Abstract
Protein arginine methyltransferase PRMT5 interacts with human SWI/SNF complexes and methylates histones H3R8 and H4R3. To elucidate the role of PRMT5 in human cancer, we analyzed PRMT5 expression in normal human B lymphocytes and a panel of lymphoid cancer cell lines as well as mantle cell lymphoma (MCL) clinical samples. We show that PRMT5 protein levels are elevated in all cancer cells, including clinical samples examined despite its low rate of transcription and messenger RNA stability. Remarkably, polysome profiling revealed that PRMT5 mRNA is translated more efficiently in Mino and JeKo MCL cells than in normal B cells, and that decreased miR-92b and miR-96 expression augments PRMT5 translation. Consequently, global methylation of H3R8 and H4R3 is increased and is accompanied by repression of suppressor of tumorigenecity 7 (ST7) in lymphoid cancer cells. Furthermore, knockdown of PRMT5 expression reduces proliferation of transformed JeKo and Raji cells. Thus, our studies indicate that aberrant expression of PRMT5 leads to altered epigenetic modification of chromatin, which in turn impacts transcriptional performance of anti-cancer genes and growth of transformed lymphoid cells.
Keywords: H3R8 and H4R3 methylation, mantle cell lymphoma, miR-92b and miR-96, PRMT5, ST7
Introduction
Human malignancies arise due to aberrant expression of genes that regulate cell growth and proliferation, and understanding the molecular mechanisms that contribute to altered gene expression is important for the design of therapies that specifically target affected central regulatory networks. Recent work has shown that chromatin-modifying enzymes, which can either activate or repress gene expression, play a crucial role in cancer etiology (Santos-Rosa and Caldas, 2005; Esteller, 2006). Chromatin can be altered by various mechanisms, including covalent modification of histones and/or DNA and non-covalent perturbation of histone–DNA contacts within nucleosomes. The net outcome of these chromatin alterations is to modulate accessibility to both repressor and activator proteins (Sif, 2004; Martin and Zhang, 2005). Several reports have shown that the interplay and crosstalk between chromatin-modifying enzymes is necessary for efficient regulation of gene expression, and that changes that affect the activity or targeting of chromatin remodelers can trigger cancer (Fischle et al, 2003; Hake et al, 2004).
Histone modifications like acetylation, methylation and ubiquitination have emerged as important marks in the control of gene expression, which can act either synergistically or antagonistically to specify transcriptional performance (Fischle et al, 2003). Both histone lysine and arginine residues can be methylated by SET proteins and protein arginine methyltransferases (PRMT), respectively, and studies by different groups have shown that methylation can either inhibit or induce transcription depending on the type and residue being modified (Bedford and Richard, 2005; Martin and Zhang, 2005). For instance, trimethylation of H3K4 is associated with active transcription and acts synergistically with H3 and H4 acetylation to activate Hoxc8 expression (Milne et al, 2002). In contrast, trimethylation of H3K9 and/or H3K27 induces gene silencing and appears to be excluded from Ubx promoter regions that harbor trimethylated H3K4 (Cao et al, 2002; Hall et al, 2002; Papp and Muller, 2006).
PRMTs are evolutionarily conserved and PRMT5 is one of the type II arginine methyltransferases that catalyze ω-NG-monomethylation and ω-NG,NG′-symmetric dimethylation (Bedford and Richard, 2005). PRMT5 interacts with a number of multisubunit complexes and is involved in a variety of cellular processes, including RNA processing, signal transduction, transcriptional regulation and germ cell development (Pollack et al, 1999; Friesen et al, 2001; Fabbrizio et al, 2002; Pal et al, 2003, 2004; Ancelin et al, 2006). Interestingly, PRMT5 can impact transcription in either a methylase-dependent or -independent manner. As a component of the androgen receptor cofactor complex, PRMT5 positively modulates androgen receptor-driven transcription independent of its methyltransferase activity (Hosohata et al, 2003); however, when PRMT5 is associated with hSWI/SNF complexes, its chromatin-modifying activity is important for histones H3 and H4 methylation, and regulation of transcription (Pal et al, 2004; Dacwag et al, 2007). Furthermore, methylation by PRMT5 can also have either a positive or negative effect on its substrates. For example, PRMT5-mediated methylation of SmD1 and SmD3 is crucial for their incorporation into snRNPs, which are involved in RNA splicing, whereas methylation of the MBD2 subunit of NURD reduces its ability to associate with methylated DNA and to repress transcription (Friesen et al, 2001; Guezennec et al, 2006; Tan and Nakielny, 2006).
Recent work indicates that PRMT5 can modulate gene expression either directly by modifying histones or indirectly by altering the activity of various transcription factors (Fabbrizio et al, 2002; Kwak et al, 2003; Pal et al, 2003, 2004; Guezennec et al, 2006; Tan and Nakielny, 2006). Direct evidence for the involvement of PRMT5 in transcriptional regulation comes from studies that clearly demonstrate that PRMT5 interacts with chromatin remodeling complexes such as BRG1/BRM-based hSWI/SNF complexes and MBD2-based NURD complexes (Pal et al, 2003, 2004; Guezennec et al, 2006). As part of these chromatin remodelers, PRMT5 is able to methylate histones H3 and H4 and repress transcription of tumor suppressor genes such as suppressor of tumorigenecity 7 (ST7) and NM23-H1, and cell cycle regulators such as CYCLIN E1, P14ARF and P16INK4a.
To assess the role of PRMT5 in human lymphoid cancers, we analyzed its expression in a panel of transformed lymphoid cells including mantle cell lymphoma (MCL) cell lines and MCL patient samples. MCL is a subtype of non-Hodgkin's B cell lymphoma characterized by the presence of t(11;14)(q13;q32) chromosomal translocation. Patients afflicted by MCL show a very poor prognosis and survival rate of 3–4 years. MCL is incurable due to the lack of effective treatments, and identification of novel therapeutic targets remains the focus of ongoing MCL research (Witzig, 2005). Using patient-derived MCL cell lines as a model system, we examined the level of PRMT5 and global methylation of histones H3R8 and H4R3, which are preferentially symmetrically methylated by PRMT5. We found that PRMT5 is highly expressed in a wide variety of lymphoid cancer cell lines, including MCL clinical samples, and consequently global symmetric methylation of H3R8 and H4R3 is also altered in transformed lymphoid cell lines and MCL clinical samples. We also show that increased PRMT5 protein expression is regulated at the translational level and correlates with transcriptional silencing of ST7. Furthermore, knockdown of PRMT5 expression interferes with growth of transformed B cells. Thus, our work identifies PRMT5 as a key chromatin regulator whose aberrant expression affects global H3R8 and H4R3 methylation, contributes to ST7 tumor suppressor gene silencing and is associated with lymphomagenesis.
Results
PRMT5 is overexpressed in lymphoid cancer cell lines and its level correlates with increased symmetric methylation of histones H3R8 and H4R3
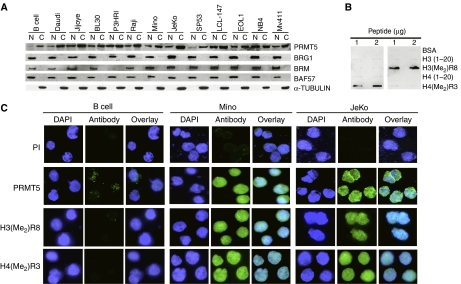
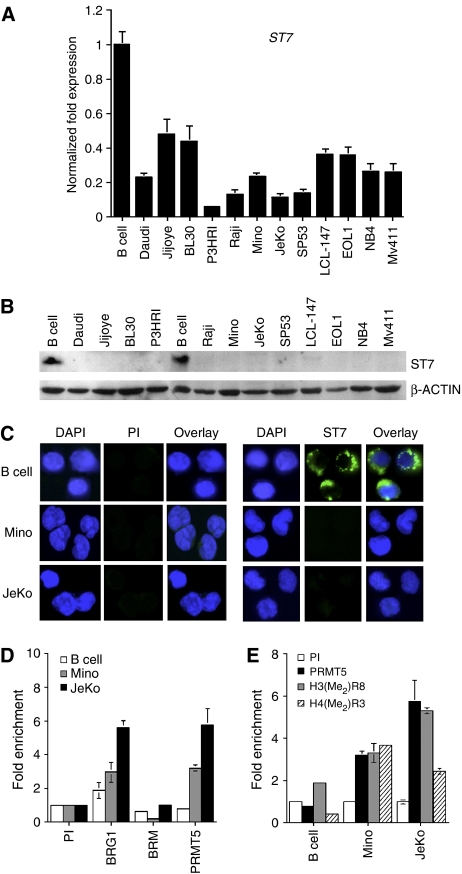
We have previously shown that PRMT5 can interact with BRG1 and BRM-based hSWI/SNF, and that overexpression of PRMT5 in an immortalized, but not transformed cell line, induces hyperproliferation and cellular transformation (Pal et al, 2004). This latter result prompted us to investigate the role of PRMT5 in human malignancies of lymphoid origin for which normal controls are readily available. Nuclear and cytosolic extracts from normal CD19+ B lymphocytes, patient-derived lymphoma (Burkitt's lymphoma: Daudi, Jijoye, BL30, P3HRI, and Raji; MCL: Mino, JeKo and SP53), in vitro EBV-transformed lymphoma (LCL-147) and leukemia cell lines (EOL1, NB4, Mv411) were analyzed by Western blotting (Figure 1A). PRMT5 protein levels were elevated in both nuclear and cytosolic fractions of transformed lymphoid cells. As PRMT5 associates with BRG1 and BRM chromatin remodeling complexes and mutations of hSWI/SNF components have been implicated in various human cancers including leukemia and lymphoma, we analyzed the levels of BRG1, BRM, and BAF57 in normal and transformed lymphoid cells. hSWI/SNF subunits were expressed in all samples except for the Burkitt's lymphoma P3HRI cell line where BRM expression was reduced. Because PRMT5 was found in both nuclear and cytosolic fractions, we measured expression of α-TUBULIN and found that it was expressed exclusively in the cytosol, indicating that the presence of PRMT5 in the nucleus is not due to cytosolic contamination.
Figure 1.
PRMT5 is overexpressed in lymphoma and leukemia cell lines. (A) Expression of PRMT5 and hSWI/SNF subunits were assessed by Western blotting using either nuclear (N) and cytosolic (C) extracts from normal CD19+ B cells, or the indicated transformed cell lines (20 μg). To discern between N and C fractions, we measured α-TUBULIN expression. (B) Anti-H3(Me2)R8 and anti-H4(Me2)R3 antibodies do not crossreact and are highly specific. Portions (1 and 2 μg) of BSA or the indicated peptides were spotted on nitrocellulose membrane, and detected using anti-H4(Me2)R3 or anti-H3(Me2)R8. (C) Immunofluorescence of PRMT5, H4(Me2)R3, and H3(Me2)R8 in normal B cells, Mino, and JeKo cells. Normal B and MCL cells were fixed and incubated with either pre-immune, or immune anti-PRMT5, anti-H3(Me2)R8, and H4(Me2)R3 antibodies. FITC-labeled goat anti-rabbit antibody was used to detect PRMT5 and modified histones, and DAPI was used to stain nuclei. Pictures were taken at × 100 magnification.
To further confirm the Western blot data, we stained normal B and patient-derived Mino and JeKo MCL cells with either pre-immune or immune anti-PRMT5 antibody (Figure 1C). A signal was detected only when immune anti-PRMT5 antibody was used, which was significantly higher in both Mino and JeKo MCL cell lines, suggesting that the level of PRMT5 protein is elevated in transformed lymphoid cells. We have previously shown that PRMT5 can methylate histones H3R8 and H4R3 in vitro, and we have linked PRMT5 recruitment to symmetric methylation of H3R8 in vivo (Pal et al, 2004). Therefore, we analyzed global H3R8 and H4R3 methylation in normal B and MCL cells using antibodies that can specifically recognize either symmetrically methylated H3R8 or H4R3 (Figure 1B, and Supplementary Figure 1A and B). Further characterization of both antibodies using whole-cell extracts revealed that the anti-H3(Me2)R8 antibody is highly specific, whereas the anti-H4(Me2)R3 antibody weakly crossreacts with four other polypeptides in addition to symmetrically methylated H4R3 (Supplementary Figure 1C and E). When anti-H3(Me2)R8 and anti-H4(Me2)R3 antibodies were used in immunoflourescence studies, they failed to detect these epigenetic marks in normal B lymphocytes; however, Mino and JeKo MCL cell lines exhibited high levels of symmetrically methylated H3R8 and H4R3 (Figure 1C). Similar results were observed when core histones isolated from normal and transformed B cells were analyzed by Western blotting using anti-H3(Me2)R8 and anti-H4(Me2)R3 antibodies (Supplementary Figure 1D and E). Collectively, these studies suggest that increased expression of PRMT5 induces symmetric methylation of H3R8 and H4R3, and is associated with MCL pathology.
PRMT5 mRNA expression and stability are altered in patient-derived Mino and JeKo MCL cell lines
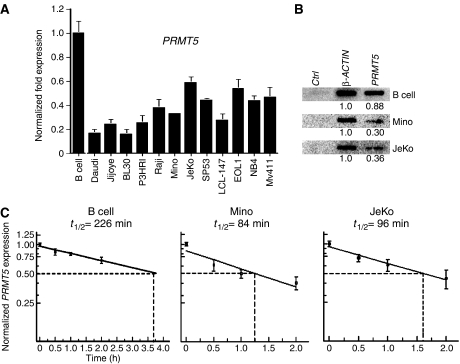
Having found that expression of PRMT5 protein is enhanced in various transformed lymphoid cell lines, we wanted to determine whether this was a direct result of increased PRMT5 mRNA expression (Figure 2A). Real-time RT–PCR revealed that, despite elevated levels of PRMT5 protein in cancer cells, the steady-state levels of PRMT5 mRNA were 2- to 5-fold lower in transformed lymphoid cells (P<10−4), suggesting that there are other mechanisms involved in upregulation of PRMT5 protein expression. To understand the underlying cause for decreased PRMT5 mRNA expression in transformed lymphoid cells, we examined the rate of PRMT5 transcription and mRNA stability in normal B cells as well as patient-derived Mino and JeKo MCL cell lines (Figures 2B and C). Consistent with the real-time RT–PCR results, nuclear run on analysis indicated that the rate of PRMT5 mRNA transcription was 2.9- and 2.4-fold lower in Mino (P=0.028) and JeKo (P=0.011) MCL cell lines, respectively, suggesting that PRMT5 mRNA transcription is more efficient in normal B lymphocytes. To measure PRMT5 mRNA stability, normal B lymphocytes as well as Mino and JeKo MCL cell lines were treated with DRB to inhibit mRNA synthesis, and PRMT5 mRNA levels were measured at various times by real-time RT–PCR. 18S rRNA was used as an internal control to normalize PRMT5 mRNA levels in these experiments, because low dose and short-time treatment with DRB does not affect RNA polymerase I transcription. We found that, in comparison to normal B cells (t1/2=226 min), the half-life of PRMT5 mRNA was reduced 2.7-fold in Mino (t1/2=84 min) and 2.3-fold in JeKo (t1/2=96 min) MCL cell lines (P=0.009). Taken together, these findings along with the observed decrease in PRMT5 mRNA transcription suggest that the observed augmentation in PRMT5 steady-state levels could be the result of either increased PRMT5 mRNA translation and/or enhanced PRMT5 protein stability in transformed MCL cell lines.
Figure 2.
Expression of PRMT5 is regulated at different levels. (A) PRMT5 mRNA expression was measured by real-time RT–PCR in normal as well as transformed B cells. The bar graph shows normalized fold expression of PRMT5 mRNA in various cell lines relative to normal B cells using GAPDH as internal control. RT–PCR analysis was performed three times in triplicates and the graph shows the average±s.d. (B) The rate of PRMT5 transcription is altered in MCL cell lines. Nuclear run on assays were conducted as described in materials and methods. Control (Ctrl) PvuII–PvuII DNA fragment of pBluescript KS(+), and β-ACTIN and PRMT5 cDNA PCR fragments were immobilized on Hybond XL membrane and detected with radiolabeled RNA isolated from the indicated cells. Signals were quantitated and expression of PRMT5 was reported relative to β-ACTIN. (C) PRMT5 mRNA is more stable in B cells. Normal and transformed B cells were treated with DRB for the indicated times, and total RNA was isolated and quantitated by real-time RT–PCR. PRMT5 mRNA expression was normalized using 18S as internal control. To calculate the half-life (t1/2) of PRMT5 mRNA, data points in each graph were generated from four distinct experiments conducted in triplicates.
Aberrant expression of miR-92b and miR-96 is associated with enhanced PRMT5 translation
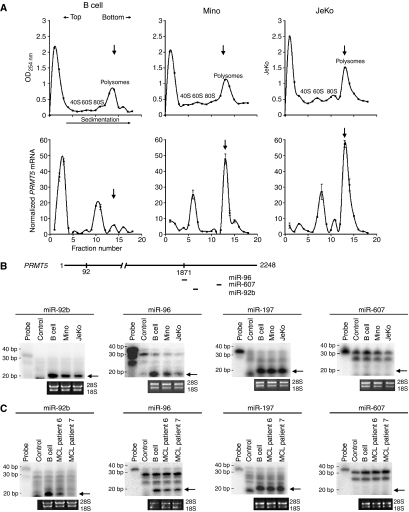
To analyze the efficiency of PRMT5 mRNA translation, we performed sucrose gradient sedimentation experiments in the presence of cycloheximide using cell lysates from normal B lymphocytes and MCL cell lines (Figure 3A). Gradient fractions were collected, and total RNA was extracted and used to measure the levels of PRMT5 mRNA by real-time RT–PCR. The amount of β-ACTIN mRNA in each fraction was used as an internal control to normalize the amount of PRMT5 mRNA present in each fraction. Sucrose gradient analysis demonstrated that, although PRMT5 mRNA was detected in polyribosomal fractions 11–18 in normal and transformed B cells, only 24% of total PRMT5 mRNA was polyribosomal in normal B lymphocytes. However, the fraction of PRMT5 mRNA associated with polyribosomes was enriched 2.2- and 2.6-fold in Mino (P<10−4) and JeKo (P=0.0009) cells, respectively. This change in the polyribosome profile of PRMT5 mRNA and the Western blot results suggest that PRMT5 mRNA translation is enhanced in transformed Mino and JeKo MCL cell lines.
Figure 3.
PRMT5 mRNA is translated more efficiently in MCL cell lines. (A) Polyribosome profiles of normal and transformed B cells. Whole-cell lysate were fractionated by 15–40% sucrose gradient sedimentation, and polyribosome profiles were determined by measuring the absorbance of each fraction at 254 nm (upper panel). Fractions representing 40S, 60S, 80S and polyribosomes are indicated. RNA isolated from each fraction was used to measure the level of PRMT5 mRNA by real-time RT–PCR, and β-ACTIN was used as an internal control to normalize PRMT5 mRNA levels in each fraction (lower panel). The amount of PRMT5 mRNA present in each fraction is reported relative to the fraction containing the lowest copy number of PRMT5 mRNA. The data points in each graph represent the average from triplicate RT–PCR reactions±s.d. Vertical arrows indicate peak polyribosome fractions. (B, C) Differential expression of PRMT5-specific miRNAs in normal and transformed B cells. Schematic representation of PRMT5 mRNA depicting the position of potential miRNA-binding sites within the 3′UTR (B, upper panel). RPA was performed on 20 μg (B) or 5 μg (C) of total RNA isolated from the indicated cells using miR-92b, miR-96, miR-197, or miR-607 probe. Probe represents 1/10th of the total amount of labeled probe used in each reaction, and control shows digestion of the probe in presence of yeast tRNA. Ethidium bromide-stained gels are included to show equal loading. Arrows show position of mature miRNAs.
To elucidate the mechanism responsible for increased translation of PRMT5, we reasoned that there might be PRMT5-specific miRNAs involved in regulating its translation. We searched the Wellcome Trust Sanger Institute miRNA registry (http://microrna.sanger.ac.uk) for the presence of potential miRNA binding site(s) in the 3′ untranslated (UTR) region of PRMT5. We analyzed expression of 11 potential PRMT5 targeting miRNAs in normal and transformed B cells using RNase protection assays (Figures 3B and C and data not shown). Of all the tested miRNAs, only miR-92b and miR-96 expression was detected; however, their expression was 2- to 2.5-fold lower in Mino (miR-92b P=0.009, miR-96 P=0.012) and JeKo (miR-92b P=0.01, miR-96 P=0.005) MCL cell lines, respectively (Figure 3B). When we analyzed the levels of an unrelated control miRNA, miR-197, we found that its expression was unaltered in normal and transformed B cells. More importantly, we also found reduced expression of miR-92b and miR-96 in MCL clinical samples 6 and 7, suggesting that aberrant expression of both miRNAs is a hallmark of MCL (Figure 3C).
Re-expression of miR-92b and miR-96 inhibits PRMT5 translation in vivo, and their binding sites are critical for translational regulation
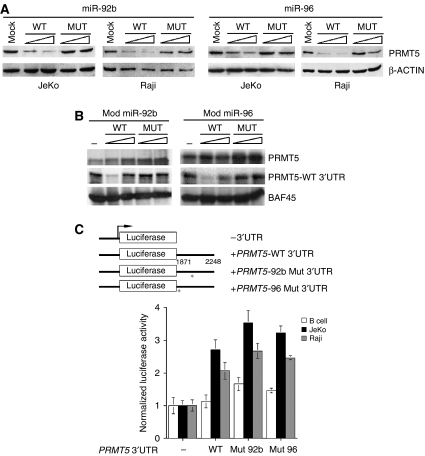
To evaluate further the effect of miR-92b and miR-96 on PRMT5 translational regulation in vivo, we electroporated JeKo cells with either wild-type or mutant miR-92b and miR-96 individually and checked for the levels of PRMT5 protein (Figure 4A). In the presence of wild-type miR-92b, PRMT5 protein expression was reduced by 80%, whereas miR-96 caused a 50% decrease in protein expression. Similarly, when we conducted the same experiment in Raji cells, PRMT5 protein levels were reduced by 60–85%, highlighting the importance of miR-92b and miR-96 expression in translational regulation of PRMT5 (Figure 4A). To verify whether reduced expression of PRMT5 is the result of translational inhibition, but not PRMT5 mRNA degradation, we measured PRMT5 mRNA steady-state levels in mock-transfected cells as well as cells transfected with wild-type and mutant miR-92b or miR-96 (Supplementary Figure 2A and B). As expected, PRMT5 mRNA levels were not significantly altered in the presence of exogenously transfected miR-92b or miR-96 in JeKo and Raji cell lines. As we have previously identified ST7 as a PRMT5 target gene, we analyzed its mRNA levels in miR-92b- or miR-96-electroporated Raji cells (Supplementary Figure 2C). In the presence of miR-92b, ST7 mRNA levels were derepressed 2.2-fold (P=0.001), whereas electroportation of miR-96 resulted in a 1.9-fold ST7 derepression (P=0.005). Taken together, these results suggest that decreased expression of PRMT5 has a direct impact on ST7 transcription.
Figure 4.
PRMT5 expression is downregulated by miR-92b and miR-96 in transformed B cells. (A) JeKo and Raji cells were electroporated with 2.5 and 5.0 μg of either wild-type or mutant miR-92b and miR-96 double-stranded RNA individually, and 20 μg of RIPA extracts were analyzed by Western blotting. (B) Effect of modified miR-92b and miR-96 on PRMT5 expression in vitro. In vitro translation was carried out in the absence or presence of increasing amounts of modified wild-type or mutant miR-92b or miR-96 using 0.25 μg of PRMT5 mRNA without 3′UTR (PRMT5) and with wild-type 3′UTR (PRMT5-WT 3′UTR). BAF45 mRNA was used as a control. (C) Normal B (25 × 106) or transformed JeKo and Raji (5 × 106) cells were electroporated in the presence of pRL-TK with either pCMV-LUC, pCMV-LUC fused to wild-type PRMT5 3′UTR, or pCMV-LUC fused to mutated miR-92b or miR-96-binding site PRMT5 3′UTR construct, and luciferase expression was measured using dual luciferase reporter assay. Luciferase activity is represented relative to pCMV-LUC for each cell line, and has been normalized using Renilla luciferase.
Next, we reconstituted miR-92b- and miR-96-mediated translational inhibition of PRMT5 in vitro. Recent work has shown that translational gene silencing by miRNAs in vitro requires the presence of a 7-methyl G cap and a poly(A) tail (Wang et al, 2006). Therefore, we conducted in vitro translation experiments using 7-methyl G-capped and poly(A)-tailed PRMT5 mRNA with and without wild-type 3′UTR. Both wild-type and mutant miR-96 failed to inhibit PRMT5 translation in vitro, suggesting that there are other components missing in the rabbit reticulocyte lysate, which might stabilize miR-96 binding to its target site (data not shown). Because miR-96 anneals to the PRMT5 3′UTR primarily through its seed sequence (+2 to +8), we assumed that the PRMT5 mRNA:miR-96 hybrid molecule would not be stable thermodynamically at 30°C (ΔG=−13.76 kcal/mol, Tm=22°C). To make up for the lack of stability, we changed six consecutive ribonucleotides (+9 to +14) in miR-96 (ΔG=−27.5 kcal/mol, Tm=44°C), and tested its ability to inhibit PRMT5 translation in vitro (Figure 4B). Modified wild–type, but not mutant miR-96, where the seed sequence (+2 to +8) has been mutated, inhibited PRMT5 translation efficiently at a 1:1 molar ratio when PRMT5 mRNA with wild-type 3′UTR was used. Translation of PRMT5 mRNA without 3′UTR and the unrelated BAF45 mRNA was unaffected. Similar results were observed when modified wild-type and mutant miR-92b were used in these in vitro translation experiments (Figure 4B).
In light of the findings, which showed that re-expression of either miR-92b or miR-96 reduces PRMT5 expression in vivo, we wanted to determine whether the binding sites of miR-92b and miR-96 were critical for translational regulation (Figure 4C). Wild-type and mutant PRMT5 3′UTRs were subcloned downstream of a CMV-driven luciferase reporter, and electroporated into either normal B lymphocytes or transformed JeKo and Raji cell lines. In the presence of wild-type PRMT5 3′UTR, luciferase expression was unchanged in B cells, whereas in JeKo and Raji cells it was enhanced 2.7- (P=0.004) and 2-fold (P=0.026), respectively, indicating that reduced expression of miR-92b and miR-96 in transformed lymphoid cells permits enhanced luciferase expression. When the miR-92b or miR-96 seed sequence binding site was mutated, luciferase expression was increased 1.5- (P=0.035) to 1.7-fold (P=0.001) in B cells, 3.2- (P=0.006) to 3.5-fold (P=0.003) in JeKo cells, and 2.5- (P=0.037) to 2.7-fold (P=0.008) in Raji cells. These results show that miR-92b and miR-96 contribute to proper regulation of PRMT5 translation.
The PRMT5 target gene ST7 is repressed in lymphoid cancer cell lines
We have previously identified ST7 as a direct target gene of PRMT5, and demonstrated using a flag-tagged PRMT5 cell line that recruitment of PRMT5 to the ST7 promoter induces symmetric methylation of histone H3R8 and is associated with ST7 transcriptional repression (Pal et al, 2004). As PRMT5 protein levels are abnormally high in an array of transformed lymphoid cell lines, we investigated whether elevated levels of PRMT5 could impact ST7 expression in these transformed cell lines. Real-time RT–PCR analysis showed that ST7 mRNA expression was reduced 2- to 12-fold in cancer cell lines (P<10−4) (Figure 5A). However, there was no correlation between PRMT5 protein levels and the extent of ST7 mRNA repression. Remarkably, when ST7 protein levels were measured by Western blotting, all transformed lymphoid cell lines lacked ST7 expression, suggesting that there is an inverse relationship between PRMT5 and ST7 protein levels (Figure 5B). To further confirm these results, we detected ST7 protein by immunofluorescence in normal B cells as well as Mino and JeKo MCL cell lines. In accord with the Western blot data, ST7 was detected in normal B cells, but not in transformed MCL cell lines (Figure 5C). It is worth noting that expression of ST7 was not detected when either nuclear or cytosolic extracts were used (data not shown); however, ST7 was readily detectable when whole-cell lysates were prepared using RIPA buffer. These results and the immunofluorescence staining pattern in normal B lymphocytes suggest that ST7 protein is insoluble and probably membrane-bound.
Figure 5.
ST7 is silenced in transformed lymphoid cell lines. (A) ST7 transcription is repressed in lymphoid cancer cell lines as determined by real-time RT–PCR. This experiment has been repeated three times in triplicates. (B) ST7 protein expression was analyzed by Western blotting using 20 μg of RIPA extracts from either normal B cells, or the indicated transformed lymphoid cell lines. β-ACTIN levels were detected to ensure equal loading. (C) Fixed normal or transformed B cells were incubated with either pre-immune or immune anti-ST7 antibody. ST7 protein was visualized using goat FITC-labeled anti-rabbit antibody, whereas nuclei were stained with DAPI. Pictures were taken at × 100 magnification. (D, E) ChIP assays were performed on crosslinked chromatin from normal or transformed B cells using either preimmune (PI) or the indicated immune antibodies, and the retained DNA was amplified by real-time PCR using ST7-specific primers and probe. Fold enrichment with each antibody was calculated relative to the PI sample. Each ChIP experiment was repeated twice in triplicates.
Expression of ST7 mRNA was inhibited in all transformed lymphoid cell lines examined, suggesting that ST7 promoter activity is altered in these cancer cell lines. To verify if PRMT5 is directly involved in inducing ST7 repression, we conducted chromatin immunoprecipitation (ChIP) experiments using anti-PRMT5-specific antibodies (Figure 5D). ChIP analysis showed that PRMT5 recruitment to the ST7 promoter was enriched 3- and 5.5-fold in transformed Mino (P=10−3) and JeKo (P=0.006) MCL cell lines, respectively. Consistent with our previous findings in NIH3T3 cells, BRG1 recruitment to the ST7 promoter was enhanced 1.6- and 3-fold in Mino (P=10−2) and JeKo (P=0.0007), respectively, whereas BRM recruitment was unaffected. These results show that the BRG1-associated PRMT5 is involved in ST7 transcriptional repression in transformed B cells. As PRMT5 can symmetrically methylate H3R8 and H4R3, we evaluated the methylation status of these residues at the ST7 promoter (Figure 5E). We found that methylation of H3R8 is augmented 1.7- (P=0.013) and 2.7-fold (P<10−4) in Mino and JeKo cells, respectively. Similarly, methylation of H4R3 is enriched 3.7- (P=0.002) and 2.5-fold (P<10−4) in these MCL cell lines. We have also analyzed BRG1, BRM, and PRMT5 recruitment as well as H3R8 and H4R3 methylation at the ST7 promoter in two other lymphoma cell lines, Daudi and Raji, and found similar results (Supplementary Figure 3). These findings show that increased expression of PRMT5 is directly involved in ST7 gene repression, and that PRMT5 recruitment as well as H3R8 and H4R3 methylation are altered in transformed lymphoid cell lines.
Expression of PRMT5 and its target gene ST7 is altered in MCL clinical samples
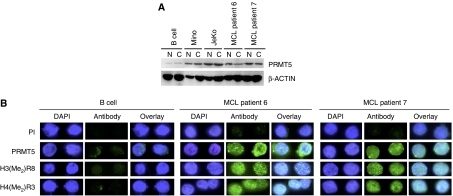
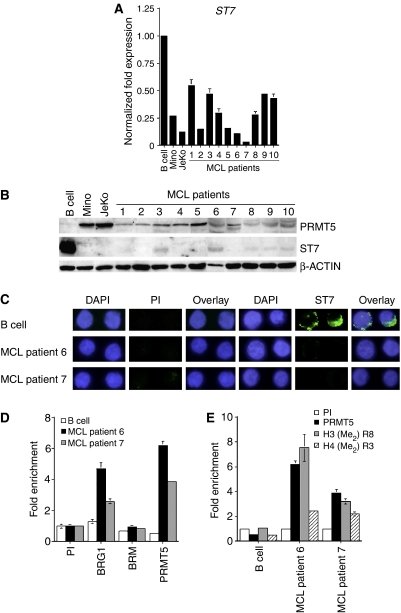
We have found that PRMT5 protein levels are aberrantly increased in a wide variety of transformed lymphoid cells, including Mino and JeKo MCL cell lines, and as a consequence ST7 expression is inhibited. To confirm our results and to assess whether the inverse relationship between PRMT5 and ST7 exists in tumors isolated from MCL patients, we first measured PRMT5 mRNA levels in several MCL clinical samples (Supplementary Figure 4). Our results demonstrate that MCL clinical samples exhibit reduced levels of PRMT5 mRNA. Furthermore, when we analyzed PRMT5 protein expression in MCL clinical samples 6 and 7, which were not limiting, we found that its levels were elevated in the nuclear and cytosolic fractions (Figure 6A). More importantly, immunofluorescence analysis revealed that PRMT5 is overexpressed and that global symmetric methylation of H3R8 and H4R3 is proportionally enhanced in MCL patient samples 6 and 7 (Figure 6B). As PRMT5 can directly inhibit ST7 transcription, we analyzed ST7 mRNA and protein expression in more MCL patient samples. Real-time RT–PCR showed that ST7 mRNA levels were reduced 2- to 30-fold (P<10−4), and Western blot analysis indicated that ST7 protein expression was severely inhibited in all MCL clinical samples examined (Figures 7A and B). In addition, immunofluorescence staining of ST7 protein in MCL samples from patients 6 and 7 confirmed the Western blot results, and showed that ST7 protein expression is inhibited in MCL clinical samples (Figure 7C). Next, we analyzed BRG1, BRM, and PRMT5 recruitment to the ST7 promoter in MCL patient samples 6 and 7 (Figure 7D). BRG1 recruitment was enriched 2- (P=0.002) to 3.6-fold (P=0.006), whereas PRMT5 recruitment was enhanced 8- (P=0.0002) to 14-fold (P=0.003) in MCL clinical samples 6 and 7. As a result of increased recruitment of BRG1-associated PRMT5 to the ST7 promoter, symmetric methylation of H3R8 was augmented 3- (P=0.003) to 7-fold (P=0.016), and H4R3 methylation was elevated 4.6- (P=0.001) to 5-fold (P=0.008) in MCL clinical samples 6 and 7 (Figure 7E). Collectively, these results demonstrate that altered expression of PRMT5 directly affects H3R8 and H4R3 global methylation and triggers changes in target gene expression which may in turn contribute to MCL.
Figure 6.
MCL patients overexpress PRMT5 protein. (A) Nuclear and cytosolic extracts from normal CD19+ B cells or MCL clinical samples 6 and 7 (20 μg) were analyzed by Western blotting using either anti-PRMT5 or control β-ACTIN antibody. (B) Immunofluorescence of normal B and transformed MCL cells from clinical samples 6 and 7 after staining with DAPI, PI, or antibodies to PRMT5, H3(Me2)R8, and H4(Me2)R3.
Figure 7.
Overexpression of PRMT5 correlates with ST7 silencing in MCL clinical samples. (A) Real-time RT–PCR analysis of ST7 mRNA expression in MCL patient samples 1–10. Expression of ST7 was normalized using GAPDH as an internal control. (B) Western blot analysis was performed on 20 μg of RIPA extracts from normal B cells, Mino, JeKo, and MCL clinical samples 1–10 using the indicated antibodies. (C) Immunofluorescence of normal B cells and MCL clinical samples 6 and 7 cells after staining with DAPI, PI, or immune anti-ST7 antibody. Pictures were taken at × 100 magnification. (D, E) ChIP was performed on crosslinked chromatin from normal B cells and MCL clinical samples 6 and 7 using either PI or the indicated immune antibodies. Immunoprecipitated DNA was amplified by real-time PCR, and the fold enrichment with each antibody was calculated relative to the PI sample.
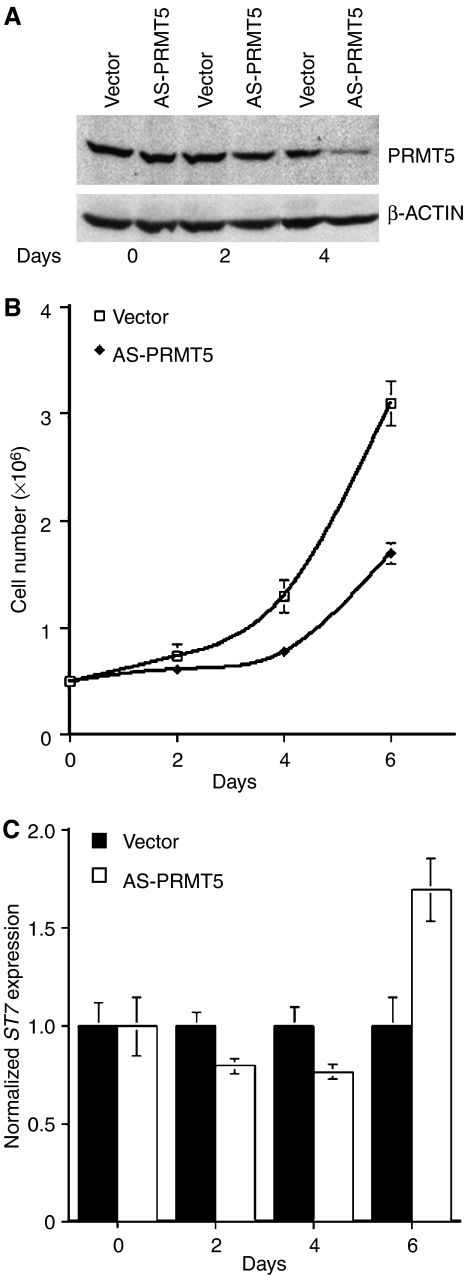
Knocking down PRMT5 alters the growth characteristics of transformed lymphoid cell lines
Previous work showed that overexpression of PRMT5 in NIH3T3 cells induces hyperproliferation, whereas knockdown of PRMT5 reduces cell growth and proliferation (Pal et al, 2004). As PRMT5 levels are high in MCL cell lines and clinical samples, we investigated whether reducing PRMT5 levels could alter MCL cell growth. To lower PRMT5 expression, JeKo cells were infected with either recombinant control or antisense (AS)-PRMT5 lentivirus, and PRMT5 protein expression was evaluated at different time points after infection (Figure 8A). PRMT5 protein levels were reduced significantly by day 4, whereas β-ACTIN expression was unaltered. When we monitored proliferation of control and AS-PRMT5 expressing JeKo cells, there was a noticeable 1.8-fold decrease in growth by day 6 (P<10−4) (Figure 8B). To evaluate whether this decrease in cell growth and proliferation was a direct result of cell death and/or slow cell cycle progression, we measured both DNA content and BrdU incorporation of control and AS-PRMT5-infected cells. Our results indicated that AS-PRMT5 cells incorporated BrdU 25% less efficiently than parental cells infected with control vector, and did not display any change in their cell cycle profile (data not shown). These results suggest that reducing expression of PRMT5 interferes with proliferation of transformed B cells.
Figure 8.
Knocking down PRMT5 expression affects growth of transformed B cells. (A) Western blot analysis was performed on 20 μg of RIPA extract from JeKo cells after infection with either vector or AS-PRMT5 lentivirus for 0, 2, and 4 days using anti-PRMT5 and control anti-β-ACTIN antibodies. (B) Proliferation of JeKo cells infected with either control vector or AS-PRMT5 lentivirus. Cells were counted every two days for 6 days, and the experiment was repeated four times in duplicates. (C) ST7 mRNA expression in lentivirus infected JeKo cells was evaluated by real-time RT–PCR at the indicated times. ST7 mRNA expression is represented relative to control vector infected JeKo cells, and is normalized to GAPDH.
We have shown that PRMT5 can directly regulate ST7 transcription. Therefore, we measured ST7 mRNA levels by real-time RT–PCR in control and AS-PRMT5-expressing JeKo cells (Figure 8C). Our results show that as PRMT5 protein levels decrease by day 6, transcription of ST7 is induced 1.7-fold (P=10−3). Despite this transcriptional derepression, ST7 protein levels did not increase, suggesting that there might be other mechanisms involved in regulating ST7 protein expression (data not shown). Similar effects on cell proliferation and ST7 mRNA expression were observed when PRMT5 was knocked down in Raji cells (Supplementary Figure 5). These findings provide more evidence that knocking down PRMT5 expression in cancer B cells impacts their growth characteristics, and that ST7 expression is regulated post-transcriptionally.
Discussion
Epigenetic modification of chromatin is the focus of intense studies, because there is a need to resolve many unanswered questions in cancer. To understand how post-translational changes of histones impact gene expression, and how perturbation of this process contributes to disease will certainly provide new avenues for cancer therapy. Our previous work showed that overexpression of PRMT5 in immortalized, but not transformed fibroblasts, induces their hyperproliferation and anchorage-independent growth, strongly suggesting that PRMT5 plays an important role in tumorigenesis (Pal et al, 2004). In this study, we showed that PRMT5 protein expression is induced in a wide variety of human lymphoid cancer cell lines, including patient-derived MCL cell lines as well as MCL clinical samples. We also linked misregulated expression of PRMT5 to aberrant expression of miR-92b and miR-96, and showed that the consequence of these changes is increased global symmetric methylation of H3R8 and H4R3. More specifically, we demonstrated that expression of the PRMT5 target gene, ST7, is inhibited in human lymphoid cancer cell lines with altered PRMT5, miR-92b and miR-96 expression. Furthermore, our studies also show that knocking down PRMT5 expression reduces proliferation of transformed lymphoid cell lines, suggesting that reduced levels of PRMT5 are critical for normal cell growth.
Aberrant expression of miR-92b and miR-96 is associated with enhanced PRMT5 translation in MCL
Analysis of nuclear and cytosolic extracts from B cells and transformed lymphoid cell lines and MCL clinical samples showed that PRMT5 protein expression is significantly higher in both compartments (Figures 1 and 6). We have also analyzed PRMT5 protein expression in various solid tumor cell lines, including glioma (U251, Gli3605), adenocarcinoma (HeLa S3, SW13), breast and lung carcinoma (BT549, A549), and hepatoma (HepG2) (Pal and Sif, unpublished), and we have found that global expression of PRMT5 protein is increased in both nucleus and cytosol in comparison to NIH3T3, Rat1a, and PC12 cell lines. Nuclear PRMT5 is associated with chromatin remodelers, including BRG1/BRM-based hSWI/SNF and NURD complexes, and is involved in transcriptional repression of cell cycle regulators and tumor suppressor genes (Pal et al, 2003, 2004; Guezennec et al, 2006). Therefore, when PRMT5 levels are elevated either by overexpression or as it is the case in transformed cancer cells, it might repress transcription of key target genes and promote tumorigenesis.
Our findings show that increased expression of PRMT5 in transformed Mino and JeKo MCL cell lines is a direct result of enhanced PRMT5 mRNA translation (Figures 2 and 3A). What is more striking is the finding that expression of two of the miRNAs predicted to bind to the PRMT5 3′UTR was reduced in patient-derived Mino and JeKo MCL cells as well as MCL clinical samples, and re-expression of miR-92b and miR-96 in two distinct lymphoma cell lines reduced PRMT5 translation in vivo (Figure 4A). Furthermore, mutation of the miR-92b or miR-96 seed sequence abolished PRMT5 translational inhibition in vivo, indicating that the observed repressive effects of miR-92b and miR-96 are specific. Our attempt to recapitulate translational inhibition of PRMT5 in vitro was not successful when wild-type miR-96 was used (data not shown). We have compensated for the lack of stability of PRMT5 mRNA:miR hybrid molecules by increasing the complementarity beyond the seed sequence, and we have been able to specifically inhibit PRMT5 translation in vitro (Figure 4B). Further support for the involvement of miR-92b and miR-96 in regulating PRMT5 translation comes from our observation that the PRMT5 3′UTR can enhance luciferase translation in transformed JeKo cells with low miR-92b and miR-96 expression (Figure 4C). The fact that transfection of either wild-type miR-92b or miR-96 resulted in efficient decrease of PRMT5 expression in JeKo and Raji cells (Figure 4A), and the finding which showed that mutation of miR-92b- or miR-96-binding site improved luciferase translation (Figure 4C), argue that both miR-92b and miR-96 play an important role in regulating PRMT5 protein expression.
Increased PRMT5 expression enhances global symmetric methylation of H3R8 and H4R3, and is accompanied by suppression of ST7
Based on our previous work and current observations, association of PRMT5 with chromatin remodelers enhances its histone methyltransferase activity, and endows it with the ability to influence target gene expression so that the net outcome is to promote cell growth and transformation (Pal et al, 2004). As PRMT5 preferentially targets histones H3R8 and H4R3, we analyzed their methylation status in transformed MCL cells and show that these sites are highly methylated in patient-derived MCL cell lines and MCL clinical samples (Figures 1C and 6B). More specifically, when we analyzed histone methylation at the ST7 promoter in MCL cells, we found that both H3R8 and H4R3 were hypermethylated (Figures 5 and 7). Whereas it is clear that recruitment of PRMT5 to the ST7 promoter enhances H3R8 and H4R3 methylation, there appears to be no correlation between the levels of histone H3R8/H4R3 methylation and ST7 transcriptional suppression (Figures 5 and 7).
It is well established that different post-translational modifications of histones can act either synergistically or antagonistically to specify transcriptional outcome (Fischle et al, 2003). For instance, histone H3K9 and H3K27 trimethylation marks colocalize at the repressed Ubx promoter (Ringrose et al, 2004). Therefore, it is possible that, in the case of ST7 transcription, there are additional epigenetic marks involved in its regulation, which may vary between cell lines and clinical samples. A second possibility is that lack of correlation between H3R8/H4R3 methylation and ST7 transcriptional suppression might arise due to differences in the expression of protein(s) that bind symmetrically methylated H3R8 and H4R3. Recent studies have clearly shown that methylated histones can be recognized and bound by specific methyl-binding protein, and that this is important for transcriptional regulation. Good examples are provided by the WDR5 protein, which can bind to dimethylated H3K4 and support transcriptional activation of HOXC8, and by HP1, which can recognize trimethylated H3K9 and induce silencing (Hall et al, 2002; Wysocka et al, 2005). Thus, it is going to be important to determine whether additional silencing epigenetic marks are present at the ST7 promoter, and whether there are specific factors that can bind symmetrically methylated H3R8 and H4R3.
We have determined that expression of ST7 is regulated both at the transcriptional and translational levels. Knocking down PRMT5 expression in transformed JeKo and Raji cells induces ST7 transcription, but does not affect ST7 protein expression (Figure 8C and Supplementary Figures 2C and 5C). Similarly, transformed lymphoid cells that express low levels of ST7 mRNA do not exhibit any detectable levels of ST7 protein (Figures 5 and 7). We have determined that decreased expression of miR-92b and miR-96 leads to enhanced PRMT5 translation. Therefore, we cannot rule out the possibility that aberrant expression of ST7-specific miRNAs might be inhibiting ST7 translation. More experiments are required to examine expression of ST7-specific miRNAs in normal and transformed B cells.
Role of PRMT5 in mantle cell lymphomagenesis
It is becoming more evident that misexpression and/or mutation of histone-modifying enzymes are associated with cancer etiology. For instance, the mixed lineage leukemia gene, MLL1, which methylates H3K4 and activates transcription, is frequently translocated in acute leukemias and as a consequence more than 50 different leukemogenic MLL fusion proteins are generated (Slany, 2005). Most of these MLL fusion proteins contribute to leukemogenesis by increasing HOX gene expression through mechanisms that involve histone hyperacetylation and H3K79 methylation (Slany, 2005). Another epigenetic mark that appears to be altered in various cancers involves the H3K27-specific methyltransferase, EZH2, which has been tightly linked to gene silencing. Overexpression of EZH2 has been documented in prostate, breast, and gastric cancers and appears to correlate with the high degree of invasiveness of tumors (Varambally et al, 2002; Kleer et al, 2003; Matsukawa et al, 2006). In addition, increased expression of EZH2 in Ramos lymphoma and prostate cancer cell lines induces their growth and hyperproliferation (Visser et al, 2001; Varambally et al, 2002). The role of histone arginine methylation in oncogenesis is not well understood.
Evidence in support of the role played by PRMT5 in cancer came from studies that showed that PRMT5 is frequently upregulated in gastric carcinoma (Kim et al, 2005). Furthermore, when MDA-MB-231 breast cancer cell line is transfected with ER-α and treated with 17β-estradiol, there is a decrease in cell proliferation, which is accompanied by reduced PRMT5 expression (Moggs et al, 2005). Our results show that PRMT5 undergoes translational upregulation in cancer cells of lymphoid origin, including MCL clinical samples. As a result, symmetric methylation of H3R8 and H4R3 is augmented. The impact of enhanced histone arginine methylation is misregulated gene expression, and in the case of the PRMT5 target gene, ST7, there is a clear inhibition of transcription, which appears to be associated with cancer cell growth. We also show that knocking down PRMT5 expression in MCL and Burkitt's lymphoma cell lines reduces cell proliferation. Thus, it appears that PRMT5 is a key histone-modifying enzyme that controls cell growth by modulating expression of target genes through histone arginine methylation.
Elevated expression of PRMT5 can impact other pathways besides methylation of histones H3R8 and H4R3. Recently, PRMT5 was shown to interact with and methylate MBD2 that is associated with the NURD complex (Guezennec et al, 2006). The consequence of this methylation is reduced MBD2 binding to methylated DNA and inability to repress transcription (Tan and Nakielny, 2006). Thus, it is possible that in addition to histone modification, PRMT5 promotes tumorigenesis by negatively modulating the activity of MBD2-based NURD complex. Therefore, it is going to be interesting to examine the methylation status of MBD2 in cancer cells that overexpress PRMT5, and to verify if expression of NURD target genes is altered in these cancer cells. Furthermore, our findings, which show that reducing expression of PRMT5 can inhibit cell proliferation, provide a good starting point to devise new strategies aimed at addressing the therapeutic relevance of targeting PRMT5.
Materials and methods
Plasmid constructions
Plasmids were constructed as described in Supplementary data.
Cell culture, B cell isolation, transfection, luciferase assay, lentivirus production, and cell infection
Normal and transformed lymphoid cells were cultured in RPMI-1640 supplemented with 10–20% FBS. Normal B cells were isolated from tonsils, obtained from Children's Hospital through the Cooperative Human Tissue Network (CHTN) as described in Supplementary Data. Normal and transformed human B lymphocytes were collected under an IRB-approved and HIPPA-compliant protocol. Details for transfection, luciferase assay, lentivirus production, and cell infection are provided in Supplementary Data.
Antibodies, Western blot, and immunofluorescence analyses
For details see Supplementary data.
Reverse transcription, real-time PCR, nuclear run on assay, polyribosome profiling, RNase protection assay (RPA), in vitro transcription, capping, polyadenylation, and translation
Detailed protocols are described in Supplementary data.
Chromatin immunoprecipitation assay
ChIP experiments were performed using soluble crosslinked chromatin from approximately 1 × 107 normal or transformed B cells as described previously, except that washing with the mixed micelle buffer was carried out twice (Pal et al, 2003, 2004). To assess recruitment to the ST7 promoter, real-time PCR was performed on 3 μl of eluted DNA in a 10 μl reaction as described in Supplementary Data.
Statistical analysis
To statistically validate data generated using multiple samples within different groups, analysis of variance (ANOVA) was used to calculate the P-value. To identify differentially expressed genes or recruitment of various chromatin remodelers between two groups, paired t-tests were used to calculate the P-value. In all cases, GraphPad Prism4 software was used to generate P-values.
Supplementary Material
Supplementary Data
Acknowledgments
We thank L Comai for providing the lentiviral expression vectors, D Schoenberg and E Murray for help with polyribosome preparation and for providing plasmid pCMV-LUC, T Ryan for technical help with tonsilar B cell isolation, and P Porcu and D Lucas for providing MCL clinical samples. We also thank D Dakhlallah, S Goel, L Wang for critical reading of the manuscript. This work was supported by National Cancer Institute grants RO1 CA116093 and PO1 CA101956, and American Cancer Society grant RSG-0418201-GMC to SS.
References
- Ancelin K, Lange UC, Hajkova P, Schneider R, Bannister AJ, Kouzaride T, Surani MA (2006) Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat Cell Biol 8: 623–630 [DOI] [PubMed] [Google Scholar]
- Bedford MT, Richard S (2005) Arginine methylation an emerging regulator of protein function. Mol Cell 18: 263–272 [DOI] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y (2002) Role of histone H3 lysine 27 methylation in polycomb -group silencing. Science 298: 1039–1043 [DOI] [PubMed] [Google Scholar]
- Dacwag CS, Ohkawa Y, Pal S, Sif S, Imbalzano AN (2007) The protein arginine methyltransferase Prmt5 is required for myogenesis because it facilitates ATP-dependent chromatin remodeling. Mol Cell Biol 27: 384–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M (2006) Epigenetics provides a new generation of oncogenes and tumour-suppressor genes. Br J Cancer 94: 179–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbrizio E, El Messaoudi S, Polanowska J, Paul C, Cook JR, Lee JH, Negre V, Rousset M, Pestka S, Le Cam A, Sardet C (2002) Negative regulation of transcription by the type II arginine methyltransferase PRMT5. EMBO Rep 3: 641–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Allis CD (2003) Histone and chromatin cross-talk. Curr Opin Cell Biol 15: 172–183 [DOI] [PubMed] [Google Scholar]
- Friesen WJ, Paushkin S, Wyce A, Massenet S, Pesiridis GS, Van Duyne G, Rappsilber J, Mann M, Dreyfuss G (2001) The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol Cell Biol 21: 8289–8300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guezennec XL, Vermeulen M, Brinkman AB, Hoeijmakers WA, Cohen A, Lasonder E, Stunnenberg HG (2006) MBD2/NuRD and MBD3/NuRD, two distinct complexes with different biochemical and functional properties. Mol Cell Biol 26: 843–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake SB, Xiao A, Allis CD (2004) Linking the epigenetic ‘language' of covalent histone modifications to cancer. Br J Cancer 90: 761–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall IM, Shankaranarayana GD, Noma K, Ayoub N, Cohen A, Grewal SI (2002) Establishment and maintenance of a heterochromatin domain. Science 297: 2232–2237 [DOI] [PubMed] [Google Scholar]
- Hosohata K, Li P, Hosohata Y, Qin J, Roeder RG, Wang Z (2003) Purification and identification of a novel complex which is involved in androgen receptor-dependent transcription. Mol Cell Biol 23: 7019–7029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, Sohn HY, Yoon SY, Oh JH, Yang JO, Kim JH, Song KS, Rho SM, Yoo HS, Kim YS, Kim JG, Kim NS (2005) Identification of gastric cancer-related genes using a cDNA microarray containing novel expressed sequence tags expressed in gastric cancer cells. Clin Cancer Res 11: 473–482 [PubMed] [Google Scholar]
- Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, Sabel MS, Livant D, Weiss SJ, Rubin MA, Chinnaiyan AM (2003) EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci USA 100: 11606–11611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak YT, Guo J, Prajapati S, Park KJ, Surabhi RM, Miller B, Gehrig P, Gaynor RB (2003) Methylation of SPT5 regulates its interaction with RNA polymerase II and transcriptional elongation properties. Mol Cell 11: 1055–1066 [DOI] [PubMed] [Google Scholar]
- Martin C, Zhang Y (2005) The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol 6: 838–849 [DOI] [PubMed] [Google Scholar]
- Matsukawa Y, Semba S, Kato H, Ito A, Yanagihara K, Yokozaki H (2006) Expression of the enhancer of zeste homolog 2 is correlated with poor prognosis in human gastric cancer. Cancer Sci 97: 484–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, Hess JL (2002) MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell 10: 1107–1117 [DOI] [PubMed] [Google Scholar]
- Moggs JG, Murphy TC, Lim FL, Moore DJ, Stuckey R, Antrobus K, Kimber I, Orphanides G (2005) Anti-proliferative effect of estrogen in breast cancer cells that re-express ERalpha is mediated by aberrant regulation of cell cycle genes. J Mol Endocrinol 34: 535–551 [DOI] [PubMed] [Google Scholar]
- Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S (2004) Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol Cell Biol 24: 9630–9645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S, Yun R, Datta A, Lacomis L, Erdjument-Bromage H, Kumar J, Tempst P, Sif S (2003) mSin3A/histone deacetylase 2- and PRMT5-containing Brg1 complex is involved in transcriptional repression of the Myc target gene cad. Mol Cell Biol 23: 7475–7487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B, Muller J (2006) Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes Dev 20: 2041–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack BP, Kotenko SV, He W, Izotova LS, Barnoski BL, Pestka S (1999) The human homologue of the yeast proteins Skb1 and Hsl7p interacts with Jak kinases and contains protein methyltransferase activity. J Biol Chem 274: 31531–31542 [DOI] [PubMed] [Google Scholar]
- Ringrose L, Ehre H, Paro R (2004) Distinct contributions of histone H3 lysine 9 and 27 methylation to locus-specific stability of polycomb complexes. Mol Cell 16: 641–653 [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Caldas C (2005) Chromatin modifier enzymes, the histone code and cancer. Eur J Cancer 41: 2381–2402 [DOI] [PubMed] [Google Scholar]
- Sif S (2004) ATP-dependent nucleosome remodeling complexes: enzymes tailored to deal with chromatin. J Cell Biochem 91: 1087–1098 [DOI] [PubMed] [Google Scholar]
- Slany RK (2005) When epigenetics kills: MLL fusion proteins in leukemia. Hematol Oncol 23: 1–9 [DOI] [PubMed] [Google Scholar]
- Tan CP, Nakielny S (2006) Control of the DNA methylation system component MBD2 by protein arginine methylation. Mol Cell Biol 26: 7224–7235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, Rubin MA, Chinnaiyan AM (2002) The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 419: 624–629 [DOI] [PubMed] [Google Scholar]
- Visser HP, Gunster MJ, Kluin-Nelemans HC, Manders EM, Raaphorst FM, Meijer CJ, Willemze R, Otte AP (2001) The Polycomb group protein EZH2 is upregulated in proliferating, cultured human mantle cell lymphoma. Br J Haematol 112: 950–958 [DOI] [PubMed] [Google Scholar]
- Wang B, Love TM, Call ME, Doench JG, Novina CD (2006) Recapitulation of short RNA-directed translational gene silencing in vitro. Mol Cell 22: 553–560 [DOI] [PubMed] [Google Scholar]
- Witzig TE (2005) Current treatment approaches for Mantle Cell Lymphoma. J Clin Oncol 23: 6409–6414 [DOI] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, Allis CD (2005) WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell 121: 859–872 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data