-isobutyl-1-methylxanthine) restored protein tyrosine phosphorylation of sNHE-null sperm cells to a pattern similar to that of wild-type spermatozoa (SI Fig. 7B). Similar results were obtained with all biologically active cAMP analogs tested in combination with 3-isobutyl-1-methylxanthine (data not shown).
sNHE-Null Spermatozoa Have Reduced Basal cAMP Levels That Are Insensitive to Bicarbonate Stimulation.
Because both the motility defect and the lack of induced protein tyrosine phosphorylation are rescued by cell-permeable cAMP analogs, we reasoned that cAMP metabolism may be impaired in spermatozoa lacking sNHE. In fact, basal cAMP concentrations were lower in sNHE-null spermatozoa compared with wild-type sperm cells (Fig. 1A). When caudal epididymal spermatozoa were diluted into bicarbonate-containing medium, the cAMP concentrations of wild-type spermatozoa rapidly increased (<1 min). Cellular cAMP concentrations subsequently decreased within 10 min (presumably due to sperm phosphodiesterase activity) but then rose again, reaching maximum levels by 60 min. (Fig. 1B). Unlike wild-type cells, cAMP concentrations in the sNHE-null sperm cells failed to increase in response to bicarbonate (Fig. 1B).
Fig. 1.
sNHE-null spermatozoa have reduced basal cAMP levels that are insensitive to bicarbonate stimulation. (A) Sperm cells from wild-type (Wt) and sNHE-null mice were collected in a bicarbonate-free medium. cAMP content was measured by using RIA. Data represent the mean ± SD (n = 4). (B) Wild-type and sNHE-null spermatozoa were diluted in a bicarbonate-containing medium and incubated at 37°C. At each indicated time point, an aliquot of sperm was collected and analyzed for cAMP content. Data represent the mean ± SD (n = 3).
Adenylyl Cyclase Activity Is Reduced in sNHE-Null Spermatozoa but Remains Sensitive to Stimulation by Bicarbonate in Broken Cell Lysates.
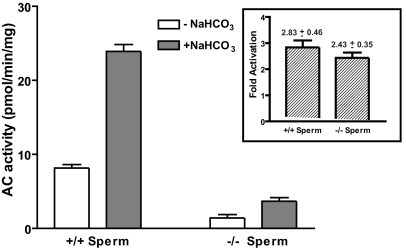
A number of causes could account for the failure of bicarbonate to elevate cAMP in sNHE-null spermatozoa, including extremely low adenylyl cyclase ATP substrate availability, defective bicarbonate transport, or abnormally high cellular phosphodiesterase activity. Null spermatozoa contain normal ATP concentrations when incubated in bicarbonate-free medium and the phosphodiesterase activity (i.e., cAMP hydrolysis) measured in vitro was lower in sNHE-null sperm cells compared with wild-type cells (SI Fig. 8 and SI Materials and Methods) indicating other causes. We then examined the in vitro adenylyl cyclase activity in broken cell lysates for wild-type and sNHE-null spermatozoa. We found that sAC activity was greatly reduced in the null cell lysates, suggesting a defect at the level of the sAC protein (Fig. 2). Interestingly, the residual sAC enzymatic activity in the cell lysates of sNHE-null spermatozoa demonstrated an ≈2-fold stimulation by bicarbonate, as was true for the sAC activity in the cell lysates of wild-type spermatozoa (Fig. 2 Inset). The equivalent sensitivity to bicarbonate stimulation of the sNHE-null sperm cell sAC activity measured in vitro contrasted with the inability of bicarbonate to elevate cAMP levels in the intact sNHE-null spermatozoa (Fig. 1B).
Fig. 2.
Bicarbonate-stimulated soluble adenylyl cyclase (AC) activity is reduced in sNHE-null spermatozoa. Whole-sperm-cell lysates from wild-type or sNHE-null mice were used to estimate in vitro adenylyl cyclase activity in the presence of 40 mM NaCl or 40 mM NaHCO3. Data represent mean ± SD (n = 4). (Inset) Fold activation by bicarbonate in each experiment was calculated. Data represent mean ± SD (n = 4).
sNHE-Null Spermatozoa Contain Undetectable Amounts of Full-Length sAC.
The reduced sAC activity in sNHE-null spermatozoa suggested that sNHE is important for the expression, stabilization, or regulation of the sAC enzyme. A Northern blot revealed no differences in the amount of sAC transcripts from sNHE-null and wild-type testes (Fig. 3A). In wild-type sperm cells, a monoclonal antibody to sAC (R21) (23) detected two major proteins: a Mr ≈180,000 full-length sAC (sACf1) and a Mr ≈50,000 truncated sAC (sACt1) (Fig. 3B), likely representing the two isoforms of sAC reported previously (9, 15). Although the sACt1 content of sNHE-null spermatozoa was unchanged relative to wild-type spermatozoa, as confirmed by using a second monoclonal antibody (R41; data not shown), the sACf1 was greatly diminished or absent. In both mouse and rat testes, alternative splicing produces two ≈5.2-kb sAC transcripts that differ by a 56-bp exon, with one transcript producing premature termination of the ORF (15). To determine whether the sAC mRNA encoding the longer sAC isoform (designated full-length) existed in the sNHE-null testes, we performed real-time PCR with a primer overlapping the exon present only in the full-length sAC cDNA and found that sNHE-null and wild-type testes contain similar levels of full-length sAC mRNA (Fig. 3C). Recently, other variants of the human sAC were reported (16). A polyclonal antibody to the human sAC revealed another truncated form of sAC (sACt2) in both wild-type and null mouse spermatozoa (Fig. 3D). The specificity of the sACt2 signal was confirmed by competition of antibody binding with a specific blocking peptide (data not shown). The intensity of sACt2 also was unchanged in the sNHE-null spermatozoa (Fig. 3D). Therefore, sNHE-null spermatozoa appear to possess normal levels of truncated sAC isoforms but lack the full-length protein. Because there is no difference in the amount of full-length sAC-encoding transcript between the null and wild-type testes, sNHE expression appears to regulate sACf1 expression at the protein level.
Fig. 3.
The sNHE-null spermatozoa contain no detectable full-length sAC protein. (A) Northern blot of total RNA (10 μg each) from wild-type, heterozygous, and sNHE-null testes with a mouse sAC cDNA probe (Upper) or a cyclophilin cDNA probe (Lower). (B) Immunoblot of total sperm cell lysate (2 × 106 cells) from wild-type or sNHE-null mice with R21 monoclonal antibody (200 ng/ml). (C) Full-length sAC transcript exists in sNHE-null testes. (Upper) Location of the sense primer (framed) relative to the sAC full-length cDNA. The gray letters show 29 of the 56 nucleotides that are missing in truncated sAC cDNA. (Lower) Relative sACfl mRNA levels in wild-type (+/+) and sNHE-null (−/−) testes determined by real-time PCR. Expression of sACfl mRNA was normalized to 18S rRNA and is reported relative to that of wild-type testis. Data represent mean ± SD (n = 3). (D) The same blot in B was stripped and reprobed with anti-human sAC polyclonal antibody (1:5,000).
Protein–Protein Interaction and Codependence of Expression for sNHE and sAC.
The requirement of sNHE for the normal expression and apparent function of sAC (Figs. 1B and 3B) suggests that sNHE and sAC may interact in spermatozoa. Therefore, their coexpression might facilitate heterologous expression. Coexpression of sACf1 did not detectably affect sNHE expression (data not shown). However, sNHE that had failed to express in past experiments was detected in HEK293F cells coexpressing sACt1, the truncated form of sAC with high specific activity (Fig. 4A, lane 2). To further enhance sNHE expression, we hypothesized that the N-terminal sequence of NHE1, which is primarily a plasma membrane protein in mammalian cells, may increase plasma membrane targeting of sNHE. Indeed, replacement of the first transmembrane domain of sNHE with the first three transmembrane domains of the mouse NHE1 produced a chimeric construct that was successfully expressed alone and even more efficiently when cotransfected with sACt1 (Fig. 4A, lanes 3 and 4).
Fig. 4.
Reciprocal facilitation of expression of sNHE and sAC in HEK293F cells. (A) Coexpression of the V5-tagged sACt1 enhanced expression of the myc-tagged full-length sNHE (sNHE) and the chimeric sNHE [N(1–3)s] in HEK293F cells as determined by immunoblot with anti-myc (1 μg/ml) (Top), anti-V5 (200 ng/ml) (Middle), and anti-tubulin (100 ng/ml) (Bottom) antibodies. (B) Wild-type and chimeric sNHE enhance V5-tagged sACfl protein expression in HEK293F cells. (Upper) Total cell lysates (80 μg per sample) were used for immunoblotting with anti-V5 antibody. (Lower) The same blot was stripped and reprobed with anti-tubulin antibody. (C) sNHE enhances V5-tagged sACt1 protein expression in HEK293F cells. (Upper) Total cell lysates (80 μg per sample) were used for immunoblotting with anti-V5 antibody. (Lower) Total cell lysates (50 μg per sample) were used for immunoblotting with anti-tubulin antibody. (B and C) The relative intensity of each sAC band was quantified with NIH ImageJ software.
sAC protein expression was also enhanced by coexpression of sNHE (Fig. 4 B and C). Both sNHE and the chimeric sNHE increased sACf1 levels (Fig. 4B, lanes 4–7), whereas coexpression of NHE1 (Fig. 4B, lanes 8–9) or another sperm-specific protein sCklfsf-2b (data not shown) did not affect sACf1 protein levels. The sACt1 protein level also was increased when the cells coexpressed sNHE protein (Fig. 4C, lanes 4 and 5). As a control, coexpression of sCKlfsf-2b (sCKlf in the Fig. 4C, lanes 6 and 7) or NHE1 (data not shown) did not affect the sACt1 protein level.
We then examined the interaction between sNHE and sAC by coimmunoprecipitation experiments using transfected HEK293F cells. The myc-tagged chimeric sNHE plasmid was cotransfected in the cells along with empty vector, V5-tagged sACf1, or V5-tagged sACt1. The immunocomplexes precipitated by anti-V5 contained myc-tagged chimeric sNHE, suggesting that both forms of sAC interact with the chimeric sNHE (Fig. 5A Left). The reciprocal coimmunoprecipitation also worked: myc-tagged chimeric sNHE protein can pull-down V5-tagged sACf1 protein (Fig. 5A Right).
Fig. 5.
sNHE and sAC coimmunoprecipitate from coexpressing cultured cells and spermatozoa. (A) Full-length chimeric sNHE protein associates with sAC proteins. (Left) myc-tagged chimeric sNHE was expressed in HEK293F cells coexpressing empty vector, V5-tagged sACfl, or V5-tagged sACt1. Total cell lysates were subject to immunoprecipitation (IP) with anti-V5 antibody, and the captured proteins were analyzed by immunoblot (IB) with anti-Myc antibody. (Right) V5-tagged sACfl was expressed in HEK293F cells coexpressing empty vector or myc-tagged chimeric sNHE. Total cell lysates were subject to immunoprecipitation with anti-myc antibody, and the captured proteins were analyzed by immunoblot with anti-V5 antibody. (B) C-terminal tail of sNHE interacts with sACt1 protein. Myc-tagged C-terminal tails of sNHE were expressed in HEK293F cells with or without V5-tagged sACt1. (Upper) Total cell lysates were immunoprecipitated with anti-V5 antibody, and the precipitated proteins were analyzed by immunoblot with anti-myc antibody. (Lower) Crude total cell lysates were analyzed by immunoblot with anti-myc antibody. (C) sNHE interacts with sAC in sperm cells. Sperm cell lysate was immunoprecipitated with anti-sNHE polyclonal IgG (lane 1) or preimmune IgG (lane 2). The precipitated proteins were analyzed by immunoblot with anti-human sAC polyclonal antibody.
We also coexpressed myc-tagged truncated sNHE proteins, sNHEC1, or sNHEC2 in HEK293F cells with V5-tagged sACt1. sNHEC1 contains the last 230 aa of sNHE and sNHEC2 contains the entire C-terminal tail of the protein plus the last two predicted transmembrane segments. Although the level of sNHEC1 protein expression is much higher than that of sNHEC2 (Fig. 5B Lower), both proteins were present in the protein complexes captured by anti-V5 antibody (Fig. 5B Upper, lanes 2 and 4), suggesting that the C-terminal tail of the sNHE protein interacts with the catalytic region of the sAC protein.
The interaction between native sNHE and sAC was then examined in sperm cell lysates. A polyclonal antibody to sNHE coimmunoprecipitated a truncated form of sAC (sACt2) and as a negative control, the preimmune IgG from the same rabbit did not (Fig. 5C). Because of the limited quantities of R21 and R41 antibodies, we were unable to confirm the interaction between native sNHE and sACf1 or sACt1.
sNHE Is a NHE.
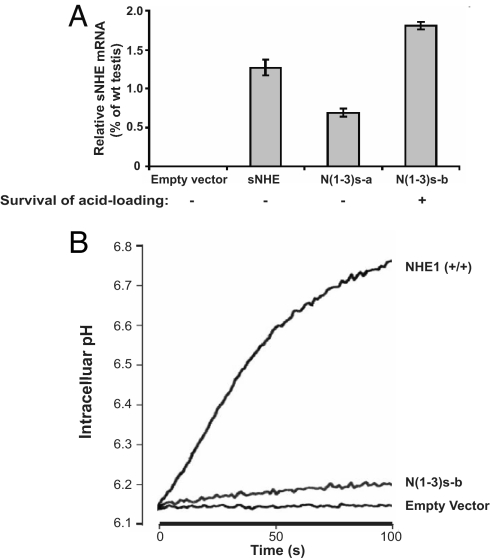
Now that we could express detectable sNHE protein in HEK293F cells (Fig. 4A) with the chimeric sNHE protein localized to the cell plasma membrane (SI Fig. 9 and SI Materials and Methods), we determined whether sNHE has NHE activity by using a NHE-null fibroblast cell line derived from NHE1−/− mice (O.W.M. and P.M., unpublished data). Expression vectors encoding sNHE or chimeric sNHE were stably transfected into these cells, and the synthesis of sNHE mRNA was confirmed by RT-PCR (data not shown) and real-time PCR (Fig. 6A). We detected evidence of protein expression after immunoprecipitation only in the stable clones with the highest level of sNHE mRNA [N (1–3)s-b; data not shown]. These cells survived 1 month of repetitive acid loading (24–26) that killed control NHE-deficient cells. The NHE activity of this stable cell line was also measured fluorimetrically by using the pH-sensitive fluorescent dye BCECF. The negative control cells showed no NHE activity, whereas the cells expressing chimeric sNHE displayed modest but consistent NHE activity (Fig. 6B).
Fig. 6.
sNHE is a NHE. (A) Detection of sNHE mRNA in stable cell lines. NHE-null cells were stably transfected with empty vector, myc-tagged sNHE, or myc-tagged chimeric sNHE constructs. sNHE mRNA in these cells was determined by real-time PCR and compared with that of wild-type mouse testis. The stable cells were acid-loaded repetitively, and the result is shown below the graph. (B) The stable cells expressing the highest sNHE message displayed NHE activity. Representative tracings of sodium-dependent pH recovery after an acid load in NHE1-expressing fibroblasts and in NHE-null fibroblasts stably transfected with either chimeric sNHE construct or empty vector. The experiment was repeated three times with three to four replicates per condition.
Discussion
The disruption of the sNHE gene causes absolute male infertility and the complete loss of sperm motility. Our new findings indicate that these effects are the consequence of an intimate interaction of sNHE and sAC in spermatozoa. The absence of sNHE severely reduces adenylyl cyclase activity in spermatozoa providing an explanation for the similarities between the sNHE and sAC knockout mouse models.
sNHE appears able to directly bind to at least three apparent sAC isoforms present in spermatozoa. The stable expression of the full-length sAC protein in mouse epididymal spermatozoa requires sNHE expression, suggesting that at least these two proteins are components of a signaling complex. This model is consistent with the colocalization and association of the sNHE and sAC orthologs in the sea urchin sperm cell (27, 28) and our own data (unpublished observations) localizing sAC on the flagellar principal piece. In contrast, two truncated sAC proteins (sACt1 and sACt2) appear to be present at comparable levels in the wild-type and sNHE-null spermatozoa. sACt2 is predicted to contain just one of the two catalytic domains necessary for cyclase activity and so is unlikely to be an active enzyme. However, as measured in vitro from overexpressing cells or testicular immunoprecipitates (9, 15), sACt1 is an active cyclase with a specific activity >10-fold higher than the full-length sAC. Despite the normal amounts of sACt1, sNHE-null spermatozoa exhibit greatly reduced adenylyl cyclase activity. One possible explanation for this observation is that the correct assembly and presence of the apparent sNHE–sACf1 complex is required for the adenylyl cyclase activity of the sACt1 isoform in epididymal spermatozoa. Alternatively, the catalytically inactive sACt2 may inhibit the cyclase activity through undefined mechanisms. Our data suggest that the in vivo regulation of the different sAC isoforms in spermatozoa is more complicated than previously appreciated and therefore remains an important topic for future study.
We have shown that the sAC activity present in both wild-type and sNHE-null sperm cell lysates is similarly responsive to bicarbonate exhibiting ≈2-fold stimulation over their respective basal specific activities. However, unlike wild-type spermatozoa, intact sNHE-null sperm cells do not elevate cAMP content in response to bicarbonate. Therefore, sNHE may directly or indirectly affect sperm cell bicarbonate transport. One hypothesis is that as CO2 diffuses into the cell and equilibrates as H+ and HCO3−, sNHE may exchange intracellular H+ for extracellular Na+ to facilitate bicarbonate uptake as reported in both NHE1 and NHE3 knockout mice (29, 30). Alternatively, it is possible that the loss of sNHE reduces the activity of a sperm bicarbonate transporter such as the Na+/HCO3− cotransporter reported in mouse and sea urchin sperm (31, 32).
Using two different methods and NHE-null fibroblasts, we have demonstrated that sNHE is a functional NHE. Because of the extremely low expression of sNHE and chimeric sNHE in this cell line, we were unable to distinguish whether sNHE has low exchange activity compared with other NHEs or simply poor surface expression and whether sAC/cAMP increases sNHE exchange activity or only its expression. Previous data suggest that mammalian sperm cell pH is predominantly set by the activity of Na+-dependent Cl−-HCO3− exchanger or Na+/HCO3− cotransporter as measured by using pH-sensitive fluorescent compounds (31, 33). We propose that sNHE may have a spatially restricted role controlling localized cellular pH changes predominantly in the flagellar principal piece that are not detected with fluorescent reporters. Therefore, sNHE may also contribute to the regulation of sAC activity as a result of the pH dependence of this enzyme (19, 34). Consistent with this postulated role for sNHE in compartmentalized flagellar pH regulation is the apparent inability of sNHE-null spermatozoa to hyperactivate after basal motility and the phosphotyrosine cascade are restored with cell-permeable cAMP analogs (our unpublished observations). Because the functional alkaline-activated CatSper channels required for motility hyperactivation are present in the sNHE-null spermatozoa, the absence of hyperactivation could represent the failure of intracellular alkalinization in the flagella of these cells (35).
In conclusion, we have presented evidence that a molecular complex including both sNHE and sAC is assembled in mouse spermatozoa. sNHE appears to modulate cellular sAC activity on multiple levels, including the stable expression of sACf1, the regulation of bicarbonate transport possibly through an associated transporter, and by modulating local pH for optimal sAC activity. In a reciprocal manner, sNHE activity and/or ion-exchange function(s) may be modulated by sAC activity through the putative cyclic nucleotide binding motif in the sNHE cytoplasmic carboxyl tail. Finally, carbonic anhydrase II binds to the cytoplasmic carboxyl tail of NHE1 (36). We propose that a similar association of carbonic anhydrase II with the sNHE cytoplasmic tail may exist. Although speculative, such a signaling complex could very efficiently regulate intracellular pH, bicarbonate, and cAMP levels, enabling the rapid and precise control of both sAC and sNHE activities to facilitate sperm motility regulation.
Materials and Methods
Immunoblot and Northern Blot.
Immunoblotting was performed as described (7). R21 and R41 antibodies were generous gifts from Lonny Levin and Jochen Buck (both from Cornell University Medical School, New York, NY). For antiphosphotyrosine immunoblotting, the sperm samples were prepared as described by Visconti et al. (20). The PY-Plus monoclonal antibody was purchased from Zymed (San Francisco, CA), anti-myc monoclonal antibody was purchased from Upstate BioTechnology (Lake Placid, NY), anti-FLAG polyclonal antibody and anti-α-tubulin antibody were purchased from Sigma (St. Louis, MO), and anti-V5 antibody was purchased from Invitrogen (Carlsbad, CA). Restore Western blot stripping buffer was purchased from Pierce (Rockford, IL). Quantification of the sAC immunoreactivity by densitometry was performed with NIH ImageJ software.
Northern blot analysis was performed as reported (37). The membrane was probed with a random-primed 32P-labeled probe (Amersham Biosciences) corresponding to nucleotides 220–2,060 of the mouse soluble adenylyl cyclase cDNA (National Center for Biotechnology Information accession no. NM_173029) or nucleotides 33–528 of the cyclophilin A cDNA (National Center for Biotechnology Information accession no. 9BC083076).
cAMP RIA.
Caudal epididymal spermatozoa were released into a modified bicarbonate-free Krebs–Ringer medium (38) at 37°C. After being washed, the cells (2 × 106 to 5 × 106 cells per sample) were incubated in the same medium with 25 mM bicarbonate for 0–90 min at 37°C under 95% air/5% CO2. The reactions were stopped at different time points by the addition of an equal volume of 1 M perchloric acid. After five freeze/thaw cycles, cAMP in the supernatant fluid (12,000 × g for 10 min) was purified and quantified by RIA (39).
In Vitro Adenylyl Cyclase Assay.
Washed caudal epididymal spermatozoa were resuspended in buffer containing 50 mM Tris·HCl (pH 7.5), 1 mM DTT, 1× protease inhibitor mixture (Roche). The cell suspension was sonicated 10 times for 10 sec with 1-min intervals on ice. After centrifugation at 13,000 × g for 10 min, the supernatant fluid was used for the adenylyl cyclase assay as described in refs. 15 and 17 with some modifications. Briefly the sperm cell lysates (10–50 μg) were incubated in a reaction buffer containing 40 mM Tris·HCl (pH 7.5), 6 mM MgCl2, 2 mM ATP, 2 mM DTT, 10 mM phosphoenol pyruvate, 3 units of pyruvate kinase, and 200 μM 3-isobutyl-1-methylxanthine with 40 mM NaHCO3 or 40 mM NaCl for 30 min at 30°C. Reactions were terminated by adding 0.5 ml of 110 mM ZnAc2, followed by addition of 0.5 ml of 110 mM Na2CO3. The samples were frozen and thawed, and the supernatant fluid (2,500 × g for 30 min) was fractionated over Dowex and alumina columns (40). cAMP in the eluant was quantified by RIA as described above. In all cases, cAMP formation was linear with time and protein concentration.
Quantitative Real-Time PCR.
Real-time PCR was performed in 20-μl reactions by using iTaq SYBR green supermix (Bio-Rad, Hercules, CA) on the Prism 7900 HT Sequence Detection System (Applied Biosystems, Foster City, CA). Signals were normalized relative to the housekeeping gene 18S-rRNA. The primers used are listed in SI Table 1. Standard curves were generated by using serially diluted plasmids containing amplified cDNAs (2 × 102 to 2 × 106 copies per reaction). The copy number of each mRNA was compared with that of wild-type mouse testis.
Plasmid Construction and Cell Transfections.
Details of the plasmids used in this study are provided in SI Table 2. For better transient expression, the inserts of pcDNA3.1/sNHE-myc and pcDNA3.1/N (1–3)sNHE-myc were subcloned into a minimal vector pCMV (41).
HEK293F cells were maintained in DMEM. NHE-null cells were maintained in DMEM/F12(50%):DMEM with low glucose (50%). All media were supplemented with 10% FBS and 1× antibiotic–antimycotic (Invitrogen). Cells were transfected with Lipofectamine 2000 (Invitrogen). Transiently transfected cells were harvested 24 or 48 h later for analysis.
Stably transfected cells were split 2 days after transfection and maintained in a medium containing Geneticin (Invitrogen) for at least 3 weeks. Individual clones were picked and analyzed. As a negative control, cells were transfected with empty pcDNA3.1(+) vector.
Immunoprecipitation.
Transfected HEK293F cells were lysed in a buffer containing 50 mM Tris·HCl (pH 7.5), 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, 0.1% SDS, and 1× complete protease inhibitor mixture (Roche). Cell lysate was first precleared with normal mouse or rabbit IgG and protein A/G PLUS-agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA) then immunoprecipitated with the appropriate antibody and protein A/G PLUS–agarose at 4°C overnight. After brief centrifugation, the beads were washed with the lysis buffer six times over 3 h. After the last wash, the proteins bound to the beads were solubilized in 2× Laemmli sample buffer (Bio-Rad). After denaturation at 65°C for 10 min, the immunoprecipitates were separated on 4–20% SDS/PAGE gradient gels (ISC Bioexpress, Kaysville, UT) and analyzed by immunoblot.
When using sperm lysate, total sperm protein (500 μg) was immunoprecipitated with 10 μg of anti-sNHE IgG. As a control, the same amount of sperm protein was incubated with 10 μg of preimmune IgG purified from the same rabbit.
Acid-Loading of Stable Cells.
Stably transfected cells were acid loaded as reported (24–26). This selection was repeated every other day over a 3- to 4-week period.
NHE Activity Assay.
NHE activity of the stably transfected cells was measured fluorimetrically by using the pH-sensitive dye BCECF-AM as described previously (42). NHE1 wild-type fibroblasts were used as a positive control.
Statistical Analysis.
Data in Fig. 1 and SI Fig. 8 were analyzed with Student's t test. All experiments were performed in accordance with the animal protocols approved by University of Texas Southwestern Medical Center Institutional Animal Care and Use Committee.
Supplementary Material
Acknowledgments
This paper is dedicated to the memory of Prof. David L. Garbers, a great scientist, mentor, and colleague. We thank Drs. Lonny Levin and Jochen Buck for providing R21 and R41 antibodies; Drs. Valeria Vasta, Donner Babcock, and Joseph Beavo for advice on the phosphodiesterase assay; Dr. Ted Chrisman for RIA advice; Dr. Weidong Geng for providing human sAC antibody; Dr. Daniel Fuster for providing pcDNA3.1/NHE1-FLAG plasmid; Dr. Timothy Megraw for help with confocal microscopy; and Dr. Donner Babcock for critically reading the manuscript and for support. This work was supported by National Institute of Health Grants HD-36022, DK-54396, and DK-48482, the Howard Hughes Medical Institute, the Charles and Jane Pak center of Mineral Metabolism and Clinical Research, and the Cecil H. and Ida Green Center for Reproductive Biology Sciences.
Abbreviations
- NHE
Na+/H+ exchanger
- sNHE
sperm-specific NHE
- sAC
soluble adenylyl cyclase
- sACt
truncated sAC
- sACfl
full-length sAC.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611296104/DC1.
References
- 1.Garcia MA, Meizel S. Mol Reprod Dev. 1999;52:189–195. doi: 10.1002/(SICI)1098-2795(199902)52:2<189::AID-MRD10>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 2.Schackmann RW, Chock PB. J Biol Chem. 1986;261:8719–8728. [PubMed] [Google Scholar]
- 3.Woo AL, James PF, Lingrel JB. Mol Reprod Dev. 2002;62:348–356. doi: 10.1002/mrd.90002. [DOI] [PubMed] [Google Scholar]
- 4.Bell SM, Schreiner CM, Schultheis PJ, Miller ML, Evans RL, Vorhees CV, Shull GE, Scott WJ. Am J Physiol. 1999;276:C788–C795. doi: 10.1152/ajpcell.1999.276.4.C788. [DOI] [PubMed] [Google Scholar]
- 5.Brett CL, Donowitz M, Rao R. Am J Physiol. 2005;288:C223–C239. doi: 10.1152/ajpcell.00360.2004. [DOI] [PubMed] [Google Scholar]
- 6.Orlowski J, Grinstein S. Pflugers Arch. 2004;447:549–565. doi: 10.1007/s00424-003-1110-3. [DOI] [PubMed] [Google Scholar]
- 7.Wang D, King SM, Quill TA, Doolittle LK, Garbers DL. Nat Cell Biol. 2003;5:1117–1122. doi: 10.1038/ncb1072. [DOI] [PubMed] [Google Scholar]
- 8.Yanagimachi R. In: The Physiology of Reproduction. Knobil E, Neill JD, editors. New York: Raven; 1994. pp. 189–317. [Google Scholar]
- 9.Buck J, Sinclair ML, Schapal L, Cann MJ, Levin LR. Proc Natl Acad Sci USA. 1999;96:79–84. doi: 10.1073/pnas.96.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Y, Cann MJ, Litvin TN, Iourgenko V, Sinclair ML, Levin LR, Buck J. Science. 2000;289:625–628. doi: 10.1126/science.289.5479.625. [DOI] [PubMed] [Google Scholar]
- 11.Hyne RV, Garbers DL. Biol Reprod. 1979;21:1135–1142. doi: 10.1095/biolreprod21.5.1135. [DOI] [PubMed] [Google Scholar]
- 12.Jaiswal BS, Conti M. Proc Natl Acad Sci USA. 2003;100:10676–10681. doi: 10.1073/pnas.1831008100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Litvin TN, Kamenetsky M, Zarifyan A, Buck J, Levin LR. J Biol Chem. 2003;278:15922–15926. doi: 10.1074/jbc.M212475200. [DOI] [PubMed] [Google Scholar]
- 14.Garbers DL, Tubb DJ, Hyne RV. J Biol Chem. 1982;257:8980–8984. [PubMed] [Google Scholar]
- 15.Jaiswal BS, Conti M. J Biol Chem. 2001;276:31698–31708. doi: 10.1074/jbc.M011698200. [DOI] [PubMed] [Google Scholar]
- 16.Geng W, Wang Z, Zhang J, Reed BY, Pak CY, Moe OW. Am J Physiol. 2005;288:C1305–C1316. doi: 10.1152/ajpcell.00584.2004. [DOI] [PubMed] [Google Scholar]
- 17.Esposito G, Jaiswal BS, Xie F, Krajnc-Franken MA, Robben TJ, Strik AM, Kuil C, Philipsen RL, Van Duin M, Conti M, et al. Proc Natl Acad Sci USA. 2004;101:2993–2998. doi: 10.1073/pnas.0400050101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hess KC, Jones BH, Marquez B, Chen Y, Ord TS, Kamenetsky M, Miyamoto C, Zippin JH, Kopf GS, Suarez SS, et al. Dev Cell. 2005;9:249–259. doi: 10.1016/j.devcel.2005.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie F, Garcia MA, Carlson AE, Schuh SM, Babcock DF, Jaiswal BS, Gossen JA, Esposito G, van Duin M, Conti M. Dev Biol. 2006;296:353–362. doi: 10.1016/j.ydbio.2006.05.038. [DOI] [PubMed] [Google Scholar]
- 20.Visconti PE, Bailey JL, Moore GD, Pan D, Olds-Clarke P, Kopf GS. Development (Cambridge, UK) 1995;121:1129–1137. doi: 10.1242/dev.121.4.1129. [DOI] [PubMed] [Google Scholar]
- 21.Visconti PE, Moore GD, Bailey JL, Leclerc P, Connors SA, Pan D, Olds-Clarke P, Kopf GS. Development (Cambridge, UK) 1995;121:1139–1150. doi: 10.1242/dev.121.4.1139. [DOI] [PubMed] [Google Scholar]
- 22.Kalab P, Visconti P, Leclerc P, Kopf GS. J Biol Chem. 1994;269:3810–3817. [PubMed] [Google Scholar]
- 23.Zippin JH, Chen Y, Nahirney P, Kamenetsky M, Wuttke MS, Fischman DA, Levin LR, Buck J. FASEB J. 2003;17:82–84. doi: 10.1096/fj.02-0598fje. [DOI] [PubMed] [Google Scholar]
- 24.Boron WF, De Weer P. J Gen Physiol. 1976;67:91–112. doi: 10.1085/jgp.67.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franchi A, Cragoe E, Jr, Pouyssegur J. J Biol Chem. 1986;261:14614–14620. [PubMed] [Google Scholar]
- 26.Sardet C, Franchi A, Pouyssegur J. Cell. 1989;56:271–280. doi: 10.1016/0092-8674(89)90901-x. [DOI] [PubMed] [Google Scholar]
- 27.Nomura M, Vacquier VD. Cell Motil Cytoskeleton. 2006;63:582–590. doi: 10.1002/cm.20147. [DOI] [PubMed] [Google Scholar]
- 28.Bookbinder LH, Moy GW, Vacquier VD. J Cell Biol. 1990;111:1859–1866. doi: 10.1083/jcb.111.5.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schultheis PJ, Clarke LL, Meneton P, Miller ML, Soleimani M, Gawenis LR, Riddle TM, Duffy JJ, Doetschman T, Wang T, et al. Nat Genet. 1998;19:282–285. doi: 10.1038/969. [DOI] [PubMed] [Google Scholar]
- 30.Good DW, Watts BA, III, George T, Meyer JW, Shull GE. Am J Physiol. 2004;287:F1244–F1249. doi: 10.1152/ajprenal.00176.2004. [DOI] [PubMed] [Google Scholar]
- 31.Demarco IA, Espinosa F, Edwards J, Sosnik J, De La Vega-Beltran JL, Hockensmith JW, Kopf GS, Darszon A, Visconti PE. J Biol Chem. 2003;278:7001–7009. doi: 10.1074/jbc.M206284200. [DOI] [PubMed] [Google Scholar]
- 32.Gunaratne HJ, Nomura M, Moy GW, Vacquier VD. Gene. 2006;375:37–43. doi: 10.1016/j.gene.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 33.Zeng Y, Oberdorf JA, Florman HM. Dev Biol. 1996;173:510–520. doi: 10.1006/dbio.1996.0044. [DOI] [PubMed] [Google Scholar]
- 34.Nomura M, Beltran C, Darszon A, Vacquier VD. Gene. 2005;353:231–238. doi: 10.1016/j.gene.2005.04.034. [DOI] [PubMed] [Google Scholar]
- 35.Qi H, Moran MM, Navarro B, Chong JA, Krapivinsky G, Krapivinsky L, Kirichok Y, Ramsey IS, Quill TA, Clapham DE. Proc Natl Acad Sci USA. 2007;104:1219–1223. doi: 10.1073/pnas.0610286104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li X, Alvarez B, Casey JR, Reithmeier RA, Fliegel L. J Biol Chem. 2002;277:36085–36091. doi: 10.1074/jbc.M111952200. [DOI] [PubMed] [Google Scholar]
- 37.Quill TA, Ren D, Clapham DE, Garbers DL. Proc Natl Acad Sci USA. 2001;98:12527–12531. doi: 10.1073/pnas.221454998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Si Y, Olds-Clarke P. Biol Reprod. 2000;62:1231–1239. doi: 10.1095/biolreprod62.5.1231. [DOI] [PubMed] [Google Scholar]
- 39.Domino SE, Tubb DJ, Garbers DL. Methods Enzymol. 1991;195:345–355. doi: 10.1016/0076-6879(91)95179-n. [DOI] [PubMed] [Google Scholar]
- 40.Johnson RA, Salomon Y. Methods Enzymol. 1991;195:3–21. doi: 10.1016/0076-6879(91)95150-i. [DOI] [PubMed] [Google Scholar]
- 41.Orlowski J. J Biol Chem. 1993;268:16369–16377. [PubMed] [Google Scholar]
- 42.Bobulescu IA, Dwarakanath V, Zou L, Zhang J, Baum M, Moe OW. Am J Physiol. 2005;289:F685–F691. doi: 10.1152/ajprenal.00447.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.