Abstract
To monitor the dissemination of resistance genes into the environment, we determined the occurrence of tetracycline resistance (Tcr) genes in groundwater underlying two swine confinement operations. Monitoring well networks (16 wells at site A and 6 wells at site C) were established around the lagoons at each facility. Groundwater (n = 124) and lagoon (n = 12) samples were collected from the two sites at six sampling times from 2000 through 2003. Total DNA was extracted, and PCR was used to detect seven Tcr genes [tet(M), tet(O), tet(Q), tet(W), tet(C), tet(H), and tet(Z)]. The concentration of Tcr genes was quantified by real-time quantitative PCR. To confirm the Tcr gene source in groundwater, comparative analysis of tet(W) gene sequences was performed on groundwater and lagoon samples. All seven Tcr genes were continually detected in groundwater during the 3-year monitoring period at both sites. At site A, elevated detection frequency and concentration of Tcr genes were observed in the wells located down-gradient of the lagoon. Comparative analysis of tet(W) sequences revealed that the impacted groundwater contained gene sequences almost identical (99.8% identity) to those in the lagoon, but these genes were not found in background libraries. Novel sequence clusters and unique indigenous resistance gene pools were also found in the groundwater. Thus, antibiotic resistance genes in groundwater are affected by swine manure, but they are also part of the indigenous gene pool.
The emergence and increasing prevalence of antibiotic-resistant bacteria that could lead to life-threatening, untreatable disease has been reported, especially in the medical field. The widespread use of antibiotics in human medicine could enhance the selection of antibiotic resistance. This is a significant issue not only with regard to human health but also for animal health and food production. Antibiotics are routinely used in the livestock industry for therapeutic (disease control), prophylactic (disease prevention), and subtherapeutic (growth promotion) purposes. During the mid-1990s, the consumption of tetracycline by farm animals in the United States was estimated at 3.5 × 106 kg/year (9). Antibiotics can select for resistant microorganisms in the gastrointestinal tracts of animals, particularly when used at subtherapeutic levels (38), providing a potential reservoir for dissemination of drug-resistant bacteria into other animals, humans, and the environment.
Although we have no direct knowledge of the ancestral tetracycline resistance gene(s), it may have evolved from housekeeping genes (7, 22) for protecting the integrity of ribosomal protein synthesis (ribosomal protection proteins [RPPs]) or maintaining intracellular ionic constituent concentrations (efflux pump proteins). These antibiotic resistance genes were then disseminated by interspecies transfer mediated by a variety of agents, such as plasmids, transposons, and bacteriophages (9, 29). Once resistance genes are introduced into the environment, they are also exposed to selective pressure, such as antibiotics produced by indigenous antibiotic producers in soil. However, selection can occur in the environment without antibiotic selective pressure (2). Gilliver et al. (12) reported that antibiotic-resistant bacteria had been found in wild rodents that had never been exposed to antibiotics. Therefore, antibiotic resistance genes might be distributed and preserved in the broader environment with or without antibiotic selective pressure.
Lagoon and pit systems are typically used for manure management in confinement animal feeding operations (CAFOs) in the United States. It is of concern that farm-generated resistant bacteria could easily migrate into soil, groundwater, and surface water via seepage from the manure lagoons and from the land application of manure. In the United States, groundwater constitutes about 40% of the water used for public supply and provides drinking water for more than 97% of the rural population (36). To assess the relationship between CAFOs and groundwater quality, the occurrence and extent of contamination from these facilities must be determined and made available. Recent monitoring studies have shown that seepage from animal waste lagoons has affected groundwater quality (17, 23). Krapac et al. (18, 19) found chemical and biological indicators of manure contamination, such as chloride and fecal enterococci, at significant levels in groundwater samples collected near CAFO lagoons and deep pits.
Recently, antibiotic resistance analysis has been employed to identify sources of fecal pollution (13, 26, 39, 40). Although traditional microbiological methods such as cultivation and phenotypic testing are commonly used, these approaches are time-consuming and biased towards the selection of bacteria that are easily grown in culture media. In addition, bacteria have been shown to readily exchange genetic information in the environment, permitting the acquisition of resistance genes or mechanisms already present in the environment and their transfer from one microbe to another (3, 11, 28). There is the potential for resistance genes, selected and excreted, to be transferred into bacteria in the soil, surface water, and groundwater around the facilities; thus, a broad range of bacteria in those environments can carry a variety of resistance genes. Traditional cultivation-based methods are not adequate to detect a broader range of resistant bacteria for monitoring the ecology of antibiotic resistance genes in the environment. In contrast, analysis of antibiotic resistance genes using molecular PCR methods can provide a rapid and sensitive method for tracking the source of fecal contamination in groundwater without cultivation. In our previous studies, using PCR methods we confirmed the distribution of tetracycline resistance (Tcr) genes in animal gastrointestinal tracts (4, 5) and in groundwater surrounding two confinement facilities (4, 8). However, these studies were based on a single sampling event; thus, long-term surveillance is necessary to assess the dynamics of resistance genes in the environment.
The aim of this study was to monitor the long-term occurrence, distribution, and quantity of Tcr genes in waste lagoons and groundwater underlying swine confinement facilities. Four Tcr genes [tet(M), tet(O), tet(Q), and tet(W)] encoding RPPs and three genes [tet(C), tet(H), and tet(Z)] encoding efflux pumps were selected, since these Tcr genes were frequently detected in our previous study (4, 8). PCR for detection of the Tcr genes was carried out at two swine confinement facilities between 2000 and 2003. The level of Tcr genes in groundwater is an important parameter for determining the magnitude of the impact of CAFOs on the environment. To quantify the seven Tcr genes in groundwater, we employed a real-time quantitative PCR (qPCR) assay that allowed precise and sensitive quantification of the target genes (21, 30, 33). In addition, comparative sequence analysis of tet(W) in lagoon and groundwater samples was carried out to determine the source of Tcr genes in groundwater.
MATERIALS AND METHODS
Sampling.
The operations and site hydrogeology of two commercial swine confinement facilities, designated site A and site C, were described previously (8, 23). Site A started operations in February of 1995 and is a 4,000-pig finishing operation (Fig. 1). The facility operates with a two-stage waste handling system in which a concrete settling basin collects most of the solids prior to the supernatant liquid passively moving into an unlined earthen lagoon. No special construction techniques were used to compact the soil during lagoon construction. The concrete settling basin is periodically pumped and the manure applied to crop fields both on-site (the field east of wells A1 and A2) and off-site (the far-northern field from well A7). Site C is a farrowing and nursery operation housing up to 2,500 sows that began operation in 1992 (Fig. 1). The facility uses a single-stage lagoon. The waste has never been applied to the crop fields surrounding the lagoon. For the purpose of therapy, preventing diseases, or growth promotion, bacitracin, chlortetracycline, and tylosin are used at site A, while chlortetracycline, penicillin, and tylosin are employed at site C.
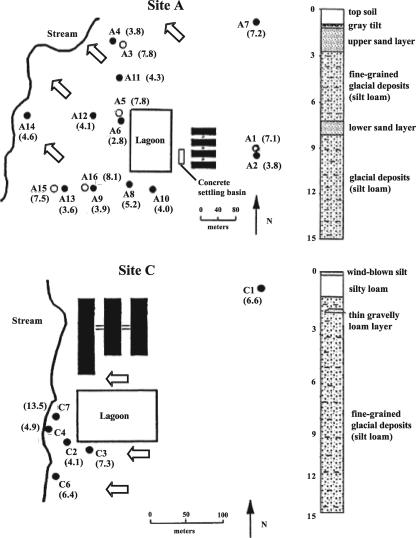
FIG. 1.
Locations of monitoring wells and direction of groundwater flow at sites A and C. Stratigraphic columns indicate the locations and characteristics of sand layers. Large open arrows indicate the direction of groundwater flow. The locations of monitoring wells are indicated by circles; open circles at site A represent nested wells screened in the deeper sand layer. Numbers in parentheses are well depths, in meters. The black rectangles represent confinement buildings.
The site hydrogeology is significantly different at sites A and C (Fig. 1). Two relatively shallow permeable sand layers underlie site A, whereas site C is underlain mainly by a silty loam material. The multiple sand layers make site A particularly susceptible to leachate migration from the lagoon, while at site C the relatively impermeable material and limited groundwater resources reduce the potential for lagoon seepage migration. A groundwater monitoring network consisting of 16 wells was installed at site A. Twelve wells were installed in the upper sand layer, and four wells were installed in the lower sand layer (Fig. 1). Six monitoring wells were installed at depths of less than 11 m at site C.
A total of 124 groundwater and 12 lagoon samples were collected between 2000 and 2003. Samples were collected at site A in April 2000, May and September 2001, January and April 2002, and March 2003. Samples were collected at site C in April 2000, May and August 2001, January and May 2002, and February 2003. Sampling procedures were as described previously (8, 17, 23). Samples were stored in clean, sterilized bottles and kept on ice in the field. Because samples were to be used for both DNA analysis and bacterial isolation (to be detailed in a future paper), samples were stored at 4°C (to limit bacterial mortality due to freezing) prior to analysis for up to 1 week.
Sample processing and DNA extraction.
Groundwater (1-liter) and lagoon (100-ml) samples were centrifuged at 17,700 × g for 20 min at 4°C. The supernatant was discarded, and the pellets were washed three times with 10 ml of phosphate-buffered saline (120 mM NaH2PO4 [pH 8.0], 0.85% NaCl). Total DNA was extracted from the pellets using a freeze-thaw method (35). Briefly, the pellets were resuspended in 400 μl of lysis solution (0.15 M NaCl, 0.1 M EDTA [pH 8.0]) containing 15 mg of lysozyme/ml and incubated at 37°C for 2 h, and then 400 μl of 0.1 M NaCl-0.5 M Tris-HCl (pH 8.0)-10% sodium dodecyl sulfate was added. Suspensions were incubated for 30 min at 37°C followed by three cycles of freezing at −80°C and thawing in a 65°C water bath. Proteinase K was added to a final concentration of 50 μg/ml, the mixture was incubated for 30 min at 37°C and centrifuged, and supernatant was collected. The crude DNA was purified with polyvinylpolypyrrolidone and Sepharose 2B (Sigma Chemical Co., St. Louis, MO), as described by Zhou et al. (41) and Miller (24), respectively. DNA concentration was measured using a DU 7500 spectrophotometer (Beckman, Fullerton, CA) and adjusted to 10 ng/μl.
PCR detection.
The class-specific primer sets used to amplify the seven Tcr genes in this study are shown in Table 1. PCR was conducted using AmpliTaq Gold DNA polymerase (Applied Biosystems, Foster City, CA). A reaction mixture containing 0.5 μM of each primer, 1.5 mM MgCl2, 0.2 mM each deoxyribonucleotide triphosphate, PCR buffer II, 1.25 U of AmpliTaq Gold DNA polymerase, and 10 ng of template DNA in a total volume of 25 μl was prepared. Typical cycle conditions were employed for the primer sets targeting tet(M), tet(O), tet(Q), tet(W), tet(C), and the 16S rRNA gene; meanwhile, a two-temperature step-down PCR cycle was used for the primer sets for tet(H) and tet(Z). The annealing temperatures used for amplification of different target genes are shown in Table 1. The temperature program for typical cycle conditions consisted of denaturation at 94°C for 10 min, followed by 40 cycles of 94°C for 30 s, annealing (Table 1) for 30 s, and extension at 72°C for 30 s and a final extension at 72°C for 10 min. The temperature program for the two-temperature step-down PCR was as follows: 94°C for 10 min, 35 [for tet(Z)] or 40 [for tet(H)] cycles of 94°C for 30 s, annealing (high temperature) (Table 1) for 30 s, and extension at 72°C for 30 s followed by 15 [for tet(Z)] or 10 [for tet(H)] cycles consisting of 94°C for 30 s, 63°C (low temperature) for 30 s, and extension at 72°C for 30 s and a final extension at 72°C for 10 min. PCR amplification was performed with a GeneAmp 2700 thermal cycler (Applied Biosystems). PCR product aliquots were analyzed by electrophoresis on 2.0% (wt/vol) agarose gel and were stained with ethidium bromide.
TABLE 1.
PCR primers used in this study
| Primera | Target gene | Sequence (5′-3′) | Target site (nucleotide of gene) | Annealing temp (°C) | Reference |
|---|---|---|---|---|---|
| TetM-FW | tet(M) | ACAGAAAGCTTATTATATAAC | 58 | 55 | 4 |
| TetM-RV | TGGCGTGTCTATGATGTTCAC | 228 | |||
| TetO-FW | tet(O) | ACGGARAGTTTATTGTATACC | 58 | 60 | 4 |
| TetO-RV | TGGCGTATCTATAATGTTGAC | 228 | |||
| TetQ-FW | tet(Q) | AGAATCTGCTGTTTGCCAGTG | 62 | 63 | 4 |
| TetQ-RV | CGGAGTGTCAATGATATTGCA | 228 | |||
| TetW-FW | tet(W) | GAGAGCCTGCTATATGCCAGC | 61 | 64 | 4 |
| TetW-RV | GGGCGTATCCACAATGTTAAC | 228 | |||
| TetW-1194RVb | CGACAGCAAAGCGGAAACA | 1212 | 67 | This study | |
| TetC-FW | tet(C) | GCGGGATATCGTCCATTCCG | 96 | 70 | 5 |
| TetC-RV | GCGTAGAGGATCCACAGGACG | 302 | |||
| TetH-FW | tet(H) | CAGTGAAAATTCACTGGCAAC | 102 | 66 | 5 |
| TetH-RV | ATCCAAAGTGTGGTTGAGAAT | 287 | |||
| TetZ-FW | tet(Z) | CCTTCTCGACCAGGTCGG | 63 | 68 | 5 |
| TetZ-RV | ACCCACAGCGTGTCCGTC | 269 | |||
| 341F | 16S rRNA | CCTACGGGAGGCAGCAG | 341 | 60 | 22 |
| 534R | TTACCGCGGCTGCTGGCAC | 534 |
All primer sets can detect a single copy of plasmid DNA containing respective target DNA fragment.
Used in source tracking analysis.
To determine the detection limit for each primer set, PCR amplification was performed on serially diluted cloned plasmids containing the respective target DNA fragments (see below). To check the negative effects of PCR-inhibiting substances in the environmental samples, two-step validation was employed. First, PCR amplification of the 16S rRNA gene (25) was performed for all samples prior to detection of Tcr genes, and strong band intensity was confirmed on the agarose gel. Then, if the samples were negative for Tcr gene detection, cloned plasmids containing the respective target DNA fragments were diluted to 10° (i.e., single) copies and used to spike those samples, and PCR amplification of the target was confirmed.
Patterns of detection of all Tcr genes from all samples were analyzed using principal components analysis (PCA). Genes detected in a sample were scored as 1, while genes not detected were scored as 0, and these presence-absence data were used for PCA. PCA was performed using the PRIMER 5 for Windows software package (10).
Standard control for real-time qPCR assay.
Plasmid DNA to be used as the standard control in PCR was obtained by PCR cloning using the primer sets shown in Table 1. Amplified target DNA of a control strain positive for relevant genes was separated on an agarose gel, and the products were excised from the gel. The gel fragments were purified with a QIAquick gel extraction kit (QIAGEN, Valencia, CA), ligated into the pGEM-T Easy vector (Promega, Madison, WI), and transformed into competent Escherichia coli JM109 cells by heat shock. Clones containing the correct insert were confirmed by PCR amplification with the appropriate primer sets. Plasmids were purified by using a QIAprep spin miniprep kit (QIAGEN), and the plasmid concentration was determined by spectroscopy. The copy number of each plasmid was calculated using the molecular weight of nucleic acid and the length (in base pairs) of the cloned plasmid.
Validation of DNA extraction and real-time qPCR assay.
DNA extraction efficiency and the accuracy of the real-time PCR assay for Tcr genes and the 16S rRNA gene were validated by using known amounts of target to spike environmental samples. E. coli cells carrying cloned plasmids for various target DNA fragments were used as the inoculum for the spiking trial. Each portion of target-carrying E. coli cells was incubated in liquid Luria-Bertani medium overnight. To determine the plasmid copy number (i.e., copy number of the target) in each culture, plasmid DNA was extracted from the overnight-incubated cultures. The plasmid copy number was calculated as described above. Each target-carrying E. coli culture was serially diluted to yield a range from 103 to 108 copies of the plasmid DNA. The dilution series of cultures containing known amounts of each target DNA fragment was spiked into 0.1 g of lagoon samples. DNA was extracted from serially diluted E. coli cultures, nonspiked lagoon samples, and target-spiked lagoon samples by the freeze-thaw method and was purified with polyvinylpolypyrrolidone and Sepharose, as described above. Purified DNA was resuspended in 50 μl of Tris-EDTA buffer. To check the recovery of the target from spiked samples, 0.5 μl of DNA was used for real-time qPCR quantification. Since the lagoon samples used in the spiking trial already contained a high copy number of the16S rRNA gene, it was impossible to spike E. coli cells to exceed the indigenous copy number of the 16S rRNA gene. Therefore, recovery of the 16S rRNA gene was determined using the DNA extracted from serially diluted E. coli cultures. The copy number of the 16S rRNA gene in E. coli cultures was calculated by the microscopic count of E. coli cells in the culture multiplied by 7, which is the reported copy number of the 16S rRNA operon in E. coli (16). The coefficient of variation (CV) for each real-time qPCR assay was determined by repeating the real-time qPCR quantification using the same sample. Overall variation of quantification was evaluated by repeating the DNA extraction and the quantification of each target.
Quantification of Tcr genes in lagoon and groundwater samples by real-time qPCR.
PCR amplifications for the quantification of Tcr genes and the 16S rRNA gene in total DNA extracted from the groundwater and lagoon samples were performed with a GeneAmp 9600 thermal cycler coupled with a GeneAmp 5700 sequence detection system (Applied Biosystems, Foster City, CA). The SYBR Green PCR Master Mix provided by Applied Biosystems was used with 10 ng of template DNA for PCR amplification. The thermal profile for all SYBR Green PCRs was the same as that for conventional PCR described above. The 10-fold dilution series of the plasmid standard for the respective genes was run along with the unknown samples for the corresponding gene controls, and each sample was duplicated in each reaction. Quantification was made using standard curves obtained from the amplification profile of known concentrations of the plasmid standard for the respective genes. All reactions were repeated in duplicate to confirm the results. Wells A6, A8, A9, A11, A12, and A16 underwent a 2-year quantitative monitoring effort for the Tcr genes, whereas wells C2, C3, C4, and C6 were monitored at site C.
The data were processed with GeneAmp 5700 SDS software (Applied Biosystems, Foster City, CA). To exclude the influence of unequal amplification efficiency among the samples, the assay values for all Tcr genes were normalized to that of the 16S rRNA gene. The normalized assay values were analyzed using the GLM (general linear models) procedure of SAS. Comparisons between means were performed using Tukey's t test at a significance level of 0.05.
Source tracking analysis of tet(W) in groundwater.
In order to identify the source of Tcr genes in groundwater, comparative sequence analysis was conducted using tet(W), which was selected based on its distribution in samples from previous work. The PCR primer TetW-1194RV, specific to tet(W), was designed to obtain a 1,152-bp fragment of tet(W) in combination with the primer TetW-FW (Table 1). Since all tet(OW) mosaic genes have a tet(O) signature sequence at the position of TetW-FW (31, 32), this primer combination allowed specific amplification of tet(W) without cross-reaction of tet(OW) mosaic genes. Samples from wells A7, A8, and A14, the lagoon and concrete settling basin at site A, wells C1, C2, and C4, and the lagoon at site C were used to construct the tet(W) clone libraries. All samples were collected in March 2003 at site A and February 2003 at site C, and DNA was extracted as described above. PCR was performed with 25 pmol of each primer and an ExTaq PCR kit (Takara Bio, Otsu, Japan) by using a final reaction mixture volume of 25 μl. Purified DNA (10 ng) was used as the template for PCR. The cycle conditions consisted of denaturation at 94°C for 5 min, followed by 35 cycles consisting of 94°C for 30 s, 67°C for 30 s, and extension at 72°C for 1 min and a final extension at 72°C for 7 min. Because of insufficient PCR product from groundwater samples, a second PCR was performed under the same conditions except that 1 μl of the PCR product from the first PCR was used as a template.
PCR products were cloned with a TOPO-TA cloning kit (Invitrogen, Carlsbad, CA), and recombinant plasmids were extracted as described above. DNA analysis for 10 recombinant plasmids from each library was performed for both strands (primers M13F and M13R) by the University of Illinois Biotechnology Center. Sequences from the tet(W) libraries were compared with the tet(W) sequences available in the GenBank database using the BLAST program. Sequences were aligned using the multiple sequence alignment software CLUSTALX version 1.83 (34). Maximum likelihood (ML) and Bayesian inference (BI) were used to infer trees and estimate clade support. ML analysis included the run of 100 resampled trees using DNAML in the PHYLIP package (distributed by J. Felsenstein, Department of Genome Sciences, University of Washington, Seattle; PHYLIP [Phylogeny Inference Package], version 3.6; http://evolution.genetics.washington.edu/phylip.html). Posterior probabilities were calculated using a Bayesian Markov chain Monte Carlo method with MrBayes version 3.1.1 (14, 27). Ten million generations were run, with sampling every 100 generations.
Nucleotide sequence accession numbers.
Nucleotide sequences from this study have been deposited in GenBank under the accession numbers DQ309594 to DQ309693.
RESULTS
DNA extraction efficiency and accuracy of real-time qPCR assay.
A summary of the validation of DNA extraction and real-time qPCR assay is shown in Table 2. The copy number of the target quantified from spiked samples showed a clear response to the increase in the amount used for spiking. The recovery of DNA was estimated by comparison between theoretical and recovered copy numbers of the target. Excluding the assays for tet(O), tet(H), and the 16S rRNA gene, DNA recovery was 71.3 to 166.3% (Table 2). The recovery for tet(O) and tet(H) was lower than that for the other Tcr genes and estimated to be between 17.7 and 40.2%, whereas the recovery of the 16S rRNA gene was higher than the theoretical number and was up to 224.3%. The CV for real-time qPCR assays was 5.0 to 25.1%, and the assay for tet(H) had a higher CV than that for the other targets. The CV for the overall assay, including the variation of DNA extraction and real-time qPCR assay, was 13.1 to 31.8%, and the assays for tet(H) and the 16S rRNA gene had higher CVs than the others (Table 2). The lower limit for accurate quantification varied between the targets; the lowest was 60 copies per reaction for tet(Z), and the highest was 910 copies per reaction for the 16S rRNA gene (Table 2). Because of differences in extraction efficiency and the presence or absence of competing templates and/or inhibitory substances, the lower limit for accurate quantification from environmental samples is different from the lower limit for detection; thus, it may be possible to detect the presence of a Tcr gene without being able to accurately determine its concentration (Table 3).
TABLE 2.
Summary of the validation of DNA extraction and real-time qPCR assays
| Target | Background (106)a | Copy no.
|
Recovery (%) | CV (%)
|
Amplification efficiency | Quantitation range (copy no.)d | ||
|---|---|---|---|---|---|---|---|---|
| Theoretical | Recovered | qPCRb | Overallc | |||||
| tet(M) | 2.13 ± 0.17 | 1.00 × 108 | 9.25 × 107 | 92.5 | 14.4 | 13.1 | 1.99 | 71-7.1 × 108 |
| 1.00 × 107 | 7.13 × 106 | 71.3 | ||||||
| tet(O) | 0.10 ± 0.01 | 1.00 × 108 | 3.75 × 107 | 17.7 | 12.0 | 13.4 | 2.30 | 370-3.7 × 108 |
| 1.00 × 107 | 1.77 × 106 | |||||||
| 1.00 × 106 | 3.04 × 105 | 30.4 | ||||||
| tet(Q) | 2.71 ± 0.43 | 1.00 × 108 | 8.68 × 107 | 86.8 | 5.0 | 16.0 | 2.10 | 240-2.4 × 108 |
| 1.00 × 107 | 9.17 × 106 | 91.7 | ||||||
| tet(W) | 1.46 ± 0.21 | 1.00 × 108 | 8.50 × 107 | 85.0 | 6.8 | 14.4 | 2.06 | 410-4.1 × 108 |
| 1.00 × 107 | 1.11 × 107 | 110.8 | ||||||
| tet(C) | 0.41 ± 0.07 | 1.00 × 108 | 1.28 × 108 | 128.0 | 13.1 | 17.6 | 1.91 | 220-2.2 × 108 |
| 1.00 × 107 | 1.50 × 107 | 150.5 | ||||||
| 1.00 × 106 | 1.51 × 106 | 150.6 | ||||||
| tet(H) | 0.25 ± 0.08 | 1.00 × 108 | 2.13 × 107 | 21.3 | 25.1 | 31.8 | 1.85 | 470-4.7 × 108 |
| 1.00 × 107 | 2.99 × 106 | 29.9 | ||||||
| 1.00 × 106 | 4.02 × 105 | 40.2 | ||||||
| tet(Z) | 0.10 ± 0.02 | 1.00 × 108 | 1.66 × 108 | 166.3 | 12.1 | 18.0 | 1.99 | 60-6.0 × 108 |
| 1.00 × 107 | 8.69 × 106 | 86.9 | ||||||
| 1.00 × 106 | 1.29 × 106 | 129.7 | ||||||
| 16S rDNA | 370.0 ± 91.7 | 2.47 × 106 | 5.54 × 106 | 224.3 | 11.0 | 24.7 | 2.13 | 910-9.1 × 108 |
| 2.47 × 105 | 2.87 × 105 | 116.3 | ||||||
| 2.47 × 104 | 5.13 × 104 | 207.7 | ||||||
Indigenous target amount in the lagoon sample used in the spiking trial.
For repeat quantifications using the same sample.
For separate repeat DNA extractions and quantifications.
Quantitation range was determined using serially diluted plasmid DNA containing respective target DNA fragment.
TABLE 3.
Quantitative monitoring of concentrations of tetracycline resistance genes in lagoon and groundwater samples between 2001 and 2003 by real-time qPCRa
| Sample | Sampling period | Relative abundance (no. of copies/106 copies of 16S rRNA gene)
|
||||||
|---|---|---|---|---|---|---|---|---|
| tet(M) | tet(O) | tet(Q) | tet(W) | tet(C) | tet(H) | tet(Z) | ||
| Site A lagoon | May 2001 | 33,167 ± 2,378 A, X | 1,819 ± 238 B, X | 19,245 ± 1,914 C, X | 2,935 ± 545 B, X | 1,402 ± 472 B, X | <470 | <60 |
| Apr. 2002 | 4,573 ± 343 A, Y | 45,460 ± 9,109 B, Y | 50,663 ± 4,034 B, Y | 19,516 ± 4,756 C, Y | 7,529 ± 922 A, X | <470 | 1,604 ± 172 A, X | |
| Mar. 2003 | 13,807 ± 586 AB, Y | 22,808 ± 2,895 A, Z | 11,729 ± 1,158 B, X | 11,832 ± 1,269 B, XY | 4,540 ± 964 BC, X | <470 | 1,532 ± 164 C, X | |
| A6 | May 2001 | <71b | <370 | <240 | <410 | 2,822 ± 673 X | <470 | <60 |
| Apr. 2002 | 2,520 ± 429 A, X | <370 | 1,859 ± 352 A, X | 467 ± 56 A, X | 10,078 ± 1,349 A, XY | <470 | <60 | |
| Mar. 2003 | 2,463 ± 190 A, X | <370 | 1,454 ± 164 A, X | <410 | 19,121 ± 831 B, Y | <470 | 145 ± 15 A | |
| A8 | May 2001 | ND | <370 | 3,622 ± 435 A, X | 1,078 ± 104 A, X | 28,081 ± 3,114 B, X | ND | 202 ± 103 A, X |
| Apr. 2002 | 6,442 ± 887 A, X | <370 | 6,225 ± 342 AB, X | 2,918 ± 217 A, X | 17,496 ± 3,062 B, X | <470 | <60 | |
| Mar. 2003 | 1,137 ± 297 A, X | <370 | 408 ± 159 A, X | <410 | <220 | <470 | 815 ± 62 A, X | |
| A9 | May 2001 | 692 ± 198 A, X | <370 | 2,416 ± 487 A, X | 451 ± 38 A, X | 56,642 ± 5,319 B, X | <470 | <60 |
| Apr. 2002 | 1,351 ± 300 A, X | <370 | 2,506 ± 488 A, X | 1,156 ± 79 A, X | 11,383 ± 651 A, Y | <470 | 807 ± 114 A, X | |
| Mar. 2003 | 8,535 ± 827 AB, X | 908 ± 97 A, X | 8,602 ± 1,127 AB, X | 4,470 ± 1,327 A, X | 14,554 ± 995 B, Y | <470 | 196 ± 11 A, X | |
| A11 | May 2001 | 1,961 ± 337 A, X | <370 | 5,626 ± 609 A, X | 1,240 ± 216 A, X | 96,156 ± 3,680 B, XZ | <470 | <60 |
| Apr. 2002 | 4,051 ± 497 A, X | 421 ± 47 A, X | 4,418 ± 754 A, X | 3,294 ± 240 A, X | 11,266 ± 2,586 A, Y | <470 | ND | |
| Mar. 2003 | 5,288 ± 364 A, X | <370 | 3,742 ± 647 A, X | 1,358 ± 118 A, X | 103,134 ± 15,344 B, Z | 3,747 ± 1,282 A, X | 2,840 ± 171 A | |
| A12 | May 2001 | <71 | <370 | <240 | 437 ± 151 A, X | 515 ± 1 A, X | <470 | <60 |
| Apr. 2002 | 830 ± 115 A, X | <370 | 724 ± 101 A, X | <410 | 1,706 ± 230 A, X | <470 | <60 | |
| Mar. 2003 | 362 ± 37 A, X | <370 | <240 | <410 | 1,934 ± 85 A, X | <470 | 448 ± 61 A | |
| A16 | May 2001 | ND | ND | ND | ND | ND | ND | ND |
| Apr. 2002 | <71 | <370 | <240 | <410 | ND | ND | 175 ± 28 X | |
| Mar. 2003 | ND | ND | <240 | <410 | ND | ND | 646 ± 14 X | |
| Site C lagoon | May 2001 | 9,658 ± 167 A, X | 2,230 ± 140 AD, X | 120,999 ± 6,510 B, X | 6,967 ± 672 AD, X | 50,616 ± 5,783 C, X | 485 ± 82 B, X | 311 ± 16 D, X |
| May 2002 | 7,816 ± 715 A, X | 10,854 ± 2,406 A, XY | 52,684 ± 3,039 B, Y | 27,200 ± 2,682 C, Y | 71,556 ± 12,655 D, Y | <470 | 604 ± 61 A, X | |
| Feb. 2003 | 9,426 ± 598 AD, X | 14,359 ± 3,282 A, Y | 27,278 ± 3,210 B, Z | 47,026 ± 4,046 C, Z | 3,241 ± 722 D, Z | <470 | 3,490 ± 279 D, X | |
| C2 | May 2001 | <71 | ND | ND | ND | ND | ND | ND |
| May 2002 | ND | <370 | ND | <410 | ND | ND | ND | |
| Feb. 2003 | <71 | <370 | <240 | <410 | 2,655 ± 155 | <470 | <60 | |
| C3 | May 2001 | <71 | <370 | <240 | <410 | 237 ± 29 | <470 | <60 |
| May 2002 | ND | ND | ND | ND | ND | ND | <60 | |
| Feb. 2003 | <71 | <370 | <240 | <410 | ND | ND | ND | |
| C4 | May 2001 | ND | ND | <240 | <410 | <220 | ND | <60 |
| May 2002 | 394 ± 64 A, X | <370 | <240 | <410 | 235 ± 16 A, X | ND | <60 | |
| Feb. 2003 | 1,231 ± 138 A, X | <370 | 1,608 ± 178 A | 515 ± 143 A, X | 1,707 ± 106 A, X | <470 | <60 | |
| C6 | May 2001 | <71 | ND | <240 | <410 | ND | ND | ND |
| May 2002 | ND | <370 | ND | <410 | ND | ND | ND | |
| Feb. 2003 | ND | <370 | ND | ND | ND | ND | ND | |
The copy numbers of the target genes were normalized to the copy number of the 16S rRNA gene. ND, not detected. Statistical significance is indicated as follows: A, B, C, and D, means in the same row with different letters differ significantly (P < 0.01) among the genes; X, Y, and Z, means within the sample in the same column with different letters differ significantly (P < 0.01) among the periods.
Below the quantification limit. See Table 2.
PCR detection of Tcr genes from lagoon and groundwater samples.
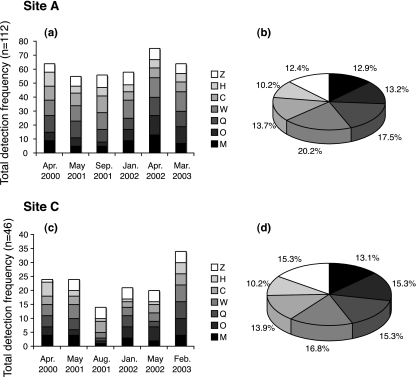
The primer sets for the Tcr genes were able to detect a single copy of the respective target gene both from pure culture (cloned plasmids) and from plasmid-spiked environmental DNA extracted from groundwater samples (data not shown). Therefore, these primers were sensitive enough to monitor the Tcr genes in groundwater. The total detection frequency of the Tcr genes in all samples within each sampling period between 2000 and 2003 is shown in Fig. 2a and c. The total detection frequency (summing the number of detections for all Tcr genes found in the lagoon and groundwater samples) was consistent among the sampling periods over the 3 years (55 to 75 detections/112 samples) at site A (Fig. 2a). At site C, the total detection frequency varied and was lowest (14 detections/46 samples) in August 2001 and highest (34 detections/46 samples) in February 2003 (Fig. 2c). There was no clear temporal pattern in total detection frequency at site C among the sampling periods. The frequency of detecting each of the Tcr genes in the samples throughout the study is shown in Fig. 2b and d. tet(Q) and tet(W) were detected slightly more often than the other tet genes at site A, while tet(H) was detected least often at both sites.
FIG. 2.
Total detection frequency (a and c) and proportion (b and d) of seven tetracycline resistance genes at two swine facilities between 2000 and 2003 determined by PCR. Total detection frequency was calculated by summing the number of positive detections of all seven tet genes found in the lagoon and groundwater samples at each sampling time. The proportion of detection frequency for each tet gene was expressed as percentage of detection frequency for each gene against total detection frequency over the 3-year monitoring period. Sample number (n) was calculated as average sample number (16.0 for site A and 6.6 for site C) multiplied by 7 (the number of targeted tet genes).
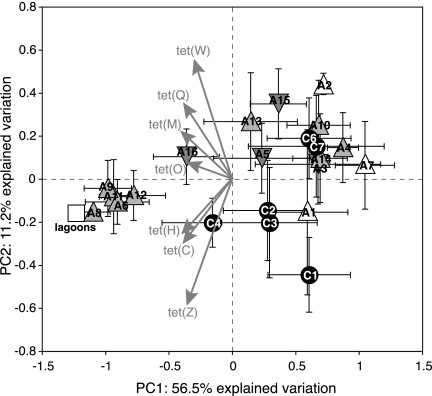
PCA based on the presence or absence of all seven Tcr genes from all well and lagoon samples (n = 135) revealed spatial variation in Tcr gene detection (Fig. 3). Because the PCA centroids were calculated from the average loading of samples on the principal axes, they are analogous to the combined detection frequencies of all Tcr genes from each sample location. All seven tet genes were detected in site A and C lagoon samples from all six sampling dates. Five wells, A6, A8, A9, A11, and A12, were also consistently positive for all seven tet genes, with gene detection frequencies ranging from 67 to 100% of samples analyzed. These wells plotted very close to the lagoon samples along the first principal axis (PC1), and the detection frequency of all tet genes was negatively correlated with PC1 (Fig. 3). Gene detection frequencies were highly variable in other wells, with some, such as A3, A4, A10, and most of the site C wells, being virtually indistinguishable from control wells A1, A2, A7, and C1 in terms of tet gene detections (Fig. 3). Wells A16 and C4 had slightly elevated gene detection frequencies in comparison with this group, but they were still distinguishable overall from lagoon samples. There was a tendency for genes encoding RPPs [tet(O), tet(M), tet(Q), tet(W)] to cooccur in samples and a tendency for efflux pump genes [tet(C), tet(H), and tet(Z)] to cooccur, and these two classes of genes were separated along PC2 (Fig. 3). Efflux pump genes were found more frequently in wells A1, C1, C2, C3, and C4, and RPPs were found more frequently in most of the wells from site A and the remaining two site C wells (Fig. 3).
FIG. 3.
PCA biplot of tetracycline resistance gene occurrence in groundwater wells and lagoon samples. Plotted points represent the centroids of the data from six sampling dates for each well, and error bars show the bivariate variance. Vectors display the loadings of tet gene presence and absence in wells on the first two principal component axes. Triangles represent wells from site A (white, background wells; light gray and upward pointing, wells in shallow sand layer; dark gray and downward pointing, wells in deep sand layer), circles represent wells from site C, and squares represent lagoon samples. For well locations, see Fig. 1.
Three-year quantitative monitoring of Tcr genes in lagoon and impacted wells.
Table 3 shows the concentration of Tcr genes in lagoon and groundwater samples collected between 2001 and 2003. Although concentrations fluctuated during the 2-year monitoring period, greater Tcr gene concentrations were detected in the lagoons than in groundwater (1.42 × 104 ± 4.18 × 103 copies per 106 16S rDNA copies versus 5.21× 103 ± 3.62× 103 copies per 106 16S rDNA copies, on average). The concentrations of genes encoding RPPs [tet(M), tet(O), tet(Q), and tet(W)] were generally greater than those of the efflux pump genes [tet(C), tet(H), and tet(Z)] in lagoon samples collected from both sites (on average, 6.5- and 1.9-fold higher at sites A and C, respectively). The Tcr gene concentrations in groundwater at site A were significantly lower than those in the lagoon; however, a remarkably higher proportion of tet(C) in wells A6, A8, A9, and A11 than in the lagoon was confirmed during the monitoring period at site A; the average concentration of tet(C) in groundwater was 5.9-fold greater than the average concentration of tet(C) in the lagoon and was 8.3- to 75.2-fold higher than the average concentration of the other Tcr genes in groundwater (Table 3). Overall Tcr gene concentrations in wells A8, A9, and A11 were 1.5- to 3.9-fold higher (on average) than those in the other selected monitoring wells at site A (Table 3). Unlike at site A, the concentrations of Tcr genes were lower than the quantification limit in almost all samples at site C. There was a significant difference in the concentration of Tcr genes in nested wells A9 and A16 at site A (Table 3; Fig. 1). Well A9, installed in the upper sand layer, contained a significant concentration of Tcr genes, whereas the Tcr gene concentration was lower than quantification limit in well A16, which was in the deeper sand layer (Table 3).
Source tracking analysis of tet(W) in groundwater.
A total of 100 clones for tet(W) from 10 libraries (A7, A8, A9, and A14 and lagoon and concrete settling basin at site A; C1, C2, and C4 and lagoon at site C; 10 clones from each library) were sequenced and subsequently subjected to phylogenetic analysis. For sequences from waste samples, clone names begin with “WS” followed by sample source (AP, solid waste in concrete settling basin at site A; AL, waste in lagoon at site A; CL, waste in lagoon at site C) and clone number. For the sequences from groundwater samples, clones are named with the prefix “GW” followed by well number and clone number.
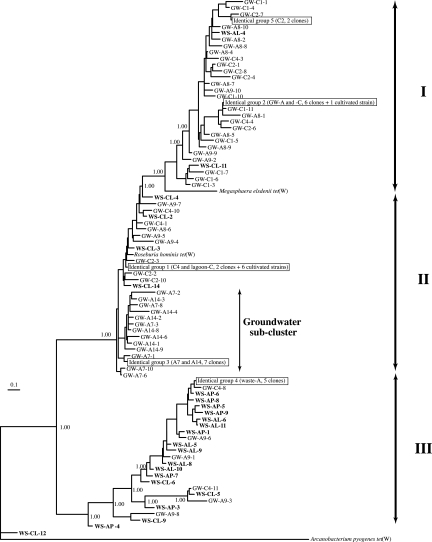
The results of phylogenetic analysis of tet(W) are shown in Fig. 4. ML and BI approaches produced similar tree topology and clade support; therefore, the result from BI is shown in Fig. 4. Among the 100 clones sequenced in this study, five sequence groups, named IG-1 to IG-5, were found. Within each IG group, all clones had identical nucleotide sequences. IG-1 contained two clones from site C (C4 and lagoon). In the 1,152-bp sequenced region, most cultivated tet(W) sequences deposited in the GenBank database belonged to IG-1. The hosts of tet(W) sequences belonging to IG-1 were as follows (with GenBank accession number in parentheses): Butyrivibrio fibrisolvens (AJ222769 and AJ427421), Clostridiaceae bacterium K10 (AY601650), Megasphaera elsdenii (AY485125), Mitsuokella multacida (AY603069), and Mitsuokella multiacidus (AJ427422). IG-2 comprised one clone from site A (A8), five clones from site C (C1, C4), and tet(W) from a Bifidobacterium sp. (AF202986). All seven clones belonging to IG-3 were retrieved from wells A7 and A14. IG-4 consisted of five clones from waste samples at site A. IG-5 contained two clones from well C2.
FIG. 4.
BI dendrogram showing the phylogenetic affiliations of tet(W) sequences from lagoon and groundwater samples. For the sequences from the waste samples, clone names begin with “WS” followed by sample source (AP, manure solids in concrete settling basin at site A; AL, waste in lagoon at site A; CL, waste in lagoon at site C) and clone number. For the sequences from groundwater samples, clones are named with the prefix “GW” followed by well number and clone number. Sequences from manure samples are shown in boldface. The number at the node indicates the probability of branching obtained by 10 million generations; 1.00 corresponds to 100% support of the branching. Bar, nucleotide substitutions per sequence position. Sequence groups IG-1 to IG-5 are boxed within the tree. Within each IG group, all clones had identical nucleotide sequences. The numbers in parentheses for IG groups indicate the number and source site of sequences belonging to each group.
The five sequence groups (IG-1 through IG-5) could be separated into three distinct clusters (I, II, and III), and the branching of clusters II and III was supported by 100% probability (Fig. 4). Twenty-two out of 30 clones from the waste at both sites were found in cluster III. Six groundwater clones from wells A9 and C4 also belonged to cluster III. All clones from distant or background wells at site A (wells A7 and A14) formed a subcluster within cluster II, whereas 12 out of 20 clones from impacted wells (wells A8 and A9) and a clone from the waste were found in cluster I (Fig. 4). Although cluster assignment differed between sites, a similar trend was observed at site C, i.e., all background well (C1) clones were found in cluster I, and five clones from waste and six clones from wells C2 and C4 belonged to cluster II (Fig. 4).
DISCUSSION
In our previous study, where only samples from a single sampling event were analyzed, tet(M), tet(O), tet(Q), tet(W), tet(C), tet(H), and tet(Z) were frequently detected in groundwater collected at sites A and C (5, 10). In the present work, all seven Tcr genes were repeatedly detected at both sites over the 3-year monitoring period without a clear temporal pattern in detection frequency or copy number of any specific gene (Fig. 2; Table 3). The variable nature of Tcr genes in these samples implies that tetracycline resistance in impacted and native microbial communities is a dynamic phenomenon subject to fluctuations in organism and gene migration, persistence of microbial populations, and/or selection pressure. In our previous study, tetracycline, chlortetracycline, and oxytetracycline residues were detected in fewer than 5% of the groundwater samples from both sites (20, 23). Tetracycline concentrations in tetracycline-positive samples were lower than 0.4 μg/liter. This concentration is low compared to the concentration for in vitro screening of resistant bacteria, where the antibiotic is used at μg/ml levels. Although low antibiotic concentrations in groundwater would likely not select for resistant bacteria, the frequency of resistance gene transfer might be enhanced at these levels (29).
To avoid underestimation of target, sufficient recovery of DNA from the samples was an important point in the environmental study. In this study, we demonstrated the satisfactorily high efficiency of our DNA extraction approach (Table 2), and the real-time qPCR assays we developed in this study produced consistent results with low variability. Therefore, the combination of DNA extraction method and real-time qPCR assays employed in this study ensured the potential for precise quantification of Tcr genes across a broad range of target copy numbers (Table 2). However, because E. coli was used in our target-spiked validation, it is possible that extraction efficiency for Tcr genes in native microbial populations may vary. There was also gene-specific variation in extraction efficiency (Table 2). The assay for tet(O) and tet(H) showed a lower recovery rate than the other target genes; therefore, the quantity of these genes could have been underestimated in this study. DNA recovery of the 16S rRNA gene showed the possibility for slight overestimation of total bacterial number in the samples. However, because 16S rRNA gene copy number was used for normalization, the relative abundances of all Tcr genes were affected equally, and therefore any overestimation of 16S rRNA gene copy number would not influence comparisons of relative Tcr gene target abundances.
Although our DNA extraction and purification methods showed the ability to provide high-quality DNA with sufficient recovery, the difference in DNA quantity and purity between the samples significantly affected the cycle threshold, which is the critical point for quantification. Therefore, experimental variability encountered with real-time qPCR should be minimized by quantifying a control gene such as a housekeeping gene (1). To compensate for differences in DNA amount and quality between different samples, we normalized the quantification values for each Tcr gene in relation to 16S rRNA gene copy number in each sample.
The relative abundance (target gene/16S rRNA gene) of tet RPP genes was greater than that of tet efflux pump genes in lagoon samples (Table 3). Similar observations were obtained for samples from swine waste treatment systems on a different set of farms, for which no groundwater samples are available (15). Here, however, the tet(C) abundance in groundwater was 5.9-fold higher (on average) than in the lagoons (Table 3). This observation suggests that differential migration or selection of Tcr genes occurred from lagoon to groundwater. The higher tet(C) abundance in groundwater can be attributed to possible greater migration of tet(C) carriers from the lagoons to groundwater, greater gene transfer of this gene from lagoon bacteria to groundwater bacteria, and/or greater persistence of migrants and indigenous tet(C) carriers in groundwater. Although the relative abundance of tet(C) was greater in groundwater, the absolute number of 16S rRNA gene copies per ml of samples was estimated to be 576.6-fold higher in lagoon samples (data not shown). Thus, the absolute copy number of tet(C) in the lagoon was likely higher than that in groundwater.
Tcr genes were continuously detected in the lagoons from both sites, and Tcr genes were detected with some frequency from all groundwater wells, even those considered to be background control wells due to their up-gradient location relative to lagoons. Nevertheless, some wells were clearly more impacted by Tcr gene contamination than others (Fig. 3). Wells A6, A8, A9, A11, and A12 were most like the lagoons in the detection frequency of Tcr genes, and these wells were consistently more likely to contain the full complement of all seven genes (Fig. 3). These wells were all situated in the shallow sand layer and located near the lagoon (Fig. 1). Wells located up-gradient or at a greater distance down-gradient and wells installed in the lower sand layer had a lower frequency of Tcr gene detection (Fig. 1 and 3). Previous work has shown that shallow groundwater down-gradient of the site A lagoon is significantly impacted by lagoon seepage, and wells A6, A8, A9, A11, and A12 contained high concentrations of chloride and ammonia (20). In fact, PC1 (Fig. 3) was highly negatively correlated with the indicators of lagoon seepage contamination measured by Krapac et al. (20): chloride (Pearson's R = −0.78), ammonia (R = −0.84), and electrical conductivity (R = −0.81). Thus, there is a correlation between chemical contamination and biological contamination of tetracycline-resistant organisms and/or their Tcr genes.
The hydrogeological characteristics of aquifers can play an important role in Tcr gene movement. The silty clay layer between the two sand layers at site A (Fig. 1) appeared to be somewhat resistant to Tcr gene movement, and nested wells installed in the lower sand had a decreased frequency of tet gene detection (Fig. 3) and a lower copy number of tet genes (Table 3) than their shallow-sand-layer counterparts. Krapac et al. (20) indicated that tetracyclines were sorbed significantly by the soil collected at site A, and silty material exhibited a much higher affinity for the tetracyclines than did the sand. Therefore, contaminants in the seepage from the lagoon may be filtered through the intervening silty loam layer, resulting in lower concentrations of antibiotics and/or Tcr genes in deeper wells. This may also explain the lower Tcr gene detection frequency (Fig. 3) and lower Tcr gene copy numbers (Table 3) observed at site C in comparison to site A, as the impermeable clay till at site C (Fig. 1) may retard the movement of contaminants from the lagoon to the groundwater. Comparison of chemical and biological contamination of groundwater at an expanded number of farms with different hydrogeological characteristics should further illustrate this relationship.
The occurrence of Tcr genes in distant background wells located up-gradient of the lagoon was confirmed in this study (Fig. 3); therefore, it was essential that the source of antibiotic resistance genes in groundwater be identified. In this study, tet(W) was chosen as a representative tetracycline resistance gene, since it was widely distributed in lagoons and polluted wells as well as background control wells (Fig. 3). The most significant finding of the comparative sequence analysis in this study is that impacted groundwater contained the gene sequences (clones GW-A9-1, GW-A9-6, and GW-C4-11) which showed more than 99.8% identity (two nucleotide substitutions in a 1,152-bp fragment) to the waste clones (Fig. 4). This result implies that antibiotic resistance genes can migrate between these connected aquatic systems. It is unclear whether the presence of antibiotic resistance genes is the result of the migration of antibiotic-resistant bacteria or transmission of resistance genes by horizontal gene transfer (3, 11, 28). Several reports indicate that E. coli can survive in anoxic groundwater after excretion from the animal (6, 37). In fact, Krapac et al. (17) previously demonstrated that groundwater collected at site A and site C contained fecal coliforms and fecal streptococci. These data indicate that bacterial migration has likely occurred to a limited extent at both sites, and it is possible that fecal bacteria contribute to dissemination of antibiotic resistance genes from manure to groundwater.
With a high posterior probability, tet(W) gene sequences from lagoon and polluted groundwater formed a cluster distinct from the known sequence group (Fig. 4), indicating that animal wastes contain a novel sequence diversity of tet(W). Although sites A and C are located 265 km from each other, tet(W) sequences from waste lagoon at both sites belonged to a unique, novel tet(W) cluster (Fig. 4). Therefore, in the context of microevolution, swine confinement facilities may have specific selection pressures. On the other hand, two distinct clusters of groundwater clones were observed, indicating that local selection pressure might exist even in nonimpacted groundwater. Focusing on site A, where the environmental impact was greater, an up-gradient clone subcluster (groundwater subcluster) was clearly isolated from the other groundwater clusters, which were comprised of clones from impacted groundwater (clusters I and II) (Fig. 4). There may be different selection pressures in impacted and nonimpacted groundwaters, and lagoon seepage might influence this. Additionally, there is evidence that site-specific differences exist within the groundwater communities at these sites: nonimpacted wells from site A tended to have a higher occurrence of RPP, while efflux pump genes tended to be more prevalent in most of the site C wells (Fig. 3). These observations concerning comparative gene diversity strongly support the presence of an indigenous tet gene pool in groundwater, and the ecological pressures maintaining these pools may operate independently of lagoon seepage.
In conclusion, animal waste seeping from unlined lagoons (20) at two swine confinement facilities had an impact on the dissemination of tetracycline resistance genes in groundwater near the facilities. Thus, these types of facilities can be reservoirs of antibiotic resistance genes. However, the magnitude and extent of antibiotic resistance gene migration resulting from lagoon seepage are likely dependent on lagoon construction and local hydrogeological conditions. The migration of Tcr genes into groundwater was confirmed by comparative sequence analysis of tet(W). In addition, novel sequence clusters of this gene and unique indigenous resistance gene pools in groundwater were also detected.
Acknowledgments
This research was supported by funding from the USDA NRI Competitive Grants Program 26.0 (award no. 2001-35102-10774 and 2005-35102-16426) and partially by Hatch funding from the Agricultural Experimental Station, University of Illinois at Urbana-Champaign.
Footnotes
Published ahead of print on 1 June 2007.
REFERENCES
- 1.Aerts, J. L., M. I. Gonzales, and S. L. Topalian. 2004. Selection of appropriate control genes to assess expression of tumor antigens using real-time RT-PCR. BioTechniques 36:84-86. [DOI] [PubMed] [Google Scholar]
- 2.Alonso, A., P. Sánchez, and J. L. Martínez. 2001. Environmental selection of antibiotic resistance genes. Environ. Microbiol. 3:1-9. [DOI] [PubMed] [Google Scholar]
- 3.Amabile-Cuevas, C. F., and M. E. Chicurel. 1992. Bacterial plasmids and gene flux. Cell 70:189-199. [DOI] [PubMed] [Google Scholar]
- 4.Aminov, R. I., J. C. Chee-Sanford, N. Garrigues, B. Teferedegne, I. J. Krapac, B. A. White, and R. I. Mackie. 2002. Development, validation, and application of PCR primers for detection of tetracycline efflux genes of gram-negative bacteria. Appl. Environ. Microbiol. 68:1786-1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aminov, R. I., N. Garrigues-Jeanjean, and R. I. Mackie. 2001. Molecular ecology of tetracycline resistance: development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins. Appl. Environ. Microbiol. 67:22-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banning, N., S. Toze, and B. J. Mee. 2002. Escherichia coli survival in groundwater and effluent measured using a combination of propidium iodide and the green fluorescent protein. J. Appl. Microbiol. 93:69-76. [DOI] [PubMed] [Google Scholar]
- 7.Baquero, F., and J. Blázquez. 1997. Evolution of antibiotic resistance. Trends Ecol. Evol. 12:482-487. [DOI] [PubMed] [Google Scholar]
- 8.Chee-Sanford, J. C., R. I. Aminov, I. J. Krapac, N. Garrigues-Jeanjean, and R. I. Mackie. 2001. Occurrence and diversity of tetracycline resistance genes in lagoons and groundwater underlying two swine production facilities. Appl. Environ. Microbiol. 67:1494-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chopra, I., and M. Roberts. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65:232-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clarke, K. R., and R. N. Gorley. 2001. PRIMER 5 for Windows: Plymouth routines in multivariate ecological research, 5.2.7 ed. PRIMER-E, Plymouth, United Kingdom.
- 11.Dzidic, S., and V. Bedekovic. 2003. Horizontal gene transfer: emerging multidrug resistance in hospital bacteria. Acta Pharmacol. Sin. 24:519-526. [PubMed] [Google Scholar]
- 12.Gilliver, M. A., M. Bennett, M. Begon, S. M. Hazel, and C. A. Hart. 1999. Enterobacteria: antibiotic resistance found in wild rodents. Nature 401:233-234. [DOI] [PubMed] [Google Scholar]
- 13.Hagedorn, C., S. L. Robinson, J. R. Filtz, S. M. Grubbs, T. A. Angier, and R. B. Reneau. 1999. Determining sources of fecal pollution in a rural Virginia watershed with antibiotic resistance patterns in fecal streptococci. Appl. Environ. Microbiol. 65:5522-5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huelsenbeck, J. P., and F. Ronquist. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 15.Jindal, A., S. Kocherginskaya, A. Mehboob, M. Robert, R. I. Mackie, L. Raskin, and J. L. Zilles. 2006. Antimicrobial use and resistance in swine waste treatment systems. Appl. Environ. Microbiol. 72:7813-7820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klappenbach, J. A., P. R. Saxman, J. R. Cole, and T. M. Schmidt. 2001. rrndb: the Ribosomal RNA Operon Copy Number Database. Nucleic Acids Res. 29:181-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krapac, I., W. S. Dey, C. A. Smyth, and W. R. Roy. 1998. Impacts of bacteria, metals, and nutrients on groundwater at two hog confinement facilities, p. 29-50. In Proceedings of Animal Feeding Operations and Groundwater: Issues, Impact, and Solutions: a Conference for the Future. National Ground Water Association, St. Louis, MO.
- 18.Krapac, I. G., W. S. Dey, W. R. Roy, B. G. Jellerichs, and C. A. Smyth. 2000. Groundwater quality near livestock manure pits, p. 710-718. In Proceedings of 8th International Symposium on Animal, Agricultural, and Food Processing Waters. American Society for Agricultural Engineering, Des Moines, IA.
- 19.Krapac, I. G., W. S. Dey, W. R. Roy, C. A. Smyth, E. Storment, S. L. Sargent, and J. D. Steele. 2002. Impacts of swine manure pits on groundwater quality. Environ. Pollut. 120:475-492. [DOI] [PubMed] [Google Scholar]
- 20.Krapac, I. G., S. Koike, M. T. Meyer, D. D. Snow, S. F. J. Chou, R. I. Mackie, W. R. Roy, and J. C. Chee-Sanford. 2004. Long-term monitoring of the occurrence of antibiotic resistance genes in groundwater near swine confinement facilities, p. 158-172. In Proceedings of 4th International Conference on Pharmaceuticals and Endocrine Disrupting Chemicals in Water. National Ground Water Association, Minneapolis, MN.
- 21.Labrenz, M., I. Brettar, R. Christen, S. Flavier, J. Bötel, and M. G. Höfle. 2004. Development and application of a real-time PCR approach for quantification of uncultured bacteria in the central Baltic Sea. Appl. Environ. Microbiol. 70:4971-4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Linares, F., and K. Sundback. 2006. Uptake of dissolved free amino acids (DFAA) by microphytobenthic communities. Aquat. Microb. Ecol. 42:175-186. [Google Scholar]
- 23.Mackie, R. I., S. Koike, I. Krapac, J. Chee-Sanford, S. Maxwell, and R. I. Aminov. 2006. Tetracycline residues and tetracycline resistance genes in groundwater impacted by swine production facilities. Anim. Biotechnol. 17:157-176. [DOI] [PubMed] [Google Scholar]
- 24.Miller, D. N. 2001. Evaluation of gel filtration resins for the removal of PCR-inhibitory substances from soils and sediments. J. Microbiol. Methods 44:49-58. [DOI] [PubMed] [Google Scholar]
- 25.Muyzer, G., E. C. De Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pillai, S. D., K. W. Widmer, K. G. Maciorowski, and S. C. Ricke. 1997. Antibiotic resistance profiles of Escherichia coli isolated from rural and urban environments. J. Environ. Sci. Health Part A 32:1665-1675. [Google Scholar]
- 27.Ronquist, F., and J. P. Huelsenbeck. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572-1574. [DOI] [PubMed] [Google Scholar]
- 28.Salyers, A. A., and C. F. Amabile-Cuevas. 1997. Why are antibiotic resistance genes so resistant to elimination? Antimicrob. Agents Chemother. 41:2321-2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salyers, A. A., N. B. Shoemaker, and L. Y. Li. 1995. In the driver's seat: the Bacteroides conjugative transposons and the elements they mobilize. J. Bacteriol. 177:5727-5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith, M. S., R. K. Yang, C. W. Knapp, Y. F. Niu, N. Peak, M. M. Hanfelt, J. C. Galland, and D. W. Graham. 2004. Quantification of tetracycline resistance genes in feedlot lagoons by real-time PCR. Appl. Environ. Microbiol. 70:7372-7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stanton, T. B., and S. B. Humphrey. 2003. Isolation of tetracycline-resistant Megasphaera elsdenii strains with novel mosaic gene combinations of tet(O) and tet(W) from swine. Appl. Environ. Microbiol. 69:3874-3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stanton, T. B., J. S. McDowall, and M. A. Rasmussen. 2004. Diverse tetracycline resistance genotypes of Megasphaera elsdenii strains selectively cultured from swine feces. Appl. Environ. Microbiol. 70:3754-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tajima, K., R. I. Aminov, T. Nagamine, H. Matsui, M. Nakamura, and Y. Benno. 2001. Diet-dependent shifts in the bacterial population of the rumen revealed with real-time PCR. Appl. Environ. Microbiol. 67:2766-2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsai, Y. L., and B. H. Olson. 1991. Rapid method for direct extraction of DNA from soil and sediments. Appl. Environ. Microbiol. 57:1070-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.U.S. Geological Survey. 1995, posting date. Ground water studies. http://water.usgs.gov/wid/html/GW.html.
- 37.Wang, G. D., T. Zhao, and M. P. Doyle. 1996. Fate of enterohemorrhagic Escherichia coli O157:H7 in bovine feces. Appl. Environ. Microbiol. 62:2567-2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wegener, H. C. 2003. Antibiotics in animal feed and their role in resistance development. Curr. Opin. Microbiol. 6:439-445. [DOI] [PubMed] [Google Scholar]
- 39.Wiggins, B. A. 1996. Discriminant analysis of antibiotic resistance patterns in fecal streptococci, a method to differentiate human and animal sources of fecal pollution in natural waters. Appl. Environ. Microbiol. 62:3997-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wiggins, B. A., R. W. Andrews, R. A. Conway, C. L. Corr, E. J. Dobratz, D. P. Dougherty, J. R. Eppard, S. R. Knupp, M. C. Limjoco, J. M. Mettenburg, J. M. Rinehardt, J. Sonsino, R. L. Torrijos, and M. E. Zimmerman. 1999. Use of antibiotic resistance analysis to identify nonpoint sources of fecal pollution. Appl. Environ. Microbiol. 65:3483-3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou, J. Z., M. A. Bruns, and J. M. Tiedje. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316-322. [DOI] [PMC free article] [PubMed] [Google Scholar]