Abstract
The exosome complex is a key component of the cellular RNA surveillance machinery and is required for normal 3′ end processing of many stable RNAs. Exosome activity requires additional factors such as the Ski or TRAMP complexes to activate the complex or facilitate substrate binding. Rrp47p promotes the catalytic activity of the exosome component Rrp6p, but its precise function is unknown. Here we show that recombinant Rrp47p is expressed as an apparently hexameric complex that specifically binds structured nucleic acids. Furthermore, pull-down assays demonstrated that Rrp47p interacts directly with the N-terminal region of Rrp6p that contains the functionally uncharacterized PMC2NT domain. Strains expressing a mutant form of Rrp6p lacking the N-terminal region failed to accumulate Rrp47p at normal levels, exhibited a slow growth phenotype characteristic of rrp47-Δ mutants and showed RNA processing defects consistent with loss of Rrp47p function. These findings suggest Rrp47p promotes Rrp6p activity by facilitating binding via the PMC2NT domain to structural elements within RNA. Notably, characterized Rrp6p substrates such as the 5.8S+30 species are predicted to contain helices at their 3′ termini, while others such as intergenic or antisense cryptic unstable transcripts could potentially form extensive double-stranded molecules with overlapping mRNAs.
INTRODUCTION
RNAs are subjected to quality control mechanisms, known collectively as RNA surveillance, which remove transcripts arising through errors in DNA replication, transcriptional fidelity, ribonucleoprotein particle assembly or RNA processing. A major component of the cellular surveillance machinery is the exosome 3′->5′ exonuclease complex, which degrades unwanted transcripts both in nuclear RNA surveillance programmes and in translation-coupled events (1–3). In addition, the exosome precisely generates the 3′ termini of mature RNAs such as 5.8S rRNA, small nucleolar RNAs (snoRNAs) that function in pre-rRNA processing and small nuclear RNAs (snRNA) that facilitate pre-mRNA splicing (4,5). The exosome also degrades the non-coding 5′ external transcribed spacer (5′ ETS) fragment that is released during pre-rRNA processing (6), and functions in normal mRNA turnover pathways (7,8).
Recent high-resolution crystal structures of exosome complexes from archaeal, yeast and human origin (9–11) have established that the conserved core complex comprises a hexameric ring of subunits homologous to Escherichia coli RNase PH, with three further proteins containing S1 and KH RNA-binding domains located at equivalent positions on one face of the ring. This arrangement of the exosome core complex is analogous to that of PNPase (12), a component of the degradosome complex that also functions in both mRNA degradation and RNA processing pathways in prokaryotes (13). The core appears to play an important structural role, with all nine components of the yeast or human complex being required for structural integrity of the complex (11,14). The eukaryotic exosome contains the salt-labile 3′->5′ exonuclease, Rrp44p/Dis3p (15), and the 3′->5′ exonuclease Rrp6p (16,17), neither of which are present in the archaeal exosome (18).
A hallmark of the exosome complex is that it requires additional proteins to facilitate its exonucleolytic activity in either RNA processing or RNA surveillance pathways. Exosome function in mRNA turnover and cytoplasmic mRNA surveillance pathways requires the associated putative GTPase Ski7p and the Ski complex, comprising the putative RNA helicase Ski2p, Ski3p and Ski8p (7,19). Nuclear RNA surveillance functions require a TRAMP complex, consisting of the putative RNA helicase Mtr4p, the poly(A) polymerase Trf4p or Trf5p, and the homologous RNA-binding proteins Air1p or Air2p (20–24). Similarly, exosome functions mediated by Rrp6p are dependent upon the small basic protein Rrp47p/Lrp1p (25–27).
Rrp6p is a member of the DEDD family of 3′->5′ exonucleases (28) that includes enzymes that act on both RNA and DNA by a hydrolytic mechanism involving two divalent metal ions (29). Rrp6p also contains a helicase and RNase D C-terminal (HRDC) domain that has been proposed to be an RNA-binding domain (30,31). However, Rrp6p itself does not show stable RNA binding in vitro (32). In addition to the catalytic and HRDC domains, Rrp6p contains a C-terminal region that contains nuclear localization signals (32) and an uncharacterized N-terminal domain denoted PMC2NT (33).
RRP6 was first characterized as a suppressor mutant of a temperature-sensitive (ts) allele of the PAP1 gene encoding the nuclear canonical poly(A) polymerase (34). Haploid yeast cells with an rrp6-Δ null allele are viable but show slow growth and a ts-lethal phenotype. Strains lacking Rrp6p have defects in stable RNA synthesis, including the accumulation of 3′ extended processing intermediates of 5.8S rRNA, snRNAs and box C/D snoRNAs (4,34) and the accumulation of polyadenylated RNAs (5,35). In addition to its role in stable RNA processing, rrp6-Δ mutants are also defective in nuclear mRNA surveillance pathways and in the retention of transcripts at the site of transcription (36, and references therein). Mutations of conserved residues in the catalytic domain of Rrp6p cause a loss of function and a cold-sensitive growth phenotype, while mutations within the HRDC domain show only mild RNA processing defects (32,37). The role of the N-terminal PMC2NT domain has not yet been addressed.
The nuclear yeast protein Rrp47p/Lrp1p was identified as a component of the exosome that remained associated with Rrp6p-containing complexes through affinity chromatography purification. Northern blot hybridization experiments and microarray analyses on RNA from rrp47-Δ strains demonstrated that Rrp47p is required at the same step in stable RNA processing pathways as Rrp6p (25,26). Subsequent studies revealed a similar relationship between Rrp47p and Rrp6p in nuclear mRNA surveillance pathways (27,38). The absence of Rrp47p has no significant effect on Rrp6p expression levels or the ability of Rrp6p to bind to the exosome (25). The similar but weaker effects on stable RNA processing observed with rrp47-Δ mutants, compared with rrp6-Δ mutants, the non-additive phenotype of rrp47-Δ rrp6-Δ double mutants and the lack of homology between Rrp47p and any characterized exonuclease led to the suggestion that Rrp47p specifically promotes the exonuclease activity of Rrp6p (25). In contrast to rrp6-Δ mutants, cells lacking Rrp47p do not show a ts-lethal growth phenotype. Therefore, Rrp6p has at least one Rrp47p-independent function that is required for optimal growth.
While Rrp47p has been shown both biochemically and genetically to function together with Rrp6p, its precise function in RNA processing and surveillance pathways is not clear. Here we report that yeast Rrp47p is a nucleic acid-binding protein that specifically binds structured nucleic acids. Furthermore, we demonstrate that Rrp47p interacts directly with the catalytic exosome component Rrp6p via its N-terminal PMC2NT domain. Yeast strains expressing a truncated version of Rrp6p lacking the N-terminal region that contains the PMC2NT domain broadly exhibit the same growth phenotypes and stable RNA processing defects as rrp47-Δ mutants, and fail to accumulate normal levels of Rrp47p. These results provide a mechanistic explanation for how Rrp47p promotes the activity of Rrp6p in degrading or processing structured substrates such as the 3′ extended ‘5.8S+30’ rRNA processing intermediate, snoRNA precursors and intergenic pol II transcripts.
MATERIALS AND METHODS
Plasmids
To express recombinant His-tagged Rrp47p, an NcoI restriction site was introduced at the initiation codon by PCR and the amplified gene was cloned into pRSETB (Invitrogen) as an NcoI–HindIII fragment (p238). The full-length recombinant GST–Rrp6p expression construct (p243) was made by successively subcloning BamHI–BamHI and KpnI–EcoRI fragments from the yeast expression construct pEG-p65 (17) (kindly provided by J.S. Butler) into pGEX-2T (GE Healthcare). Digestion of p243 with KpnI, treatment with T4 DNA polymerase, digestion with StuI and religation generated the rrp6Δ212-721 construct (p245). Digestion of p243 with BglII and religation generated the rrp6Δ42-450 construct (p246). To express the PMC2NT domain of Rrp6p as a GST fusion protein, the N-terminal region of RRP6 encoding residues 13–102 was amplified from genomic yeast DNA by PCR and cloned into pGEX-2T as a BamHI–EcoRI fragment, yielding p249. GST fusions of extended variants of the PMC2NT domain (P176X, p256; L197X, p257) were generated by site-directed mutagenesis of p245 using the Quick-change mutagenesis kit (Stratagene). To express Rrp6p lacking the N-terminal region in E. coli, p243 was digested with KpnI, treated with T4 polymerase and digested with EcoRI. The released fragment was cloned into pGEX-2T after digestion with BamHI, treatment with klenow and subsequent digestion with EcoRI. The incurred frameshift at the BamHI–KpnI junction was then removed by deleting the 3′ C residue of the BamHI site by site-directed mutagenesis, as above, generating the rrp6Δ1-212 construct (p266). To express an rrp6 mutant lacking the N-terminal region in yeast, the 1.8 kb KpnI–EcoRI fragment of RRP6 was cloned in frame behind two tandem copies of the z domain of Staphylococcus aureus protein A and expression of the fusion protein was driven from the RRP4 promoter (39). Specifically, the KpnI–EcoRI region was recovered as a blunt end fragment from p243 by treatment with T4 DNA polymerase and the klenow fragment of E. coli DNA polymerase, and blunt-cloned into the EcoRI site at the end of the zz epitope tag, generating the construct p260 (zz-rrp6Δ1-213). To express the N-terminal region of Rrp6p in yeast, the BamHI–EcoRI fragment of plasmid p245 encompassing the truncated RRP6 coding region was cloned into pEG-KT (40).
Yeast strains and media
The wild-type, rrp6-Δ::TRP1Kl (16) and rrp47-Δ::kanr null mutants, and the epitope-tagged rrp47-zz::HIS5Sp strain (25) used in this study were derivatives of BMA38 (Mata ade2-1 his3-Δ200 leu2-3,112 trp1-1 ura3-1 can1-100) (41). The rrp47-zz::HIS5Sp allele had previously been shown to be fully functional (25). The rrp6-TAP derivative of BY4741 (Mata his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) was obtained from Open Biosystems. This strain showed vigorous growth at 37°C, demonstrating that the TAP-tagged Rrp6p protein is functional. The rrp6-TAP rrp47-Δ::kanr and rrp47-zz rrp6-Δ::TRP1Kl strains were generated by PCR-mediated mobilization of the rrp47-Δ::kanr or rrp47-zz::HIS5Sp allele and correct integration at the RRP47 locus was confirmed by PCR. Yeast strains expressing the zz-rrp6Δ1-213 fusion allele, the corresponding, fully functional full-length Rrp6p derivative (16) or the GAL-regulated GST-rrp6Δ212-712 fusion protein were generated by plasmid transformation of suitable strains. Yeast transformations with plasmids or PCR-amplified DNA were performed using standard techniques. Transformants were selected by growth on suitable selective media and integrants were screened by PCR on genomic DNA.
Strains were routinely grown in minimal media containing 2% glucose or galactose, 0.67% yeast nitrogen base and appropriate supplements. For spot growth assays, the relative concentration of URA+ viable cells in saturated precultures was first determined by plating 100 μl of a 106 dilution on solid glucose-based medium lacking uracil and counting the number of colonies obtained. Ten-fold serial dilutions of standardized precultures were prepared and 5 μl aliquots were applied to the surface of solid growth medium and incubated at 26 and 34°C for 4 days.
Expression and purification of recombinant proteins
The E. coli strain BL21(DE3)LysS was transformed with plasmids encoding full-length or truncated GST-Rrp6p polypeptides and grown up at 30°C in LB medium containing ampicillin and chloramphenicol to an OD600 of 0.5. Transformants expressing His(6)-Rrp47p were grown up at 37°C. Expression was induced by adding IPTG to 0.5 mM and the cultures were incubated for a further 4 h before harvesting the cells. Cell lysates were prepared by sonication in 20 mM HEPES pH 7.6, 300 mM NaCl, 10 mM imidazole pH 7.6 and clarified by centrifugation at 15 000g for 30 min. Clarified lysates were mixed with pre-washed Ni-NTA superflow (Qiagen) or glutathione-sepharose (GE Healthcare) resin and after extensive washing with lysis buffer, the bound proteins recovered by elution in lysis buffer containing 250 mM imidazole or 20 mM reduced glutathione. His(6)-Rrp47p was further purified by ion exchange chromatography and gel filtration. The eluate from the Ni-NTA affinity chromatography was diluted 10-fold with 20 mM HEPES pH 7.6 300 mM NaCl to reduce the imidazole concentration and then mixed with SP-sepharose resin. Bound His(6)-Rrp47p was eluted with cell lysis buffer containing 500 mM NaCl. After concentration under vacuum, the eluate was resolved through a 25 ml superdex 200 column on an ÄKTA purifier system (GE Healthcare) using an elution buffer of 20 mM HEPES pH 7.6, 300 mM NaCl. Concentrations of protein samples were determined by Bradford assay.
Protein and RNA analyses
Recombinant protein-binding assays were performed by mixing lysates from cells expressing one protein with pull-downs of the partner protein on glutathione-sepharose or Ni-NTA superflow beads. After extensively washing the beads in lysis buffer, retained proteins were eluted, resolved by SDS–PAGE and visualized by staining with Coomassie blue G250 or transferred to nylon membrane and decorated with penta-His monoclonal antibodies (Qiagen) or anti-GST antiserum (Sigma). Yeast cell lysates were made in TBS containing 1 mM PMSF by glass bead extraction and clarified by centrifugation at 30 000g for 20 min. Yeast fusion proteins carrying two copies of the z domain of protein A from S. aureus, alone or within the larger TAP tag, were analysed by western blot using peroxidase/anti-peroxidase (PAP) antibody (Sigma). A custom-ordered rabbit polyclonal antiserum raised against recombinant His(6)-Rrp47p (Eurogentec) was used at a dilution of 1:5000 for western analyses.
Total cellular RNA was recovered from cells harvested at early log growth in glucose-based minimal medium by phenol/GTC extraction using glass beads (25). RNA was resolved through 8% polyacrylamide gels, transferred to Hybond N+ membrane (GE Healthcare) and hybridized with radiolabelled transcript-specific oligonucleotide probes to detect specific RNAs. The probes used in this study were specific for the following RNAs: snR38 (oligo 272), gagaggttacctattattacccattcagacagggataactg; 5.8S (oligo 236), gcgttgttcatcgatgc; ITS2 (oligo 237), tgagaaggaaatgacgct; SCR1 (oligo 242), aaggacccagaactaccttg.
Protein–nucleic acid-binding assays
Band-shift assays were performed on His(6)-Rrp47p using EcoRI-digested pBluscript II KS (+) (Stratagene) or the synthetic homopolymeric RNAs poly(A) and poly(A)–poly(U) (Sigma). Twenty-microlitre serial dilutions of Rrp47p purified from cell lysates by Ni-NTA superflow chromatography were prepared in EMSA buffer (10 mM Tris–HCl pH 8, 20 mM KCl, 2 mM MgCl2, 0.1 mM EDTA, 7% glycerol) and mixed with nucleic acids on ice for 20 min. One microlitre of 30% glycerol-containing bromophenol blue was then added and the bound and non-bound nucleic acid fractions were resolved by electrophoresis through 0.5% agarose gels in 0.5× TBE at 5 V/cm. Resolved nucleic acids were then transferred to Hybond N+ membrane and detected by hybridization with radiolabelled T7 primer (aatacgactcactataggg) to detect plasmid DNA, or with oligo(dT) to visualize poly(A).
For the on-bead-binding assays, recombinant proteins or protein complexes were purified on Ni-NTA or glutathione-sepharose beads, as described above. The beads were then equilibrated in EMSA buffer containing 150 mM NaCl and incubated with radiolabelled ligand in 50 μl of the same binding buffer for 30 min on ice. After binding, the supernatant was removed and the beads were washed three times in binding buffer. The amount of ligand retained on the beads was determined by measuring the Cherenkov radiation in a Beckman Coulter LS650 scintillation counter. Ligands were labelled at their 5′ hydroxyl group with γ[32P]-ATP using polynucleotide kinase. The ligands used were poly(A), an ∼40-nt-long synthetic oligodeoxynucleotide (o280, cggaacctcttgactttgagctctatgcacaattcagcttatc), a gel-purified 40-nt-long DNA marker (Promega), E. coli tRNAPhe (Sigma) and a CIP-treated ∼0.6-kb long restriction fragment.
Bioinformatics
Rrp6p homologous sequences were identified by BLAST searches of the EMBL non-redundant database using WU-BLASTp2 (http://dove.embl-heidelberg.de/Blast2/). Multiple sequence alignments was performed using ClustalW (http://www.ebi.ac.uk/clustalw/index.html) and viewed using Jalview (42). Protein structure predictions were performed using PHYRE (http://www.sbg.bio.ic.ac.uk/~phyre/).
RESULTS
Rrp47p is expressed as a multimeric protein complex
To purify yeast Rrp47p for in vitro binding assays, the protein was expressed in E. coli as a 25.5 kDa His(6)-tagged fusion and purified to near homogeneity from cell lysates by affinity chromatography using Ni-NTA superflow beads (Figure 1A). The recombinant protein was further purified by ion exchange chromatography over SP-sepharose beads and subjected to gel filtration analysis. A single peak was observed on the gel filtration column trace (Figure 1B) that corresponded to recombinant Rrp47p by SDS–PAGE analysis. The elution volume of Rrp47p gave an apparent molecular weight of 152 kDa (mean and median averages of 10 independent experiments), indicative of a hexameric complex. We conclude that Rrp47p is expressed as a multimeric protein complex that is most probably hexameric.
Figure 1.
Rrp47p is a multimeric complex. (A) Recombinant His(6)-Rrp47p purification. Samples were resolved through a 15% SDS–PAGE gel and visualized with Coomassie blue G250. Lane 1, non-induced cell extract; lane 2, cell extract after 4 h induction; lane 3, Ni-NTA superflow eluate; lane 4, SP-sepharose non-bound fraction, lane 5, SP-sepharose eluate; lane 6, peak fraction from the superdex 200 GF column. The positions of molecular weight markers (in kDa) are indicated on the left. (B) Gel filtration analysis of the SP-sepharose retained fraction. The A280 profile is shown, together with the elution volumes of the markers thyroglobulin (670 kDa), γ-globulin (158 kDa), ovalbumin (44 kDa), myoglobin (17 kDa) and vitamin B12 (1.3 kDa). The calibration curve obtained from the molecular weight markers is shown.
Rrp47p binds nucleic acids
The murine Rrp47p homologue C1D has been reported to have in vitro DNA-binding activity (43) and previous analyses suggested that yeast Rrp47p promotes the exonuclease activity of Rrp6p by facilitating recruitment of RNA substrates (25). We therefore tested recombinant Rrp47p for DNA and RNA binding in a conventional electrophoretic mobility shift assay (EMSA).
Incubation of plasmid DNA with Rrp47p caused a clear retardation at protein concentrations of ∼1 μM in the EMSA (Figure 2A, upper panel). Increased protein concentrations generated lower mobility complexes (lanes 8 and 9), most probably due to multiple Rrp47p complexes associated with the DNA. No retardation was observed in control experiments where GST protein was incubated with plasmid DNA under the same conditions (Figure S1).
Figure 2.
Rrp47p is a nucleic acid-binding protein. (A) DNA EMSA. Upper panel, 50 ng linearized plasmid was incubated with 2-fold increasing amounts of IMAC-purified, recombinant Rrp47p over the indicated concentration range. Free and bound forms were resolved by agarose gel electrophoresis and detected by Southern blot hybridization (see Materials and Methods section). Lower panel, EMSA in the presence of competitor RNA. Lane 1, free DNA substrate; lane 2, DNA and Rrp47p; lane 3, DNA and mock-purified protein from a pre-induced culture; lane 4, DNA, Rrp47p and 10 μg poly(A); lane 5, DNA, Rrp47p and 10 μg poly(A)–poly(U). (B) Titration of competitor RNA in EMSA. Mixes contained 50 ng DNA, 5 μM Rrp47p and 2-fold increasing amounts of RNA over the indicated concentration range. Upper panel, poly(A); Lower panel, poly(A)–poly(U). (C) RNA EMSA with poly(A) (left) or poly(A)–poly(U) (right). One hundred nanogram of RNA was incubated 2-fold increasing amounts of Rrp47p over the indicated range. The mixtures were resolved by agarose gel electrophoresis and the poly(A) RNA was detected by northern blot hybridization.
We then used this DNA EMSA to test RNAs for their ability to act as competitor substrates. Incubation in the presence of poly(A) RNA had little inhibitory effect on the formation of protein–DNA complexes (Figure 2A, lower panel), even at 200-fold excess over the plasmid DNA (the faster migrating material in lane 4 migrates with protein–DNA complexes seen in the absence of competitor, e.g. upper panel, lane 9). However, incubation in the presence of the double-stranded RNA poly(A)–poly(U) inhibited DNA binding completely. Titration experiments revealed that poly(A)–poly(U) was a very efficient binding competitor, negating DNA binding at approximately equimolar concentrations (Figure 2B). Competitive inhibition was also observed in the presence of the natural heteropolymeric RNAs tRNAPhe from E. coli and total cellular RNA from yeast (Figure S2). In contrast, single-stranded DNA oligonucleotides failed to compete for binding with Rrp47p (Figure S2).
Having established that RNA is an effective competitor to DNA binding by Rrp47p, we then addressed the ability of the protein to interact with RNA directly. EMSA experiments were performed with increasing amounts of purified Rrp47p in the presence of poly(A) or poly(A)–poly(U) RNA and the poly(A) component was detected by northern hybridization using an oligo(dT) probe. Consistent with the competitive inhibition EMSA experiments, incubation of poly(A)–poly(U) with Rrp47p caused a reduced electrophoretic mobility at protein concentrations of ∼1 μM (Figure 2C). As in the DNA-binding experiments, slower migrating complexes were observed at higher protein concentrations. No gel retardation was observed upon incubation of Rrp47p with poly(A) RNA.
The EMSA results were corroborated using an on-bead-binding assay. Double-stranded DNA and tRNA were retained on beads coated with His(6)-Rrp47p, while there was no significant binding of poly(A) or single-stranded DNA oligonucleotide (Figure S3). We conclude that Rrp47p binds to double-stranded DNA or structured RNA with comparable affinity, with an apparent dissociation constant in the 1 μM range, and shows no detectable interaction with single-stranded nucleic acids.
Rrp47p directly binds Rrp6p in vitro
Rrp47p has previously been shown to be a component of the Rrp6p-containing exosome complex (25,26) but previous studies have not addressed whether the proteins physically interact. To demonstrate a putative direct interaction, Rrp6p was expressed in E. coli as an N-terminal GST fusion protein and assayed for its ability to bind His(6)-Rrp47p in pull-down assays (see Materials and Methods section). The His tag antibody detected an ∼25 kDa protein in pull-downs of lysates containing GST-Rrp6p and His(6)-Rrp47p (Figure 3A). No signal was observed if lysate from cells expressing GST was used, or if bound GST-Rrp6p was incubated with lysate from cells transformed with the control vector pRSETB. Specific binding between His(6)-Rrp47p and GST-Rrp6p was also observed in the reciprocal pull-down experiment on Ni-NTA beads (data not shown). We conclude that Rrp47p interacts directly with Rrp6p in the absence of other exosome proteins.
Figure 3.
Rrp47p binds directly to the N-terminal region of Rrp6p. (A) Rrp47p co-purifies with Rrp6p in pull-down assays. Glutathione-sepharose beads were incubated first with lysate containing recombinant GST–Rrp6p or GST and then with lysate from cells expressing His(6)-Rrp47p or containing the control vector pRSETB. Eluates were resolved through a 10% SDS–PAGE gel and analysed by western blotting using an antibody specific for the His tag. O denotes the top of the resolving gel; X marks the position of the bromophenol blue tracking dye. (B) Schematic of the domain structure of Rrp6p. PMC2NT, exonuclease (EXO) and HRDC domains are indicated as boxes. Deletions within Rrp6p constructs made in this study are indicated by broken lines. (C and D) Western analyses of eluates from pull-down assays performed using different Rrp6p constructs, as in A. Upper panels show western blots of eluates using an anti-GST antibody; lower panels show corresponding results with an anti-His antibody. Multiple bands are visible with the anti-GST antibody due to proteolytic degradation of Rrp6p. The electrophoretic migration of the full-length Rrp6p protein constructs are indicated to the right of the panels.
To map the Rrp47p-binding site within the 84 kDa Rrp6p protein, deletion constructs of Rrp6p were made (Figure 3B) and their ability to interact with His(6)-Rrp47p was compared with that of full-length Rrp6p in pull-down assays. The GST-Rrp6Δ212-721 mutant contains the N-terminal region of Rrp6p without the catalytic domain, the HRDC domain and the C-terminal domain bar 12 residues at the extreme C-terminus. In contrast, the GST-Rrp6Δ42-450 construct contains the complete C-terminal domain and most of the HRDC domain, but not the catalytic domain and the N-terminal region bar the extreme N- terminus. His(6)-Rrp47p was retained on beads charged with full-length GST-Rrp6p or the GST-Rrp6Δ212-721 mutant (Figure 3C, lanes 1 and 2) but no binding was observed with the GST-Rrp6Δ42-450 construct (Figure 3C, lane 3). A GST-Rrp6p fusion lacking only residues 1–212 (GST-Rrp6Δ1-212) also failed to show Rrp47p-binding activity (Figure 3D). Although the expression levels of the N-terminal deletion constructs were low, compared with fragments containing the N-terminal region, there was absolutely no detectable binding of His(6)-Rrp47p after prolonged exposure times while binding to the full-length construct was easily detectable. We conclude that the N-terminal 212 residues of Rrp6p are both necessary and sufficient for binding to Rrp47p in vitro.
Analysis of the PMC2NT domain
To analyse the N-terminal 212 residues of Rrp6p in more detail, homologues were identified by BLAST searches and the sequences aligned using ClustalW (Figure S4). All Rrp6p homologues contained a common N-terminal domain that is not found in other subgroups of the DEDD family of 3′->5′ exonucleases. Previous bioinformatics analyses (33) suggested that residues 13–102 of Rrp6p constitute an independent domain, denoted PMC2NT (shown schematically in Figure 4). Protein structure prediction programmes indicated that the PMC2NT domain is largely α-helical in nature, while residues 103–212 have low structural content. However, a recent structural analysis of residues 129–536 of yeast Rrp6p (37) revealed four short α-helices at positions 132–135, 162–165, 184–189 and 208–210 (denoted α1–α4 in Figure 4 and Figure S4). The multiple sequence alignment (Figure S4) indicates that the regions of the protein between α1 and α2 (residues 129–162 in Saccharomyces cerevisiae Rrp6p), and around α3 (residues 179–197) contain a number of highly conserved residues.
Figure 4.
Residues C-terminal to the PMC2NT domain contribute to Rrp47p interaction. (A) Schematic of the N-terminal region of Rrp6p. The location of the PMC2NT domain and the four short α-helices α1–α4 are shown. Residues P176 and L197 define the C-termini of truncated Rrp6p constructs. These residues are mutated to stop codons and therefore not the C-terminal residues. (B) Pull-down assay of His(6)-Rrp47p with GST-tagged N-terminal Rrp6p fragments. Eluates from glutathione-sepharose beads were resolved by SDS–PAGE and assayed by western blot analyses with primary antisera against GST (upper panel) or the His tag (lower panel). Electrophoretic mobilites of protein molecular weight markers (in kDa) are given on the left.
To assay whether the PMC2NT domain was sufficient for Rrp47p binding, we expressed residues 13–102 of Rrp6p as a GST fusion protein in E. coli and assayed the recombinant protein for Rrp47p-binding activity, as above. As shown in Figure 4B, the PMC2NT domain of Rrp6p alone is sufficient for Rrp47p binding. However, increased levels of His(6)-Rrp47p were observed in pull-downs of Rrp6p fusion proteins that extend up to P176 or L197 (compare Figure 4, lane 2 with lanes 3–5). This effect is not simply due to differences in the expression levels of the different Rrp6p constructs, since the fusion protein extended to L197 is expressed at lower levels than the PMC2NT fusion (Figure 4B upper panel, lanes 2 and 4). We conclude that the PMC2NT domain is sufficient for binding with Rrp47p, and that residues between 103 and 197 contribute to this interaction.
Rrp47p binds both nucleic acid and Rrp6p concomitantly
To address whether Rrp47p binding to Rrp6p and nucleic acid is mutually exclusive or can occur simultaneously, we carried out RNA- and DNA-binding assays on pull-downs of Rrp6p/Rrp47p complexes. Glutathione-sepharose beads were charged with either the N-terminal Rrp6p expression construct (GST-Rrp6Δ212-721) or GST and then incubated with lysate containing His(6)-Rrp47p or a control extract. The beads were then incubated with radiolabelled nucleic acids (tRNAPhe or a DNA restriction fragment), the non-bound fraction removed and the amount of bound ligand quantified (Figure 5). Binding of both tRNAPhe and DNA was observed for the assembled Rrp6p–Rrp47p complex. No significant binding was observed in the absence of His(6)-Rrp47p, consistent with the lack of nucleic acid-binding activity of GST or GST-Rrp6Δ212-721 (Figure S1). Moreover, binding was dependent upon the presence of the truncated GST-Rrp6p fusion. We conclude that Rrp6p-associated Rrp47p can form a stable complex with structured nucleic acids.
Figure 5.
Rrp47p interacts with nucleic acid when bound to Rrp6p. Glutathione-sepharose beads charged with GST or a GST fusion protein containing the N-terminal region of Rrp6p (GST-Rrp6Δ212-721) were incubated with lysate containing His(6)-Rrp47p or a control extract (pRSETB). After washing the beads, protein complexes were incubated with radiolabelled E. coli tRNAPhe or a DNA restriction fragment. The non-bound material was removed and bound tRNA or DNA was determined by quantifying the associated Cherenkov radiation. (A) Levels of tRNA retained. (B) Levels of DNA retained. The results shown are the average of three independent assays, the error bars indicating the associated range.
The N-terminal region of Rrp6p is required for normal levels of Rrp47p
There is no available data regarding Rrp47p expression levels, although the levels of several other exosome components such as Rrp6p have been determined (44). We therefore compared the expression levels of Rrp47p and Rrp6p proteins fused to the same epitope (two copies of the z domain of S. aureus protein A) by western blot analysis. We also addressed whether the expression level of Rrp47p was dependent upon Rrp6p.
Western blot analyses revealed that the expression levels of Rrp47p-zz and Rrp6p-TAP in yeast cells were comparable (Figure 6A, compare lanes 1 and 3). The expression level of Rrp47p–zz was reduced to very low levels in the absence of Rrp6p (Figure 6A, lanes 1 and 2). In contrast, the expression of Rrp6p-TAP was not significantly sensitive to the level of Rrp47p (Figure 6A, and 25). We conclude that Rrp6p is required for the normal accumulation of Rrp47p. Since Rrp47p binds directly to the N-terminal region of Rrp6p in vitro (Figures 3 and 4), we reasoned that its interaction with Rrp6p was required for the accumulation of Rrp47p in the cell. To test this hypothesis, we determined Rrp47p-zz expression levels in yeast rrp6 deletion mutants specifically lacking the N-terminal region of the protein (rrp6Δ1-213) or comprising essentially just the N-terminal region (rrp6Δ212-721). The rrp6Δ1-213 mutant was expressed as a zz tag fusion to allow a direct comparison of the relative expression levels of Rrp6p and Rrp47p proteins, while the rrp6Δ212-721 mutant was expressed as a GST fusion to test for association with Rrp47p in pull-down assays. The GST-Rrp6Δ212-721 protein was expressed in yeast under the control of the inducible GAL promoter, allowing repression during growth in glucose-based glucose-based medium and induction during growth in galactose-based medium.
Figure 6.
Rrp47p interacts with the N-terminal region of Rrp6p in yeast. (A–D) Western analyses of cell extracts from strains expressing epitope-tagged forms of Rrp47p and/or Rrp6p. Extracts were resolved through 10% SDS–PAGE gels, protein transferred to western blots and the fusion proteins detected using PAP or GST-specific antisera. Relevant strain genotypes are indicated above each lane. The rrp47-zz, rrp6-TAP and zz-rrp6 alleles encode full-length fusion proteins, whereas the zz-Rrp6Δ1-213 and the GST-Rrp6Δ212-721 proteins are truncated Rrp6p variants. The GST fusion proteins were expressed under the control of the GAL promoter; panel D shows analyses of extracts from strains grown in glucose-based media (glc) and in galactose-based media (gal). The upper panel in D shows a western analysis using the GST-specific antibody. The centre panel in D shows a western analysis of the same samples using the PAP antibody. Bands corresponding to the detected fusion proteins are indicated on the right. SDS–PAGE analyses (lower panels) indicate the relative loading of each extract. (E) Western analysis of lysates from isogenic wild-type and rrp47-Δ strains, using a His(6)-Rrp47p antiserum. Proteins were resolved through a 15% SDS–PAGE gel. The migration of size markers (in kDa) is indicated on the left. A strong band of the expected size is detected in the wild-type lysate (lane 1) and absent in the rrp47-Δ extract (lane 2). (F) Western analyses of the bound fractions of extracts from strains expressing GST (lane 1) or GST-Rrp6Δ212-721 (lane 2) after incubation with glutathione-sepharose beads. The upper panel shows a blot probed with the GST-specific antibody, the lower panel shows a blot probed with the His(6)-Rrp47p antiserum.
Rrp6p was expressed at similar levels as a C-terminal TAP tag or an N-terminal zz tag (Figure 6B), and the N-terminal deletion did not significantly affect the expression level of the protein (Figure 6C). As observed above, a very low level of expression of Rrp47p-zz was detected in the rrp6-Δ strain (Figure 6C, lane 1). Expression of full-length zz-Rrp6p resulted in the accumulation of a band of identical electrophoretic mobility to Rrp47p-zz (Figure 6C, compare lanes 1 and 2). The observed levels of this band were comparable to that of zz-Rrp6p and a band of this size was not observed in strains expressing zz-Rrp6p as the only epitope-tagged protein (Figure 6B, lane 2). In contrast, this protein did not accumulate in strains expressing Rrp6p that lacked the N-terminal region (Figure 6C, lane 3). Rrp47p-zz levels in an rrp6-Δ strain were significantly increased upon induction of GST-Rrp6Δ212-721 (Figure 6D, compare lanes 5 and 6) but not in the vector control (Figure 6D, lanes 3 and 4). We conclude that the N-terminal region of Rrp6p is necessary and sufficient for normal expression levels of Rp47p in yeast.
An interaction between Rrp47p and the N-terminal region of Rrp6p in yeast extracts was demonstrated in pull-down assays. Lysate from strains expressing either GST or the GST-Rrp6pΔ211-721 fusion were incubated with glutathione-sepharose beads and the eluates analysed on western blots using the GST-specific antibody or a rabbit polyclonal antiserum raised against recombinant His(6)-Rrp47p. Rrp47p was detected in the pull-down from the strain expressing the GST-Rrp6Δ212-721 fusion but not the GST control (Figure 6F). We conclude that the N-terminal region of Rrp6p is sufficient for Rrp47p interaction in yeast cell extracts.
The N-terminal region of Rrp6p is required for normal growth
Yeast rrp6-Δ mutants have a ts-lethal growth phenotype and show slow growth at permissive temperatures (34), while rrp47-Δ mutants exhibit a more moderate non-conditional growth defect. To analyse the contribution of the N-terminal region of Rrp6p to the function of the protein in vivo, we first assayed the growth of cells expressing the truncated Rrp6p protein. Yeast rrp6-Δ strains transformed with plasmids expressing the mutant zz-rrp6Δ1-213 allele, the full-length zz-Rrp6p protein or the empty cloning vector were grown on selective solid growth medium or in liquid culture and their growth characteristics were compared with that of an isogenic rrp47-Δ mutant and the parental wild-type strain.
Spot growth assays revealed that both the full-length zz-Rrp6p protein and the truncated zz-rrp6Δ1-213 allele complemented the ts-lethal growth phenotype of the rrp6-Δ mutant (Figure 7A). To compare the growth phenotypes of the zz-rrp6Δ1-213 and rrp47-Δ strains further, we directly measured their growth rates at 30°C, the standard laboratory growth temperature for S. cerevisiae, and compared them to rrp6-Δ and wild-type strains (Figure 7B). The doubling times of the wild-type strain and the rrp6-Δ mutant complemented by the full-length zz–Rrp6p construct were 175 min and 163 min, respectively. Growth of the rrp47-Δ and zz-rrp6Δ1-213 mutants was comparably impaired, with doubling times of 200 min and 223 min, respectively. The rrp6-Δ mutant grew considerably more slowly, with a doubling time of 319 min. We conclude that the zz-rrp6Δ1-213 allele encodes a functional Rrp6p protein that supports growth at a level close to that observed upon loss of Rrp47p function.
Figure 7.
Loss of the N-terminal region of Rrp6p elicits a slow growth phenotype comparable to rrp47-Δ mutants. Growth of isogenic rrp6-Δ, zz-rrp6, zz-rrp6Δ1-213, rrp47-Δ and wild-type strains were compared on selective solid or liquid growth medium. (A) Spot growth assays. Ten-fold serial dilutions of yeast precultures were incubated on solid medium at 26°C (permissive for growth of rrp6-Δ strains) and 34°C (non-permissive for growth of rrp6-Δ strains) and the plates photographed after 4 days. (B) Growth curves. Cultures grown at 30°C were maintained in the exponential growth phase by dilution in pre-warmed medium. The measured increase in OD600, expressed as log10 values, is plotted against time.
The N-terminal region of Rrp6p is required for 5.8S rRNA and snoRNA processing
Yeast strains lacking Rrp6p or Rrp47p accumulate 3′ extended forms of 5.8S rRNA and snoRNAs (4,5,25,34). To test the role of the N-terminal region of Rrp6p in stable RNA processing pathways, total cellular RNA isolated from the zz-rrp6Δ1-213 mutant, the isogenic wild-type strain, a strain expressing full-length zz-Rrp6p and rrp6-Δ or rrp47-Δ null mutants was analysed by northern blot hybridization using oligonucleotide probes complementary to the mature 5.8S rRNA, 3′ extended 5.8S rRNA species, and the intron-encoded box C/D snoRNA snR38.
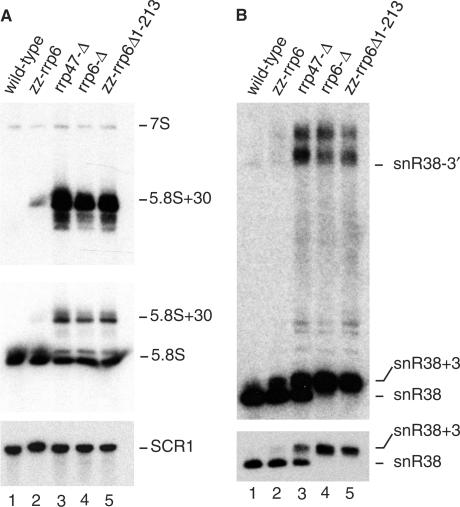
The zz-rrp6Δ1-213 mutant exhibited a 5.8S rRNA processing phenotype that was comparable to that of rrp6-Δ and rrp47-Δ mutants (Figure 8A, lanes 3–5), with a slight increase in the level of 7S pre-rRNA, the 3′ extended precursor to 5.8S rRNA, compared to the wild-type strain, and a large accumulation of the ‘5.8S+30’ processing intermediate. A much weaker accumulation of the band corresponding to the 5.8S+30 intermediate was observed in cells expressing the full-length zz-Rrp6p epitope-tagged protein (Figure 8A, lane 2), demonstrating that the defect observed in the mutant lacking the N-terminal region is not due to the presence of the epitope tag. The zz-rrp6Δ1-213 mutant also accumulated snR38 species that were ∼3 nt longer than the mature RNA (denoted snR38+3) or extended to the 3′ splice site (denoted snR38-3′). A similar defect was also observed for U14 snoRNA (data not shown). The snoRNA processing phenotype of rrp6-Δ mutants is more exacerbated than that of rrp47-Δ mutants, as demonstrated by the complete block to final trimming of snR38 (Figure 8B, compare lanes 3 and 4, and 25). The snR38-processing defect observed in the zz-rrp6Δ1-213 mutant matched that observed in the rrp6-Δ null mutant. We conclude that loss of the N-terminal domain of Rrp6p elicits stable RNA-processing phenotypes that are consistent with a loss of Rrp47p function. However, loss of the N-terminal region of Rrp6p has a more severe effect on the protein's function than simply abrogating its interaction with Rrp47p.
Figure 8.
The N-terminal region of Rrp6p is required for 5.8S rRNA and snoRNA processing. Northern blot analyses of RNA in the zz-rrp6Δ1-213 mutant. Total cellular RNA from yeast cultures was resolved through 8% denaturing acrylamide gels and analysed by northern blot hybridization using probes specific to 5.8S rRNA and snR38 RNA species. Relevant strain genotypes are indicated above each lane. Detected RNA species are indicated to the right of each panel. (A) Hybridization with probes to 5.8S rRNA species. Upper panel, hybridization with a probe (237) specific for 3′ extended 5.8S rRNA species. Centre panel hybridization with a probe (236) specific for the mature 5.8S rRNA. 5.8S rRNA is expressed as a predominant short form and a less abundant long form. Lower panel hybridization with a probe (242) complementary to the RNA pol III transcript SCR1, which serves as a loading control. (B) Hybridization with a probe (272) specific for snR38. Lower panel, exposure showing the mature snR38 and a species with a short 3′ extension (snR38 + 3). Upper panel, longer exposure to reveal 3′ extended precursors. The snR38 species extended at the 3′ end to the 3′ splice site is indicated as snR38-3′.
DISCUSSION
To test an earlier model that Rrp47p functions as an exosome cofactor by docking the Rrp6p enzyme onto the 3′ end of RNA substrates (25), we assayed Rrp47p for its ability to bind RNA and to interact with Rrp6p. Recombinant Rrp47p was shown to bind concomitantly to both RNA and Rrp6p. The region of Rrp6p involved in Rrp47p binding was mapped to the functionally uncharacterized N-terminal PMC2NT domain (33) and we therefore addressed the contribution of this domain to Rrp6p activity in vivo.
Rrp47p bound poly(A)–poly(U) and tRNAPhe but did not bind poly(A) RNA, and bound to structured RNA or double-stranded DNA with comparable affinity in EMSA analyses. Rrp47p is not related in sequence to any characterized double-stranded nucleic acid-binding protein (45) and therefore represents a novel class of proteins with this activity. Analysis of the protein sequence using the structure prediction algorithms PHYRE (http://www.sbg.bio.ic.ac.uk/~phyre/) and PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred/) suggest that Rrp47p and its homologues consist of an α-helical N-terminal region, followed by an unstructured region of variable length which has a basic stretch of residues at the C-terminus (50% lysine or arginine residues in the final 22 amino acids of S. cerevisiae Rrp47p). Residues within this lysine-rich region might contribute to nucleic acid binding (46) but the nature of the specificity for double-stranded nucleic acids is unclear at present. Rrp47p may potentially form a collar around double-stranded DNA or RNA, analogous to some members of the six-membered ring ATPase family (47). Consistent with our results, the Rrp47p homologue C1D has recently been reported to bind structured RNAs such as poly(G) and tRNA in vitro (48).
The RNA-binding activity of Rrp47p is most probably of key importance to its function as an exosome cofactor. The 3′ termini of 5.8S rRNA and snoRNA precursors that accumulate in rrp6-Δ and rrp47-Δ strains are predicted to be involved in imperfect double-stranded structures (49,50). In addition, recent genome-wide expression studies have revealed extensive transcription of both intergenic regions and antisense mRNA (51–53), which could hybridize with mRNA to form stretches of double-stranded RNA. Such intergenic transcripts are nuclear exosome substrates (20) and are degraded in an Rrp47p-dependent manner (27). Furthermore, recombinant Rrp6p is highly active on poly(A) or A/U-rich RNAs in vitro but degrades structured substrates rather poorly (11,17). Rrp47p has also been implicated in processes of DNA repair (38,54) and telomere length control (55), which may reflect its ability to bind DNA.
Previous estimates suggest Rrp6p is present at ∼2000 copies per cell (44). Our western analyses revealed comparable expression levels of Rrp47p and Rrp6p. The organization of Rrp47p into hexameric complexes suggests that the majority of Rrp6p–exosome complexes in the cell do not contain Rrp47p, a notion supported by the stoichiometric levels of Rrp47p and Rrp6p in non-fractionated exosome complexes (25). One possibility is that Rrp6p-containing exosome complexes lacking Rrp47p are required for the Rrp47p-independent function of Rrp6p that is important for optimal cell growth.
Recombinant protein studies and analyses of yeast extracts showed that Rrp47p interacts with the ∼200-residue long N-terminal region of Rrp6p. While the PMC2NT domain of Rrp6p (residues 13–102) was sufficient for Rrp47p interaction, binding was improved with extended constructs. Sequence alignments and structural studies (37) reveal that this region includes a semi-conserved loop flanked by short α-helices (P132-L166). We propose that this loop structure promotes binding of Rrp47p to the PMC2NT domain, either by stabilizing the fold of the PMC2NT domain or by providing additional points of interaction. Furthermore, expression of an rrp6 mutant lacking the N-terminal region correlated with loss of Rrp47p function in vivo by three independent criteria: (i) the depletion of Rrp47p expression levels; (ii) a reduction of cell growth rate; and (iii) the accumulation of 5.8S rRNA and snoRNA processing intermediates. C1D has recently been shown to interact with both RNA and PM/Scl-100 (48), the human homologue of Rrp6p, demonstrating that this mode of exosome regulation has been conserved from yeast to humans.
Deletion of the N-terminal region had a greater effect on Rrp6p function than simply preventing interaction with Rrp47p. Loss of the N-terminal region of Rrp6p may cause subtle structural defects within the protein; it has been proposed that this region may be important to anchor the HRDC domain to the exonuclease domain (37). Alternatively, additional factors involved in snoRNA maturation such as Bcd1p (26) might also interact with this portion of Rrp6p.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Scot Butler for the GST-Rrp6p expression construct pEG-p65, and Guillaume Hautbergue and Stefan Milson for help with the FPLC analyses. This work was funded by the BBSRC, Yorkshire Cancer Research and the Royal Society. Funding to pay the Open Access publication charges for this article was provided by the BBSRC.
Conflict of interest statement. None declared.
REFERENCES
- 1.Vasudevan S, Peltz SW. Nuclear mRNA surveillance. Curr. Opin. Cell. Biol. 2003;15:332–337. doi: 10.1016/s0955-0674(03)00051-6. [DOI] [PubMed] [Google Scholar]
- 2.Houseley J, LaCava J, Tollervey D. RNA quality control by the exosome. Nat. Rev. Mol. Cell. Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- 3.Reinisch KM, Wolin SL. Emerging themes in non-coding RNA quality control. Curr. Opin. Struct. Biol. 2007;17:1–6. doi: 10.1016/j.sbi.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 4.Allmang C, Kufel J, Chanfreau G, Mitchell P. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 1999;18:5399–5410. doi: 10.1093/emboj/18.19.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Hoof A, Lennertz P, Parker R. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell. Biol. 2000;20:441–452. doi: 10.1128/mcb.20.2.441-452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de la Cruz J, Kressler D, Tollervey D, Linder P. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J. 1998;17:1128–1140. doi: 10.1093/emboj/17.4.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacobs Anderson JS, Parker R. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen C-Y, Gherzi R, Ong S-E, Chan EL, Raijmakers R, Pruijn GJM, Stoeklin G, Moroni C, Mann M, et al. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 9.Büttner K, Wening K, Hopfner K-P. Structural framework for the mechanism of archeal exosome in RNA processing. Mol. Cell. 2005;20:461–471. doi: 10.1016/j.molcel.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 10.Lorentzen E, Conti E. Structural basis of 3′ end RNA recognition and exoribonucleolytic cleavage by an exosome RNase PH core. Mol. Cell. 2005;20:473–481. doi: 10.1016/j.molcel.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 11.Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 12.Symmons MF, Jones GH, Luisi BF. A duplicated fold is the structural basis for polynucleotide phosphorylase catalytic activity, processivity, and regulation. Structure. 2000;8:1215–1226. doi: 10.1016/s0969-2126(00)00521-9. [DOI] [PubMed] [Google Scholar]
- 13.Carpousis AJ. The Escherichia coli RNA degradosome: structure, function and relationship in other ribonucleolytic multienzyme complexes. Biochem. Soc. Trans. 2002;30:150–155. [PubMed] [Google Scholar]
- 14.Dziembowski A, Lorentzen E, Conti E, Séraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat. Struct. Mol. Biol. 2007;14:15–22. doi: 10.1038/nsmb1184. [DOI] [PubMed] [Google Scholar]
- 15.Mitchell P, Petfalski E, Schevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic processing complex containing multiple 3′->5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 16.Allmang C, Petfalski E, Podtelejnikov A, Mann M, Tollervey D, Mitchell P. The yeast exosome and human PM-Scl are related complexes of 3′->5′ exonucleases. Genes Dev. 1999;13:2148–2158. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burkard KTD, Butler JS. A nuclear 3′-5′ exonuclease involved in mRNA degradation interacts with poly(A) polymerase and the hnRNA protein Npl3p. Mol. Cell. Biol. 2000;20:604–616. doi: 10.1128/mcb.20.2.604-616.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koonin EV, Wolf YI, Aravind L. Prediction of the archaeal exosome and its connections with the proteasome and the translation and transcription machineries by a comparative-genomic approach. Genome Res. 2001;11:240–252. doi: 10.1101/gr.162001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Hoof A, Frischmeyer PA, Dietz HC, Parker R. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science. 2002;295:2262–2264. doi: 10.1126/science.1067272. [DOI] [PubMed] [Google Scholar]
- 20.Wyers F, Rougemaille M, Badis G, Rousselle J-C, Dufour M-E, Boulay J, Régnault B, Devaux F, Namane A, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 21.LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 22.Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Houseley J, Tollervey D. Yeast Trf5p is a nuclear poly(A) polymerase. EMBO Rep. 2005;7:205–211. doi: 10.1038/sj.embor.7400612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Egecioglu DE, Henras AK, Chanfreau G. Contributions of Trf4p- and Trf5p-dependent polyadenylation to the processing and degradative functions of the yeast nuclear exosome. RNA. 2006;12:26–32. doi: 10.1261/rna.2207206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitchell P, Petfalski E, Houalle R, Podtelejnikov A, Mann M, Tollervey D. Rrp47p is an exosome-associated protein required for the 3′ processing of stable RNAs. Mol. Cell. Biol. 2003;23:6982–6992. doi: 10.1128/MCB.23.19.6982-6992.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peng W-T, Robinson MD, Mnaimneh S, Krogan NJ, Cagney G, Morris Q, Davierwala AP, Grigull J, Yang X, et al. A panoramic view of yeast noncoding RNA processing. Cell. 2003;113:919–933. doi: 10.1016/s0092-8674(03)00466-5. [DOI] [PubMed] [Google Scholar]
- 27.Arigo JT, Eyler DE, Carroll KL, Corden JL. Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol. Cell. 2006;23:841–851. doi: 10.1016/j.molcel.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 28.Moser MJ, Holley WR, Chatterjee A, Mian IS. The proofreading domain of Escherichia coli DNA polymerase I and other DNA and/or RNA exonuclease domains. Nucleic Acids Res. 1997;25:5110–5118. doi: 10.1093/nar/25.24.5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beese LS, Steitz TA. Structural basis for the 3′-5′ exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J. 1991;10:25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morozov V, Mushegian AR, Koonin EV, Bork P. A putative nucleic acid-binding domain in Bloom's and Werner's syndrome helicases. Trends Biochem. Sci. 1997;22:417–418. doi: 10.1016/s0968-0004(97)01128-6. [DOI] [PubMed] [Google Scholar]
- 31.Liu Z, Macias MJ, Bottomley MJ, Atier G, Linge JP, Nilges M, Bork P, Sattler M. The three-dimensional structure of the HRDC domain and implications for the Werner and Bloom syndrome proteins. Structure. 1999;7:1557–1566. doi: 10.1016/s0969-2126(00)88346-x. [DOI] [PubMed] [Google Scholar]
- 32.Phillips S, Butler JS. Contribution of domain structure to the RNA 3′ end processing and degradation functions of the nuclear exosome subunit Rrp6p. RNA. 2003;9:1098–1107. doi: 10.1261/rna.5560903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Staub E, Fiziev P, Rosenthal A, Hinzmann B. Insights into the evolution of the nucleolus by an analysis of its protein domain repertoire. Bioessays. 2004;26:567–581. doi: 10.1002/bies.20032. [DOI] [PubMed] [Google Scholar]
- 34.Briggs MW, Burkard KTD, Butler JS. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8S rRNA 3′ end formation. J. Biol. Chem. 1998;273:13255–13263. doi: 10.1074/jbc.273.21.13255. [DOI] [PubMed] [Google Scholar]
- 35.Kuai L, Fang F, Butler JS, Sherman F. Polyadenylation of rRNA in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2004;101:8581–8586. doi: 10.1073/pnas.0402888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomsen R, Libri D, Boulay J, Rosbash M, Jensen TH. Localisation of nuclear retained mRNAs in Saccharomyces cerevisiae. RNA. 2003;9:1049–1057. doi: 10.1261/rna.5170303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Midtgaard SF, Assenholt J, Jonstrup AT, Van LB, Jensen TH, Brodersen DE. Structure of the nuclear exosome component Rrp6p reveals an interplay between the active site and the HRDC domain. Proc. Natl Acad. Sci. USA. 2006;103:11898–11903. doi: 10.1073/pnas.0604731103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hieronymus H, Yu MC, Silver PA. Genome-wide mRNA surveillance is coupled to mRNA export. Genes Dev. 2004;18:2652–2662. doi: 10.1101/gad.1241204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitchell P, Petfalski E, Tollervey D. The 3′ end of yeast 5.8S rRNA is generated by an exonuclease processing mechanism. Genes Dev. 1996;10:502–513. doi: 10.1101/gad.10.4.502. [DOI] [PubMed] [Google Scholar]
- 40.Mitchell DA, Marshall TK, Deschenes RJ. Vectors for the inducible overexpression of glutathione S-transferase fusion proteins in yeast. Yeast. 1993;9:715–722. doi: 10.1002/yea.320090705. [DOI] [PubMed] [Google Scholar]
- 41.Baudin AO, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- 43.Nehls P, Keck T, Greferath R, Spiess E, Glaser T, Rothbarth K, Stammer H, Werner D. cDNA cloning, recombinant expression and characterization of polypeptides with exceptional DNA affinity. Nucleic Acids Res. 1998;26:1160–1166. doi: 10.1093/nar/26.5.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ghaemmaghami S, Huh W-K, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 45.Saunders LR, Barber GN. The dsRNA binding protein family: critical roles, diverse cellular functions. FASEB J. 2003;17:961–983. doi: 10.1096/fj.02-0958rev. [DOI] [PubMed] [Google Scholar]
- 46.Wilson SA, Brown EC, Kingsman AJ, Kingsman SM. TRIP: a novel double stranded RNA binding protein which interacts with the leucine rich repeat of Flightless I. Nucleic Acids Res. 1998;26:3460–3467. doi: 10.1093/nar/26.15.3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patel SS, Picha KM. Structure and function of hexameric helicases. Ann. Rev. Biochem. 2000;69:651–697. doi: 10.1146/annurev.biochem.69.1.651. [DOI] [PubMed] [Google Scholar]
- 48.Schilders G, van Dijk E, Pruijn GJM. C1D and hMtr4p associate with the human exosome subunit PM/Scl-100 and are involved in pre-rRNA processing. Nucleic Acids Res. 2007;35:2564–2572. doi: 10.1093/nar/gkm082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Villa T, Ceradini F, Bozzoni I. Identification of a novel element required for processing of intron-encoded box C/D small nucleolar RNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 2000;20:1311–1320. doi: 10.1128/mcb.20.4.1311-1320.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yeh L-CC, Lee JC. Structural analysis of the internal transcribed spacer 2 of the precursor ribosomal RNA from Saccharomyces cerevisiae. J. Mol. Biol. 1990;211:699–712. doi: 10.1016/0022-2836(90)90071-S. [DOI] [PubMed] [Google Scholar]
- 51.Yelin R, Dahary D, Sorek R, Levanon EY, Goldstein O, Shoshan A, Diber A, Biton S, Tamir Y, et al. Widespread occurence of antisense transcription in the human genome. Nat. Biotechnol. 2003;21:379–386. doi: 10.1038/nbt808. [DOI] [PubMed] [Google Scholar]
- 52.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 53.David L, Huber W, Granovskaia M, Toedling J, Palm CJ, Bofkin L, Jones T, Davis RW, Steinmetz LM. A high-resolution map of transcription in the yeast genome. Proc. Natl Acad. Sci. USA. 2006;103:5320–5325. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Erdemir T, Bilican B, Cagatay T, Goding C, Yavuzer U. Saccharomyces cerevisiae C1D is implicated in both non-homologous DNA end joining and homologous recombination. Mol. Microbiol. 2002;46:947–957. doi: 10.1046/j.1365-2958.2002.03224.x. [DOI] [PubMed] [Google Scholar]
- 55.Askree SH, Yehuda T, Smolikov S, Gurevich R, Hawk J, Coker C, Krauskopf A, Kupiec M, McEachern MJ. A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc. Natl Acad. Sci. USA. 2004;101:8658–8663. doi: 10.1073/pnas.0401263101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.