Abstract
We investigated autotrophic anaerobic ammonium-oxidizing (anammox) biofilms for their spatial organization, community composition, and in situ activities by using molecular biological techniques combined with microelectrodes. Results of phylogenetic analysis and fluorescence in situ hybridization (FISH) revealed that “Brocadia”-like anammox bacteria that hybridized with the Amx820 probe dominated, with 60 to 92% of total bacteria in the upper part (<1,000 μm) of the biofilm, where high anammox activity was mainly detected with microelectrodes. The relative abundance of anammox bacteria decreased along the flow direction of the reactor. FISH results also indicated that Nitrosomonas-, Nitrosospira-, and Nitrosococcus-like aerobic ammonia-oxidizing bacteria (AOB) and Nitrospira-like nitrite-oxidizing bacteria (NOB) coexisted with anammox bacteria and accounted for 13 to 21% of total bacteria in the biofilms. Microelectrode measurements at three points along the anammox reactor revealed that the NH4+ and NO2− consumption rates decreased from 0.68 and 0.64 μmol cm−2 h−1 at P2 (the second port, 170 mm from the inlet port) to 0.30 and 0.35 μmol cm−2 h−1 at P3 (the third port, 205 mm from the inlet port), respectively. No anammox activity was detected at P4 (the fourth port, 240 mm from the inlet port), even though sufficient amounts of NH4+ and NO2− and a high abundance of anammox bacteria were still present. This result could be explained by the inhibitory effect of organic compounds derived from biomass decay and/or produced by anammox and coexisting bacteria in the upper parts of the biofilm and in the upstream part of the reactor. The anammox activities in the biofilm determined by microelectrodes reflected the overall reactor performance. The several groups of aerobic AOB lineages, Nitrospira-like NOB, and Betaproteobacteria coexisting in the anammox biofilm might consume a trace amount of O2 or organic compounds, which consequently established suitable microenvironments for anammox bacteria.
Anaerobic ammonium oxidation (anammox) is a microbiological oxidation of ammonium, with nitrite as the electron acceptor and dinitrogen gas as the main product, and is mediated by a group of deep-branching Planctomycete-like bacteria (12, 39). Anammox bacteria have been detected in different wastewater treatment facilities and environments in the world (for a review, see reference 34). Currently, four genera of anammox bacteria have been reported and named, including Brocadia, Kuenenia, Scalindua, and Anammoxoglobus (14, 34).
The anammox process is a new and promising alternative to conventional nitrogen removal processes. A better understanding of the ecophysiology (i.e., microbial community structure and in situ activity) of anammox bacteria in complex biofilms is essential for implementing the anammox process as a manageable and reliable nitrogen removal process in wastewater treatment. However, related information is limited because anammox bacteria are strict anaerobic autotrophs and thus have not yet been isolated in pure culture.
The community structures and in situ activities of nitrifying biofilms (23, 36), sulfate-reducing biofilms (11, 24), and phosphate-removing biofilms (10) have been investigated extensively by using a combined microelectrode and fluorescence in situ hybridization (FISH) approach. However, only a few studies have been carried out to study in situ anammox activity, including a lab-scale anammox reactor study and a study with subtropical mangrove sediments, using microelectrodes (20, 22). Recent molecular biological techniques have also been used to analyze in situ microbial community structures in various oxygen-limited anammox reactors and have revealed the coexistence of anammox bacteria and aerobic ammonia-oxidizing bacteria (AOB) and/or nitrite-oxidizing bacteria (NOB) in biofilms or aggregates (22, 32, 41, 45). As far as we know, however, a comparative analysis between in situ anammox activity and community structure (spatial distribution) in anammox biofilms has never been reported so far. A better understanding of reactor performance, including the distribution of anammox activities along the reactor and the microbial community structures (the spatial organization of anammox bacteria and other coexisting bacteria) in biofilms, will lead to the future optimization and efficient design of the anammox process.
The objective of this study was therefore to directly relate the in situ spatial organization of anammox bacteria and other coexisting bacteria to the in situ anammox activity in autotrophic anammox biofilms grown in an anaerobic fixed-bed column reactor. To achieve this objective, we directly measured concentration profiles of O2, NH4+, NO2−, and NO3− at three points along the anammox reactor by using microelectrodes. This was done under realistic operating conditions (i.e., water flow, water chemistry, temperature, and so on). The in situ spatial organization of anammox bacteria and coexisting bacteria in the anammox biofilms was analyzed by FISH and phylogenetic analysis based on small-subunit-rRNA gene sequencing.
MATERIALS AND METHODS
Biofilm samples.
Anaerobic ammonium-oxidizing (anammox) biofilms were cultured with a synthetic nutrient medium in an anaerobic fixed-bed column reactor equipped with three ports (inner diameter, 1 cm) for microelectrode measurement, as shown in Fig. 1. The anammox biomass was obtained from a fixed-bed biofilm column reactor, which was developed previously in our laboratory (44), and then directly inoculated as the source for the anammox biofilm. The reactor volume was 150 cm3 (length, 28 cm; diameter, 2.6 cm). The temperature was maintained at 37°C. Nonwoven fabric sheets comprised of polyester (thickness, 0.8 cm; length, 25 cm; width, 2 cm) (Japan Vilene Co., Ltd., Tokyo, Japan) were used for carrier materials of biofilms. The total projection area of the biofilm carrier materials was 143 cm2. The hydraulic retention time of the reactors was fixed at 0.7 to 1.0 h. The synthetic nutrient medium was composed of 5.5 mM (NH4)2SO4, 5 to 7 mM NaNO2, 1.0 mM KHCO3, 0.2 mM KH2PO4, 1.2 mM MgSO4·7H2O, 1.2 mM CaCl2·2H2O, and 1 ml of trace element solutions I and II, as described by Van de Graaf et al. (45). The medium was flushed with N2 gas for at least 1 h to achieve a concentration of dissolved oxygen (DO) below 0.8 mg/liter. The pH of the medium was adjusted in the range of 6.2 to 6.5 with 1 N H2SO4. The microelectrode measurements and biofilm samplings were carried out after 74 days of operation.
FIG. 1.
Schematic drawing of anaerobic fixed-bed glass column reactor with three ports (diameter, 1 cm) for liquid sampling and microelectrode measurements. P1, P2, P3, and P4 indicate the sampling points of anammox biofilms for FISH and phylogenetic analyses.
DNA extraction and PCR amplification.
Total DNA was extracted from the anammox biomass taken from the biofilm at P1 (see below) and from detached biomass in the effluent after 74 days of operation by using a Fast DNA spin kit (Bio 101; Qbiogene Inc., Carlsbad, CA) as described in the manufacturer's instructions. The effluent water samples containing detached biomass were collected in sterile 500-ml bottles and then centrifuged (10 min, 10,000 × g) to recover the anammox biomass. The pellets were resuspended in 2 ml of distilled water. 16S rRNA gene fragments from the isolated total DNA extracted from the anammox biomass at P1 (biofilm) were amplified with Taq DNA polymerase (TaKaRa Bio Inc., Ohtsu, Japan) by using Planctomycetales-specific primer set pla46f and 1492r as described by Tsushima et al. (44). The PCR conditions targeted for anammox bacteria were as follows: 5 min of initial denaturation at 94°C, followed by 25 cycles of 1 min at 94°C, 1 min at 50°C, and 70 s at 72°C. Final extension was carried out for 4 min at 72°C. 16S rRNA gene fragments from the isolated total DNA extracted from the detached biomass in the effluent were amplified with Taq DNA polymerase (TaKaRa Bio Inc.) by using bacterial primer set 11f and 1492r as described by Kindaichi et al. (15). The PCR conditions targeted for bacteria were as follows: 5 min of initial denaturation at 94°C and 20 cycles of 30 s at 94°C, 30 s at 50°C, and 1 min at 72°C. Final extension was carried out for 4 min at 72°C. For both PCRs, the reaction was performed with a total volume of 50 μl and 1 μg of DNA was added as template DNA. The PCR products were electrophoresed in a 1% (wt/vol) agarose gel.
Cloning and sequencing of 16S rRNA gene and phylogenetic analysis.
PCR products were ligated into a pCR-XL-TOPO vector and transformed into One Shot Escherichia coli cells following the manufacturer's instructions (TOPO XL PCR cloning kit; Invitrogen, Carlsbad, CA), and then clone libraries were constructed. Nucleotide sequencing was performed with an automatic sequencer (ABI Prism 3100 Avant genetic analyzer; Applied Biosystems, Foster City, CA). The almost-full-length sequences (>1,400 bp) obtained were checked for chimeric artifacts by the CHECK_CHIMERA program in the Ribosomal Database Project (19) and compared with similar sequences of the reference organisms by a BLAST search (1). Sequences with 97% or greater similarity were grouped into operational taxonomic units (OTUs) by the Similarity Matrix program from the Ribosomal Database Project (19). The sequences were aligned with the CLUSTAL W package (42). Phylogenetic trees were constructed by MEGA2 software with the neighbor-joining method (30) and the maximum parsimony method. Bootstrap resampling analysis for 1,000 replicates was performed to estimate the confidence of the tree topologies.
Fixation and cryosectioning of biofilm samples.
After microelectrode measurements, the biofilm samples were obtained from four points (P1, -2, -3, and -4) in the fixed-bed column reactor, as shown in Fig. 1. The biofilm samples were fixed in a 4% paraformaldehyde solution for 8 h at 4°C and were embedded in Tissue-Tek OCT compound (Sakura Finetek, Torrance, CA) overnight to allow the OCT compound to infiltrate into the biofilm, as described previously (23). After rapid freezing at −20°C, 30-μm-thick vertical slices were prepared with a cryostat (Reichert-Jung Cryocut 1800; Leica, Bensheim, Germany).
Oligonucleotide probes and FISH.
The 16S rRNA-targeted oligonucleotide probes used in this study are listed in Table 1. To detect all bacteria, probes EUB338, EUB338II, and EUB338III were used in an equimolar mixture (EUB338mix) (4). The probes were labeled with fluorescein isothiocyanate (FITC) or tetramethylrhodamine 5-isothiocyanate (TRITC). In situ hybridization was performed according to the procedure described by Okabe et al. (23). Simultaneous hybridizations with probes requiring different stringency conditions were performed by using a successive hybridization procedure; hybridization with the probe requiring higher stringency was performed first, and then hybridization with the probe requiring lower stringency was performed (46). A model LSM510 confocal laser scanning microscope (CLSM; Carl Zeiss, Oberkochen, Germany) equipped with an Ar ion laser (488 nm) and a HeNe laser (543 nm) was used. Image combining and processing were performed with the standard software package provided by Zeiss, as described previously (15, 23, 25). For quantitative determination of the microbial composition in the biofilm, the surface fractions of the specific probe-hybridized cell area and the EUB338mix probe-hybridized cells (total biomass) were determined after simultaneous in situ hybridizations with various probe sets. The average surface fraction was determined from at least 10 randomly chosen CLSM projection images obtained from each of the duplicate biofilm samples by using image analysis software provided by Zeiss (26). Similarly, the relative abundance of anammox bacteria in the upper (0 to 1 mm from the surface), middle (1 to 3 mm), and deeper (3 to 4 mm) parts of the biofilm was determined.
TABLE 1.
16S rRNA-targeted oligonucleotide probes used in this study
| Probe | Sequence (5′ to 3′) | FA (%)a | Specificity | Reference |
|---|---|---|---|---|
| EUB338 | GCTGCCTCCCGTAGGAGT | 0-50 | Most bacteria | 2 |
| EUB338 II | GCAGCCACCCGTAGGTGT | 0-50 | Planctomycetales | 4 |
| EUB338 III | GCTGCCACCCGTAGGTGT | 0-50 | Verrucomicrobiales | 4 |
| Amx820 | AAAACCCCTCTACTTAGTGCCC | 40 | “Candidatus Brocadia anammoxidans,” “Candidatus Kuenenia stuttgartiensis” | 33 |
| Nmo218 | CGGCCGCTCCAAAAGCAT | 35 | Nitrosomonas oligotropha lineage | 10 |
| NmV | TCCTCAGAGACTACGCGG | 35 | Nitrosococcus mobilis lineage | 13 |
| Nse1472 | ACCCCAGTCATGACCCCC | 50 | Nitrosomonas europea, Nitrosomonas halophila, and Nitrosomonas eutropha | 13 |
| Nsv443 | CCGTGACCGTTTCGTTCCG | 30 | Nitrosospira spp. | 21 |
| Ntspa662 | GGAATTCCGCGCTCCTCT | 35 | Genus Nitrospira | 5 |
| Comp Ntspa662b | GGAATTCCGCTCTCCTCT | Competitor for Ntspa662 | 5 | |
| NIT3 | CCTGTGCTCCATGCTCCG | 40 | Nitrobacter spp. | 47 |
| Comp NIT3c | CCTGTGCTCCAGGCTCCG | Competitor for NIT3 | 47 |
FA, formamide concentration in the hybridization buffer.
Unlabeled probe Ntspa662 was used as a competitor to enhance specificity.
Unlabeled probe NIT3 was used as a competitor to enhance specificity.
Microelectrode measurements.
Concentration profiles in the biofilm were measured using Clark-type microelectrodes for O2 (29) and LIX-type microelectrodes for NH4+, NO2−, NO3−, and pH (7). The microelectrodes were prepared, calibrated, and operated as described by Okabe et al. (23) and Satoh et al. (31). The microelectrodes were inserted through ports A, B, and C in the top of the reactor at 74 days of operation (Fig. 1). At least three profiles in the biofilm were measured for each chemical species at each point.
Total production and consumption rates of NH4+, NO2−, and NO3− were calculated from the concentration profiles by using Fick's first law of diffusion, i.e., J = −D(dCs/dz), where D is the molecular diffusion coefficient in the liquid phase and dCs/dz is the measured concentration gradient of each solute in the boundary layer at the biofilm-liquid interface. Furthermore, the net specific consumption or production rates of NH4+, NO2−, and NO3− were calculated from the mean concentration profiles by using Fick's second law of diffusion as described by Lorenzen et al. (18). Molecular diffusion coefficients at 37°C of 1.47 × 10−5 cm2 s−1 for NH4+, 1.30 × 10−5 cm2 s−1 for NO2−, and 1.30 × 10−5 cm2 s−1 for NO3− were used for the calculations (3).
Analytical procedure.
To analyze the concentrations of NH4+, NO2−, and NO3− in the reactor, water samples were obtained from P2, P3, and P4 by using a syringe and from inflow and outflow lines (Fig. 1). The concentrations of NH4+, NO2−, and NO3− were determined using an ion chromatograph (DX-100; Dionex, Sunnyvale, CA) equipped with an IonPac CS3 cation column and IonPac AS9 anion column. The samples were filtered through 0.2-μm-pore-size membranes (Advantec Co. Ltd., Tokyo, Japan) before the analysis. The dissolved organic carbon (DOC) concentration was measured by using a TOC analyzer (TOC-5000A; Shimadzu, Kyoto, Japan) after filtration with 0.45-μm-pore-size membranes (Advantec, Tokyo, Japan).
Nucleotide sequence accession numbers.
The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA gene sequences of the six OTUs used for the phylogenetic tree analysis are AB290144 to AB290148 and AB302409.
RESULTS AND DISCUSSION
Reactor performance.
The total inorganic nitrogen removal rate, which was defined as the sum of the concentrations of NH4+, NO2−, and NO3− in the reactor, gradually increased to 250 μmol cm−3 reactor day−1, with a nitrogen removal efficiency of more than 70%, within the initial 2 months. The maximum total inorganic nitrogen removal rate increased to 385 μmol cm−3 reactor day−1, with >95% nitrogen removal efficiency, after 74 days of operation. Thereafter, the nitrogen removal rate became stable.
Average concentrations of NH4+, NO2−, NO3−, and DOC in the influent and effluent and at P2, P3, and P4 are listed in Table 2. The concentrations of NH4+ and NO2− significantly decreased from the influent to P2, whereas the concentration of NO3− and the pH increased. In contrast, the concentrations of NH4+ and NO2− slightly decreased from P2 to P4 in the reactor, below which points these concentrations tended to increase slightly. This indicates that the anammox activity was high in the inlet zone (up to P2) of the column reactor and decreased along the flow direction, and no activity was observed in the outlet zone. The DOC concentration steadily increased, from 0.33 ± 0.02 mM in the influent to 0.64 ± 0.02 mM in the effluent. It is noted that the fresh medium contained a DOC concentration of 0.33 mM, which was derived from EDTA in the trace element solutions I and II.
TABLE 2.
Average NH4+, NO2−, NO3−, and DOC concentrations and pH values in the anammox reactor
| Compound or parameter | Concn (mM) or value (pH)a
|
||||
|---|---|---|---|---|---|
| Influent | P2 | P3 | P4 | Effluent | |
| NH4+ | 5.5 ± 0.1 | 1.8 ± 0.6 | 1.4 ± 0.6 | 1.2 ± 0.7 | 1.5 ± 0.8 |
| NO2− | 6.3 ± 0.6 | 0.9 ± 0.3 | 0.7 ± 0.4 | 0.3 ± 0.2 | 0.5 ± 0.3 |
| NO3− | 0.0 ± 0.0 | 1.1 ± 0.1 | 1.2 ± 0.1 | 1.2 ± 0.2 | 1.3 ± 0.1 |
| DOC | 0.33 ± 0.02 | 0.47 ± 0.02 | 0.60 ± 0.03 | 0.64 ± 0.02 | 0.64 ± 0.03 |
| pH | 6.3 ± 0.2 | 7.6 ± 0.4 | 7.7 ± 0.5 | 7.8 ± 0.5 | 8.0 ± 0.7 |
The values are means ± standard deviations (n = 6).
Phylogenetic analysis of anammox biofilms.
Thirty-six clones were randomly selected from a clone library constructed from the detached biomass with the bacterial primer set, partial sequences (approximately 500 bp) were analyzed, and the clones were grouped into five OTUs on the basis of >97% sequence similarity. No chimeric sequences were observed in the clone library. The nearly complete sequences of five OTUs were analyzed and used to generate phylogenetic trees (Fig. 2). It is noted that there were no differences in the tree topologies of both trees, generated by using the neighbor-joining and the maximum parsimony methods (Fig. 2A and B). The sequence of the OTU HU1 (24 of 36 clones) was closely related to that of the uncultured clone KU1 (AB054006), with 97% sequence similarity. This clone was obtained from a continuous-flow fixed-bed reactor operated in Japan, in which a nonwoven biomass carrier was used as the biomass carrier and denitrifying sludge was seeded. The sequence similarity of the OTU HU1 to the closest proposed anammox bacterium, “Candidatus Brocadia anammoxidans,” was only 95% (Fig. 2A). Furthermore, the sequence similarities of the OTU HU1 to other proposed anammox bacteria, namely, “Candidatus Brocadia fulgida,” “Candidatus Jettenia asiatica,” “Candidatus Kuenenia stuttgartiensis,” “Candidatus Scalindua brodae,” and “Candidatus Anammoxoglobus propionicus,” were 94, 91, 92, 90, and 92%, respectively. According to Stackebrandt and Goebel (38), the dominant anammox-like bacterium detected in this study is most likely a member of a novel species of the genus “Brocadia.”
FIG. 2.
Phylogenetic trees showing the positions of clones retrieved from the detached biomass (OTUs HU1 to HU5) and biofilm (OTU Biofilm-PU13) in the reactor after 74 days of operation in relation to previously published anammox-like bacterial sequences (A) and betaproteobacterial sequences (B). The trees were generated by using 1,429 bp (A) and 1,462 bp (B) of the 16S rRNA gene and the neighbor-joining method. Bars = 2% sequence divergence. The values at the nodes are bootstrap values (1,000 resampling analyses). The GenBank/EMBL/DDBJ accession numbers are also indicated.
The sequences of the OTUs HU2 (7 of 36 clones), HU3 (2 of 36 clones), HU4 (2 of 36 clones), and HU5 (1 of 36 clones) belonged to the Betaproteobacteria (Fig. 2B). The sequences of the OTUs HU3 and HU5 were affiliated with the Thauera spp. and Acidovorax spp., respectively, with 98% sequence similarity. Thauera spp. and Acidovorax spp. are capable of denitrification (9, 17). Tsushima et al. have reported the coexistence of Acidovorax sp. with anammox bacteria in anammox enrichment cultures (43). Furthermore, some members of the Betaproteobacteria present in autotrophic nitrifying biofilms utilized the organic compounds derived from nitrifying bacteria (26). Therefore, it could be speculated that these coexisting putative heterotrophs utilize by-products derived from anammox and/or coexisting nitrifying bacteria as a carbon source for denitrification. Further studies focusing on in situ ecophysiology of these bacteria are needed to clarify microbial interactions between these putative heterotrophs and anammox bacteria in the anaerobic anammox biofilm.
Sixteen clones were randomly selected from a clone library constructed from the biofilm with the Planctomycetales-specific primer set, partial sequences (approximately 500 bp) were analyzed, and all clones were grouped into one OTU on the basis of >97% sequence similarity. No chimeric sequences were observed in the clone library. The nearly complete sequence of the OTU was analyzed and included in the phylogenetic tree (Fig. 2A). The sequence of the OTU Biofilm-PU13 (16 of 16 clones) was also closely related to the OTU HU1, with 99% sequence similarity (Fig. 2A).
Biofilm structure and spatial organization of anammox biofilms.
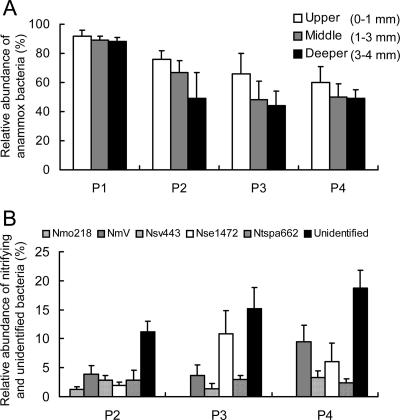
Biofilm cross sections revealed a heterogeneous structure of the anammox biofilm, consisting of densely and tightly packed biomass and interstitial voids (Fig. 3A and B). FISH of vertical cross sections of the anammox biofilm indicated that anammox bacteria hybridized with the probe Amx820 were present throughout the anammox biofilm (Fig. 3A and 4A). The relative abundance of anammox bacteria was >90% throughout the biofilm at P1 (inlet side) but decreased with the flow direction of the reactor and with biofilm depth (Fig. 4A), corresponding to the overall reactor performance (Table 2). One possible explanation is that the biomass in the deeper part of the biofilm tends to decay and liberate organic matter, probably due to substrate transport limitation in the biofilms, which promotes the growth of other heterotrophic bacteria and/or inhibits the growth of anammox bacteria.
FIG. 3.
(A) CLSM image of a vertical section (30 μm thick) of the anammox biofilm at P1 after 74 days of operation after FISH with FITC-labeled EUB338mix probe (green) and TRITC-labeled Amx820 probe (red). Yellow signals result from binding of both probes to one cell, indicating anammox bacteria. Bar = 100 μm. (B) Phase-contrast image of the vertical section in panel A. The biofilm surface is at the top of the images.
FIG. 4.
Relative abundance of anammox bacteria at different depths in the anammox biofilm (A) and of nitrifying and unidentified bacteria in the upper part of the biofilm (>1,000 μm) (B). Relative abundance is shown as the percentage of each probe signal in a microscopic field compared with the EUB338mix probe signal. The number of unidentified bacteria was calculated as the difference between the EUB338mix signal and the total signal from each specific probe. P1, P2, P3, and P4 refer to the biofilm sampling points, as shown in Fig. 1. The error bars indicate standard deviations.
Anammox bacteria accounted for only 60% of total bacteria hybridized with the probe EUB338mix in the upper layer (0 to 1 mm) of the biofilm at P4 (Fig. 4A). In the anammox biofilm, several groups of nitrifying bacteria (aerobic AOB plus NOB) that were hybridized with probes Nmo218, Nse1472, NmV, Nsv443, and Ntspa662 were detected mainly in the upper part of the biofilm (>1,000 μm). No hybridization signal with the probe NIT3, specific for the genus Nitrobacter, was observed in the biofilms. These results indicate that the aerobic AOB belonging to at least four phylogenetically different lineages, Nitrosomonas europaea, Nitrosomonas oligotropha, Nitrosococcus mobilis, and Nitrosospira spp., were present. Furthermore, Nitrospira sp. was the dominant NOB in the anammox biofilm. Collectively, these nitrifying bacteria accounted for 13 to 21% of total bacteria (Fig. 4B), among which the probe Ntspa662-hybridized Nitrospira-like NOB accounted for 2 to 3%. One possible explanation of the presence of AOB is that some AOB can reduce nitrite to N2O or NO gases under oxygen-limiting conditions (27, 28). Another possible reason is that since nitrification perhaps occurred in the inlet zone (up to P2) of the reactor because a trace amount of O2 was present in the medium (<0.8 mg/liter) (37), nitrifying bacteria present in the P1 biofilm could detach and then reattach to the biofilms or accumulate downstream in the reactor. In addition, for AOB, the FISH signal intensity (i.e., ribosome content) does not decrease significantly during periods of starvation. Several studies have also reported the coexistence of anammox bacteria and Nitrosomonas-like AOB (12, 32, 45) or Nitrospira-like NOB (8).
We demonstrated the in situ spatial organization of the anammox bacteria and coexisting bacteria in the anammox biofilms. The coexisting bacteria (nitrifying and putative heterotrophic bacteria) were also detected at the upper layer of the biofilm and distributed around the anammox bacterial clusters (Fig. 5). Some of them were present inside the anammox bacterial clusters (Fig. 5). Since the sum of the relative abundances of anammox bacteria, aerobic AOB, and Nitrospira-like NOB was 81 to 89%, the other (e.g., putative heterotrophic) bacteria were present in the anammox biofilm. In addition, FISH results revealed that some filamentous bacteria hybridized with only the probe EUB338mix (not identified at present) were present around the anammox bacterial clusters (data not shown). The relative abundance of the unidentified other bacteria (defined as total bacteria hybridized with the probe EUB338 mix [the sum of anammox bacteria, aerobic AOB, and Nitrospira-like NOB]) increased with the flow direction (Fig. 4B). The coexisting nitrifying and putative heterotrophic bacteria in the anammox biofilm might consume a trace amount of O2 or organic by-products of anammox bacteria, which consequently prevented O2 inhibition and accumulation of organic waste products in the biofilm. However, an excess growth of the coexisting bacteria may negatively affect the activity of anammox bacteria. Further studies are needed to clarify the ecophysiological roles and functions of these coexisting bacteria in the anammox biofilm.
FIG. 5.
CLSM images showing the in situ spatial organization of anammox bacteria and coexisting bacteria (nitrifying and putative heterotrophic bacteria) in the anammox biofilm. All red signals show FISH with the TRITC-labeled probe Amx820 (A to D). Green signals show FISH with the FITC-labeled probe EUB338mix at P2 (A) and P3 (B), with probe NmV at P2 (C), and with probe Ntspa662 at P2 (D). Bars = 10 μm. Yellow signals result from binding of both probes to one cell, indicating anammox bacteria (A and B). The biofilm surface is at the top of the images.
Concentration profiles in anammox biofilms.
Steady-state concentration profiles of O2, NH4+, NO2−, and NO3− in the anammox biofilms were measured directly at three points (i.e., P2, P3, and P4) under realistic operating conditions (i.e., water flow, water chemistry, temperature, and so on) (Fig. 6A to C). The O2 concentration was under the detection limit (ca. 1 μM) throughout the biofilms at all three points. Both NH4+ and NO2− concentrations decreased and the NO3− concentration slightly increased in the upper 600 μm of the biofilms at P2 and P3, indicating the occurrence of anammox in the biofilms. In contrast, the NH4+ concentration increased within the biofilm at P4, probably due to mineralization of organic compounds and biomass. The pH value for P2 and P3 biofilms increased in the zones where anammox reactions occurred (Fig. 6A and B). These anammox activities determined with microelectrodes were consistent with the overall reactor performance (Table 2).
FIG. 6.
Steady-state concentration profiles of O2 (closed circles), NH4+ (gray squares), NO2− (open circles), and NO3− (triangles) and pH profiles (crosses) in the anammox biofilms measured at P2 (A), P3 (B), and P4 (C), as shown in Fig. 1. Spatial distributions of the estimated specific consumption rates of NH4+ (gray bars), NO2− (open circles), and NO3− (black bars) at P2, P3, and P4 are shown in panels D, E, and F, respectively. Negative values indicate production rates. The surface of the biofilm was at a depth of 0 μm. The error bars indicate standard deviations (n = 3).
The spatial distributions of net specific NH4+, NO2−, and NO3− consumption rates showed that the anammox reaction was restricted to the upper 1,300 and 800 μm of P2 and P3 biofilms, respectively (Fig. 6D and E), although sufficient concentrations of NH4+ and NO2− were still present in the deeper part of the biofilms (Fig. 6A and B). The low anammox activity in the deeper part of the biofilms was probably not due to the substrate limitations, because the Km values for NO2− for anammox bacteria have been reported to be ≤7 μM (40), ≤3 μM (6), and 14 to 23 μM (22) and the Km values for NH4+ for anammox bacteria have been reported to be ≤7 μM (40) and ≤50 μM (6).
No anammox reaction was observed at P4 (Fig. 6F), although the anammox bacteria were detected at a higher abundance (Fig. 4). DOC concentrations (Table 2) and the relative abundance of the unidentified other bacteria (Fig. 4B) slightly increased at this point. These results indicate that heterotrophic denitrification using DOC as the electron donor increased with the flow direction. A possible explanation for the lower anammox activity at P4 could be an inhibitory effect of the organic compounds derived from biomass decay and/or produced by anammox and coexisting nitrifying bacteria in the upper parts of the biofilm and in the upstream region of the reactor. Tsushima et al. have reported that the anammox activity decreased when the DOC concentration increased from 0.5 to 0.8 mM in the enrichment batch cultures (43). Moreover, the addition of the supernatants of anammox enrichment cultures significantly decreased the anammox activities in the batch cultures (43). It is therefore important to identify these organic compounds and evaluate their inhibitory effects for maintaining high anammox activity throughout the reactor.
Total rates of NH4+ consumption, NO2− consumption, and NO3− production in the anammox biofilms, determined for the boundary layer at the biofilm-liquid interface, are summarized in Table 3. The total rates of NH4+ consumption in this anammox biofilm (0.30 and 0.68 μmol cm−2 h−1) were higher than aerobic NH4+ oxidation rates detected in nitrifying biofilms (16, 23, 36). The total rates of NO2− consumption (0.15 to 0.64 μmol cm−2 h−1) detected in this biofilm were also higher than the NO2− uptake rates (0.05 to 0.15 μmol cm−2 h−1) of CANON aggregates, as determined by microelectrodes (22). Such a high anammox rate was attributed mainly to the high concentration of biomass (ca. 2.4 g volatile suspended solids per reactor) retained in the reactor. The much higher solubility of NO2− as an electron acceptor for anammox bacteria than that of O2 as an electron acceptor for aerobic AOB should be advantageous for a high NH4+ oxidation rate. In addition, it is possible that dominant anammox bacteria in this biofilm have high specific anammox activity because the anammox bacteria in this biofilm are most likely a novel species. The reported stoichiometric ratio of NO2− consumption to NH4+ consumption for anammox was 1.31 (45), whereas the ratio was 0.94 at P2 and 1.17 at P3 in this study (Table 3). This is partly because partial nitrification occurred in the inlet zone, using a small amount of O2 entering into the reactor. Another possibility is that the Nitrosomonas-like AOB could oxidize NH4+ anaerobically when nitrogen dioxide (NO2) gas was present (35). If this anaerobic ammonia oxidation by AOB occurs, the stoichiometry of the converted NO2−/NH4+ ratio becomes lower than 1.31. Further studies on the presence of nitrogen oxides in this reactor are needed to understand nitrogen removal mechanisms by coculture of anammox bacteria and aerobic AOB in biofilms.
TABLE 3.
Total rates of NH4+ consumption, NO2− consumption, and NO3− production in anammox biofilms
| Parameter | Total ratea (μmol cm−2 h−1)
|
||
|---|---|---|---|
| P2 | P3 | P4 | |
| NH4+ consumption | 0.68 ± 0.34 | 0.30 ± 0.07 | −0.22 ± 0.02 |
| NO2− consumption | 0.64 ± 0.46 | 0.35 ± 0.16 | 0.15 ± 0.10 |
| NO3− production | 0.33 ± 0.24 | 0.15 ± 0.02 | 0.09 ± 0.05 |
The values are means ± standard deviations (n = 3).
Concluding remarks.
In conclusion, this study provided new and important information on distributions of in situ anammox activities along an anaerobic fixed-bed column reactor as well as with biofilm depth and on the in situ spatial organization of anammox and coexisting bacteria in the anammox biofilms. FISH and phylogenetic analyses revealed that several groups of aerobic AOB, Nitrospira-like NOB, Betaproteobacteria, unidentified bacteria, and novel anammox-like bacteria coexisted in the anammox biofilms. The anammox activity decreased with the flow direction, probably due to accumulation of organic compounds (by-products), even though the relative abundance of anammox bacteria was high (60 to 92%). To investigate whether the organic compounds produced by the anammox bacteria are utilized by coexisting heterotrophic bacteria, the microautoradiography-FISH technique should be applied directly in the future.
Acknowledgments
This research was partially supported by a Grant for Research and Technology Development on Waste Management (K1627) from the Ministry of the Environment and by the Steel Industry Foundation for Advancement of Environmental Protection Technology, the Electric Technology Research Foundation of Chugoku, the Foundation of River & Watershed Environment Management, and the Kurita Water and Environment Foundation. Ikuo Tsushima was financially supported by the 21st Century Center of Excellence (COE) program Sustainable Metabolic System of Water and Waste for Area-Based Society from the Ministry of Education, Science and Culture of Japan.
Footnotes
Published ahead of print on 25 May 2007.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrussow, L. 1969. Diffusion, p. 513-727. In H. Borchers, H. Hauser, K. H. Hellwege, K. Schafer, and E. Schmidt (ed.), Landolt-Bornstein zahlenwerte und functionen, vol. II/5a. Springer, Berlin, Germany. [Google Scholar]
- 4.Daims, H., A. Brühl, R. Amann, K.-H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 5.Daims, H., J. L. Nielsen, P. H. Nielsen, K.-H. Schleifer, and M. Wagner. 2001. In situ characterization of Nitrospira-like nitrite-oxidizing bacteria active in wastewater treatment plants. Appl. Environ. Microbiol. 67:5273-5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dalsgaard, T., and B. Thamdrup. 2002. Factors controlling anaerobic ammonium oxidation with nitrite in marine sediments. Appl. Environ. Microbiol. 68:3802-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Beer, D., A. Schramm, C. M. Santegoeds, and M. Kühl. 1997. A nitrite microsensor for profiling environmental biofilms. Appl. Environ. Microbiol. 63:973-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Egli, K., F. Bosshard, C. Werlen, P. Lais, H. Siegrist, A. J. B. Zehnder, and J. R. van der Meer. 2003. Microbial composition and structure of a rotating biological contactor biofilm treating ammonium-rich wastewater without organic carbon. Microbiol. Ecol. 45:419-432. [DOI] [PubMed] [Google Scholar]
- 9.Etchebehere, C., and J. Tiedje. 2005. Presence of two different active nirS nitrite reductase genes in a denitrifying Thauera sp. from a high-nitrate-removal-rate reactor. Appl. Environ. Microbiol. 71:5642-5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gieseke, A., U. Purkhold, M. Wagner, R. Amann, and A. Schramm. 2001. Community structure and activity dynamics of nitrifying bacteria in a phosphate-removing biofilm. Appl. Environ. Microbiol. 67:1351-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ito, T., S. Okabe, H. Satoh, and Y. Watanabe. 2002. Successional development of sulfate-reducing bacterial populations and their activities in a wastewater biofilm growing under microaerophilic conditions. Appl. Environ. Microbiol. 68:1392-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jetten, M. S. M., M. Strous, K. T. van de Pas-Schoonen, J. Schalk, U. G. J. M. van Dongen, A. A. van de Graaf, S. Logemann, G. Muyzer, M. C. M. van Loosdrecht, and J. G. Kuenen. 1999. The anaerobic oxidation of ammonium. FEMS Microbiol. Rev. 22:421-437. [DOI] [PubMed] [Google Scholar]
- 13.Juretschko, S., G. Timmermann, M. Schmid, K.-H. Schleifer, A. Pommerening-Röser, H. P. Koops, and M. Wagner. 1998. Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl. Environ. Microbiol. 64:3042-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kartal, B., J. Rattray, L. A. van Niftrik, J. van de Vossenberg, M. C. Schmid, R. I. Webb, S. Schouten, J. A. Fuerst, J. S. Damsté, M. S. M. Jetten, and M. Strous. 2007. Candidatus “Anammoxoglobus propionicus” a new propionate oxidizing species of anaerobic ammonium oxidizing bacteria. Syst. Appl. Microbiol. 30:39-49. [DOI] [PubMed] [Google Scholar]
- 15.Kindaichi, T., T. Ito, and S. Okabe. 2004. Ecophysiological interaction between nitrifying bacteria and heterotrophic bacteria in autotrophic nitrifying biofilms as determined by MAR-FISH. Appl. Environ. Microbiol. 70:1641-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kindaichi, T., Y. Kawano, T. Ito, H. Satoh, and S. Okabe. 2006. Population dynamics and in situ kinetics of nitrifying bacteria in autotrophic nitrifying biofilms as determined by real-time quantitative PCR. Biotechnol. Bioeng. 94:1111-1121. [DOI] [PubMed] [Google Scholar]
- 17.Kniemeyer, O., C. Probian, R. Roselló-Mora, and J. Harder. 1999. Anaerobic mineralization of quaternary carbon atoms: isolation of denitrifying bacteria on dimethylmalonate. Appl. Environ. Microbiol. 65:3319-3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lorenzen, J., L. H. Larsen, T. Kjær, and N. P. Revsbech. 1998. Biosensor detection of the microscale distribution of nitrate, nitrate assimilation, nitrification, and denitrification in a diatom-inhabited freshwater sediment. Appl. Environ. Microbiol. 64:3264-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maidak, B. L., G. L. Olsen, N. Larsen, R. Overbeek, M. J. McCaughey, and C. R. Woese. 1997. The RDP (Ribosomal Database Project). Nucleic Acids Res. 25:109-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meyer, R. L., N. Risgaard-Petersen, and D. E. Allen. 2005. Correlation between anammox activity and microscale distribution of nitrite in a subtropical mangrove sediment. Appl. Environ. Microbiol. 71:6142-6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mobarry, B. K., M. Wagner, V. Urbain, B. E. Rittmann, and D. A. Stahl. 1996. Phylogenetic probes for analyzing abundance and spatial organization of nitrifying bacteria. Appl. Environ. Microbiol. 62:2156-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nielsen, M., A. Bollmann, O. Sliekers, M. Jetten, M. Schmid, M. Strous, I. Schmidt, L. H. Larsen, L. P. Nielsen, and N. P. Revsbech. 2005. Kinetics, diffusional limitation and microscale distribution of chemistry and organisms in a CANON reactor. FEMS Microbiol. Ecol. 51:247-256. [DOI] [PubMed] [Google Scholar]
- 23.Okabe, S., H. Satoh, and Y. Watanabe. 1999. In situ analysis of nitrifying biofilms as determined by in situ hybridization and the use of microelectrodes. Appl. Environ. Microbiol. 65:3182-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okabe, S., T. Itoh, H. Satoh, and Y. Watanabe. 1999. Analyses of spatial distributions of sulfate-reducing bacteria and their activity in aerobic wastewater biofilms. Appl. Environ. Microbiol. 65:5107-5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okabe, S., T. Kindaichi, T. Ito, and H. Satoh. 2004. Analysis of size distribution and areal cell density of ammonia-oxidizing bacterial microcolonies in relation to substrate microprofiles in biofilms. Biotechnol. Bioeng. 85:86-95. [DOI] [PubMed] [Google Scholar]
- 26.Okabe, S., T. Kindaichi, and T. Ito. 2005. Fate of 14C-labeled microbial products derived from nitrifying bacteria in autotrophic nitrifying biofilms. Appl. Environ. Microbiol. 71:3987-3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poth, M., and D. D. Focht. 1985. 15N kinetic analysis of N2O production by Nitrosomonas europaea: an examination of nitrifier denitrification. Appl. Environ. Microbiol. 49:1134-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Remde, A., and R. Cornad. 1990. Production of nitric oxide in Nitrosomonas europaea by reduction of nitrite. Arch. Microbiol. 154:187-191. [Google Scholar]
- 29.Revsbech, N. P. 1989. An oxygen microelectrode with a guard cathode. Limnol. Oceanogr. 34:474-478. [Google Scholar]
- 30.Saito, N., and M. Nei. 1987. The neighbor-joining method: a new method for constructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 31.Satoh, H., S. Okabe, Y. Yamaguchi, and Y. Watanabe. 2003. Evaluation of the impact of bioaugmentation and biostimulation by in situ hybridization and microelectrode. Water Res. 37:2206-2216. [DOI] [PubMed] [Google Scholar]
- 32.Schmid, M., U. Twachtmann, M. Klein, M. Strous, S. Juretschko, M. S. M. Jetten, J. W. Metzger, K.-H. Schleifer, and M. Wagner. 2000. Molecular evidence for genus level diversity of bacteria capable of catalyzing anaerobic ammonium oxidation. Syst. Appl. Microbiol. 23:93-106. [DOI] [PubMed] [Google Scholar]
- 33.Schmid, M., S. Schmitz-Esser, M. Jetten, and M. Wagner. 2001. 16S-23S rDNA intergenic spacer and 23S rDNA of anaerobic ammonium-oxidizing bacteria: implications for phylogeny and in situ detection. Environ. Microbiol. 3:450-459. [DOI] [PubMed] [Google Scholar]
- 34.Schmid, M. C., B. Maas, A. Dapena, K. van de Pas-Schoonen, J. van de Vossenberg, B. Kartal, L. van Niftrik, I. Schmidt, I. Cirpus, J. G. Kuenen, M. Wagner, J. S. Sinninghe Damste, M. Kuypers, N. P. Revsbech, R. Mendez, M. S. Jetten, and M. Strous. 2005. Biomarkers for in situ detection of anaerobic ammonium-oxidizing (anammox) bacteria. Appl. Environ. Microbiol. 71:1677-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidt, I., C. Hermelink, K. van de Pas-Schoonen, M. Strous, H. J. Op den Camp, J. G. Kuenen, and M. S. M. Jetten. 2002. Anaerobic ammonia oxidation in the presence of nitrogen oxides (NOx) by two different lithotrophs. Appl. Environ. Microbiol. 68:5351-5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schramm, A., L. H. Larsen, N. P. Revsbech, R. I. Amann, and K.-H. Schleifer. 1996. Structure and function of a nitrifying biofilm as determined by in situ hybridization and the use of microelectrodes. Appl. Environ. Microbiol. 62:4641-4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sliekers, A. O., S. C. M. Haaijer, M. H. Stafsnes, J. G. Kuenen, and M. S. M. Jetten. 2005. Competition and coexistence of aerobic ammonium- and nitrite-oxidizing bacteria at low oxygen concentrations. Appl. Microbiol. Biotechnol. 68:808-817. [DOI] [PubMed] [Google Scholar]
- 38.Stackebrandt, E., and B. M. Goebel. 1994. A place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 39.Strous, M., J. A. Fuerst, E. H. Kramer, S. Logemann, G. Muyzer, K. T. van de Pas-Schoonen, R. Webb, J. G. Kuenen, and M. S. Jetten. 1999. Missing lithotroph identified as new planctomycete. Nature 400:446-449. [DOI] [PubMed] [Google Scholar]
- 40.Strous, M., J. G. Kuenen, and M. S. M. Jetten. 1999. Key physiology of anaerobic ammonium oxidation. Appl. Environ. Microbiol. 65:3248-3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Third, K. A., A. O. Sliekers, J. G. Kuenen, and M. S. M. Jetten. 2001. The CANON system (completely autotrophic nitrogen-removal over nitrite) under ammonium limitation: interaction and competition between three groups of bacteria. Syst. Appl. Microbiol. 24:588-596. [DOI] [PubMed] [Google Scholar]
- 42.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsushima, I., T. Kindaichi, and S. Okabe. 2007. Quantification of anaerobic ammonium-oxidizing bacteria in enrichment cultures by real-time PCR. Water Res. 41:785-794. [DOI] [PubMed] [Google Scholar]
- 44.Tsushima, I., Y. Ogasawara, T. Kindaichi, H. Satoh, and S. Okabe. 2007. Development of high-rate anaerobic ammonium-oxidizing (anammox) biofilm reactors. Water Res. 41:1623-1634. [DOI] [PubMed] [Google Scholar]
- 45.Van de Graaf, A. A., P. de Bruijn, L. A. Robertson, M. S. M. Jetten, and J. G. Kuenen. 1996. Autotrophic growth of anaerobic ammonium-oxidizing micro-organisms in a fluidized bed reactor. Microbiology 142:2187-2196. [Google Scholar]
- 46.Wagner, M., R. I. Amann, P. Kampfer, B. Assmus, A. Hartmann, P. Hutzler, N. Springer, and K.-H. Schleifer. 1994. Identification and in situ detection of gram-negative filamentous bacteria in activated sludge. Syst. Appl. Microbiol. 17:405-417. [Google Scholar]
- 47.Wagner, M., G. Rath, H. P. Koops, J. Flood, and R. Amann. 1996. In situ analysis of nitrifying bacteria in sewage treatment plants. Water Sci. Technol. 34:237-244. [Google Scholar]