Abstract
DNA polymerase III is one of the five eubacterial DNA polymerases that is responsible for the replication of DNA duplex. Among the ten subunits of the DNA polymerase III core enzyme, the alpha subunit catalyzes the reaction for polymerizing both DNA strands. In this study, we extracted genomic sequences of the alpha subunit from 159 sequenced eubacterial genomes, and carried out sequence-based phylogenetic and structural analyses. We found that all eubacterial genomes have one or more alpha subunits, which form either homodimers or heterodimers. Phylogenetic and domain structural analyses as well as copy number variations of the alpha subunit in each bacterium indicate the classification of alpha subunit into four basic groups: polC, dnaE1, dnaE2, and dnaE3. This classification is of essence in genome composition analysis. We also consolidated the naming convention to avoid further confusion in gene annotations.
Key words: DNA polymerase III alpha subunit, dnaE, polC
Introduction
DNA polymerases play central roles in DNA replication and repair, whose fidelity is the primary source of DNA sequence variations. Among the five eubacterial DNA polymerases (I-V) 1., 2., 3., 4., polymerase III is responsible for catalyzing DNA polymerization in replication, and the rest are involved in subsidiary roles in replication (I) and repair (I, II, IV, and V) 1., 2., 3., 4., 5., 6.. DNA polymerase III is a ten-component complex consisting of the replicase (α, ε, and θ), the clamp loader or gamma complex (γ, δ, δ’, ζ, χ and ψ), and the sliding clamp (β2) 2., 7., 8., 9., 10., 11., 12., 13., 14., 15., 16., 17.. The alpha subunit works as a dimer at the replication fork and plays a central role in the complex. However, its classification remains controversial and ambiguous due to occasional redundancy and seemingly unrelated groupings, which are mainly resulted from the horizontal gene transfer among prokaryotes. The alpha subunit has been roughly classified into polC and dnaE groups 18., 19., 20., 21., 22.. A “confused dnaE” (23) and “a split of dnaE” (into dnaE1 and dnaE2) (24), as well as the integrated dnaE1 and dnaE2 in several cyanobacterial species were noticed previously (25). When two intact dnaE genes were detected, they were usually named as dnaE1 and dnaE2 in an arbitrary way 25., 26.. No detailed analysis has been carried out to classify these important molecules.
In this study, we collected DNA sequences from 159 representative eubacterial genomes and extracted their alpha subunit genes according to their annotations, followed by thorough searches with the BLAT tools under variable parameters. After that, we compiled amino acid sequences of these genes and studied the enzyme systematically at three different levels, including their overall phylogeny, domain structures, and identifiable motifs. As a result, we classified all the alpha subunit genes into four groups.
Results
The distribution of the alpha subunit genes per genome among the 159 eubacterial genomes is quite variable (Table S1). According to our statistics, 77 (48.4%) genomes have one single alpha subunit gene, 75 (47.2%) genomes have two alpha subunit genes, and the remaining seven (4.4%) genomes (Agrobacterium tumefaciens C58 UWash, Bacillus subtilis, Clostridium acetobutylicum, Sinorhizobium meliloti, Staphylococcus epidermidis RP62A, Streptomyces coelicolor, and Symbiobacterium thermophilum IAM14863) each contain three alpha subunit genes. Therefore, these genomes have 248 alpha subunit genes in total. We examined these genes carefully and did not find frame shifts, stop codons, or other anomalies. We believe that they are not pseudogenes although we lack empirical data to establish their exact functionality.
Phylogenetic analysis
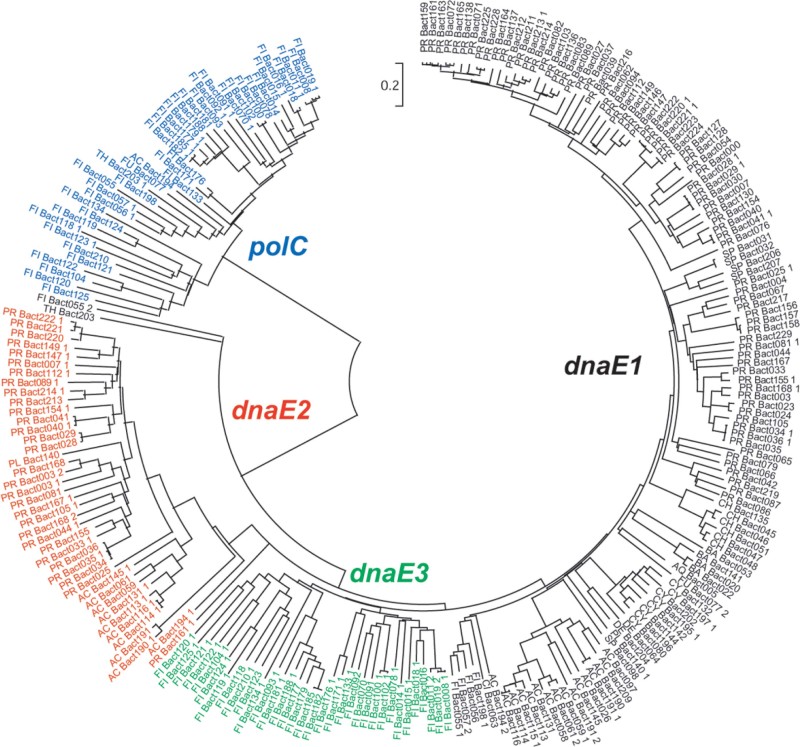
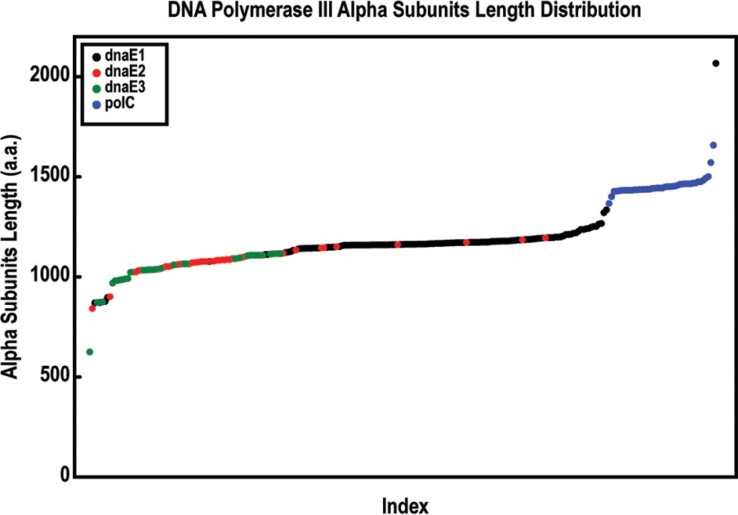
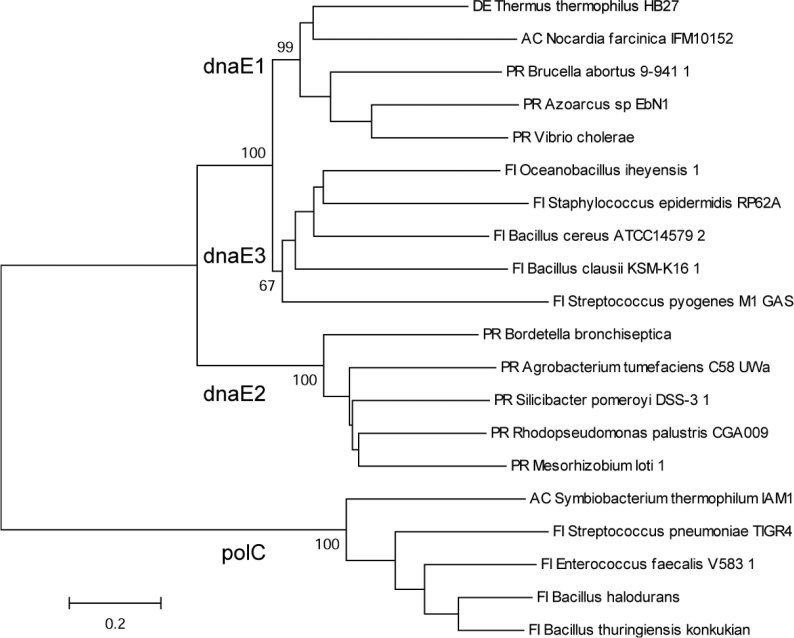
We tried several methods (neighbor-joining, minimum evolution, and maximum parsimony) to assess the phylogeny of the alpha subunit genes and achieved similar results. Based on the phylogenetic analysis, we classified the 248 genes into four simple groups (Figure 1), namely polC, dnaE1, dnaE2, and dnaE3, regardless whether some of the bacteria have multiple alpha subunit genes or not. The polC group is rather distinct from the other three, in which 41 polC genes are clustered together, forming a single group. The lengths of these genes in this group range from 1,367 to 1,658 amino acids (a.a.), with an average of 1,455 a.a., which are longer than those of the other three groups in general. The dnaE genes are divided into three groups according to the cluster results. The lengths of these three groups are quite different: dnaE1 is the longest, with an average length of 1,174 a.a., whereas dnaE2 and dnaE3 are shorter, with average lengths of 1,078 and 1,033 a.a., respectively (Figure 2). To demonstrate the unequivocal relationship, we selected five alpha subunits randomly from each of the four groups, and grouped the subset of twenty alpha subunits into four clusters with significant bootstrap values (Figure 3).
Fig. 1.
Phylogeny of the alpha subunit gene groups. Genomes are labeled in abbreviated characters for the phylum names (Table S1). The Arabian number after the becterium number denotes whether it is classified based on the first alpha subunit gene or other co-existed isoforms. For instance, FI Bact181 1 stands for the Firmicute bacterium No. 181 in the collection, which is classified based on its first alpha subunit gene. AC, Actinobacteria; AQ, Aquificae; BA, Bacteroidetes/Chlorobi group; CH, Chlamydiae/Verrucomicrobia group; CI, Chloroflexi; CY, Cyanobacteria; DE, Deinococcus-Thermus; FI, Firmicutes; FU, Fusobacteria; PL, Planctomycetes; PR, Proteobacteria; TH, Thermotogae; SP, Spirochaetes.
Fig. 2.
The length distribution of all alpha subunit genes in the collection. Among the 248 genes, there are 129 dnaE1 genes (average length of 1,174 a.a.), 44 dnaE2 genes (average length of 1,078 a.a.), 34 dnaE3 genes (average length of 1,033 a.a.), and 41 polC genes (average length of 1,455 a.a.). The shortest sequence is a dnaE3 gene found in Onion yellows phytoplasma (Bact134; 625 a.a.). The longest one is a single dnaE1 gene from Thermus thermophilus HB27 (Bact204; 2,067 a.a.).
Fig. 3.
Phylogeny of four alpha subunit groups from a partial and random selection. Five alpha subunits are randomly selected from each group. The same nomenclature is used as in Figure 1. The trees are constructed with the program MEGA (V3.1) by neighbor-joining with Kimura 2-parameter distance (scale bar). The reliability of the branching orders is estimated by bootstrapping (500 replicates), and bootstrap values (percentage after 500 iterations) for major branches are shown.
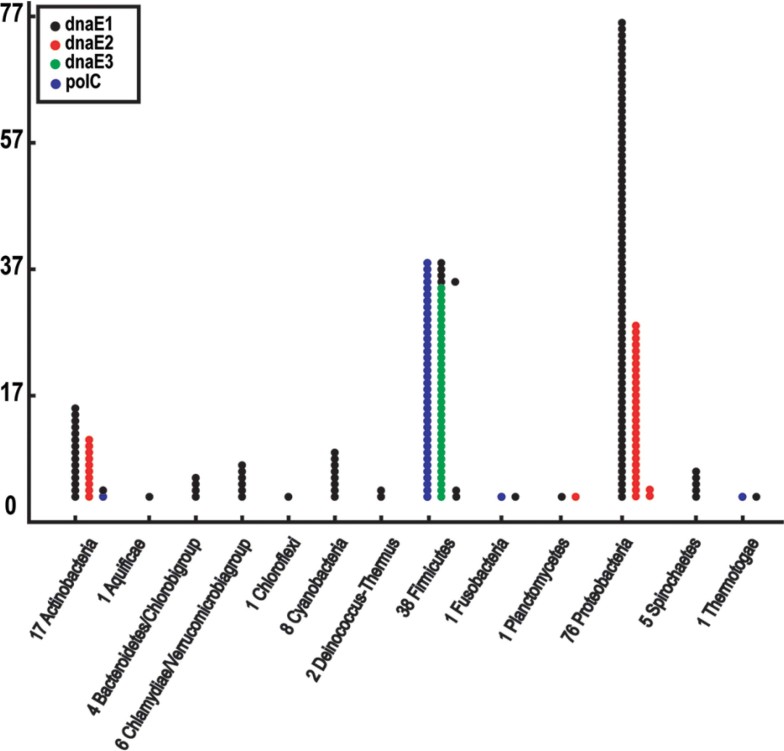
In addition to sequence-based phylogeny, there is ample taxonomic evidence that supports our four group scheme. Firstly, polC and dnaE3 frequently appear together (27) and work on the leading and the lagging strand, respectively, catalyzing the chemical reaction of DNA replication 20., 21.. In contrast, dnaE1 and dnaE2 in general do not appear together in our collection so that dnaE1 could work on both the leading and the lagging strand. Secondly, in the case of dnaE2, it pairs with dnaE1 primarily; however, in very rare occasions it works together with polC (we found only one example of S. thermophilum IAM14863 in the 159 bacteria). It seems that dnaE2 is not able to form a functional homodimer since it always pairs with either dnaE1 or polC, even though some of the bacteria have more than one copy of the gene. Thirdly, the distribution of dnaE1 homodimer and polC/dnaE3 heterodimer is restricted to taxonomic schemes. For instance, polC/dnaE3 combination is dominant in phylum Firmicutes, whereas dnaE1 is mostly found in non-Firmicute bacteria. In contrast, dnaE2 is not related to bacterial taxonomy so that it has been found in different phyla, such as Actinobacteria and Proteobacteria (Figure 4). Fourthly, even for bacteria in the same genus, some have dnaE2 while others do not. For instance, in Proteobacteria, a fraction possesses only dnaE1 whereas others have both dnaE1 and dnaE2; a similar classification result has also been observed in Actinobacteria (data not shown). The best example is the genus Vibrio that has four species sequenced; two of them, Vibrio vulnificus CMCP6 and Vibrio parahaemolyticus, have dnaE1 and dnaE2, whereas the other two, Vibrio fischeri ES114 and Vibrio cholerae, do not have dnaE2. Finally, it is rather curious that how dnaE1 functions in a genome that also has dnaE2, since dnaE1 can form functional homodimers by itself. From the distribution pattern of dnaE2, we believe that the heterodimer of dnaE1 and dnaE2 overrides dnaE1 homodimer when forming the replicase complex.
Fig. 4.
Alpha subunits in different bacterial phyla. One solid circle represents one alpha subunit. We mark the bacterial genome numbers together with phylum names; for example, “17 Actinobacteria” represents 17 bacterial genomes included in Actinobacteria.
Domain analysis
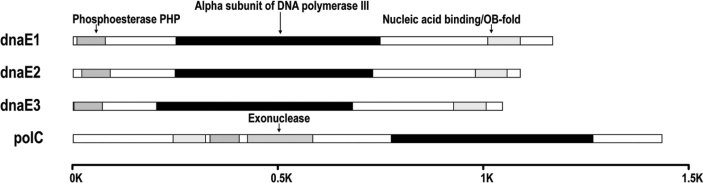
In this part of analysis, we investigated what domain structures and their function classes separate the four alpha subunit groups. We used InterPro (28), a popular database for protein annotations, which integrates information for protein families and functional domains as well as incorporates efforts from different data-mining and data-curating projects, such as PROSITE, PRINTS, Pfam, and ProDom. We initially identified 49 domains in the alpha subunit genes among the eubacterial genomes. However, upon scrutiny on the sequences, we learnt that most of these domains are rare and only shared by less than five genomes. After removing the rare domains, we have 11 domains left. We then classified them into five basic classes based on their functions and positions in the four groups of alpha subunit genes (Table 1). The first class contains two exonuclease domains, IPR006054 (DNA polymerase III and epsilon subunit) and IPR006055 (exonuclease), which are located at the same position in all polC genes and appear to be the same sequences. In fact, the function of epsilon subunit is its exonuclease activity so that it is class-specific. The second class is the alpha subunit domains composed of IPR006308 (DNA polymerase III, alpha subunit, and Gram-positive type), IPR004805 (DNA polymerase III and alpha subunit), and IPR011708 (bacterial DNA polymerase III and alpha subunit); two of them are class-specific, IPR006308 to polC and IPR004805 to dnaE, and one, IPR011708, appears common to all alpha subunit genes. The third class is the phosphoesterase PHP domains, including IPR003141 (phosphoesterase PHP N-terminal) and IPR004013 (PHP C-terminal). This class is universal to all alpha subunit genes but becomes ambiguous in a minor fraction of them. The fourth class is the nucleic acid-binding/OB-fold domains consisting of IPR004365 (nucleic acid binding, OB-fold, and tRNA/helicasetype) and IPR008994 (nucleic acid-binding and OB-fold). Some of these domains become more ambiguous among the genomes. The fifth class has two domains, IPR001093 (IMP dyhydrogenase/GMP reductase) and IPR012337 (polynucleotidyl transferase and ribonuclease H fold); the former appears universal and the latter seems polC-specific, although the sequence-based detection of these domains is usually poor for this class of domains.
Table 1.
Domain Analysis of the Four Groups of Alpha Subunit Genes
| Class | Domain | Domain annotation | polC | dnaE1 | dnaE2 | dnaE3 |
|---|---|---|---|---|---|---|
| (41) | (129) | (44) | (34) | |||
| 1 | IPR006054 | DNA polymerase III, epsilon subunit | 100% | 0 | 0 | 0 |
| IPR006055 | Exonuclease | 100% | 0 | 0 | 0 | |
| 2 | IPR006308 | DNA polymerase III, alpha subunit, Gram-positive type | 100% | 0 | 0 | 0 |
| IPR004805 | DNA polymerase III, alpha subunit | 0 | 100% | 100% | 97% | |
| IPR011708 | Bacterial DNA polymerase III, alpha subunit | 100% | 100% | 100% | 100% | |
| 3 | IPR003141 | Phosphoesterase PHP N-terminal | 95% | 95% | 95% | 85% |
| IPR004013 | PHP C-terminal | 93% | 99% | 75% | 76% | |
| 4 | IPR004365 | Nucleic acid binding, OB-fold, tRNA/helicase-type | 88% | 88% | 95% | 74% |
| IPR008994 | Nucleic acid-binding, OB-fold | 60% | 56% | 55% | 68% | |
| 5 | IPR001093 | IMP dehydrogenase/GMP reductase | 33% | 25% | 23% | 33% |
| IPR012337 | Polynucleotidyl transferase, ribonuclease H fold | 55% | 0 | 0 | 0 | |
We also examined the relative positions of the identified domains in protein alignments (Figure 5; the last class was ignored in the display). The alpha subunit domains are universal but their relative positions are distinct in dnaE (in the middle) and polC (in the C-terminal portion). This most lengthy domain class, with nearly 500 a.a. in length, occupies the core of the alpha subunit gene and manifests DNA-dependent 5’-to-3’ polymerase activity (29). The other two universal domain classes are the phosphoesterase PHP domains and the nucleic acid binding/OB-fold domains. They are shorter than the alpha subunit domains, having a length of nearly 100 a.a. The positions of these domains are conserved only within polC and dnaE. Comparatively, the exonuclease domain class is the most distinctive, for it is polC-specific and positioned in the middle of the gene with a length of nearly 200 a.a., which has 3’-to-5’ proofreading exonuclease activity (21). Since genomes without polC always have an equivalent activity fulfilled by dnaQ, the exonuclease activity seems essential for the DNA polymerase III core enzyme 11., 21..
Fig. 5.
Domain alignments of the four groups of alpha subunit genes. InterProScan has been used to identify the domains. Scale bar marks the length measured by amino acids.
The lengths of dnaE1, dnaE2, and dnaE3 domains showed great differences in their sequences (Table 2). We analyzed the three dnaE groups to search for the common ancestor for each dnaE group, believing that dnaE1 is older than dnaE2 since it is much longer than dnaE2 and dnaE3. When deciphering three shared domains (IPR003141, IPR004365, and IPR011708) of each dnaE group, we found that their lengths are all longer in dnaE1 than in the other two dnaE groups, especially domain IPR011708, whose length is 839 a.a. in dnaE1, compared with 563 a.a. in dnaE2 and 528 a.a. in dnaE3. We suspect that the longer dnaE1 domains may form secondary structures for special functional requirement.
Table 2.
Average Lengths of three Domains of the dnaE Groups
| Group | Domain length (a.a.) |
||
|---|---|---|---|
| IPR003141 | IPR004365 | IPR011708 | |
| dnaE1 | 134 | 110 | 839 |
| dnaE2 | 105 | 83 | 563 |
| dnaE3 | 68 | 81 | 528 |
Motif analysis
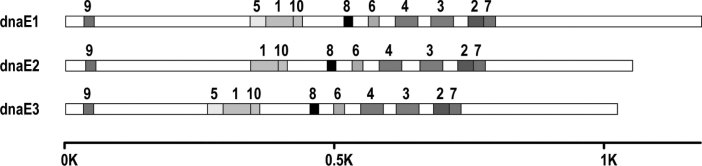
To find further differences among the dnaE groups, we analyzed the protein motifs, which are subsets of domain structures, by using the MEME tool for discovering motifs or highly conserved regions with its default settings 30., 31.. We detected ten motifs among the dnaE genes, nine of which are shared by all members (Figure 6). Eight of the nine motifs were found in the alpha subunit domains while one (motif 9) was found in the phosphoesterase PHP domains. Motif 5 is the only one standing out that is different from the other nine motifs in its distribution among the genomes; it was identified in dnaE1 and dnaE3 genes but rarely in dnaE2 genes. Detailed analysis revealed that motif 5 is part of the alpha subunit domain and has a unique amino acid sequence, “LD-VINQMGFPGYFLIVMEFIQWSKDNGIP”, which is almost undetectable in dnaE2 genes. No recognizable motif was found in the nucleic acid-binding/OB-fold domains with similar parameters in the same way. In addition, we further investigated individual portions of the alpha subunit genes by varying MEME parameters but still failed in finding any meaningful motifs in the phosphoesterase PHP domains. However, this procedure paid off in finding another motif unique to dnaE2 in the nucleic acid-binding/OB-fold domains, which is a short sequence, “VVHLVAQRLED”, at the end of this domain (data not shown). Collectively, dnaE1 and dnaE3 share a common motif in the alpha subunit domain, while dnaE2 has a unique motif in its nucleic acid-binding/OB-fold domain.
Fig. 6.
Protein motifs among the three groups of dnaE genes. The MEME tool has been employed for this analysis. Each motif describes a pattern of a fixed width, and no gaps are allowed in MEME motifs. MEME numbers the motifs consecutively starting from 1 as it found them, and the most statistically significant (low E-value) motifs are usually found first. Relative to dnaE1 and dnaE3, dnaE2 appears to miss motif 5. Scale bar indicates amino acid positions.
Discussion
Our results clearly pointed out two important observations about the grouping of alpha subunit genes; one is the existence of the four distinct groups in eubacterial genomes, and the other is that the grouping scheme always has exceptions albeit mostly minor. We do believe that the grouping scheme is related to the nucleotide compositional dynamics of prokaryotic genomes, which exceeds taxonomy, so that it breaks the boundaries of the bacterial taxa, even within a genus. For instance, polC is characteristic in phylum Firmicutes and co-exists with dnaE3 (89% in Firmicutes), except in four genomes in class Clostridia of Firmicutes where polC co-exists with dnaE1. Having detected these four dnaE1 genes, we found that they are clustered together in dnaE1 group and are homologous to the dnaE1 gene of S. thermophilum IAM14863 in phylum Actinobacteria. Meanwhile, phylogenetic analysis (Figure 1) showed that the polC gene of S. thermophilum IAM14863 is homologous to the polC genes of the above four genomes in Clostridia and also homologous to that of one genome in phylum Fusobacteria. The dnaE1 gene of S. thermophilum IAM14863 is also different from those of other bacteria in Actinobacteria; it is clustered with the dnaE1 gene of Dehalococcoides ethenogenes 195 (Bact063) in phylum Chloroflexi (which contains only one dnaE1) and the four dnaE1 genes of the genomes in Clostridia, whereas the other dnaE1 genes in Actinobacteria are clustered together. These results of the detection of homologous dnaE1 and polC genes in distant species suggests that horizontal gene transfer occurs frequently among prokaryotic genomes (32).
This co-existence between polC and dnaE3/dnaE1 suggests that polC can form heterodimers with both dnaE1 and dnaE3 but prefers dnaE3 in most of the cases. Besides the bacteria in Firmicutes, three bacteria in other phyla also possess polC: Fusobacterium nucleatum in Fusobacteria and Thermotoga maritima in Thermotogae both harbor polC and dnaE1. The third one, S. thermophilum IAM14863 in Actinobacteria, is even more complex, which contains one polC, one dnaE1, and one dnaE2; however, it seems that dnaE2 may pair with dnaE1 to form a heterodimer just like other bacteria in Actinobacteria. Alternatively, polC might be a latecomer that has not yet made its signature visible in this genome. It would be interesting to know what is going on in this bacterium in terms of DNA replication.
PolC and dnaE1 play a dominant role in the replication in Firmicutes and non-Firmicutes, respectively. DnaE3 is present only together with polC in Firmicutes. DnaE2 is present only in the dnaE1/dnaE2 type of bacteria (mostly Actinobacteria and Proteobacteria), and pairs with dnaE1, while very rarely with polC, and never dnaE3 (at least in our current collection), despite the fact that horizontal gene transfer is frequent. Another observable fact is the compatibility among these genes. For instance, dnaE2 and dnaE3 are never found in the same genome so that they should be incompatible.
We also reviewed the literature to see what has been noticed in alpha subunit diversity by other investigators. It is clear that the difference of dnaE genes in E. coli and B. subtilis was described previously (20). Our analysis indicated that the dnaEBS gene discovered in B. subtilis actually belongs to the dnaE3 group in our study. It is reported that dnaE genes contain two different isoforms; one is encoded by an intact gene, the other is encoded by a split dnaE1 and dnaE2 interrupted by an intein sequence (24). We identified these “split dnaE genes” in four Cyanobacteria genomes within our dataset: Nostoc sp., Synechococcus elongatus PCC 6301, Synechocystis sp. PCC6803, and Thermosynechococcus elongates. The four genes are all members of the dnaE1 group. The dnaE2 genes found in other studies 25., 26. are exclusive members of our dnaE2 group although they were assigned with different functions.
Conclusion
We have classified eubacterial alpha subunit genes into four different groups: polC, dnaE1, dnaE2, and dnaE3. DnaE1 functions as homodimers and the rest form heterodimers. Phylogenetic analysis have suggested that dnaE and polC might have diverged from the same ancestor and the former further split into three subclasses. Two isoforms of dnaE1 form a homodimer that catalyzes DNA replication. DnaE2 might have subsequently lost a small but significant motif and is flexible to dimerize with dnaE1 and polC. DnaE3 is the least flexible so that it pairs only with polC, becoming Firmicute-specific as a result. We hope that our consolidated naming convention will be used in future gene annotations for eubacterial genomes.
Materials and Methods
We extracted alpha subunit genes from 191 bacterial sequences deposited in GenBank (May 19, 2005; ftp.ncbi.nlm.nih.gov/genomes/Bacteria/), which cover 159 species, and classified them according to the NCBI taxonomy database. For convenience, we numbered the bacteria in an alphabetic order and assigned abbreviations (Table S1). For those taxonomic groups with more than one genome sequenced, we randomly selected one to represent the species. Since incomplete annotation may lead to oversight, we also searched each genome with the extracted sequence with the BLAT tools (33). We detected the alpha subunit genes that are diverged in sequences and lengths, which are far shorter than the average sizes. As a result, we obtained 248 alpha subunit genes from the 159 genomes. For phylogenetic analysis, we used ClustalW for amino acid sequence alignments (34) and MEGA (bootstrap neighbor-joining method) (35) for phylogeny. The InterProScan (28) and MEME 30., 31. tools were used for domain and motif analyses.
Authors’ contributions
XQZ carried out sequence extraction, sequence analysis, and drafted the manuscript. JFH performed the sequence collection and biological information classification. JY designed and supervised the project as well as revised the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors have declared that no competing interests exist.
Acknowledgements
We thank Zhang Zhang and Xiang-Jun Tian for their constructive comments on this manuscript. This work was supported by a grant from the Chinese Academy of Sciences awarded to JY (KSCX2-SW-331).
Supporting Online Material
http://evolution.genomics.org.cn/supp/supp.htm
Table S1
References
- 1.Goodman M.F. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu. Rev. Biochem. 2002;71:17–50. doi: 10.1146/annurev.biochem.71.083101.124707. [DOI] [PubMed] [Google Scholar]
- 2.Lewin B. Oxford University Press; Oxford, UK: 2004. Genes VIII. [Google Scholar]
- 3.Lehman I.R. Discovery of DNA polymerase. J. Biol. Chem. 1989;278:34733–34738. doi: 10.1074/jbc.X300002200. [DOI] [PubMed] [Google Scholar]
- 4.Tang M. UmuD’2C is an error-prone DNA polymerase, Escherichia coli pol V. Proc. Natl. Acad. Sci. USA. 1999;96:8919–8924. doi: 10.1073/pnas.96.16.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kobayashi S. Fidelity of Escherichia coli DNA polymerase IV. Preferential generation of small deletion mutations by dNTP-stabilized misalignment. J. Biol. Chem. 2002;277:34198–34207. doi: 10.1074/jbc.M204826200. [DOI] [PubMed] [Google Scholar]
- 6.Patel P.H. Prokaryotic DNA polymerase I: evolution, structure, and “base flipping” mechanism for nucleotide selection. J. Mol. Biol. 2001;308:823–837. doi: 10.1006/jmbi.2001.4619. [DOI] [PubMed] [Google Scholar]
- 7.McHenry C.S., Crow W. DNA polymerase III of Escherichia coli. Purification and identification of subunits. J. Biol. Chem. 1979;254:1748–1753. [PubMed] [Google Scholar]
- 8.Kelman Z., O’Donnell M. DNA polymerase III holoenzyme: structure and function of a chromosomal replicating machine. Annu. Rev. Biochem. 1995;64:171–200. doi: 10.1146/annurev.bi.64.070195.001131. [DOI] [PubMed] [Google Scholar]
- 9.Herendeen D.R., Kelly T.J. DNA polymerase III: running rings around the fork. Cell. 1996;84:5–8. doi: 10.1016/s0092-8674(00)80069-0. [DOI] [PubMed] [Google Scholar]
- 10.McHenry C., Kornberg A. DNA polymerase III holoenzyme of Escherichia coli. Purification and resolution into subunits. J. Biol. Chem. 1977;252:6478–6484. [PubMed] [Google Scholar]
- 11.Scheuermann R.H., Echols H. A separate editing exonuclease for DNA replication: the epsilon subunit of Escherichia coli DNA polymerase III holoenzyme. Proc. Natl. Acad. Sci. USA. 1984;81:7747–7751. doi: 10.1073/pnas.81.24.7747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maki H. The polymerase subunit of DNA polymerase III of Escherichia coli. I. Amplification of the dnaE gene product and polymerase activity of the alpha subunit. J. Biol. Chem. 1985;260:12982–12986. [PubMed] [Google Scholar]
- 13.Maki H., Kornberg A. The polymerase subunit of DNA polymerase III of Escherichia coli. II. Purification of the alpha subunit, devoid of nuclease activities. J. Biol. Chem. 1985;260:12987–12992. [PubMed] [Google Scholar]
- 14.Maki H., Kornberg A. Proofreading by DNA polymerase III of Escherichia coli depends on cooperative interaction of the polymerase and exonuclease subunits. Proc. Natl. Acad. Sci. USA. 1987;84:4389–4392. doi: 10.1073/pnas.84.13.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maki H. DNA polymerase III holoenzyme of Escherichia coli. IV. The holoenzyme is an asymmetric dimer with twin active sites. J. Biol. Chem. 1988;263:6570–6578. [PubMed] [Google Scholar]
- 16.Stukenberg P.T. Mechanism of the sliding beta-clamp of DNA polymerase III holoenzyme. J. Biol. Chem. 1991;266:11328–11334. [PubMed] [Google Scholar]
- 17.Onrust R. Assembly of a chromosomal replication machine: two DNA polymerases, a clamp loader, and sliding clamps in one holoenzyme particle. J. Biol. Chem. 1995;270:13366–13377. doi: 10.1074/jbc.270.22.13366. [DOI] [PubMed] [Google Scholar]
- 18.Inoue R. Genetic identification of two distinct DNA polymerases, DnaE and PolC, that are essential for chromosomal DNA replication in Staphylococcus aureus. Mol. Genet. Genomics. 2001;266:564–571. doi: 10.1007/s004380100564. [DOI] [PubMed] [Google Scholar]
- 19.Barnes M.H. DNA polymerases of low-GC gram-positive eubacteria: identification of the replication-specific enzyme encoded by dnaE. J. Bacteriol. 2002;184:3834–3838. doi: 10.1128/JB.184.14.3834-3838.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dervyn E. Two essential DNA polymerases at the bacterial replication fork. Science. 2001;294:1716–1719. doi: 10.1126/science.1066351. [DOI] [PubMed] [Google Scholar]
- 21.Bruck I., O’Donnell M. The DNA replication machine of a gram-positive organism. J. Biol. Chem. 2000;275:28971–28983. doi: 10.1074/jbc.M003565200. [DOI] [PubMed] [Google Scholar]
- 22.Bruck I. The essential C family DnaE polymerase is error-prone and efficient at lesion bypass. J. Biol. Chem. 2003;278:44361–44368. doi: 10.1074/jbc.M308307200. [DOI] [PubMed] [Google Scholar]
- 23.Wu H. Protein trans-splicing by a split intein encoded in a split DnaE gene of Synechocystis sp. PCC6803. Proc. Natl. Acad. Sci. USA. 1998;95:9226–9231. doi: 10.1073/pnas.95.16.9226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang Y.P., Ito J. DNA polymerase C of the thermophilic bacterium Thermus aquaticus: classification and phylogenetic analysis of the family C DNA polymerases. J. Mol. Evol. 1999;48:756–769. doi: 10.1007/pl00006520. [DOI] [PubMed] [Google Scholar]
- 25.Galhardo R.S. An SOS-regulated operon involved in damage-inducible mutagenesis in Caulobacter crescentus. Nucleic Acids Res. 2005;33:2603–2614. doi: 10.1093/nar/gki551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boshoff H.I. DnaE2 polymerase contributes to in vivo survival and the emergence of drug resistance in Mycobacterium tuberculosis. Cell. 2003;113:183–193. doi: 10.1016/s0092-8674(03)00270-8. [DOI] [PubMed] [Google Scholar]
- 27.Koonin E.V., Bork P. Ancient duplication of DNA polymerase inferred from analysis of complete bacterial genomes. Trends Biochem. Sci. 1996;21:128–129. [PubMed] [Google Scholar]
- 28.Apweiler R. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 2001;29:37–40. doi: 10.1093/nar/29.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewin B. Oxford University Press; Oxford, UK: 2000. Genes VII. [Google Scholar]
- 30.Grundy W.N. ParaMEME: a parallel implementation and a web interface for a DNA and protein motif discovery tool. Comput. Appl. Biosci. 1996;12:303–310. doi: 10.1093/bioinformatics/12.4.303. [DOI] [PubMed] [Google Scholar]
- 31.Grundy W.N. Meta-MEME: motif-based hidden Markov models of protein families. Comput. Appl. Biosci. 1997;13:397–406. doi: 10.1093/bioinformatics/13.4.397. [DOI] [PubMed] [Google Scholar]
- 32.Lawrence J.G., Ochman H. Molecular archaeology of the Escherichia coli genome. Proc. Natl. Acad. Sci. USA. 1998;95:9413–9417. doi: 10.1073/pnas.95.16.9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kent W.J. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ebedes J., Datta A. Multiple sequence alignment in parallel on a workstation cluster. Bioinformatics. 2004;20:1193–1195. doi: 10.1093/bioinformatics/bth055. [DOI] [PubMed] [Google Scholar]
- 35.Kumar S. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1