Abstract
Lethal congenital contractural syndrome type 2 (LCCS2) is an autosomal recessive neurogenic form of arthrogryposis that is associated with atrophy of the anterior horn of the spinal cord. We previously mapped LCCS2 to 6.4 Mb on chromosome 12q13 and have now narrowed the locus to 4.6 Mb. We show that the disease is caused by aberrant splicing of ERBB3, which leads to a predicted truncated protein. ERBB3 (Her3), an activator of the phosphatidylinositol-3-kinase/Akt pathway—regulating cell survival and vesicle trafficking—is essential for the generation of precursors of Schwann cells that normally accompany peripheral axons of motor neurons. Gain-of-function mutations in members of the epidermal growth-factor tyrosine kinase–receptor family have been associated with predilection to cancer. This is the first report of a human phenotype resulting from loss of function of a member of this group.
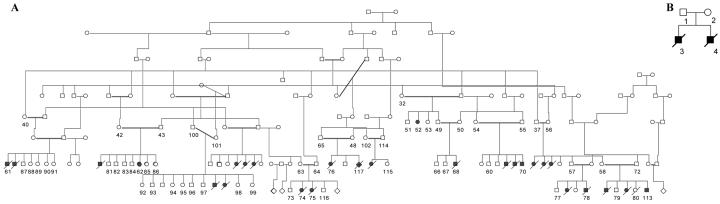
Lethal congenital contractual syndrome type 2 (LCCS2 [MIM 607598]), a neonatally lethal form of arthrogryposis presenting in two Israeli-Bedouin kindreds1 (families A and B) (fig. 1), is characterized by multiple joint contractures, anterior horn atrophy in the spinal cord, and a unique feature of a markedly distended urinary bladder. The phenotype suggests a spinal cord neuropathic etiology.2 Through genomewide linkage analysis, we previously mapped this syndrome to 6 cM (6.4 Mb) on chromosome 12q13 between markers D12S325 and D12S1072 (maximum LOD score of Zmax=9.23 at θ=0),1
Figure 1. .
Israeli-Bedouin kindreds affected with LCCS2. Blackened and unblackened symbols represent affected and unaffected individuals, respectively. The numbers denote individuals whose DNA samples were analyzed. A, Pedigree of family A. B, Pedigree of family B.
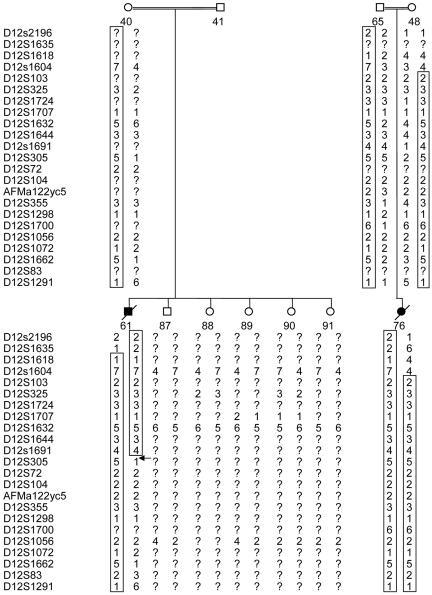
We now have narrowed the disease-associated locus, using additional markers between and flanking D12S325 and D12S1072. Eleven microsatellite markers were selected from public genome databases (Marshfield, Généthon, and deCODE Genetic maps): D12S1712, D12S398, D12S103, D12S1691, D12S72, D12S104, AFMa122yc5, D12S329, D12S375, D12S1052, and D12S1662. The lower boundaries were determined on the basis of analysis of individual 61 (fig. 1). This individual was homozygous for the common haplotype between D12S1635 and D12S305. As seen in figure 2, on the basis of his and his mother’s haplotypes and of the results for markers D12S305, D12S1072, D12S1662, and D12S1291, we could assume that a crossing-over event had occurred proximal to marker D12S305 in the affected individual. Thus, the region of homozygosity common to affected individuals in both families was narrowed to 5.3 cM, corresponding to 4.6 Mb between markers D12S325 and D12S305. Markers D12S1712, D12S398, D12S329, D12S375, D12S1052, and D12S1662 were demonstrated to be outside the defined boundaries of the region of homozygosity (results not shown). The possibility that the disease-associated locus is between D12S305 and D12S1072 cannot be ruled out, but it is less likely, since this would require two crossing-over events occurring in individual 61 (fig. 2).
Figure 2. .
Linkage results for chromosome 12 of two nuclear families of family A. The haplotype showing the homozygosity region is boxed. The arrow indicates a crossing-over event. The lower boundary was determined on the basis of analysis of individual 61.
In an attempt to identify the specific molecular defect leading to the phenotype, we sequenced 61 of the 162 genes in the linkage interval (according to Map Viewer, build 36, and University of California–Santa Cruz [UCSC] Genome Browser). Blood samples were obtained with informed consent after approval was received from the Soroka University Medical Center institutional review board. DNA was isolated from whole blood, tissue samples, or cultured cells, with use of the PUREGENE DNA Isolation Kit per the manufacturer’s instructions (Gentra Systems). RNA was extracted from Epstein-Barr virus–transformed lymphoblastoid cells generated from affected individuals, with use of standard methods. The coding sequences were amplified from genomic DNA or cDNA with PCR primers designed using the Primer3 program. PCR products were subjected to agarose-gel electrophoresis and gel extraction (QIAGEN), followed by sequencing with either the forward or reverse primer on an ABI PRISM 377 DNA Sequencer (Applied Biosystems). Results were analyzed using ChromasPro software, and the obtained DNA sequences were compared with published sequences by use of BLAST.
Genes selected to be analyzed were of several categories: genes that were previously known to be expressed in the spinal cord or to take part in neurogenesis; genes that encode proteins that participate in the function of cells of the neural system, such as NEUROD4, GEFT, and TAC3; and genes known to be involved in development of the nervous system, such as NAB2, GDF11, and several HOXC genes. Another form of autosomal recessive arthrogryposis, arthrogryposis–renal dysfunction–cholestasis syndrome (ARC [MIM 208085]), is caused by mutations in the VPS33B gene.3 VPS33B encodes an Sec1/Munc-18 (SM) protein that belongs to a subfamily of class C vacuolar sorting proteins (VPSs)3,4 that are essential for intracellular membrane-fusion reactions and are associated with severe vacuolar protein–sorting and –morphology defects.5 We therefore also sequenced genes within the LCCS2 linkage interval that are known to be associated with vesicle transport, such as COPZ1, DNAJC14, and OS9. No mutations were found in the coding sequence or within the intron-exon boundaries of any of the genes sequenced (listed in table 1).
Table 1. .
Candidate Genes in the LCCS2 Interval[Note]
| Position (bp) |
Gene | Type of DNA Sequenceda |
| 52618843 | HOXC13 | C |
| 52634981 | HOXC12 | G |
| 52634981 | HOXC12 | C |
| 52653196 | HOXC11 | C |
| 52665221 | HOXC10 | C |
| 52680144 | HOXC9 | C |
| 52689157 | HOXC8 | C |
| 52696940 | HOXC4 | C |
| 52708467 | HOXC6 | C |
| 52713099 | HOXC5 | C |
| 53005178 | COPZ1 | C |
| 53229671 | PDE1B | C |
| 53259302 | PPP1R1A | C |
| 53699889 | NEUROD4 | G |
| 54396088 | BLOC1S1 | C |
| 54423335 | GDF11 | G |
| 54437325 | DNAJC14 | C |
| 54498121 | ORMDL2 | C |
| 54611213 | DGKA | C |
| 54654027 | RAB5B | C |
| 54704791 | ZNFN1A4 | C |
| 54760154 | ERBB3 | G |
| 54784692 | PA2G4 | C |
| 54798297 | NP_116175.1 | C |
| 54838413 | MYL6 | C |
| 54884555 | RNF41 | C |
| 54904418 | NP_076973.1 | C |
| 54910805 | SLC39A5 | C |
| 54918270 | Q8NB46 | C |
| 54990484 | TMEM4 | C |
| 54996988 | USP52 | C |
| 55097177 | TIMELESS | C |
| 55131149 | MIP | G |
| 55148618 | Q8WZ21 | C |
| 55151009 | GLS2 | C |
| 55343403 | TEBP | C |
| 55392484 | NACA | C |
| 55443375 | HSD17B6 | C |
| 55674497 | ADMR | C |
| 55690053 | TAC3 | C |
| 55708569 | MYO1A | G |
| 55769157 | NAB2 | G |
| 55775462 | STAT6 | C |
| 55808543 | LRP1 | C |
| 55923509 | STAC3 | C |
| 56114810 | INHBC | G |
| 56135363 | INHBE | G |
| 56140201 | GLI1 | C |
| 56152314 | ARHGAP9 | C |
| 56196640 | DDIT3 | C |
| 56210202 | DCTN2 | G |
| 56230114 | KIF5A | G |
| 56271253 | PIP5K2C | C |
| 56305945 | GALGT | C |
| 56374171 | OS9 | G |
| 56425051 | SAS | C |
| 56428272 | CDK4 | C |
| 56477704 | AVIL | C |
| 56503617 | CTDSP2 | C |
| 56621627 | NP_150592.1 | C |
| 57008431 | ENK1 | C |
Note.— The candidate genes in the interval were arranged in the table by their location on the chromosome, according to Map Viewer, build 36, and UCSC Genome Browser.
G indicates that genomic DNA was sequenced. C indicates that cDNA was sequenced.
In a study6(in this issue) done in parallel with the one described here, we demonstrated through genomewide linkage analysis that a similar form of autosomal recessive lethal congenital contractural syndrome (LCCS3) without the neurogenic bladder is caused by a mutation in PIP5K1C, which encodes phosphatidylinositol-4-phosphate 5-kinase, type I, gamma (PIPKIγ). Because the phenotypes of LCCS2 and LCCS3 are similar, we sequenced genes in the LCCS2 interval that are related to PIP5K1C. PIP5K1C is involved in signal transduction through the phosphatidylinositol signaling pathway; PIP5K1C catalyzes the phosphorylation of phosphatidylinositol-4-phosphate (PI4P) to generate phosphatidylinositol-4,5-bisphosphate (PIP2),7,8 a substrate for a variety of enzymes and an essential regulator of many cellular processes.9 PIP2 in turn is phosphorylated by phosphatidylinositol-3-kinase (PI3K) to produce a lipid second messenger, phosphatidylinositol-3,4,5-triphosphate (PIP3).10,11 Two genes within the LCCS2 locus were chosen on the basis of their association with the phosphatidylinositol pathway: PIP5K2C and ERBB3 (GenBank accession number NM_001982). PIP5K2C catalyzes the phosphorylation of phosphatidylinositol-5-phosphate (PI5P) to generate PIP2,7,8 and ERBB3 binds p85, a regulatory unit of PI3K.12
Whereas no mutation was found in PIP5K2C or in the coding sequence of ERBB3, an A→G substitution 8 bp 5′ of exon 11 (IVS10-8A→G) of ERBB3 was identified in all LCCS2-affected patients of family A (fig. 3A and 3C). An identical homozygous substitution was found in the affected individual of family B. Primers used to amplify the exons and flanking intron sequences of ERBB3 from genomic DNA in LCCS2-affected patients are listed in table 2.
Figure 3. .
Sequence analysis of the ERBB3 gene in an LCCS2-affected individual. A, Homozygosity for the G→A substitution in intron 10 (affected individual 61). B, Insertion (8 bp) in the cDNA sequence at position 1183 of the ERBB3 gene. chr = Chromosome; ex = exon. C, Schematic representation of the chromosomal locus, the genomic DNA of ERBB3, and the IVS10-8A→G mutation. D, The splice variation occurring in affected individuals. E, Schematic illustration of the protein structure of ErbB3 (according to InterPro) and the predicted mutant truncated protein (395FS399Stop). RLD = Recep_L_domain; FU = Furin-like domain.
Table 2. .
Primers Used for Sequencing the Genomic DNA of ERBB3
| Primer Sequence(5′→3′) |
||
| Primer | Forward | Reverse |
| ERBB3_ex1 | TCTTGCCTCGATGTCCTAGC | TGAGACCGTGCCCATACC |
| ERBB3_ex2 | AATTTGTGTCCAGCCCTCAG | AGCCCTGAAGAAGGAAGGAG |
| ERBB3_ex3 | AATGCCTGAAGGAGGAGAGG | TGGGTCATCATCAAGGAGGT |
| ERBB3_ex4 | TGGATGGGTGGAGAGGTAAG | ACTCTGGTTTGGTGGGACTG |
| ERBB3_ex5–7 | CAAGCCTTTCTTAGCCCTGA | GATTGATTGCACCATTGCAC |
| ERBB3_ex6 | ATGAGCAGTGGCCTCAGAAT | CCCCTACCTCCTCCTCAAAG |
| ERBB3_ex8–9 | AGGGATCTAGGGTGGCAGAT | CTGGGGTCTAAGGAGGGAAG |
| ERBB3_ex10–12 | ATCCCAGCCATTTCTCCAAG | GTCCTTTCAGCCCTCTCCTC |
| ERBB3_ex11 | CACTGGCATAAATTGCGGTA | CATTGCCTGAAGATGCCATT |
| ERBB3_ex12 | GGGATTCCCTCTCAGTGGAT | GTCCTTTCAGCCCTCTCCTC |
| ERBB3_ex13–15 | AGGCTGGAAGCAGTAACGAG | CCAGCTGGGAGTGATTCTTG |
| ERBB3_ex16–7 | CCTCTGCTGTCCAAGCTCTC | GGAAAGAGGAGGGGAGTGAC |
| ERBB3_ex18–20 | TCCAGGACTAACGGTGCTTC | CCCCCAGACAAGCAGTTCT |
| ERBB3_ex20 | CAGGGTCCTGTGCTTCTCAG | CCCCCAGACAAGCAGTTCT |
| ERBB3_ex21 | GATTTGGGGGAATTCCAGAT | ATCTGCTTGCCTCTGGAAGA |
| ERBB3_ex22 | CGGCATGCATCTGTACTCC | GAAGTGAAGGGAAGGCAGAA |
| ERBB3_ex23 | ACCTTTTGAGACCCCCTCTT | AGAGGCGGAGGTTGTAGTGA |
| ERBB3_ex24–26 | GTCTCAAAGTCCCGACCTCA | TCCTAAAACCCAGCCTCTCA |
| ERBB3_ex27 | GGCGACAAGAACAAGACTCC | GGGGAAAGAATTCGAGGAAA |
| ERBB3_ex28 | CCCCTACCCTCATGAAGTTCT | GCCATTAAATGCTCCCTGAG |
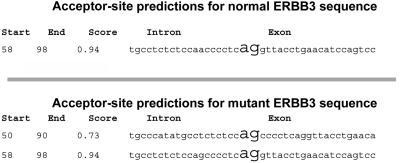
The A→G mutation in ERBB3 abolishes an MmeI restriction site. Restriction-analysis screening for the mutation in all available family members (of families A and B) and in 170 additional unrelated Bedouin individuals was performed (ERBB3 exon 11 was amplified using PCR primers ex11F [5′-CAC TGG CAT AAA TTG CGG TA-3′] and ex11R [5′-CAT TGC CTG AAG ATG CCA TT-3′]). All obligate carriers in families A and B were shown to be heterozygous for the mutation. The mutation segregated with the disease in 67 affected and unaffected individuals in extended families A and B, whereas none of 170 unrelated Bedouins carried the mutation (data not shown). To evaluate the effects of the mutation on the splice site, in silico analysis was done using the Neural Networks Splice Site Prediction software (Berkeley Drosophila Genome Project). Although the mutation did not alter the original acceptor site, an alternative acceptor splice site was predicted (fig. 4). The alternative splice site was confirmed in vitro through sequence analysis of cDNA from LCCS2-affected patients, which showed that the IVS10-8A→G mutation resulted in aberrant alternative splicing, causing an insertion of 8 bases between exons 10 and 11 (fig. 3B and 3D). In all four affected individuals from whom fibroblasts were available, the sole transcript found was the one with the aberrant splicing. The primers used for PCR amplification and for sequencing were CDS_ex11F (5′-GTG GAC TCG AGC AAC ATT GA-3′) and CSD_ex11R (5′-TAC GCC CAG CAC TAA TTT CC-3′). The insertion in position 1183 of the ERBB3 mRNA resulted in a frameshift leading to missense coding beginning at amino acid 395 and an early stop codon generating a 399-aa protein in place of the native 1,341-aa protein (fig. 3E).
Figure 4. .
Splice-site prediction with use of the Neural Networks Splice Site Prediction software (Berkeley Drosophila Genome Project). An alternative additional acceptor splice site was predicted for the ERBB3-mutated sequence.
ERBB3 encodes an epidermal growth factor (EGF) receptor, one of four transmembrane glycoprotein EGF receptor tyrosine kinases: ErbB1 (also called “EGFR” or “HER1”), ErbB2 (HER2 or Neu), ErbB3 (HER3), and ErbB4 (HER4). Members of this family play a key role in regulation of mammalian cell survival, proliferation, adhesion, and differentiation. Overexpression of ErbB receptors has been reported in various human cancers. To date, no human phenotype has been associated with loss of function of any of these receptors. ErbB receptors contain an extracellular region, a transmembrane domain, and a cytoplasmic tyrosine kinase domain. The putative protein structure of ErbB3, according to InterPro (EMBL-EBI), is illustrated in figure 3E. The truncated ERBB3 protein lacks part of the extracellular region, the transmembrane domain, the entire tyrosine kinase domain, and the proline-rich carboxy-terminal tail. An alternative transcriptional splice variant encodes a short isoform of the ErbB3 protein, isoform s, which lacks the transmembrane region and is secreted outside the cell. This isoform acts to regulate the long isoform 1.13 The mutation demonstrated in the LCCS2-affected patients affects only isoform 1, leaving the secreted isoform (isoform s) unaffected. Activation of the ErbB kinases requires binding of a ligand of the EGF or heregulin (also called “Neuregulin” [NRG]) family. NRGs are predominantly expressed in the embryonic CNS and peripheral nervous systems.14 In response to ligand-specific binding, the ErbB receptors form homo- or heterodimers, which in turn initiate tyrosine kinase activity in the intracellular domain by recruiting a different set of Src homology 2 (SH2) domain–containing proteins, which lead to a signaling pathway. ErbB3 is known to heterodimerize with ErbB2 (Her2), causing NRG-dependent activation of the PI3K pathway. The ErbB2/ErbB3 dimer is unique in that ErbB2 has the tyrosine kinase activity, whereas ErbB3 (whose tyrosine kinase domain is inactive) does the ligand binding through its extracellular domain. The mutation found in patients with LCCS2 abrogates the ERBB3 domains known to bind both ERBB2 and the p85 regulatory unit of PI3K. Thus, the LCCS2 mutation is expected to abolish the PI3K-activating function of the ErbB2/ErbB3 dimer.
In mice, ErbB receptors mediate cellular growth and differentiation in multiple tissues, both in the developing embryo and in the adult. Null mutations of any of the ErbB family members result in embryonic lethality in mice, with defects observed in organs, including the brain, heart, skin, lung, and gastrointestinal tract, depending on the receptor affected.15 The human disease caused by a homozygous defect in ERBB3 is reminiscent of the phenotype of the null mutant of its mouse orthologue. Most ErbB3-knockout embryos die before birth. The few that develop to term are slightly reduced in size, do not move or react to tactile stimulation, fail to breathe, and die shortly after birth. Homozygous ErbB3-mutant embryos lack Schwann-cell precursors and Schwann cells that normally accompany peripheral axons of sensory and motor neurons. It should be noted that NRG is highly expressed in Schwann cells. Thus, the specific effect on Schwann cells seen in ErbB3-null mutant mice is in agreement with the known NRG-mediated action of ERBB2-ERBB3 dimers in activating the PI3K pathway.
Neuronal apoptosis has been clearly demonstrated in the LCCS2 phenotype,2 as well as in the homologous Erbb3−/− mouse model16; in Erbb3−/− mice, the initial development of motor neurons and sensory neurons of dorsal root ganglia occurs as it should, but, at later stages, most motor neurons and sensory neurons in dorsal root ganglia undergo cell death. By embryonic day 18.5, the number of motor neurons in the ventral horn of the spinal cord is markedly reduced,16 in line with the anterior-horn atrophy in the human LCCS2 phenotype.2 PI3K phosphorylates PIP2 to produce PIP3, a second messenger that is essential for the translocation of Akt to the plasma membrane. The PI3K/Akt pathway, a critical survival pathway in neurons and in most cell types, is regulated by ErbB2/ErbB3 heterodimers through heregulin.17 Thus, the neuronal apoptosis seen in the human and mouse ErbB3 defects is likely mediated through downregulation of the Akt pathway. In yeast, in vivo conversion of the essential PIP2 pool into PIP3 by PI3K impairs yeast growth by altering morphogenesis and vesicular trafficking.18 We have shown6 that LCCS3, with a phenotype similar to that of LCCS2, is caused by a mutation in PIP5K1C that is essential for the synthesis of PIP2—the substrate of PI3K. Thus, defects in the phosphatidylinositol pathway, altering neuronal concentrations of PIP3 or its precursor PIP2, can lead to defective synaptic vesicular trafficking and enhanced neuronal apoptosis, culminating in a human phenotype of congenital arthrogryposis.
Acknowledgment
We sincerely thank the Kahn Family Foundation for Humanitarian Support for making this study possible.
Web Resources
The accession number and URLs for data presented herein are as follows:
- Berkeley Drosophila Genome Project, http://www.fruitfly.org/seq_tools/splice-instrucs.html (for the Neural Networks Splice Site Prediction software)
- BLAST, http://www.ncbi.nlm.nih.gov/blast/
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (forERBB3 [accession number NM_001982])
- InterPro, http://www.ebi.ac.uk/interpro/ (for the EMBL-EBI ERBB3 protein structure)
- Map Viewer, http://www.ncbi.nlm.nih.gov/mapview (for build 36)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for LCCS2 and ARC)
- Primer3, http://www.genome.wi.mit.edu/cgi-bin/primer/primer3.cgi (for primer design)
- UCSC Genome Browser, http://www.genome.ucsc.edu/
References
- 1.Narkis G, Landau D, Manor E, Elbedour K, Tzemach A, Fishelson M, Geiger D, Ofir R, Carmi R, Birk OS (2004) Homozygosity mapping of lethal congenital contractural syndrome type 2 (LCCS2) to a 6 cM interval on chromosome 12q13. Am J Med Genet A 130:272–276 10.1002/ajmg.a.30266 [DOI] [PubMed] [Google Scholar]
- 2.Landau D, Mishori-Dery A, Hershkovitz R, Narkis G, Elbedour K, Carmi R (2003) A new autosomal recessive congenital contractural syndrome in an Israeli Bedouin kindred. Am J Med Genet A 117:37–40 10.1002/ajmg.a.10894 [DOI] [PubMed] [Google Scholar]
- 3.Gissen P, Johnson CA, Morgan NV, Stapelbroek JM, Forshew T, Cooper WN, McKiernan PJ, Klomp LW, Morris AA, Wraith JE, et al (2004) Mutations in VPS33B, encoding a regulator of SNARE-dependent membrane fusion, cause arthrogryposis-renal dysfunction-cholestasis (ARC) syndrome. Nat Genet 36:400–404 10.1038/ng1325 [DOI] [PubMed] [Google Scholar]
- 4.Lo B, Li L, Gissen P, Christensen H, McKiernan PJ, Ye C, Abdelhaleem M, Hayes JA, Williams MD, Chitayat D, et al (2005) Requirement of VPS33B, a member of the Sec1/Munc18 protein family, in megakaryocyte and platelet α-granule biogenesis. Blood 106:4159–4166 10.1182/blood-2005-04-1356 [DOI] [PubMed] [Google Scholar]
- 5.Carim L, Sumoy L, Andreu N, Estivill X, Escarceller M (2000) Cloning, mapping and expression analysis of VPS33B, the human orthologue of rat Vps33b. Cytogenet Cell Genet 89:92–95 10.1159/000015571 [DOI] [PubMed] [Google Scholar]
- 6.Narkis G, Ofir R, Landau D, Manor E, Volokita M, Hershkowitz R, Elbedour K, Birk OS (2007) Lethal contractural syndrome type 3 (LCCS3) is caused by a mutation in PIP5K1C, which encodes PIPKIγ of the phophatidylinsitol pathway. Am J Hum Genet 81:530–539 (in this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boronenkov IV, Anderson RA (1995) The sequence of phosphatidylinositol-4-phosphate 5-kinase defines a novel family of lipid kinases. J Biol Chem 270:2881–2884 10.1074/jbc.270.7.2881 [DOI] [PubMed] [Google Scholar]
- 8.Loijens JC, Anderson RA (1996) Type I phosphatidylinositol-4-phosphate 5-kinases are distinct members of this novel lipid kinase family. J Biol Chem 271:32937–32943 10.1074/jbc.271.51.32937 [DOI] [PubMed] [Google Scholar]
- 9.Doughman RL, Firestone AJ, Anderson RA (2003) Phosphatidylinositol phosphate kinases put PI4,5P2 in its place. J Membr Biol 194:77–89 10.1007/s00232-003-2027-7 [DOI] [PubMed] [Google Scholar]
- 10.Vanhaesebroeck B, Leevers SJ, Ahmadi K, Timms J, Katso R, Driscoll PC, Woscholski R, Parker PJ, Waterfield MD (2001) Synthesis and function of 3-phosphorylated inositol lipids. Annu Rev Biochem 70:535–602 10.1146/annurev.biochem.70.1.535 [DOI] [PubMed] [Google Scholar]
- 11.Halstead JR, Jalink K, Divecha N (2005) An emerging role for PtdIns(4,5)P2-mediated signalling in human disease. Trends Pharmacol Sci 26:654–660 10.1016/j.tips.2005.10.004 [DOI] [PubMed] [Google Scholar]
- 12.Hellyer NJ, Cheng K, Koland JG (1998) ErbB3 (HER3) interaction with the p85 regulatory subunit of phosphoinositide 3-kinase. Biochem J 333:757–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee H, Akita RW, Sliwkowski MX, Maihle NJ (2001) A naturally occurring secreted human ErbB3 receptor isoform inhibits heregulin-stimulated activation of ErbB2, ErbB3, and ErbB4. Cancer Res 61:4467–4473 [PubMed] [Google Scholar]
- 14.Alroy I, Yarden Y (1997) The ErbB signaling network in embryogenesis and oncogenesis: signal diversification through combinatorial ligand-receptor interactions. FEBS Lett 410:83–86 10.1016/S0014-5793(97)00412-2 [DOI] [PubMed] [Google Scholar]
- 15.Cho HS, Leahy DJ (2002) Structure of the extracellular region of HER3 reveals an interdomain tether. Science 297:1330–1333 10.1126/science.1074611 [DOI] [PubMed] [Google Scholar]
- 16.Riethmacher D, Sonnenberg-Riethmacher E, Brinkmann V, Yamaai T, Lewin GR, Birchmeier C (1997) Severe neuropathies in mice with targeted mutations in the ErbB3 receptor. Nature 389:725–730 10.1038/39593 [DOI] [PubMed] [Google Scholar]
- 17.Li BS, Ma W, Jaffe H, Zheng Y, Takahashi S, Zhang L, Kulkarni AB, Pant HC (2003) Cyclin-dependent kinase-5 is involved in neuregulin-dependent activation of phosphatidylinositol 3-kinase and Akt activity mediating neuronal survival. J Biol Chem 278:35702–35709 10.1074/jbc.M302004200 [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez-Escudero I, Roelants FM, Thorner J, Nombela C, Molina M, Cid VJ (2005) Reconstitution of the mammalian PI3K/PTEN/Akt pathway in yeast. Biochem J 390:613–623 10.1042/BJ20050574 [DOI] [PMC free article] [PubMed] [Google Scholar]