Abstract
Considerable progress has been made recently on solution NMR studies of multi-transmembrane helix membrane protein systems of increasing size. Careful correlation of structure with function has validated the physiological relevance of these studies in detergent micelles. However, larger micelle and bicelle systems are sometimes required to stabilize the active forms of dynamic membrane proteins, such as the bacterial small multidrug resistance transporters. Even in these systems with aggregate molecular weights well over 100 kDa, solution NMR structural studies are feasible – but challenging.
Keywords: Micelle, Bicelle, Structure, Smr, Multidrug resistance
1. Introduction
Despite recent increases in the number of membrane protein structures solved annually, the gap between soluble and membrane protein structures continues to increase, and new technologies for tackling membrane protein structures are sorely needed. Even among the membrane proteins of known structure, small, highly membrane-embedded proteins are underrepresented, presumably due to their lack of extramembrane domains that could make crystal contacts necessary for 3D crystal formation [1]. Since these are the systems most amenable to structural characterization by NMR, and since methods for expressing, reconstituting and obtaining assignments and structural constraints are continuously improving, it is very likely that solution NMR will play an important part in overcoming the current bottleneck in membrane protein structure determination.
There have recently been a number of excellent reviews on membrane protein NMR [2–5], and we will therefore not try to give an exhaustive overview of the field but mainly focus on two important steps in the structure determination of membrane proteins by solution state NMR: sample preparation, including choice of membrane mimic, and obtaining assignments and structural constraints for α-helical membrane protein complexes at the upper currently accessible size limit. In addition to reviewing recent advances in the field, we will focus on our experience with the Staphylococcal multidrug resistance transporter (Smr). Smr is a member of the small multidrug-resistance transporter family of bacterial proteins which utilizes the proton gradient across the membrane to extrude a number of lipophilic, mostly cationic drugs in an antiporter manner [6]. It is functional as a dimer with 4 transmembrane helices (TMHs) per monomer [7,8]. In spite of numerous efforts, no X-ray crystal structure of any Smr homologue is currently available, and the highest resolution structure is a 7 Å cryo-EM 3D reconstruction map [9]. We are working towards solving the NMR structure of Smr in order to better understand drug selectivity and transport mechanism and to find ways of inhibiting this transporter.
2. Membrane protein solubilization
In order to study an integral membrane protein by solution-state NMR, it must be extracted from its native lipid membrane and solubilized in a form that allows for sufficiently fast isotropic tumbling in solution. The most important consideration in choosing such a solubilization system is to ensure that the protein retains its native conformation under the chosen experimental conditions. We will therefore discuss a number of membrane mimics that have been successfully used in NMR studies of integral membrane proteins in light of this requirement, and highlight our own experiences made in the study of the highly dynamic 8 TMH protein Smr.
Detergent micelles are the most commonly used solubilization agents for membrane protein solution NMR studies. So far, no generally applicable rules have been found for the choice of detergent, and optimal solubilization conditions must be worked out for each membrane protein of interest. Detergents that have been successfully used for solution NMR studies of membrane proteins containing more than a single TMH are summarized in Table 1. In addition to these studies of individual proteins, extensive work has also been conducted on assessing membrane protein purification and detergent solubilization methods in light of their ability to produce high quality solution NMR spectra for larger sets of proteins [10–12]. While our own extensive detergent screen for five membrane proteins (containing 1, 2 or 4 TMHs) indicated that the best spectra in all cases were obtained in the lysolipid detergent lysopalmitoyl-phospatidylglycerol (LPPG) [10], two other studies on larger sets of proteins did not reveal a single universally favorable detergent [11,12]. Taken together, the results of these surveys identify a useful subset of the available detergents for testing, but stress the importance of conducting a screen for each individual protein. As already mentioned, the detergent should not only yield good quality NMR spectra, but even more importantly it should be able to solubilize the protein of interest in its native conformation. The following few examples will illustrate this second point.
Table 1.
Detergents used in solution NMR studies of integral membrane proteins. In all these examples, at least partial assignments have been obtained in the given detergents. Abbreviations used are DPC (dodecylphosphocholine), SDS (sodium dodecylsulfate), DHPC (dihexanoylphosphatidylcholine), LDAO (lauryldimethylamine-N-oxide), βOG (n-octyl-β-D-glucopyranoside), DM (n-decyl-β-D-maltopyranoside), DDM (n-dodecyl-β-D-maltopyranoside), CYFOS-7 (7-cyclohexyl-1-heptylphosphocholine).
In the case of an enzyme, the presence of the protein in a native conformation can easily be confirmed by a functional assay. For the enzyme diacylglycerol kinase (DAGK), which has been studied extensively in the Sanders laboratory, a functional detergent screen revealed that the best combination of enzymatic activity and NMR characteristics was achieved in DPC [13]. The short sample lifetimes of a few days to a week in DPC at NMR concentrations were overcome by the use of a thermostable mutant of DAGK [14], which yielded NMR samples that were stable over much longer periods of time and allowed essentially complete backbone assignments to be made [15].
Another enzyme that has been extensively studied by NMR is the β-barrel integral membrane protein PagP. Kay and co-workers initially determined its global fold by solution NMR in both DPC and βOG, and found that the protein adopted a very similar conformation in both detergents, especially in the transmembrane β-barrel [16], pointing to the relevance of the solved structures. However, neither DPC nor βOG supports enzymatic activity of PagP. A subsequent crystal structure of the enzyme in LDAO also displayed essentially the same global fold, and moreover revealed a bound molecule of detergent in the putative active site [17]. It was therefore assumed that the lack of enzymatic activity in DPC and βOG was due to detergent binding in the active site, and indeed the enzyme activity was recovered in CYFOS-7, an analogue of DPC with a bulky cyclohexyl moiety in its hydrocarbon tail that should prevent its entry into the active site. NMR studies in this detergent revealed two distinct dynamic states in the protein, one more mobile state that is suggested to allow substrate entry into the active site, and a more rigid state that is essential for catalysis [18].
For the potassium channel KcsA, a functional assay is not available in detergent micelles, as selective potassium flow can only be measured across a sealed bilayer. However, Bax and co-workers took advantage of a large body of knowledge about this protein to ensure the functional relevance of their KcsA preparation in SDS micelles used for NMR assignments [19] and dynamics measurements [20]. KcsA is known to exist in its functional tetrameric oligomerization state in SDS micelles [21] and to be gated by pH with the intracellular C-terminal region of the protein representing the proton interaction site [22]. It has also been observed that the narrowest part of the ion conduction pore, the so-called selectivity filter, adopts a collapsed non-conductive conformation at 3 mM K+ concentration in a crystal structure [23]. Bax and co-workers have indeed observed chemical shift changes in the selectivity filter upon titration with K+ and found the dissociation constant to be ~ 3 mM [19]. Chemical shift changes that were induced by lowering of the sample pH could be mapped mostly to the C-terminal region of the protein [19], in agreement with the previous data [22].
Recently, Chou and co-workers solved the structure of the ζζ dimer transmembrane portion (ζζTM) by solution-state NMR [24]. Since the disulfide-linked dimer could only be reconstituted in SDS, DPC was then titrated into the sample until no more chemical shift changes were observed in an HSQC spectrum (~5:1 DPC:SDS). Complete exchange into DPC was impossible due to sample aggregation in the absence of any SDS. The validity of the dimer structure was corroborated by extensive mutational analysis of residues in ζζTM. The dimer formation ability of mutants was assayed in the context of the full-length protein in vivo, and an excellent correlation was found between residues involved in the dimer interface in the NMR structure and residues disrupting dimer formation in vivo [24].
3. Smr – a cautionary tale about the importance of a functional assay
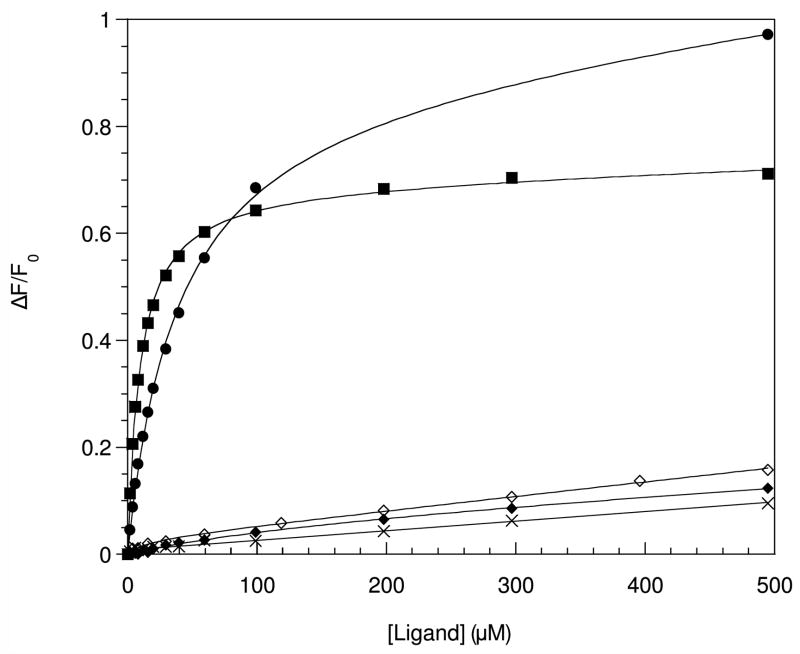
In our own work with Smr we had originally identified LPPG as the detergent giving the best spectral quality and sample lifetimes [10]. Equilibrium sedimentation analysis was performed to confirm the presence of Smr in its native dimeric oligomerization state in LPPG micelles [25]. In addition, preliminary paramagnetic spin-labeling studies had shown the protein to form a compact helical bundle, and experiments with a water-soluble paramagnetic broadening agent revealed solvent-exposed residues consistent with the proposed membrane topology [10]. We therefore used the standard TROSY-based triple-resonance suite of experiments to make full backbone and Cβ assignments [25], and found that the chemical shift deviations were consistent with 4 individual α-helices. In spite of all these indicators of a properly folded protein, we performed a functional assay to absolutely ascertain the presence of Smr in its native conformation in LPPG micelles. A ligand binding assay was carried out using the intrinsic fluorescence of the single tryptophan 62 residue as a reporter to monitor the binding of the drug tetraphenylphosphonium (TPP). In this assay, no specific binding could be detected to Smr in LPPG micelles (Figure 1), indicating that although Smr forms a compact dimeric α-helical bundle in this detergent, this conformation does not fully correspond to the protein’s native tertiary structure. In order to study Smr in a functionally relevant form, we conducted binding assays in a number of detergents and, among all tested detergents, found reproducible ligand binding only in DDM (and to a lesser extent in DM). In addition to simple detergent micelles, we also assayed phospholipid bicelles and found reproducible ligand binding in dimyristoyl-/ dihexanoylphospatidylcholine (DMPC/DHPC) bicelles with DMPC to DHPC molar ratios (q) of between 0.5 and 0.25. As expected, the TPP binding was pH-dependent, and specific binding was essentially abolished at pH 6.0 [26] (Figure 1). In addition to TPP, we also investigated Smr’s ability to bind methyl viologen, which has been shown to be transported by Smr’s Escherichia coli homologue EmrE [27]. We found that it also binds to bicelle-solubilized Smr with a higher dissociation constant of 28 μM as compared to 3.5 μM for TPP (Figure 1), as expected based on competition assays carried out with EmrE [27]. We also determined that the protein was present in its native dimeric oligomerization state in bicelles by equilibrium sedimentation analysis [26]. These results confirm the presence of Smr in its native conformation in bicelles and validate NMR structural studies carried out in this system.
Figure 1.
Ligand binding activity of Smr in different sample conditions. The relative fluorescence change upon addition of ligand is plotted, with the solid lines representing the best fit of the binding function to the data. Solid squares and diamonds represent TPP binding to Smr in bicelles at pH 8.0 and 6.0, respectively. Solid circles denote methyl viologen binding to Smr in bicelles at pH 8.0. Open diamonds and crosses denote the fluorescence changes upon addition of TPP to Smr in β-nonylglucoside and LPPG, respectively.
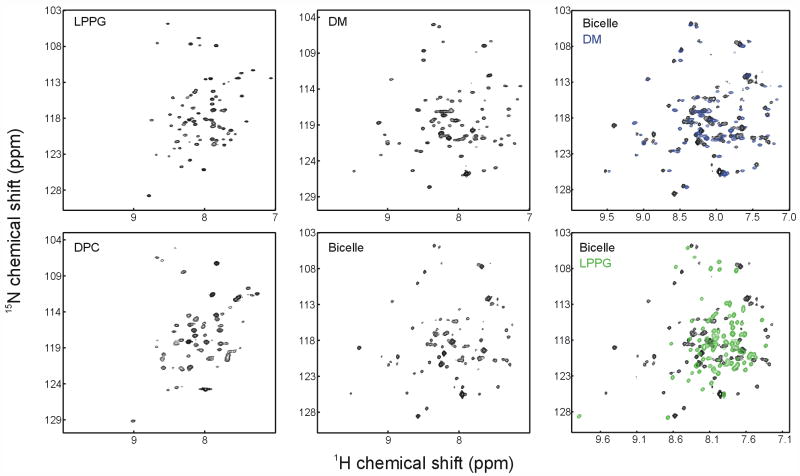
A comparison of 1H-15N-TROSY spectra of Smr in two detergents that do not support TPP binding (LPPG and DPC) as well as in two systems that do (DM and bicelles) is shown in Figure 2. It can immediately be noted that the dispersion of crosspeaks in the proton dimension is significantly wider for the two systems that support binding (DM and bicelles) compared to LPPG and DPC. Also, the general features of the spectra in DM and bicelles are extremely similar, and many crosspeaks can be related from one spectrum to the other (Figure 2). Between LPPG and DPC on the other hand, the main similarity is the lack of proton dispersion, and individual peaks cannot be easily correlated from one spectrum to another. This suggests that despite significant differences in the chemical environments of DM micelles and DMPC/DHPC bicelles, Smr adopts a single (native) conformation in both membrane mimics, as shown by the similarities of the TROSY spectra. Less stabilizing detergents like LPPG and DPC on the other hand are not able to support this native conformation, and the differences between spectra in those two detergents indicate that Smr can adopt different non-native conformations depending on the solubilizing detergent.
Figure 2.
1H-15N TROSY spectra of Smr in different detergents and bicelles. The left four spectra are recorded in different detergents and bicelles as indicated. The two right panels show overlays of the spectrum in bicelles with spectra in either DM or LPPG.
Although NMR sample lifetimes of Smr in bicelles were longer than those in DM (which is why we chose to work in bicelles over DM), continuous protein precipitation nevertheless led to sample halftimes of only 3–4 days for q 0.33 bicelles. Sample lifetimes could be modestly extended by increasing the long-chain to short-chain molar ratios in the bicelles, but this comes at the cost of NMR spectral quality. One pathway for protein aggregation is the loss of phospholipid through ester hydrolysis. This problem can be solved by using the ether-linked analogues of DMPC and DHPC, 1,2-di-O-tetradecyl-sn-glycero-3-phosphocholine (DoMPC) and 1,2-di-O-hexyl-sn-glycero-3-phosphocholine (DoHPC) [28]. Another way of increasing sample lifetimes is by doping the bicelles with a fraction of negatively charged phospholipids [29,30], as this should increase the repulsion between bicelles and therefore minimize opportunities for protein aggregation. In general, incorporating at least a small fraction of charged detergent or lipid into micelles or bicelles appears helpful in extending sample lifetimes for solution NMR. Triba et al. have adopted this approach and used ether-linked phospholipid analogs doped with a small fraction of negatively charged phosphatidylserine (PS) lipids to stabilize their preparation of a β-barrel membrane protein in bicelles for NMR studies [31]. We therefore prepared Smr samples in 27:9:3:1 DoHPC, DoMPC, DHPS, DMPS bicelles, which lead to a doubling of sample half-life times to about 7 days, enough for the collection of 3D double- and triple-resonance experiments [26].
4. Bicelles – a generally applicable solubilization system for solution-state NMR?
The success with functional reconstitution of Smr suggests that bicelles may be a generally useful solubilization system for the study of membrane proteins in their native form. Small isotropic bicelles are the most membrane-like environment in which solution NMR experiments are still possible. Nanodiscs [32] may provide similar benefits, but to our knowledge no one has yet reported their use for solution NMR of a membrane protein. The great advantage of these systems is that they provide a layer of long-chain phospholipids surrounding the protein. The presence of lipids has been shown to be essential both for supporting the native function [33–36] as well as the crystallization [37–40] of many solubilized membrane proteins. In addition to our own functional studies on Smr, bicelles have also been reported to support native-like enzymatic activity in DAGK [41]. Bicelles may therefore represent a useful starting point for the study of membrane proteins of unknown function, for example in a structural genomics project. As shown in Figure 2, the appearance of the TROSY spectra of Smr in DM and bicelles, which both support ligand binding, are very similar. In the absence of a functional assay one might reconstitute the protein of interest into bicelles and use the appearance of the TROSY spectrum in this system as a criterion to evaluate the ability of other detergents to support the protein’s native conformation. Along these lines, Opella and co-workers have recently used similarities between HSQC spectra in bicelles and SDS micelles to suggest that the structure of MerF in SDS corresponds to the protein’s native state [42].
So far, bicelles have mostly been used in solution NMR for the study of small membrane-inserting and membrane-associated peptides (see [43] for a recent review). The recent solution NMR structure of the Bnip3 transmembrane domain dimer (2 TMH total) [44] is the first example of a solution NMR structure in bicelles of a membrane protein with more than a single TMH, and proves that NMR structures can indeed be obtained for more complex membrane proteins in bicelles.
Another intriguing aspect of bicelles is that larger bicelles (q ~ 3–3.5) spontaneously align in a magnetic field and are very successfully used in solid-state NMR studies of membrane proteins ([45–48], and [43] for a review). It is therefore tempting to think of bicelles as a template for combined solution/solid-state NMR studies for the structure determination of membrane proteins. It has recently been shown that a combined use of structural restraints from solution NMR in micelles and solid-state NMR in aligned lipid bilayers can indeed improve the quality of a membrane protein NMR structure, as long as the protein adopts the same conformation in micelles and bilayers [49,50]. Using bicelles of different size, solution- and solid-state type NMR experiments could be carried out in a very similar environment, therefore increasing the probability of having the protein in the exactly same conformation for the solution- and solid-state type experiments. Solution-state experiments could then be used to obtain resonance assignments and limited structural constraints, and solid-state PISEMA-type experiments ([51], and reviewed in [52,53]) would provide information on the orientation of the transmembrane α-helices.
Several methods have been employed to reconstitute membrane proteins into bicelles. One can: i) solubilize the protein in short-chain lipid micelles and then add the long-chain component, ii) reconstitute the protein into long-chain lipid vesicles and redisolve it in the short-chain component, or iii) directly reconstitute the protein into bicelles from detergent solution, lyophilized protein or organic solvents. Triba et al. have reconstituted the transmembrane part of OmpA into bicelles by adding the protein in DoHPC micelles to dry powder of the appropriate long-chain phospholipids in the desired ratio, and inducing bicelle formation by four vortex/freeze/thaw cycles [31]. Park et al. prepared bicelle samples of the G protein-coupled receptor CXCR1 by first reconstituting the protein into lipid vesicles (at a 1:15 w/w protein:lipid ratio). An appropriate amount of DoHPC solution was then added to the proteoliposomes that had been pelleted by ultracentrifugation, and the bicelles formed after vortexing and equilibration at room temperature [46]. Czerski and Sanders reconstituted DAGK in bicelles by adding concentrated protein in DM micelles into preformed bicelle solutions such that the DM was diluted to below its CMC [41]. Bocharov et al. mixed lyophilized peptide with a DMPC/DHPC aqueous suspension and achieved bicelle formation through several vortex/freeze/thaw cycles [44]. For incorporation of Smr into bicelles, the purified protein in 1:1 chloroform/methanol with added short- and long-chain lipids of interest is thoroughly dried to a thin film under argon, followed by resolubilization in an aqueous buffer of choice [26].
5. Structural study of helical membrane proteins in bicelles by solution NMR
Bicelles clearly make an excellent membrane mimic for membrane proteins, but just how feasible are solution structural studies of multiple TMH membrane proteins in these large assemblies? For micelle-solubilized membrane proteins up to about 40 kDa in protein molecular weight and about 80 kDa in total mass for the protein-micelle complex [15,16,24,54–57], the methods developed for large globular proteins – perdeuterated 13C15N labeled protein samples, TROSY-based triple resonance assignment methods, and constraints based on chemical shifts for backbone torsion angles, backbone and methyl NOEs, paramagnetic relaxation enhancements (PREs) from site-directed spin-labeling, and residual dipolar couplings (RDCs) – have sufficed, if barely, for structural study by solution NMR. Recent reviews [3,5] describe the application of these methods. For larger ensembles such as KcsA in SDS micelles (~100 kDa) [19] and Smr in bicelles (~150 kDa), more significant modifications in approach appear necessary.
6. Backbone assignments for large membrane protein systems
Most TROSY-based 3D assignment experiments perform poorly for perdeuterated membrane protein systems tumbling as particles larger than 100 kDa. For Smr in bicelles, only the trHNCO, trHNCA, and 1H-15N-NOESY-TROSY yielded cross-peaks for all residues in the protein. All other triple resonance experiments along with the TROSY-NOE-TROSY performed much more poorly, showing only 5–15% of the expected cross-peaks, with these representing mainly terminal and loop residues. Even for the successful experiments, high field strengths (800–900 MHz), cryoprobes, perdeuteration, and long acquisition times (7–10 days) were required for complete data sets. Without complementary intra- and inter-residue data sets, alternative procedures must be used to assign backbone resonances. Promising methods include i) combined use of the trHNCA and NOESY-TROSY for helical segments, as employed for MISTIC [56] and KcsA [19], ii) amino acid type-specific labeling, most powerfully implemented as a combinatorial approach [58], and iii) “protonless” 13C-detected experiments, for which a full suite of assignment experiments have been developed [59].
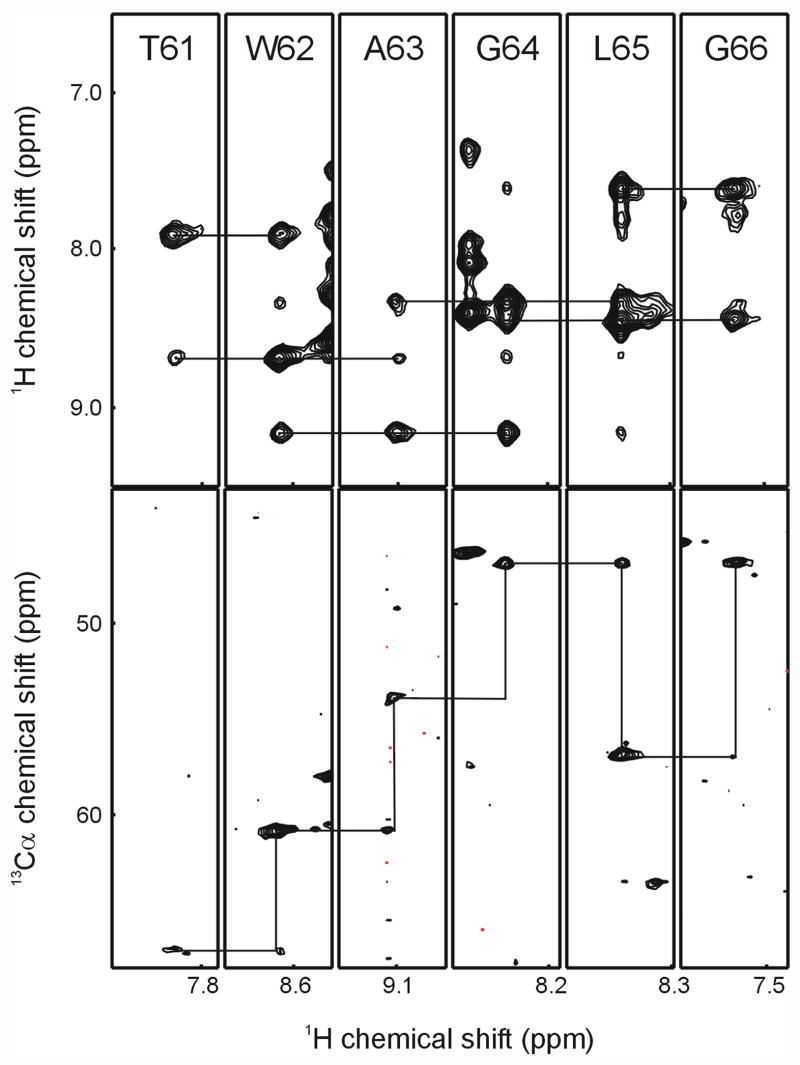
For highly α-helical membrane proteins showing strong HNi-HNi±1 NOEs, the Riek and Bax groups have shown that it is possible to make significant backbone assignments using just the HNCA and NOESY-HSQC (or HMQC) data sets [19,56]. In the case of Smr in bicelles, a NOESY-TROSY experiment collected with a 125 ms mixing time gave strong HNi-HNi±1 NOEs, and in many cases HNi-HNi±2 NOE cross-peaks were also observable, greatly assisting the sequential assignment process. Hence the NOESY-TROSY was used to find sequentially connected spin systems, and the HNCA was then used to confirm those connections and to assign possible amino acid types for individual spin systems. This approach was relatively successful, yielding unambiguous assignments for 55% of the backbone [26], and is illustrated with a sample assignment stretch in Figure 3. Overlap in the NOESY spectrum, missing Cαi-1 cross-peaks in the HNCA, and Cα shifts that were not sufficiently diagnostic for a single amino acid type precluded further assignments from these two data sets alone. Type specific labeling with 15N amino acids that show little isotopic scrambling in E. coli overcomes amino acid type ambiguity to provide safe anchor points for mapping an assigned series of resonances onto the membrane protein sequence [15]. A more extensive combinatorial scheme making use of both 15N and 13C’ labeled amino acids in a cell-free synthesis system was first used by Parker et al. [60], and a similar approach in combination with the standard triple-resonance assignment experiments was used to obtain nearly complete assignments for a 24 kDa membrane protein fragment [58].
Figure 3.
Representative sequential strips from the NOESY-TROSY (top panel) and TROSY-HNCA (bottom panel) experiments. In the NOESY-TROSY strips, the connectivities from each amide peak to its i±1 NOE cross-peaks are shown. In the TROSY-HNCA, the connectivities from i to i-1 peak to i peak in the preceding residue’s strip are indicated.
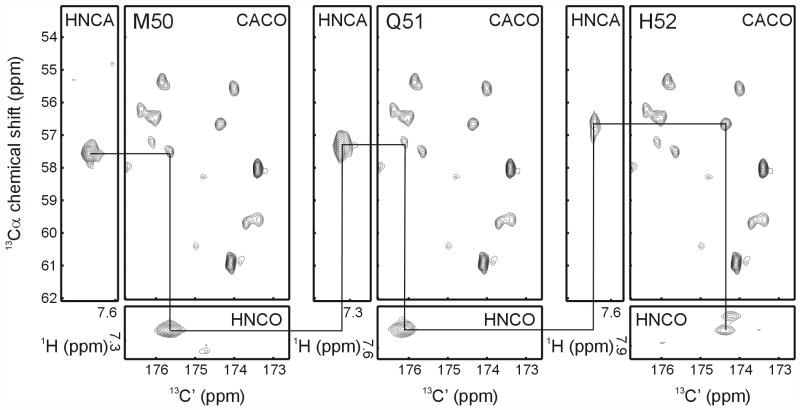
Another potential assignment strategy uses 13C-detected experiments [59]. This approach was developed mainly to overcome extremely rapid proton transverse relaxation rates in paramagnetic proteins, but as pointed out by the authors, it should also be applicable to large diamagnetic proteins where the transverse relaxation rates of carbons (lower γ) can be considerably slower than those of protons. The Bertini group has developed a full suite of 2D and 3D assignment experiments [59] that are most effectively used on cryogenic probes optimized for 13C detection sensitivity. To evaluate their usefulness for large membrane protein samples, we began with the 2D CACO experiment. This experiment correlates the intra-residue Cα and C’, and should provide assignment connections by linking the C’ of the inter-residue trHNCO with the intra-residue Cα of the trHNCA. The resulting data were quite promising. Over 80% of the CACO correlations could be resolved, and used in combination with the trHNCA and trHNCO to make assignments, as shown in Figure 4. With the carbon T1 values of nearly 10 sec for Smr in bicelles, the acquisition time for this 2D experiment was nearly three days. Such lengthy recycle delays make 3D experiments prohibitively long, but could be reduced significantly by the addition of paramagnetic agents, which have been shown to reduce carbon T1’s with little effect on their T2’s [61], and the hybrid approach demonstrated in a recent “out and stay” c-trHNCO from the Clore group [62], where relaxation delays are governed by amide proton relaxation rather than that of the detected 13C.
Figure 4.
Using the 13C-detect CACO experiment for making sequential backbone assignments. Starting from the i Cα chemical shift determined in the trHNCA experiment, the i CαC’ cross-peak in the CACO experiment is found, which then allows identification of the i+1 cross-peak in the trHNCO experiment through correlation of the C’ chemical shifts.
7. Structural constraints for large membrane protein systems
The accessible structural constraints for larger helical membrane protein systems will generally parallel those available for large perdeuterated globular proteins. Local backbone distance constraints can be readily obtained from amide NOEs, and phi/psi dihedral angle constraints from backbone chemical shifts [63]. Useful long range constraints of modest precision can be derived using PREs from site-directed spin-labels [64] as described in detail for OmpA in DPC micelles [65]. Long-range amide-amide NOEs appear to be quite rare in multi-TMH membrane proteins. Long-range NOEs from selectively protonated methyl groups are also unlikely to be generally useful as structural constraints for these proteins – the number of methyl groups is typically extremely large, and the chemical shift dispersion in both 1H and 13C is quite low, as pointed out by Sanders [3], and borne out by assignments of other helical membrane proteins in our lab. However, a more targeted use of methyl group NOEs and chemical shift changes can be quite informative, as was shown in the study of charybdotixin binding to KcsA [66].
RDCs measured in partially aligned samples [57,67–71] are expected to play a major role in defining structures and conformational changes in large membrane proteins. Here the challenge has been to induce long-term partial alignment without significantly decreasing the membrane protein concentration or increasing the linewidths of its amide resonances. Of the commonly used alignment media, only stressed polyacrylamide gels [69,72,73] are compatible with high concentrations of detergents or isotropic bicelles. Uncharged or charged gels have been used for partial alignment of small to medium sized membrane proteins [24,42,57,70,71], but have been less successful for larger membrane protein systems [2,3,74] where reduced protein concentrations, interactions with the gel, and short sample lifetimes have limited their use to measuring 1H15N couplings in 2D experiments. An alignment method based on DNA nanotubes was reported recently that overcomes all of these limitations [75]. The nanotubes are resistant to detergent and do not reduce achievable protein concentrations, permitting 1H15N and 13Cα1Hα dipolar couplings to be measured for two oligomeric membrane proteins. Another approach is to incorporate a lanthanide ion binding site into the protein, and use the incorporated ion to align the protein. This can be achieved by adventitious ion binding [76], addition of a metal binding motif (e.g. an EF-hand [77]), or incorporation of single cysteines into the protein and subsequent modification with a thiol-reactive EDTA derivative [78,79]. We have found the thiol-reactive EDTA derivatives to be the most versatile method [74], allowing placement of the metal at different locations in the protein, typically at single cysteines that were already engineered into the sequence for spin-label modification. Since the alignment tensor varies with different lanthanides [80], multiple orientations can be obtained by varying the identity of the metal used. Pseudocontact shifts and PREs from the metals also make useful distance constraints. Like DNA nanotube induced alignment, the lanthanide method overcomes the difficulties encountered with stressed gels, permitting RDCs for multiple internuclear vectors to be readily measured for membrane proteins [74], with the additional advantage of being able to induce different orientations with different metals.
8. Conclusions and prospects
Advances in expression and labeling [81,82], instrumentation, reconstitution [4,12], and adaptation of the latest solution NMR approaches to membrane proteins have recently led to a number of multiple TMH membrane protein structures being solved by NMR, and to complete or nearly complete backbone assignments for some impressively large systems. The largest systems studied to date have either been unusually stable (KcsA) or engineered to be unusually stable (DAGK) [14] – suggesting that a generally applicable strategy for selecting thermally stable membrane proteins could play an important role in making them more amenable to structural characterization. When this is not possible, and for membrane proteins such as Smr that must maintain access to multiple conformations for functional reasons, more highly stabilizing but larger membrane mimics such as bicelles do appear to both support function and permit structural characterization by solution NMR. And in cases where function cannot be readily measured, comparison of 1H-15N spectra of the target protein in bicelles with spectra of the protein in other potential detergents should help identify solution conditions leading to natively folded protein.
Acknowledgments
This work would not have been possible without instrument time and support from the New York Structural Biology Center (NYSBC). We would like to thank Drs. S. Bhattacharia, S. Shekar and S. Cahill for help with NMR experiments. Financial support was received from the NIH (NYSBC: GM66354; MG: GM72085) and from the Swiss National Science Foundation and the Swiss Foundation for medical-biological Grants (SFP).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gao FP, Cross TA. Recent developments in membrane-protein structural genomics. Genome Biol. 2005;6:244. doi: 10.1186/gb-2005-6-13-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hwang PM, Kay LE. Solution structure and dynamics of integral membrane proteins by NMR: a case study involving the enzyme PagP. Methods Enzymol. 2005;394:335–350. doi: 10.1016/S0076-6879(05)94013-5. [DOI] [PubMed] [Google Scholar]
- 3.Sanders CR, Sönnichsen F. Solution NMR of membrane proteins: practice and challenges. Magn Reson Chem. 2006;44:S24–40. doi: 10.1002/mrc.1816. [DOI] [PubMed] [Google Scholar]
- 4.Tian C, Karra MD, Ellis CD, Jacob J, Oxenoid K, Sönnichsen F, Sanders CR. Membrane protein preparation for TROSY NMR screening. Methods Enzymol. 2005;394:321–334. doi: 10.1016/S0076-6879(05)94012-3. [DOI] [PubMed] [Google Scholar]
- 5.Tamm LK, Liang B. NMR of membrane proteins in solution. Prog Nucl Mag Res Sp. 2006;48:201–210. [Google Scholar]
- 6.Grinius LL, Goldberg EB. Bacterial multidrug resistance is due to a single membrane protein which functions as a drug pump. J Biol Chem. 1994;269:29998–30004. [PubMed] [Google Scholar]
- 7.Butler PJG, Ubarretxena-Belandia I, Warne T, Tate CG. The Escherichia coli multidrug transporter EmrE is a dimer in the detergent-solubilised state. J Mol Biol. 2004;340:797–808. doi: 10.1016/j.jmb.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 8.Elbaz Y, Steiner-Mordoch S, Danieli T, Schuldiner S. In vitro synthesis of fully functional EmrE, a multidrug transporter, and study of its oligomeric state. Proc Natl Acad Sci USA. 2004;101:1519–1524. doi: 10.1073/pnas.0306533101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ubarretxena-Belandia I, Baldwin JM, Schuldiner S, Tate CG. Three-dimensional structure of the bacterial multidrug transporter EmrE shows it is an asymmetric homodimer. EMBO J. 2003;22:6175–6181. doi: 10.1093/emboj/cdg611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krueger-Koplin RD, Sorgen PL, Krueger-Koplin ST, Rivera-Torres IO, Cahill SM, Hicks DB, Grinius L, Krulwich TA, Girvin ME. An evaluation of detergents for NMR structural studies of membrane proteins. J Biomol NMR. 2004;28:43–57. doi: 10.1023/B:JNMR.0000012875.80898.8f. [DOI] [PubMed] [Google Scholar]
- 11.Columbus L, Lipfert J, Klock H, Millett I, Doniach S, Lesley SA. Expression, purification, and characterization of Thermotoga maritima membrane proteins for structure determination. Protein Sci. 2006;15:961–975. doi: 10.1110/ps.051874706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Page RC, Moore JD, Nguyen HB, Sharma M, Chase R, Gao FP, Mobley CK, Sanders CR, Ma L, Sönnichsen FD, Lee S, Howell SC, Opella SJ, Cross TA. Comprehensive evaluation of solution nuclear magnetic resonance spectroscopy sample preparation for helical integral membrane proteins. J Struct Funct Genomics. 2006;7:51–64. doi: 10.1007/s10969-006-9009-9. [DOI] [PubMed] [Google Scholar]
- 13.Vinogradova O, Sönnichsen F, Sanders CR. On choosing a detergent for solution NMR studies of membrane proteins. J Biomol NMR. 1998;11:381–386. doi: 10.1023/a:1008289624496. [DOI] [PubMed] [Google Scholar]
- 14.Zhou Y, Bowie JU. Building a thermostable membrane protein. J Biol Chem. 2000;275:6975–6979. doi: 10.1074/jbc.275.10.6975. [DOI] [PubMed] [Google Scholar]
- 15.Oxenoid K, Kim HJ, Jacob J, Sönnichsen FD, Sanders CR. NMR assignments for a helical 40 kDa membrane protein. J Am Chem Soc. 2004;126:5048–5049. doi: 10.1021/ja049916m. [DOI] [PubMed] [Google Scholar]
- 16.Hwang PM, Choy WY, Lo EI, Chen L, Forman-Kay JD, Raetz CRH, Privé GG, Bishop RE, Kay LE. Solution structure and dynamics of the outer membrane enzyme PagP by NMR. Proc Natl Acad Sci U S A. 2002;99:13560–13565. doi: 10.1073/pnas.212344499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahn VE, Lo EI, Engel CK, Chen L, Hwang PM, Kay LE, Bishop RE, Privé GG. A hydrocarbon ruler measures palmitate in the enzymatic acylation of endotoxin. EMBO J. 2004;23:2931–2941. doi: 10.1038/sj.emboj.7600320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hwang PM, Bishop RE, Kay LE. The integral membrane enzyme PagP alternates between two dynamically distinct states. Proc Natl Acad Sci USA. 2004;101:9618–9623. doi: 10.1073/pnas.0402324101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chill JH, Louis JM, Miller C, Bax A. NMR study of the tetrameric KcsA potassium channel in detergent micelles. Protein Sci. 2006;15:684–698. doi: 10.1110/ps.051954706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chill JH, Louis JM, Baber JL, Bax A. Measurement of 15N relaxation in the detergent-solubilized tetrameric KcsA potassium channel. J Biomol NMR. 2006;36:123–136. doi: 10.1007/s10858-006-9071-4. [DOI] [PubMed] [Google Scholar]
- 21.Heginbotham L, Odessey E, Miller C. Tetrameric stoichiometry of a prokaryotic K+ channel. Biochemistry. 1997;36:10335–10342. doi: 10.1021/bi970988i. [DOI] [PubMed] [Google Scholar]
- 22.Cortes DM, Cuello LG, Perozo E. Molecular architecture of full-length KcsA: role of cytoplasmic domains in ion permeation and activation gating. J Gen Physiol. 2001;117:165–180. doi: 10.1085/jgp.117.2.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Y, Morais-Cabral JH, Kaufman A, MacKinnon R. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 Å resolution. Nature. 2001;414:43–48. doi: 10.1038/35102009. [DOI] [PubMed] [Google Scholar]
- 24.Call ME, Schnell JR, Xu C, Lutz RA, Chou JJ, Wucherpfennig KW. The structure of the ζζ transmembrane dimer reveals features essential for its assembly with the T cell receptor. Cell. 2006;127:355–368. doi: 10.1016/j.cell.2006.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poget SF, Krueger-Koplin ST, Krueger-Koplin RD, Cahill SM, Chandra Shekar S, Girvin ME. NMR assignment of the dimeric S. aureus small multidrug-resistance pump in LPPG micelles. J Biomol NMR. 2006;36(Suppl 5):10. doi: 10.1007/s10858-005-5346-4. [DOI] [PubMed] [Google Scholar]
- 26.Poget SF, Cahill SM, Girvin ME. Isotropic bicelles stabilize the functional form of a small multidrug-resistance pump for NMR structural studies. J Am Chem Soc. 2007;129:1232–1233. doi: 10.1021/ja0679836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yerushalmi H, Lebendiker M, Schuldiner S. EmrE, an Escherichia-coli 12-kDa multidrug transporter, exchanges toxic cations and H+ and is soluble in organic solvents. J Biol Chem. 1995;270:6856–6863. doi: 10.1074/jbc.270.12.6856. [DOI] [PubMed] [Google Scholar]
- 28.Ottiger M, Bax A. Bicelle-based liquid crystals for NMR-measurement of dipolar couplings at acidic and basic pH values. J Biomol NMR. 1999;13:187–191. doi: 10.1023/a:1008395916985. [DOI] [PubMed] [Google Scholar]
- 29.Vold RR, Prosser RS, Deese AJ. Isotropic solutions of phospholipid bicelles: a new membrane mimetic for high-resolution NMR studies of polypeptides. J Biomol NMR. 1997;9:329–335. doi: 10.1023/a:1018643312309. [DOI] [PubMed] [Google Scholar]
- 30.Losonczi JA, Prestegard JH. Improved dilute bicelle solutions for high-resolution NMR of biological macromolecules. J Biomol NMR. 1998;12:447–451. doi: 10.1023/a:1008302110884. [DOI] [PubMed] [Google Scholar]
- 31.Triba MN, Zoonens M, Popot JL, Devaux PF, Warschawski DE. Reconstitution and alignment by a magnetic field of a beta-barrel membrane protein in bicelles. Eur Biophys J. 2006;35:268–275. doi: 10.1007/s00249-005-0014-x. [DOI] [PubMed] [Google Scholar]
- 32.Civjan NR, Bayburt TH, Schuler MA, Sligar SG. Direct solubilization of heterologously expressed membrane proteins by incorporation into nanoscale lipid bilayers. Biotechniques. 2003;35:556–60. 562–3. doi: 10.2144/03353rr02. [DOI] [PubMed] [Google Scholar]
- 33.Walsh JP, Bell RM. sn-1,2-Diacylglycerol kinase of Escherichia coli. Structural and kinetic analysis of the lipid cofactor dependence. J Biol Chem. 1986;261:15062–15069. [PubMed] [Google Scholar]
- 34.Zimmer J, Doyle DA. Phospholipid requirement and pH optimum for the in vitro enzymatic activity of the E. coli P-type ATPase ZntA. Biochim Biophys Acta. 2006;1758:645–652. doi: 10.1016/j.bbamem.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 35.Lee S, Lee A, Chen J, MacKinnon R. Structure of the KvAP voltage-dependent K+ channel and its dependence on the lipid membrane. Proc Natl Acad Sci USA. 2005;102:15441–15446. doi: 10.1073/pnas.0507651102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shanmugavadivu B, Apell H, Meins T, Zeth K, Kleinschmidt JH. Correct folding of the beta-barrel of the human membrane protein VDAC requires a lipid bilayer. J Mol Biol. 2007;368:66–78. doi: 10.1016/j.jmb.2007.01.066. [DOI] [PubMed] [Google Scholar]
- 37.Zhang H, Kurisu G, Smith JL, Cramer WA. A defined protein-detergent-lipid complex for crystallization of integral membrane proteins: The cytochrome b6f complex of oxygenic photosynthesis. Proc Natl Acad Sci USA. 2003;100:5160–5163. doi: 10.1073/pnas.0931431100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jidenko M, Nielsen RC, Sorensen TL, Moller JV, le Maire M, Nissen P, Jaxel C. Crystallization of a mammalian membrane protein overexpressed in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2005;102:11687–11691. doi: 10.1073/pnas.0503986102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Long SB, Campbell EB, Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 40.Guan L, Smirnova IN, Verner G, Nagamoni S, Kaback HR. Manipulating phospholipids for crystallization of a membrane transport protein. Proc Natl Acad Sci USA. 2006;103:1726. doi: 10.1073/pnas.0510922103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Czerski L, Sanders CR. Functionality of a membrane protein in bicelles. Anal Biochem. 2000;284:327–333. doi: 10.1006/abio.2000.4720. [DOI] [PubMed] [Google Scholar]
- 42.Howell SC, Mesleh MF, Opella SJ. NMR structure determination of a membrane protein with two transmembrane helices in micelles: MerF of the bacterial mercury detoxification system. Biochemistry. 2005;44:5196–5206. doi: 10.1021/bi048095v. [DOI] [PubMed] [Google Scholar]
- 43.Prosser RS, Evanics F, Kitevski JL, Al-Abdul-Wahid MS. Current applications of bicelles in NMR studies of membrane-associated amphiphiles and proteins. Biochemistry. 2006;45:8453–8465. doi: 10.1021/bi060615u. [DOI] [PubMed] [Google Scholar]
- 44.Bocharov EV, Pustovalova YE, Pavlov KV, Volynsky PE, Goncharuk MV, Ermolyuk YS, Karpunin DV, Schulga AA, Kirpichnikov MP, Efremov RG, Maslennikov IV, Arseniev AS. Unique dimeric structure of BNip3 transmembrane domain suggests membrane permeabilization as a cell death trigger. J Biol Chem. 2007;282:16256–16265. doi: 10.1074/jbc.M701745200. [DOI] [PubMed] [Google Scholar]
- 45.De Angelis AA, Howell SC, Nevzorov AA, Opella SJ. Structure determination of a membrane protein with two trans-membrane helices in aligned phospholipid bicelles by solid-state NMR spectroscopy. J Am Chem Soc. 2006;128:12256–12267. doi: 10.1021/ja063640w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park SH, Prytulla S, De Angelis AA, Brown JM, Kiefer H, Opella SJ. High-resolution NMR spectroscopy of a GPCR in aligned bicelles. J Am Chem Soc. 2006;128:7402–7403. doi: 10.1021/ja0606632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park SH, De Angelis AA, Nevzorov AA, Wu CH, Opella SJ. Three-dimensional structure of the trans-membrane domain of Vpu from HIV-1 in aligned phospholipid bicelles. Biophys J. 2006 doi: 10.1529/biophysj.106.087106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dürr UHN, Yamamoto K, Im S, Waskell L, Ramamoorthy A. Solid-state NMR reveals structural and dynamical properties of a membrane-anchored electron-carrier protein, cytochrome b(5) J Am Chem Soc. 2007;129:6670–6671. doi: 10.1021/ja069028m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Franzin CM, Gong X, Thai K, Yu J, Marassi FM. NMR of membrane proteins in micelles and bilayers: the FXYD family proteins. Methods. 2007;41:398–408. doi: 10.1016/j.ymeth.2006.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Franzin CM, Teriete P, Marassi FM. Structural similarity of a membrane protein in micelles and membranes. J Am Chem Soc. 2007;129:8078–8079. doi: 10.1021/ja0728371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu CH, Ramamoorthy A, Opella SJ. High resolution heteronuclear dipolar solid-state NMR spectroscopy. J Magn Reson A. 1994;109:270–272. [Google Scholar]
- 52.Opella SJ, Marassi FM. Structure determination of membrane proteins by NMR spectroscopy. Chem Rev. 2004;104:3587–3606. doi: 10.1021/cr0304121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramamoorthy A, Wei Y, Lee D. PISEMA solid-state NMR spectroscopy. Ann Rep NMR Spectrosc. 2004;52:1–52. [Google Scholar]
- 54.Fernández C, Hilty C, Wider G, Güntert P, Wüthrich K. NMR structure of the integral membrane protein OmpX. J Mol Biol. 2004;336:1211–1221. doi: 10.1016/j.jmb.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 55.Arora A, Abildgaard F, Bushweller JH, Tamm LK. Structure of outer membrane protein A transmembrane domain by NMR spectroscopy. Nature Struct Biol. 2001;8:334–338. doi: 10.1038/86214. [DOI] [PubMed] [Google Scholar]
- 56.Roosild TP, Greenwald J, Vega M, Castronovo S, Riek R, Choe S. NMR structure of Mistic, a membrane-integrating protein for membrane protein expression. Science. 2005;307:1317–1321. doi: 10.1126/science.1106392. [DOI] [PubMed] [Google Scholar]
- 57.Oxenoid K, Chou JJ. The structure of phospholamban pentamer reveals a channel-like architecture in membranes. Proc Natl Acad Sci USA. 2005;102:10870–10875. doi: 10.1073/pnas.0504920102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trbovic N, Klammt C, Koglin A, Löhr F, Bernhard F, Dötsch V. Efficient strategy for the rapid backbone assignment of membrane proteins. J Am Chem Soc. 2005;127:13505. doi: 10.1021/ja0540270. [DOI] [PubMed] [Google Scholar]
- 59.Bermel W, Bertini I, Felli IC, Piccioli M, Pierattelli R. C-13-detected protonless NMR spectroscopy of proteins in solution. Prog Nucl Magn Reson Sp. 2006;48:45. [Google Scholar]
- 60.Parker MJ, Aulton-Jones M, Hounslow AM, Craven CJ. A combinatorial selective labeling method for the assignment of backbone amide NMR resonances. J Am Chem Soc. 2004;126:5020–5021. doi: 10.1021/ja039601r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eletsky A, Moreira O, Kovacs H, Pervushin K. A novel strategy for the assignment of side-chain resonances in completely deuterated large proteins using C-13 spectroscopy. J Biomol NMR. 2003;26:179. doi: 10.1023/a:1023572320699. [DOI] [PubMed] [Google Scholar]
- 62.Hu K, Vögeli B, Clore GM. Spin-state selective carbon-detected HNCO with TROSY optimization in all dimensions and double echo-antiecho sensitivity enhancement in both indirect dimensions. J Am Chem Soc. 2007;129:5484–5491. doi: 10.1021/ja067981l. [DOI] [PubMed] [Google Scholar]
- 63.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 64.Battiste JL, Wagner G. Utilization of site-directed spin labeling and high-resolution heteronuclear nuclear magnetic resonance for global fold determination of large proteins with limited nuclear overhauser effect data. Biochemistry. 2000;39:5355–5365. doi: 10.1021/bi000060h. [DOI] [PubMed] [Google Scholar]
- 65.Liang B, Bushweller JH, Tamm LK. Site-directed parallel spin-labeling and paramagnetic relaxation enhancement in structure determination of membrane proteins by solution NMR spectroscopy. J Am Chem Soc. 2006;128:4389–4397. doi: 10.1021/ja0574825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu L, Sun C, Song D, Shen J, Xu N, Gunasekera A, Hajduk PJ, Olejniczak ET. Nuclear magnetic resonance structural studies of a potassium channel-charybdotoxin complex. Biochemistry. 2005;44:15834–15841. doi: 10.1021/bi051656d. [DOI] [PubMed] [Google Scholar]
- 67.Tolman JR, Flanagan JM, Kennedy MA, Prestegard JH. Nuclear magnetic dipole interactions in field-oriented proteins: information for structure determination in solution. Proc Natl Acad Sci USA. 1995;92:9279–9283. doi: 10.1073/pnas.92.20.9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tjandra N, Bax A. Direct measurement of distances and angles in biomolecules by NMR in a dilute liquid crystalline medium. Science. 1997;278:1111–1114. doi: 10.1126/science.278.5340.1111. [DOI] [PubMed] [Google Scholar]
- 69.Cierpicki T, Bushweller JH. Charged gels as orienting media for measurement of residual dipolar couplings in soluble and integral membrane proteins. J Am Chem Soc. 2004;126:16266. doi: 10.1021/ja046054g. [DOI] [PubMed] [Google Scholar]
- 70.Cierpicki T, Liang B, Tamm LK, Bushweller JH. Increasing the accuracy of solution NMR structures of membrane proteins by application of residual dipolar couplings. High-resolution structure of outer membrane protein A. J Am Chem Soc. 2006;128:6947–6951. doi: 10.1021/ja0608343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chou JJ, Kaufman JD, Stahl SJ, Wingfield PT, Bax A. Micelle-induced curvature in a water-insoluble HIV-1 Env peptide revealed by NMR dipolar coupling measurement in stretched polyacrylamide gel. J Am Chem Soc. 2002;124:2451. doi: 10.1021/ja017875d. [DOI] [PubMed] [Google Scholar]
- 72.Chou JJ, Gaemers S, Howder B, Louis JM, Bax A. A simple apparatus for generating stretched polyacrylamide gels, yielding uniform alignment of proteins and detergent micelles. J Biomol NMR. 2001;21:382. doi: 10.1023/a:1013336502594. [DOI] [PubMed] [Google Scholar]
- 73.Tycko R, Blanco F, Ishii Y. Alignment of biopolymers in strained gels: A new way to create detectable dipole-dipole couplings in high-resolution biomolecular NMR. J Am Chem Soc. 2000;122:9341. [Google Scholar]
- 74.Kamen DE, Cahill SM, Girvin ME. Multiple alignment of membrane proteins for measuring residual dipolar couplings using lanthanide ions bound to a small metal chelator. J Am Chem Soc. 2007;129:1846–1847. doi: 10.1021/ja067089e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Douglas SM, Chou JJ, Shih WM. DNA-nanotube-induced alignment of membrane proteins for NMR structure determination. Proc Natl Acad Sci U S A. 2007;104:6644–6648. doi: 10.1073/pnas.0700930104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Veglia G, Opella SJ. Lanthanide ion binding to adventitious sites aligns membrane proteins in micelles for solution NMR spectroscopy. J Am Chem Soc. 2000;122:11733–11734. [Google Scholar]
- 77.Ma C, Opella SJ. Lanthanide ions bind specifically to an added “EF-hand” and orient a membrane protein in micelles for solution NMR spectroscopy. J Magn Reson. 2000;146:381–384. doi: 10.1006/jmre.2000.2172. [DOI] [PubMed] [Google Scholar]
- 78.Dvoretsky A, Gaponenko V, Rosevear PR. Derivation of structural restraints using a thiol-reactive chelator. FEBS Lett. 2002;528:192. doi: 10.1016/s0014-5793(02)03297-0. [DOI] [PubMed] [Google Scholar]
- 79.Ikegami T, Verdier L, Sakhaii P, Grimme S, Pescatore B, Saxena K, Fiebig KM, Griesinger C. Novel techniques for weak alignment of proteins in solution using chemical tags coordinating lanthanide ions. J Biomol NMR. 2004;29:349. doi: 10.1023/B:JNMR.0000032611.72827.de. [DOI] [PubMed] [Google Scholar]
- 80.Barbieri R, Bertini I, Cavallaro G, Lee Y, Luchinat C, Rosato A. Paramagnetically induced residual dipolar couplings for solution structure determination of lanthanide binding proteins. J Am Chem Soc. 2002;124:5587. doi: 10.1021/ja025528d. [DOI] [PubMed] [Google Scholar]
- 81.Schwarz D, Klammt C, Koglin A, Löhr F, Schneider B, Dötsch V, Bernhard F. Preparative scale cell-free expression systems: New tools for the large scale preparation of integral membrane proteins for functional and structural studies. Methods. 2007;41:355–369. doi: 10.1016/j.ymeth.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 82.Kainosho M, Torizawa T, Iwashita Y, Terauchi T, Mei Ono A, Güntert P. Optimal isotope labelling for NMR protein structure determinations. Nature. 2006;440:52–57. doi: 10.1038/nature04525. [DOI] [PubMed] [Google Scholar]
- 83.MacKenzie KR, Prestegard JH, Engelman DM. A transmembrane helix dimer: structure and implications. Science. 1997;276:131–133. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]
- 84.Oxenoid K, Sönnichsen FD, Sanders CR. Topology and secondary structure of the N-terminal domain of diacylglycerol kinase. Biochemistry. 2002;41:12876–12882. doi: 10.1021/bi020335o. [DOI] [PubMed] [Google Scholar]
- 85.Fernández C, Adeishvili K, Wüthrich K. Transverse relaxation-optimized NMR spectroscopy with the outer membrane protein OmpX in dihexanoyl phosphatidylcholine micelles. Proc Natl Acad Sci U S A. 2001;98:2358–2363. doi: 10.1073/pnas.051629298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Johansson MU, Alioth S, Hu K, Walser R, Koebnik R, Pervushin K. A minimal transmembrane beta-barrel platform protein studied by nuclear magnetic resonance. Biochemistry. 2007;46:1128–1140. doi: 10.1021/bi061265e. [DOI] [PubMed] [Google Scholar]
- 87.Malia TJ, Wagner G. NMR structural investigation of the mitochondrial outer membrane protein VDAC and its interaction with antiapoptotic Bcl-xL. Biochemistry. 2007;46:514–525. doi: 10.1021/bi061577h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schubert M, Kolbe M, Kessler B, Oesterhelt D, Schmieder P. Heteronuclear multidimensional NMR spectroscopy of solubilized membrane proteins: resonance assignment of native bacteriorhodopsin. Chembiochem. 2002;3:1019–1023. doi: 10.1002/1439-7633(20021004)3:10<1019::AID-CBIC1019>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 89.Takeuchi K, Takahashi H, Kawano S, Shimada I. Identification and characterization of the slow-exchanging pH-dependent conformational rearrangement in KcsA. J Biol Chem. 2007;282:15179–15186. doi: 10.1074/jbc.M608264200. [DOI] [PubMed] [Google Scholar]