Abstract
DNA polymerase γ (pol γ) is the sole DNA polymerase devoted to mitochondrial DNA (mtDNA) replication. We have characterized the molecular and physiological effects of over-expression of the catalytic subunit of pol γ, pol γ-α, in the nervous system of Drosophila melanogaster using the upstream activation sequence (UAS)/yeast transcriptional activator by binding to UAS (GAL4) system. Tissue-specific over-expression of pol γ-α was confirmed by immunoblot analysis, whereas the very low levels of endogenous protein are undetectable in UAS or GAL4 control lines. The transgenic flies over-expressing pol γ-α in the nervous system showed a moderate increase in pupal lethality, and a significant decrease in the median life span of adult flies. Moreover, these flies displayed a decrease in the rate of synthesis of mtDNA, which is accompanied by a significant mtDNA depletion, and a corresponding decrease in the levels of mitochondrial transcription factor A (mtTFA). Biochemical analysis showed an oxidative phosphorylation (OXPHOS) defect in transgenic flies, which were more susceptible to oxidative stress. Although we did not detect apoptosis in the nervous system of adult transgenic flies, brains of larvae over-expressing pol γ-α showed evidence of increased cell death that correlates with the observed phenotypes. Our data establish an animal model that mimics some of the features of human mtDNA depletion syndromes.
Keywords: apoptosis, DNA polymerase γ, Drosophila melanogaster, mitochondria, oxidative phosphorylation, oxidative stress
Cellular energy requirements vary in the different tissues of an organism and in response to different physiological circumstances. This differential demand is accompanied by changes in mitochondrial proliferation and/or differentiation, a process known as mitochondrial biogenesis (Garesse and Vallejo 2001). Animal mitochondria contain a double-stranded circular DNA genome that is replicated, transcribed and translated within the organelle. It encodes only 37 genes: 22 tRNAs, 2 rRNAs and 13 polypeptides that are components of the oxidative phosphorylation (OXPHOS) system (Attardi and Schatz 1988; Garesse 1988). The limited coding capacity of mitochondrial DNA (mtDNA) requires that most of the mitochondrial proteins, including the majority of the proteins involved in OXPHOS and all of the components involved in mtDNA maintenance and expression are encoded in the nucleus. These proteins are synthesized by cytoplasmic ribosomes and are imported into mitochondria by a complex machinery located in the inner and outer mitochondrial membranes (Pfanner and Wiedemann 2002). Therefore, mitochondrial biogenesis requires the coordinated expression of two distinct genomes, nuclear and mitochondrial (Kelly and Scarpulla 2004).
Mitochondrial proliferation requires mtDNA replication, and the key replicative enzyme is DNA polymerase γ [pol γ; mtDNA polymerase (EC: 2.7.7.7)]. The mitochondrial replicase has been characterized biochemically in several organisms including Drosophila and mammals (Kaguni 2004). Pol γ comprises a catalytic subunit or subunit α, pol γ-α, containing both 5′->3′ DNA polymerase and 3′->5′ exonuclease activities, and an accessory subunit or subunit β, pol γ-β, which contributes to the catalytic efficiency and structural integrity of the holoenzyme. In Drosophila melanogaster, the two subunits of pol γ are encoded by genes that are located in a highly compact complex (Lefai et al. 2000a) and at the transcriptional level, their expression is regulated differentially during embryogenesis (Lefai et al. 2000b). Pol γ-α mRNA is stored in the egg and its level decreases sharply as development proceeds. In contrast, the maternal storage of pol γ-β mRNA is lower and its concentration reaches the maximum level at the time that replication of mtDNA resumes, suggesting a role in the control of mtDNA replication.
We have previously over-expressed in Schneider cells the catalytic subunit of Drosophila pol γ, and found that mitochondrial function is unaffected with regard to growth and mtDNA replication in the transformed cells. In contrast, constitutive over-expression in the whole animal produces a variety of effects ranging from morphological abnormalities to pupal lethality (Lefai et al. 2000c). Furthermore, in molecular terms, the excess of the pol γ-α subunit induces mtDNA depletion, suggesting that mtDNA replication is imbalanced in the transgenic animals.
Mitochondrial dysfunction has been reported as causing or contributing to a wide variety of degenerative diseases mainly affecting brain, heart and muscle (DiMauro and Schon 2003). Of particular interest are the mtDNA depletion syndromes (MDDS), a group of encephalomyopathies that are caused by mutations in nuclear genes required to maintain nucleotide pools in mitochondria (Salviati et al. 2002) or genes involved in mtDNA replication and maintenance (Van Goethem et al. 2001). Whereas mtDNA depletion has been associated with neurological defects, data regarding disruption of mitochondrial function specifically in neurons is lacking.
To characterize further the consequences of an excess of pol γ-α in Drosophila, we over-expressed specifically the catalytic subunit of D. melanogaster DNA polymerase γ in the nervous system of flies, one of the target tissues of MDDS, using the upstream activation sequence (UAS)/yeast transcriptional activator by binding to UAS (GAL4) system. Over-expression in the nervous system decreased mtDNA replication rate, thereby causing mtDNA depletion, reduced OXPHOS activity and increased sensitivity to oxidative stress. Furthermore, flies with an excess of pol γ-α showed an increase in apoptosis in the nervous system of larvae, a moderate lethality at the end of the pupal stage and a reduction in the median life span of adult flies.
Materials and methods
Generation of transgenic lines and fly stocks
The plasmid pUAST-γ125 containing the complete pol γ-α cDNA subcloned in pUAST was injected into y−w− embryos according to standard procedures (Spradling and Rubin 1982) to give the UAS-DNA polymerase γ subunit α (PolG) transgenic fly lines (Lefai et al. 2000c). Transformants with w+ eye color were mapped and rendered homozygous using the double balancer stock w; CyO/If; MKRS/TM2. The fly lines UAS-green fluorescent protein (GFP) {w; P[w+(GFPUAS)]/CyO}, and 1407-GAL4 {w; P[w+(GAL41407)]} were obtained from Ginés Morata and Sonsoles Campuzano, respectively. To over-express pol γ-α in the nervous system, we used the UAS/GAL4 system established by Brand and Perrimon (1993): a homozygous GAL4 line (1407-GAL4) was crossed with homozygous UAS-PolG lines. For all of the experiments, at least two independent lines of transgenic flies were tested and were found to give similar results.
Survival curves
Flies were maintained at 25°C and 60% humidity on standard medium (Calleja et al. 1993) unless otherwise specified. For longevity experiments, a cohort of male flies (20 per vial) were transferred to fresh medium every 3 or 4 days and scored for survivors. Several independent experiments were carried out with ~60 flies per experiment. The starting population for each genotype was between 200 and 400 animals. The median life span was calculated as the age in days required to reach 50% survivorship, and the maximum life span was calculated as 10%.
Pupal survival
Pupal survival was measured by the frequency of pupal eclosion. Adult flies were removed from the vials after 12 h. After 20 days, non-eclosed pupae were assumed to be dead, and all pupal cases on the side of the vials were scored as either eclosed (empty) or dead. The percentage of decrease of pupal survival was calculated from the data of Table 1.
Table 1.
Pupal survival. Decreased pupal survival in transgenic flies over-expressing DNA polymerase γ-α under control of the 1407-GAL4 driver. The percentage of pupae that eclosed is shown
| n | Pupal survival (%) | Decreasea (%) | ||
|---|---|---|---|---|
| UAS-PolGI × | (−) | 209 | 75 | |
| 1407-GAL4 | 319 | 50 | 33 | |
| UAS-PolGIII-1 × | (−) | 247 | 66 | |
| 1407-GAL4 | 239 | 37 | 44 | |
| UAS-PolGIII-2 × | (−) | 180 | 80 | |
| 1407-GAL4 | 259 | 51 | 36 |
Measurement of pupal lethality was as described in ‘Materials and methods’; n is the total number of pupae; (−) indicates no cross.
Pupal survival decrease or increase of pupal lethality.
Immunoblot analysis
Protein extracts from flies of each genotype were separated by 10% (w/v) sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) according to Laemmli (1970). Proteins were transferred to polyvinylidene difluoride membrane (Immobilon-P, Millipore, Madrid, Spain), probed with rabbit antiserum against the different proteins, and detected with an ECL kit (Amersham, Barcelona, Spain) using horseradish peroxidase-labeled goat anti-rabbit antibody. The polyclonal antibodies against D. melanogaster proteins used were anti-pol γ-α (Wang et al. 1997), anti-mtTFA (mitochondrial transcription factor A or Tfam) (Goto et al. 2001), anti-nuclear-encoded mitochondrial ATPase subunit α (α-ATPase) (Garesse, unpublished results), antibody against cleaved human caspase 3 (Cell Signaling Technology, Danvers, MA, USA) that cross-reacts with activated Drosophila caspase 3 Drice (Yu et al. 2002), and anti-Grim (Claveria et al. 1998).
Southern analyses
Total DNA from flies was extracted as described previously (Calleja et al. 1993), and Southern blot analysis was performed using standard procedures. DNA was digested with NsiI, electrophoresed in a 0.6% (w/v) agarose gel and transferred to Zeta-Probe membrane (Bio-Rad, Madrid, Spain). The mtDNA probe was a PCR-amplified region of D. melanogaster mtDNA between nucleotide positions 12502 and 12943 (Garesse 1988). The nuclear DNA (nDNA) probe was a fragment of 459 bp of D. melanogaster 18S rDNA amplified by PCR [nucleotide positions 488–946 (Tautz et al. 1988)]. The mtDNA and nDNA probes were radiolabeled by random priming (Random Prime Labeling System, Amersham). The nylon filters were pre-hybridized, hybridized with the probes, washed and exposed with intensifying screens at −70°C. The relative amounts of mtDNA and nDNA were determined by densitometric analysis of autoradiographs.
In organello mitochondrial DNA replication
Isolation of mitochondria and analysis of mtDNA was carried out as described previously (Enriquez et al. 1994), except that adult flies were used. Flies (~1 g) were placed on ice, suspended at a ratio of 4 mL/g flies in homogenization medium (10 mmol/L Tris-HCl pH 7.4, 0.32 mol/L sucrose and 1 mmol/L EDTA), and homogenized with five strokes at 600 rpm in a glass Potter-Elvehjem homogenizer with a motor-driven Teflon pestle. The homogenate was centrifuged for 5 min at 1000 g at 4°C to pellet unbroken tissue, cell debris and nuclei. The supernatant was centrifuged for 10 min at 12 000 g at 4°C to pellet mitochondria. The mitochondrial pellet was washed four times in homogenization medium. The mitochondrial protein concentration was determined by the method of Waddel (Waddel and Hill 1956). The mitochondrial fraction was resuspended at a final concentration of 2 mg/mL in incubation buffer that contained 10 mmol/L Tris-HCl pH 7.4, 25 mmol/L sucrose, 75 mmol/L sorbitol, 100 mmol/L KCl, 10 mmol/L K2HPO4, 50 μmol/L EDTA, 5 mmol/L MgCl2, 1 mg/mL bovine serum albumin, 1 mmol/L ADP, 10 mmol/L glutamate, 2.5 mmol/L malate, 50 μmol/L dATP, 50 μmol/L dGTP, 50 μmol/L 2′-deoxythymidine 5′-triphosphate (dTTP) and 2.5 mCi/mL [α-32P]deoxycytidine triphosphate (dCTP) (800 Ci/mmol). Incubation was performed in a rotary shaker at 37°C. At the indicated times, aliquots (50 μL) were centrifuged, washed twice and the mtDNA was extracted. Nucleic acids were analyzed by electrophoresis in a 0.6% (w/v) agarose gel after digestion with DNase-free RNase and the restriction enzyme BglII. The gel was stained with ethidium bromide, photographed under UV light and then dried and exposed for autoradiography at −70°C.
Respiratory chain enzyme activities
Assays for rotenone-sensitive NADH cytochrome c reductase [(EC: 1.6.99.3), complexes I+III] (Ferguson et al. 2005) were measured by monitoring the reduction of cytochrome c at 550 nm. Total activity was measured in the presence of NADH, cytochrome c, and the mitochondrial protein. The rotenone-resistant activity was measured in the presence of rotenone, and then subtracted from the total NADH cytochrome c reductase activity to yield the activity of the rotenone-sensitive cytochrome c reductase.
Cytochrome c oxidase [(EC: 1.9.3.1), complex IV] was determined spectrophotometrically by the decrease in absorbance at 550 nm of reduced cytochrome c in presence of the mitochondrial protein. Reduced cytochrome c was freshly prepared before each experiment by adding a few grains of sodium borohydride to a 10 g/L solution of the pigment in 10 mmol/L potassium phosphate buffer (pH 7.0). Addition of 0.1 mol/L HCl stabilized the reduced cytochrome c, and the excess borohydride was removed by centrifugation at 12 000 g for 4 min.
Citrate synthase (EC: 4.1.3.7) was measured by monitoring the change in absorbance at 412 nm caused by the reaction of 5,5′-dithiobis(2-nitrobenzoic acid) with the free coenzyme A formed by the condensation of acetyl-CoA with oxalacetate in the presence of the mitochondrial protein. All activities (complexes I+III, complex IV and citrate synthase) were calculated as nmol/min per mg of protein.
Oxidative stress assay
Oxidative stress was assayed in terms of resistance to paraquat feeding (Cvejic et al. 2004) as follows: males and females, 4–5 days of age, were starved for 6 h by keeping them in vials (20 flies per vial) with filters soaked in water. Flies were then transferred to vials with filters soaked in 1% (w/v) sucrose and 20 mmol/L paraquat (Sigma, Madrid, Spain) and kept at 25°C. The flies were scored for survival at 48 h. The average and standard deviation was calculated for 15 vials of flies.
Immunohistochemistry
Apoptosis was visualized using a commercial rabbit antibody against activated Drosophila caspase 3 Drice. Brains from late third instar larvae were dissected in chilled phosphate buffered saline and fixed with 4% (v/v) paraformaldehyde for 25 min at 25°C. Blocking was performed with 1% (w/v) bovine serum albumin, 0.3% (v/v) Triton X-100 in phosphate buffered saline for 1 h, followed by incubation with primary antibody for at least 5 h at 4°C and four washes with blocking buffer. Samples were then incubated with the appropriate fluorescent secondary antibody for 2 h at 25°C. Brains and imaginal discs were washed with blocking buffer and mounted in Vectashield (Vector laboratories, Burlingame, CA, USA). Confocal images were taken in a laser MicroRadiance microscope (Bio-Rad) and subsequently processed using Adobe Photoshop (Adobe, San Jose, CA, USA).
Statistical analysis
The survival curves were fitted to the three-parametric Gompertz equation (y = a * exp(−exp (−(x −x0)/b))) using the SigmaPlot 8.0 software (SSI, San Jose, CA, USA). The median and maximum life span were calculated from these fitted curves. The results were compared using the Student’s t test. p values of less than 0.05 were considered statistically significant.
Results
Over-expression of the catalytic subunit of pol γ in the nervous system of D. melanogaster increases pupal lethality and reduces significantly the median life span of adult flies
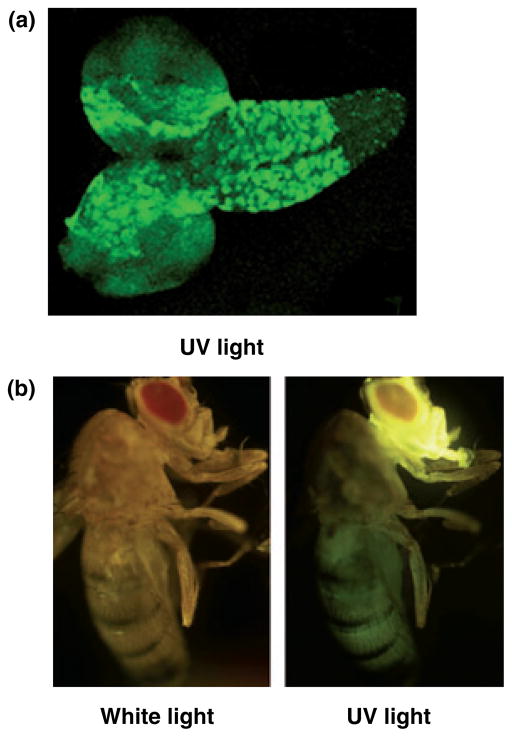
We reported previously that over-expression of the catalytic subunit of Drosophila pol γ has no phenotypic consequences in Schneider cells, but induces pupal lethality when it is constitutively over-expressed in the whole animal. To evaluate further the effects of an excess of pol γ-α in different tissues of the fly, we restricted over-expression to the nervous system using the UAS-GAL4 system. To do so, we constructed three independent UAS transgenic lines, two with the complete pol γ-α cDNA construct integrated into chromosome III (UAS-PolGIII-1 and UAS-PolGIII-2), and one in chromosome X (UAS-PolGI). We used the GAL4 line 1407 (1407-GAL4) to drive expression of the transcriptional regulator GAL4 early in neuroblasts (stage 10/11), and subsequently in most neurons of the CNS and in all neurons of the peripheral nervous system (Luo et al. 1994). The pattern of expression of the 1407 driver in larvae and adult flies was confirmed using a UAS reporter line containing the GFP cDNA integrated into chromosome II (UAS-GFP). As expected, GFP was expressed at a high level in the larval brain (Fig. 1a) and in the adult head, with a lower level of expression in the rest of the body (Fig. 1b).
Fig. 1.
Expression pattern of the 1407-GAL4 line revealed by crossing with UAS-GFP. (a) Brain with the ventral nerve chord to the right and the brain hemispheres to the left from third instar larvae. (b) Adult fly.
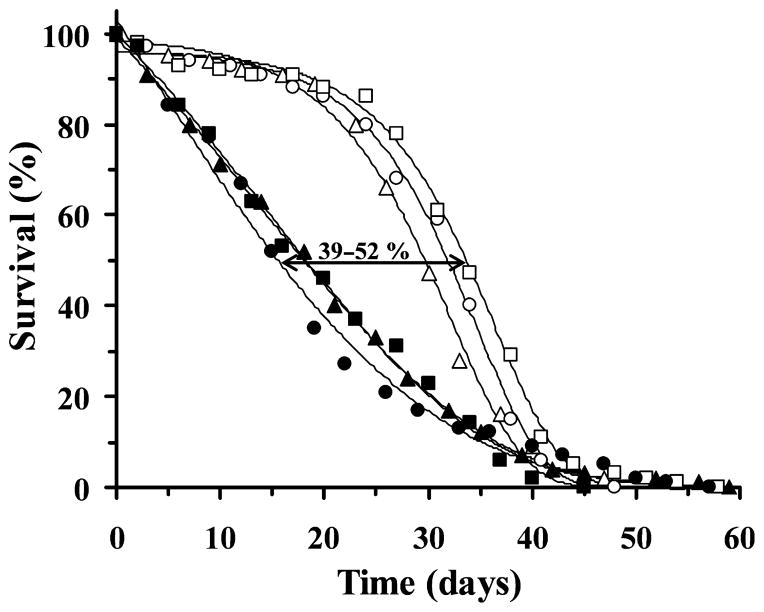
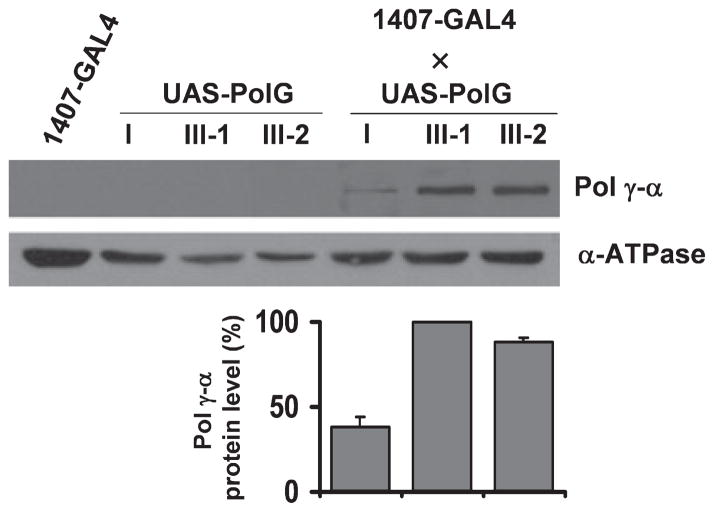
When we crossed the UAS-PolG lines with 1407-GAL4 stocks to obtain the 1407-GAL4 × UAS-PolG combination in the F1 generation, we found that pupal eclosion was moderately reduced as compared to wild type (ranging between 33–44% in the different crosses) (Table 1). This prompted us to evaluate the survival rate of control flies and flies over-expressing pol γ-α from the F1 populations (Fig. 2). Males were selected to avoid the complications arising from the substantial mitochondrial content in ovaries. We determined the median and maximum life span from the survival curves in Fig. 2 (Table 2). The median life span of the three UAS-PolG lines was 29.6 days (UAS-PolGI), 31.9 days (UAS-PolGIII-1), and 33.6 days (UAS-PolGIII-2), similar to that of the 1407-GAL4 stock (31.0 days, data not shown). However, the 1407-GAL4 × UAS-PolG transgenic flies showed a significant decrease in the median life span ranging from 39% to 52% (Fig. 2 and Table 2). The median life span in crosses of the three UAS-PolG lines was 18.0 days (1407-GAL4 × UAS-PolGI), 15.4 days (1407-GAL4 × UAS-PolGIII-1) and 18.2 days (1407-GAL4 × UAS-PolGIII-2). However, the maximum life span (calculated as the average life span of the last 10% of flies) was not statistically different between transgenic and control populations (Table 2). To demonstrate that the observed phenotype was because of the over-expression of the pol γ-α gene, we determined the steady-state level of pol γ-α protein by immunoblot analysis (Fig. 3). The expression of the protein in total protein extracts of adult flies was slightly higher for the UAS-PolGIII-1 cross as compared to UAS-PolGI, in agreement with its stronger phenotype (52% vs. 39% reduction of median life span); pol γ-α was not detected in the parental lines because of the very low level of expression of the endogenous gene (Fig. 3). Furthermore, the over-expressed protein was detected in the mitochondrial fraction (data not shown), indicating that it is properly targeted to the organelle as had been demonstrated previously (Lefai et al. 2000c). As a control, the same membranes were probed with a specific antiserum against the α subunit of the Drosophila F1-H+-ATPase complex (α-ATPase).
Fig. 2.
Longevity curves of adult males over-expressing pol γ-α in the nervous system. Survival curves of UAS-PolGI (open triangles), UAS-PolGIII-1 (open circles) and UAS-PolGIII-2 lines (open squares), and males expressing pol γ-α (closed symbols as above) in the nervous system. Results are expressed as a percentage of flies alive at each day of age. Median life spans of the pol γ-α expressing lines range from 39% to 52% of the control lines, as shown by the double arrow. The survival curves were fitted to Gompertz equation.
Table 2.
Life span analysis. Increased mortality in transgenic flies over-expressing DNA polymerase γ-α under control of the 1407-GAL4 driver. Maximum (10% survival) and median (50% survival) life span are shown
| n | Median life span (days) | Decrease (%) | Maximum life span (days) | Decrease (%) | ||
|---|---|---|---|---|---|---|
| UAS-PolGI × | (−) | 232 | 29.6 ± 1.2 | 38.1 ± 2.8 | ||
| 1407-GAL4 | 197 | 18.0 ± 2.3 | 39 (p < 0.0001) | 36.5 ± 1.2 | 4 (p ~ 0.3340) | |
| UAS-PolGIII-1 × | (−) | 428 | 31.9 ± 1.4 | 39.8 ± 3.1 | ||
| 1407-GAL4 | 346 | 15.4 ± 1.2 | 52 (p < 0.0001) | 35.0 ± 2.5 | 12 (p ~ 0.0525) | |
| UAS-PolGIII-2 × | (−) | 203 | 33.6 ± 2.1 | 41.7 ± 3.5 | ||
| 1407-GAL4 | 189 | 18.2 ± 3.1 | 46 (p < 0.0001) | 36.1 ± 3.2 | 13 (p ~ 0.0562) |
The mean ± SD (days) is calculated from the data in Fig. 2; p value is determined by Student’s t test; n is the total number of flies from four independent experiments; (−) indicates no cross.
Fig. 3.
Over-expression of pol γ-α protein in the nervous system. Upper panel, mitochondrial proteins isolated from adult flies were detected by immunoblot analysis using rabbit antiserum directed against Drosophila melanogaster pol γ-α subunit or nuclear-encoded mitochondrial ATPase subunit α (α-ATPase). Lower panel, steady-state levels of pol γ-α protein normalized to the α-ATPase subunit. Data are means ± SD.
Pol γ-α over-expression in the nervous system induces mtDNA depletion by decreasing the rate of mtDNA replication
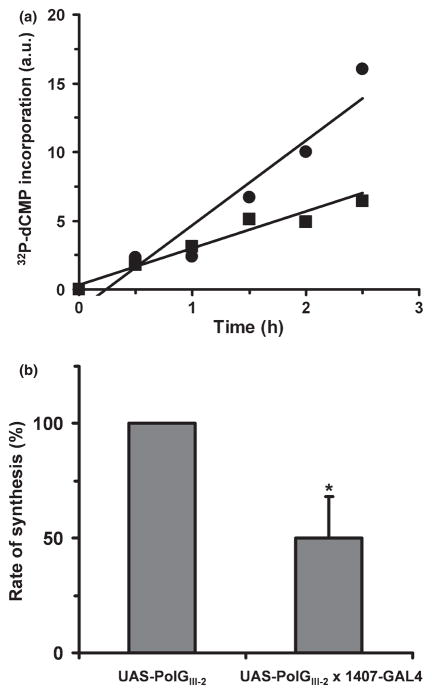
Because pol γ is the replicative mtDNA polymerase, accumulation of pol γ-α in the mitochondrial matrix might affect either the rate or mechanism of mtDNA replication. To evaluate this possibility, we examined DNA replication in organello by measuring the capacity of isolated mitochondria to incorporate [32P]-deoxycytidine monophosphate (dCMP) into mtDNA over a time course of incubation (Fig. 4a). Gel electrophoresis and autoradiography of the products of DNA synthesis showed only a single fragment of the expected size (~20 kb), indicating that the overall integrity of mtDNA is not affected (data not shown). DNA synthesis was higher in the control line (UAS-PolGIII-2) as compared to the pol γ-α over-expression line (1407-GAL4 × UAS-PolGIII-2), and the data show that the rate of mtDNA synthesis is decreased ~50% as a result of nervous-system over-expression (Fig. 4b).
Fig. 4.
Time course of mitochondria DNA (mtDNA) synthesis in organello. (a) Incorporation of [α-32P]dCMP into mtDNA at various times of incubation of isolated mitochondria at 37°C as described under Materials and methods. Mitochondria were isolated from UAS-PolGIII-2 (circles) and UAS-PolGIII-2 × 1407-GAL4 flies (squares). a.u.: arbitrary units. (b) Rate of synthesis of mtDNA derived from the data shown in (a). Data are means ± SD. *Statistical comparison was made using the Student’s t test, p < 0.003.
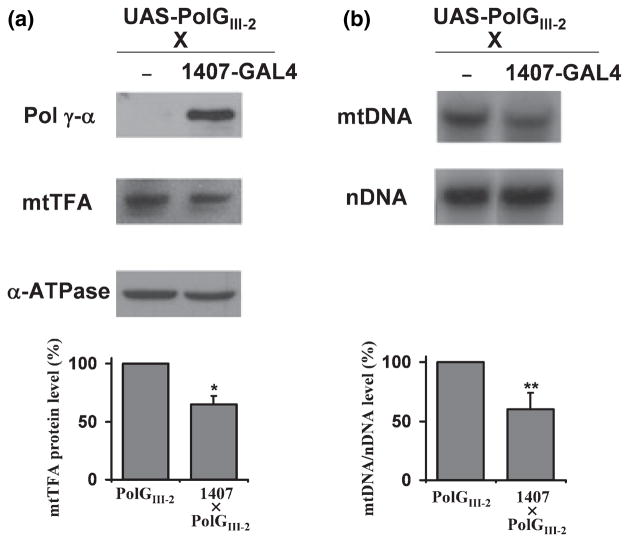
A reduced rate of mtDNA replication could affect the steady-state level of mtDNA. mtTFA is required for mtDNA maintenance, and its level correlates well with mtDNA content (Larsson et al. 1994, 1998). mtTFA knock-out mice exhibit embryonic lethality concomitant with depletion of mtDNA (Larsson et al. 1998), and recent genetic evidence supports a novel role for mtTFA in regulation of mtDNA copy number in mammals (Ekstrand et al. 2004). To assess the effect of over-expression of pol γ-α on mtTFA level, an immunoblot analysis was performed using antibodies against pol γ-α, mtTFA and α-ATPase, and the level of mtTFA protein was normalized relative to the level of the α-ATPase protein (Fig. 5a). This showed a ~45% decrease in the level of mtTFA in the 1407-GAL4 × PolGIII-2 flies.
Fig. 5.
pol γ-α over-expression induces mitochondrial DNA (mtDNA) depletion in the nervous system. (a) Immunoblot analysis of nuclear-encoded mitochondrial proteins. Upper panel, mitochondrial proteins isolated from adult flies were detected by immunoblotting with antibodies against Drosophila melanogaster pol γ-α, mitochondrial transcription factor A (mtTFA) and nuclear-encoded mitochondrial ATPase subunit α (α-ATPase). Lower panel, steady-state levels of mtTFA protein were normalized to the α-ATPase subunit. Data are means ± SD. (b) Southern blot analysis. Upper panel, total DNA was extracted from adult flies, digested with NsiI, electrophoresed, transferred to a nylon membrane and hybridized with a mtDNA fragment (mtDNA) and a fragment of the 18S rRNA gene (nDNA). Lower panel, quantitation of mtDNA/nDNA levels. Data are means ± SD. Statistical comparison was made using the Student’s t test, *p < 0.007, **p < 0.02.
To investigate a corresponding mtDNA depletion, mtDNA content was examined by Southern blot analysis after digestion of total DNA with NsiI, which cuts only once in D. melanogaster mtDNA. The blot was hybridized with a radiolabeled mtDNA-specific probe and a fragment of nuclear-encoded 18S rDNA, and the mtDNA/nDNA hybridization ratio was used to quantitate mtDNA (Fig. 5b). We observed that the mtDNA/nDNA ratio decreased ~40% in 1407-GAL4 × PolGIII-2 flies as compared to controls. In composite, these data demonstrate that over-expression of the pol γ-α subunit in the nervous system produces mtDNA depletion.
Mitochondrial DNA depletion reduces OXPHOS activity and resistance to oxidative stress
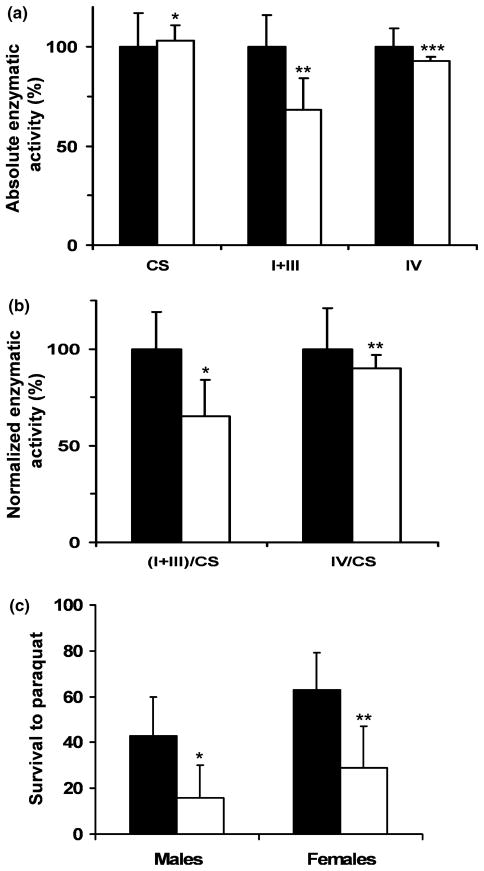
To analyze the effect of mtDNA depletion on the mitochondrial respiratory chain (OXPHOS system), the activities of rotenone-sensitive NADH cytochrome c reductase (complexes I+III) and cytochrome c oxidase (complex IV) were assayed in isolated mitochondria. The measurement of citrate synthase activity was carried out as a control for differences in mitochondrial purity or content. As shown in Fig. 6a, mitochondrial complex I+III activity was decreased markedly in the mitochondria of transgenic flies, corresponding to 32% of the mean of the control flies (50 ± 12 vs. 74 ± 12 nmol/min per mg protein, p < 0.026), but the specific activity of complex IV was not decreased significantly (7%, p ~ 0.25). The activity of the mitochondrial marker citrate synthase was not affected by over-expression of pol γ-α. Thus, there was a significant decrease in the activity of complex I+III normalized with citrate synthase activity of 35% between control and transgenic flies, and a moderate decrease of 10% in complex IV activity (Fig. 6b), which represents a specific mtDNA depletion-associated decline in the activity of the OXPHOS system.
Fig. 6.
Transgenic flies over-expressing pol γ-α display oxidative phosphorylation deficiency and a higher susceptibility to reactive oxygen species. (a) Absolute activities of citrate synthase, complex I+III and complex IV. The activities were assayed per milligram of protein (nmol/min per mg of protein) on mitochondria from control flies UAS-PolGIII-2 (black) and flies over-expressing pol γ-α [1407-GAL4 × UAS-PolGIII-2] (white), and are represented as percentage of the control value. Data are means ± SD. Statistical comparison was made using the Student’s t test, *p ~ 0.785, **p < 0.026, ***p ~ 0.250. (b) Normalized activities of complexes. Complex I+III and complex IV activities were normalized by citrate synthase activity. Student’s t test, *p < 0.035, **p ~ 0.602. (c) Resistance to the reactive oxygen species-generating agent paraquat. Survival of male and female flies (4–5 days of age) over-expressing pol γ-α (1407-GAL4 × UAS-PolGIII-2) (white) and control flies (UAS-PolGIII-2) (black) was measured 48 h post-treatment with 20 mmol/L paraquat at 25°C. Student’s t test, *p < 0.019, **p < 0.015.
Because deficiencies of the OXPHOS system have been associated with an accumulation of oxidative injuries (Esposito et al. 1999), we examined the effects of oxidative stress after over-expression of the pol γ-α subunit. Oxidative stress was induced in 4–5 day old flies by feeding paraquat as a ROS (reactive oxygen species)-generating agent, and survival was evaluated 48 h post-treatment. We found that over-expression of the pol γ-α subunit in the nervous system reduces resistance of transgenic flies to oxidative stress: control flies were more resistant to paraquat than flies over-expressing pol γ-α subunit (Fig. 6c). At day two, 27% more control (UAS-PolGIII-2) male flies were alive after exposure to paraquat than the pol γ-α over-expression (1407-GAL4 × UAS-PolGIII-2) male flies. Over-expression of the pol γ-α subunit also induced a 34% decrease in survival of female flies.
Transgenic flies over-expressing pol γ-α showed increased apoptosis in the larval brain but not in adults
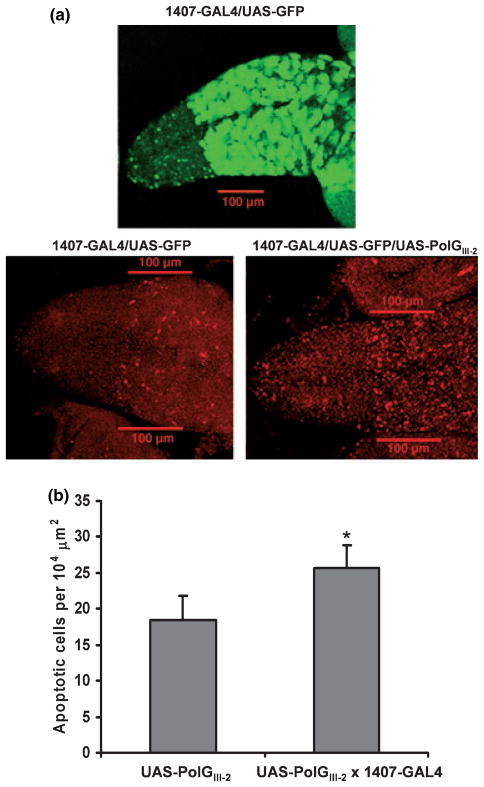
Oxidative stress causes cellular damage and subsequent cell death (oxidative stress-induced apoptosis), especially in organs such as the brain (Sanvicens et al. 2006). Moreover, compounds that inhibit mtDNA replication and deplete mtDNA also induce apoptosis (Mansouri et al. 2003). We evaluated the level of apoptosis in heads of adult flies by immunoblot analysis using an antibody against the proapoptotic protein Grim, and an antibody that recognizes the activated Drosophila effector caspase Drice. Both proteins were undetectable in control (UAS-PolGIII-2) flies, indicating a low level or absence of apoptosis in the nervous system of adult flies. In heads of flies over-expressing the pol γ-α subunit (1407-GAL4 × UAS-PolGIII-2) the Grim and Drice proteins were also undetectable (data not shown). Because apoptosis plays an important role during development, we measured the level of the active form of the caspase Drice in brains of larvae over-expressing the pol γ-α subunit (Fig. 7). To confirm the domain of expression driven by the 1407-GAL4 line we crossed flies carrying UAS-GFP with the lines carrying UAS-PolGIII-2 inserted to obtain a UAS-GFP/UAS-PolGIII-2 stock. Figure 7a shows a representative experiment, in which a significant increase in the level of apoptosis is observed in cells expressing pol γ-α, with a roughly 40% increase in the cells suffering apoptosis. Quantitative analysis indicates an increase in cells suffering apoptosis in brain larvae over-expressing pol γ-α (25.7 ± 3.1) as compared to controls (18.5 ± 3.3) (Fig. 7b).
Fig. 7.
Mitochondrial DNA depletion in the nervous system increases the level of apoptosis in larval brain. (a) Brains were dissected from third instar larvae over-expressing pol γ-α (1407-GAL4/UAS-GFP/ UAS-POLGIII-2) and control larvae (1407-GAL4/UAS-GFP) as described under Materials and methods. The domain of over-expression directed by the 1407-GAL4 driver was monitored by GFP fluorescence (green; upper panel). Apoptosis was detected using an antibody that detects specifically activated caspase 3 Drice (red; lower panel). The bar represents 100 μm. (b) Quantitation of the numbers of apoptotic cells in (a) lower panel. Data are means ± SD. Statistical comparison was made using the Student’s t test: UAS-PolGIII-2 (n = 18) versus UAS-PolGIII-2 × 1407-GAL4 (n = 30), *p < 0.0001.
Discussion
We have developed a system for tissue-specific depletion of mtDNA in flies by over-expression of pol γ-α in the nervous system using the UAS/GAL4 system. This fly model for mtDNA depletion shows tissue-specific mtDNA depletion in the nervous system, OXPHOS defects, increased sensitivity to oxidative stress and a disease-specific mortality rate.
We have shown previously that over-expression of pol γ-α in UAS-PolG transgenic flies using a constitutive GAL4 driver induces a significant mtDNA depletion that causes morphological abnormalities and pupal lethality (Lefai et al. 2000c). In sharp contrast, the same level of over-expression in Schneider cells had no phenotypic consequences, indicating that an excess of pol γ-α is compatible with cell growth and viability, at least in cell culture. To gain insight into the pathogenic role of pol γ-α over-expression in the whole animal, we characterized the molecular and physiological effects of over-expression of Drosophila pol γ-α in a tissue particularly dependent on mitochondrial function, the nervous system. To avoid potential variations because of chromosome position effects (Spradling et al. 1995), we analyzed three independent transgenic UAS-PolG lines with the construct inserted at different chromosomal locations. The physiological behavior of the three resulting homozygous UAS-PolG transgenic lines was similar to a wild-type line with regard to fertility, pupal lethality and median life span. In transgenic lines over-expressing the pol γ-α subunit in the nervous system, pupal eclosion was only moderately decreased over controls when compared with the low pupal eclosion or high pupal lethality observed previously when pol γ-α was constitutively over-expressed (Lefai et al. 2000c). These data suggest that the high pupal lethality was because of disruption of mitochondrial function in other tissue(s).
The nervous system in both flies and nematodes has been implicated as a critical tissue in life span determination (Braeckman et al. 2001). Over-expression of pol γ-α in the nervous system reduces significantly the median life span, likely because of an increase in the mortality rate of the population. However, the maximum life span is not changed significatively, indicating that there is no variation in the rate of aging. Such survival curves are typical of populations in which an increase in mortality because of disease obscures the effects of aging; these populations are characterized by a rapid mortality of young members with a parallel decrease in the median life span, but without affecting the maximum life span.
In molecular terms, a primary effect induced by the over-expression of pol γ-α was a substantial reduction in the rate of mtDNA synthesis that produced a decrease in the mtDNA level. It is important to note, that the real values of mtDNA replication rate, OXPHOS activities and steady-state levels of mtTFA and mtDNA in the nervous system of transgenic flies must be much lower than those shown in Figs 4, 5 and 6, because the latter correspond to values in the whole organism. Several drugs that inhibit mtDNA replication also deplete mtDNA, such as tacrine (Mansouri et al. 2003) and nalidixic acid (Lawrence et al. 1993). Similarly, transient expression of a polymerase-deficient human pol γ-α in cultured cells caused a reduction in the level of mtDNA (Spelbrink et al. 2000). Although multiple mechanisms are likely involved in inducing mtDNA depletion, it is clear in our study that an excess of pol γ-α subunit interferes with the process of mtDNA replication. One possible explanation is that excess pol γ-α sequesters other mtDNA replication proteins and inhibits their function in mtDNA replication complexes. In this regard, functional interaction between pol γ and mitochondrial single-stranded DNA-binding protein (mtSSB) enhances greatly the overall activity of pol γ (Farr et al. 1999), and low levels of mtSSB result in growth defects and in the depletion of mtDNA (Farr et al. 2004). Another possible explanation is that in the transgenic flies over-expressing pol γ-α, the catalytic core itself (Lewis et al. 1996) participates in mtDNA replication, but its lower activity and processivity (Wang and Kaguni 1999) results in a reduced rate of replication. These two hypotheses would be compatible with the aforementioned lack of effect in Schneider cells if the mechanism of mtDNA replication or its regulation is different in cultured cells as compared to the whole organism. Thus, if the steady-state of the replication accessory factors (pol γ-β, mtSSB, etc.) is higher in cells than in flies, the effect of the over-expression of pol γ-α may be masked. Moreover, different topological forms of mtDNA in Drosophila embryos, adults and Schneider cells have been described (Rubenstein et al. 1977), and two different modes of mtDNA replication that might be involved in the changes of mtDNA copy number have been suggested (Holt et al. 2000). Whatever the biochemical mechanism, it is clear that an imbalance in the amount of pol γ-α protein in the nervous system has significant physiological consequences.
In individuals with mtDNA depletion, the enzymatic activities of the mitochondrial respiratory chain complexes containing mtDNA-encoded subunits (complexes I, III and IV) are reduced (Mandel et al. 2001). In this study, we found a significant decrease in the normalized complexes (I+III)/citrate synthase activity, and a mild decrease of normalized complex IV activity in mitochondria from flies over-expressing pol γ-α in the nervous system. The decrease in these activities must result from a loss in the relative abundance of subunits encoded by mtDNA as consequence of the mtDNA depletion. mtDNA depletion results in various types of enzyme deficiencies (Sarzi et al. 2007), and although most patients with mtDNA depletion present reduced absolute activity values of mtDNA-encoded complexes (complexes I, III, IV and/or V), others can exhibit different combinations of enzyme deficiencies. The respiratory chain complex thresholds in mitochondria of Drosophila subobscura have been determined (Farge et al. 2002). Threshold values of 70% were measured for complex I, thus is, a 70% inhibition of complex I activity was required to induce inhibition of both mitochondrial respiration and ATP synthesis. In contrast, the complex III activity threshold was 20%. Although the decrease of complex I+III activity in the present study was of 35%, as mentionated before, the real values of OXPHOS activities in the nervous system of transgenic flies must be much lower, probably below the thresholds for these complexes. Thus, our study shows that, in flies over-expressing pol γ-α in the nervous system, severe mtDNA depletion occurs and as a consequence, the activity of the mitochondrial electron transport chain (OXPHOS system) as measured by complex I+III activity is reduced.
The transgenic flies over-expressing pol γ-α in the nervous system are more susceptible to oxidative damage than control flies. Thus, mtDNA depletion and OXPHOS defects are associated with a higher sensitivity to oxidative damage, suggesting a possible mechanism for the observed decrease in the median life span of the transgenic flies. Neuronal sensitivity to oxidative stress contributes significantly to limiting life span (Wolkow 2002). Reactive oxygen species are widely recognized as an important contributor to the aging process, and it has been proposed that an increase in resistance to oxidative stress contributes to longevity (Sohal 2002). In agreement with these ideas, transgenic and mutant flies with increased life span also showed enhanced resistance to oxidative stress (Parkes et al. 1998; Mourikis et al. 2006). Furthermore, various single gene mutations that affect nervous system activity in Drosophila (Kaplan and Trout 1969; Mourikis et al. 2006), as well as mutations affecting antioxidant systems, lead to shortened life span (Sohal 2002). Moreover, the contribution of different tissues to life span is not equal, and manipulation of free radical scavenging enzymes in the nervous system of Drosophila extends longevity, whereas the same manipulation in muscle has no effect (Parkes et al. 1999).
The nervous system of adult flies over-expressing pol γ-α did not show signs of apoptosis. However, a moderate but significant increase in the level of apoptosis in the nervous system of larvae over-expressing pol γ-α was detected, suggesting that pathological programmed cell death in the brain from the larval stage may contribute to the phenotype observed in the flies with mtDNA depletion. In Drosophila, although newly eclosed adults show extensive cell death in the nervous system within the first 12 h of adulthood, apoptosis associated specifically with the adult nervous system has not been shown to play a prominent role in fly aging (Zheng et al. 2005). This observation suggests either a differential response or a higher tolerance to oxidative damage during normal aging rather than the absence of apoptosis (Zheng et al. 2005), because in several models of neurodegenerative diseases apoptosis has been detected in the nervous system of adult flies (Kretzschmar et al. 1997; Finley et al. 2003; Ghosh and Feany 2004). One possible explanation for the apparent absence of apoptosis in the nervous system of transgenic adult flies is that apoptosis occurs in the nervous system but is not detected using antibodies against caspases. Thus, it has been suggested that the pathway leading to cell death may be different in respiratory chain-deficient cardiomyocytes and neurons (Sorensen et al. 2001). These authors generated MILON mice by disruption of the mtTFA gene in cortical neurons, which showed abundant TUNEL-reactive neuron cells. However, these neurons did not express activated caspase 3 or caspase 7 despite the massive amount of cell death. Furthermore, the levels of mRNAs for Bax and Bcl-xL were unchanged, and DNA fragmentation was only observed by using a sensitive PCR assay. Another possibility is that no apoptosis occurs, but neuronal cells suffer rapid degeneration. It has been shown in Drosophila that stress events that initiate apoptosis in tissues where the death pathway is blocked generate undead cells that can induce the oncogenic transformation of neighboring cells (Perez-Garijo et al. 2005).
Our work suggests that functional mitochondria are necessary for normal brain function, and that abnormal brain function deriving from the larval stage may be responsible for the progressive brain deterioration and death in the adult mutants. Thus, during their shortened life, the transgenic flies undergo more rapidly a series of events that are normally associated with advancing age. This conclusion is supported by a study in Coenorhabditis elegans that proposes that the developing animal has a regulatory system that monitors mitochondrial activity early in life and in response, establishes rates of mitochondrial respiration, behavior, and aging that persist during adulthood (Dillin et al. 2002).
The over-expression of pol γ-α in the Drosophila nervous system mimics some of the findings observed in human mtDNA depletion diseases. MDDS (Moraes et al. 1991) are autosomal recessive mitochondrial abnormalities characterized by a variable decrease of mtDNA in one or more tissues during development in the absence of additional mutations or rearrangements. This results in neonatal or childhood organ failure and lethality. Pedigree analysis and somatic cell genetics have demonstrated that all forms of mtDNA depletion are because of mutations in nuclear genes encoding factors involved in the control of mtDNA replication or deoxyribonucleotide metabolism. A preliminary study reported a markedly-deficient activity of pol γ in tissues from patients with mtDNA depletion (Naviaux et al. 1999). The hepatocerebral form of mtDNA depletion syndrome has been associated with mutation in the nuclear-encoded mitochondrial deoxyguanosine kinase gene (Salviati et al. 2002), the subunit β of the ADP-forming succinyl-CoA synthetase ligase gene (Elpeleg et al. 2005), and recently the MPV17 (Spinazzola et al. 2006) gene. This form is characterized by fasting hypoglycemia and episodic acute hepatic failure, and patients show a neonatal or an early infantile presentation, with a rapid progression to death within less than 1 year of age (Carrozzo et al. 2003). In Drosophila, survival curves provide a direct measure of mortality rate. In our model, the flies over-expressing pol γ-α in the nervous system develop a profound tissue-specific mtDNA depletion and respiratory chain deficiency by affecting the rate of mtDNA replication. The decrease of mtDNA expression is associated with an increase in oxidative stress susceptibility and lethality in adult flies. The neurodegenerative process is manifested by substantial apoptosis in the nervous system of larvae and a moderate lethality at the end of the pupal stage. Thus, over-expression of Drosophila pol γ-α in the nervous system provides an animal model for the human MDDS, opening new avenues to find treatments that decrease the effects and revert the consequences of the disease.
Acknowledgments
We would like to thank Alicia Torrado for valuable comments on the manuscript. This research was supported by European Union Project QLG1-CT-2001-00966, Ministerio de Ciencia y Tecnologia, Spain (Grants BMC01-1525 and BFU2004-04591) and Instituto de Salud Carlos III, Redes de centros RCMN (C03/08) and Temáticas (G03/011) (to RG) and National Institutes of Health Grant GM45295 (to LSK).
Abbreviations used
- α-ATPase
nuclear-encoded mitochondrial ATP synthase subunit α
- GAL4
yeast transcriptional activator by binding to UAS
- GFP
green fluorescent protein
- MDDS
mtDNA depletion syndromes
- mtDNA
mitochondrial DNA
- mtSSB
mitochondrial single-stranded DNA-binding protein
- mtTFA
mitochondrial transcription factor A
- nDNA
nuclear DNA
- OXPHOS
oxidative phosphorylation
- pol γ-α
DNA polymerase γ subunit α
- PolG
DNA polymerase γ
- UAS
upstream activation sequence
References
- Attardi G, Schatz G. Biogenesis of mitochondria. Annu Rev Cell Biol. 1988;4:289–353. doi: 10.1146/annurev.cb.04.110188.001445. [DOI] [PubMed] [Google Scholar]
- Braeckman BP, Houthoofd K, Vanfleteren JR. Insulin-like signaling, metabolism, stress resistance and aging in Caenorhabditis elegans. Mech Ageing Dev. 2001;122:673–693. doi: 10.1016/s0047-6374(01)00222-6. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a mean of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Calleja M, Peña P, Ugalde C, Ferreiro C, Marco R, Garesse R. Mitochondrial DNA remains intact during Drosophila aging, but the levels of mitochondrial transcripts are significantly reduced. J Biol Chem. 1993;268:18891–18897. [PubMed] [Google Scholar]
- Carrozzo R, Bornstein B, Lucioli S, et al. Mutation analysis in 16 patients with mtDNA depletion. Hum Mutat. 2003;21:453. doi: 10.1002/humu.9135. [DOI] [PubMed] [Google Scholar]
- Claveria C, Albar JP, Serrano A, Buesa JM, Barbero JL, Martinez-A C, Torres M. Drosophila grim induces apoptosis in mammalian cells. EMBO J. 1998;17:199–208. doi: 10.1093/emboj/17.24.7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvejic S, Zhu Z, Felice SJ, Berman Y, Huang XY. The endogenous ligand Stunted of the GPCR Methuselah extends lifespan in Drosophila. Nat Cell Biol. 2004;6:540–546. doi: 10.1038/ncb1133. [DOI] [PubMed] [Google Scholar]
- Dillin A, Hsu AL, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298:2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- DiMauro S, Schon EA. Mitochondrial respiratory-chain diseases. N Engl J Med. 2003;348:2656–2668. doi: 10.1056/NEJMra022567. [DOI] [PubMed] [Google Scholar]
- Ekstrand MI, Falkenberg M, Rantanen A, Park CB, Gaspari M, Hultenby K, Rustin P, Gustafsson CM, Larsson NG. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum Mol Genet. 2004;13:935–944. doi: 10.1093/hmg/ddh109. [DOI] [PubMed] [Google Scholar]
- Elpeleg O, Miller C, Hershkovitz E, Bitner-Glindzicz M, Bondi-Rubinstein G, Rahman S, Pagnamenta A, Eshhar S, Saada A. Deficiency of the ADP-forming succinyl-CoA synthase activity is associated with encephalomyopathy and mitochondrial DNA depletion. Am J Hum Genet. 2005;76:1081–1086. doi: 10.1086/430843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enriquez JA, Ramos J, Perez-Martos A, Lopez-Perez MJ, Montoya J. Highly efficient DNA synthesis in isolated mitochondria from rat liver. Nucleic Acids Res. 1994;22:1861–1865. doi: 10.1093/nar/22.10.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito LA, Melov S, Panov A, Cottrell BA, Wallace DC. Mitochondrial disease in mouse results in increased oxidative stress. Proc Natl Acad Sci USA. 1999;96:4820–4825. doi: 10.1073/pnas.96.9.4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farge G, Touraille S, Debise R, Alziari S. The respiratory chain complex thresholds in mitochondria of a Drosophila subobscura mutant strain. Biochimie. 2002;84:1189–1197. doi: 10.1016/s0300-9084(02)00038-x. [DOI] [PubMed] [Google Scholar]
- Farr CL, Wang Y, Kaguni LS. Functional interactions of mitochondrial DNA polymerase and single-stranded DNA binding protein: template-primer DNA binding and initiation and elongation of DNA strand synthesis. J Biol Chem. 1999;274:14779–14785. doi: 10.1074/jbc.274.21.14779. [DOI] [PubMed] [Google Scholar]
- Farr CL, Matsushima Y, Lagina AT, III, Luo N, Kaguni LS. Physiological and biochemical defects in functional interactions of mitochondrial DNA polymerase and DNA-binding mutants of single-stranded DNA-binding protein. J Biol Chem. 2004;279:17047–17053. doi: 10.1074/jbc.M400283200. [DOI] [PubMed] [Google Scholar]
- Ferguson M, Mockett RJ, Shen Y, Orr WC, Sohal RS. Age-associated decline in mitochondrial respiration and electron transport in Drosophila melanogaster. Biochem J. 2005;390:501–511. doi: 10.1042/BJ20042130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley KD, Edeen PT, Cumming RC, Mardahl-Dumesnil MD, Taylor BJ, Rodriguez MH, Hwang CE, Benedetti M, McKeown M. blue cheese mutations define a novel, conserved gene involved in progressive neural degeneration. J Neurosci. 2003;23:1254–1264. doi: 10.1523/JNEUROSCI.23-04-01254.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garesse R. Drosophila melanogaster mitochondrial DNA: gene organization and evolutionary considerations. Genetics. 1988;118:649–663. doi: 10.1093/genetics/118.4.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garesse R, Vallejo CG. Animal mitochondrial biogenesis and function: a regulatory cross-talk between two genomes. Gene. 2001;263:1–16. doi: 10.1016/s0378-1119(00)00582-5. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Feany MB. Comparison of pathways controlling toxicity in the eye and brain in Drosophila models of human neurodegenerative diseases. Hum Mol Genet. 2004;13:2011–2018. doi: 10.1093/hmg/ddh214. [DOI] [PubMed] [Google Scholar]
- Goto A, Matsushima Y, Kadowaki T, Kitagawa Y. Drosophila mitochondrial transcription factor A (d-TFAM) is dispensable for the transcription of mitochondrial DNA in Kc167 cells. Biochem J. 2001;354:243–248. doi: 10.1042/0264-6021:3540243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt IJ, Lorimer HE, Jacobs HT. Coupled leading- and lagging-strand synthesis of mammalian mitochondrial DNA. Cell. 2000;100:515–524. doi: 10.1016/s0092-8674(00)80688-1. [DOI] [PubMed] [Google Scholar]
- Kaguni LS. DNA polymerase gamma, the mitochondrial replicase. Annu Rev Biochem. 2004;73:293–320. doi: 10.1146/annurev.biochem.72.121801.161455. [DOI] [PubMed] [Google Scholar]
- Kaplan WD, Trout WE., 3rd The behavior of four neurological mutants of Drosophila. Genetics. 1969;61:399–409. doi: 10.1093/genetics/61.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly DP, Scarpulla RC. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004;18:357–368. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- Kretzschmar D, Hasan G, Sharma S, Heisenberg M, Benzer S. The swiss cheese mutant causes glial hyperwrapping and brain degeneration in Drosophila. J Neurosci. 1997;17:7425–7432. doi: 10.1523/JNEUROSCI.17-19-07425.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Larsson NG, Oldfors A, Holme E, Clayton DA. Low levels of mitochondrial transcription factor A in mitochondrial DNA depletion. Biochem Biophys Res Commun. 1994;200:1374–1381. doi: 10.1006/bbrc.1994.1603. [DOI] [PubMed] [Google Scholar]
- Larsson NG, Wang JM, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh GS, Clayton DA. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- Lawrence JW, Darkin-Rattray S, Xie F, Neims AH, Rowe TC. 4-Quinolones cause a selective loss of mitochondrial DNA from mouse L1210 leukemia cells. J Cell Biochem. 1993;51:165–174. doi: 10.1002/jcb.240510208. [DOI] [PubMed] [Google Scholar]
- Lefai E, Fernandez-Moreno MA, Kaguni LS, Garesse R. The highly compact structure of the mitochondrial DNA polymerase genomic region of Drosophila melanogaster: functional and evolutionary implications. Insect Mol Biol. 2000a;9:315–322. doi: 10.1046/j.1365-2583.2000.00191.x. [DOI] [PubMed] [Google Scholar]
- Lefai E, Fernandez-Moreno MA, Alahari A, Kaguni LS, Garesse R. Differential regulation of the catalytic and accessory subunit genes of Drosophila mitochondrial DNA polymerase. J Biol Chem. 2000b;275:33123–33133. doi: 10.1074/jbc.M003024200. [DOI] [PubMed] [Google Scholar]
- Lefai E, Calleja M, Ruiz de Mena I, Lagina AT, III, Kaguni LS, Garesse R. Overexpression of the catalytic subunit of DNA polymerase γ results in depletion of mitochondrial DNA in Drosophila melanogaster. Mol Gen Genet. 2000c;264:37–46. doi: 10.1007/s004380000301. [DOI] [PubMed] [Google Scholar]
- Lewis DL, Farr CL, Wang Y, Lagina AT, III, Kaguni LS. Catalytic subunit of mitochondrial DNA polymerase from Drosophila embryos. Cloning, bacterial overexpression, and biochemical characterization. J Biol Chem. 1996;271:23389–23394. doi: 10.1074/jbc.271.38.23389. [DOI] [PubMed] [Google Scholar]
- Luo L, Liao YJ, Jan LY, Jan YN. Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Gene Dev. 1994;8:1787–1802. doi: 10.1101/gad.8.15.1787. [DOI] [PubMed] [Google Scholar]
- Mandel H, Szargel R, Labay V, et al. The deoxyguanosine kinase gene is mutated in individuals with depleted hepatocerebral mitochondrial DNA. Nat Genet. 2001;29:337–341. doi: 10.1038/ng746. [DOI] [PubMed] [Google Scholar]
- Mansouri A, Haouzi D, Descatoire V, Demeilliers C, Sutton A, Vadrot N, Fromenty B, Feldmann G, Pessayre D, Berson A. Tacrine inhibits topoisomerases and DNA synthesis to cause mitochondrial DNA depletion and apoptosis in mouse liver. Hepatology. 2003;38:715–725. doi: 10.1053/jhep.2003.50353. [DOI] [PubMed] [Google Scholar]
- Moraes CT, Shanske S, Tritschler HJ, Aprille JR, Andreetta F, Bonilla E, Schon EA, DiMauro S. mtDNA depletion with variable tissue expression: a novel genetic abnormality in mitochondrial diseases. Am J Hum Genet. 1991;48:492–501. [PMC free article] [PubMed] [Google Scholar]
- Mourikis P, Hurlbut GD, Artavanis-Tsakonas S. Enigma, a mitochondrial protein affecting lifespan and oxidative stress response in Drosophila. Proc Natl Acad Sci USA. 2006;103:1307–1312. doi: 10.1073/pnas.0510564103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naviaux RK, Nyhan WL, Barshop BA, Poulton J, Markusic D, Karpinski NC, Haas RH. Mitochondrial DNA polymerase g deficiency and mtDNA depletion in a child with Alpers’ syndrome. Ann Neurol. 1999;45:54–58. doi: 10.1002/1531-8249(199901)45:1<54::aid-art10>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Parkes TL, Elia AJ, Dickinson D, Hilliker AJ, Phillips JP, Boulianne GL. Extension of Drosophila lifespan over-expression of human SOD1 in motorneurons. Nat Genet. 1998;19:171–174. doi: 10.1038/534. [DOI] [PubMed] [Google Scholar]
- Parkes TL, Hilliker AJ, Phillips JP. Motorneurons, reactive oxygen, and life span in Drosophila. Neurobiol Aging. 1999;20:531–535. doi: 10.1016/s0197-4580(99)00086-x. [DOI] [PubMed] [Google Scholar]
- Perez-Garijo A, Martin FA, Struhl G, Morata G. Dpp signaling and the induction of neoplastic tumors by caspase-inhibited apoptotic cells in Drosophila. Proc Natl Acad Sci USA. 2005;102:17664–17669. doi: 10.1073/pnas.0508966102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfanner N, Wiedemann N. Mitochondrial protein import: two membranes, three traslocases. Curr Opin Cell Biol. 2002;14:400–411. doi: 10.1016/s0955-0674(02)00355-1. [DOI] [PubMed] [Google Scholar]
- Rubenstein JL, Brutlag D, Clayton DA. The mitochondrial DNA of Drosophila melanogaster exists in two distinct and stable superhelical forms. Cell. 1977;12:471–482. doi: 10.1016/0092-8674(77)90123-4. [DOI] [PubMed] [Google Scholar]
- Salviati L, Sacconi S, Mancuso M, et al. Mitochondrial DNA depletion and dGK gene mutations. Ann Neurol. 2002;52:311–317. doi: 10.1002/ana.10284. [DOI] [PubMed] [Google Scholar]
- Sanvicens N, Gomez-Vicente V, Messeguer A, Cotter TG. The radical scavenger CR-6 protects SH-SY5Y neuroblastoma cells from oxidative stress-induced apoptosis: effect on survival pathways. J Neurochem. 2006;98:735–747. doi: 10.1111/j.1471-4159.2006.03914.x. [DOI] [PubMed] [Google Scholar]
- Sarzi E, Bourdon A, Chretien D, Zarhrate M, Corcos J, Slama A, Cormier-Daire V, de Lonlay P, Munnich A, Rotig A. Mitochondrial DNA depletion is a prevalent cause of multiple respiratory chain deficiency in childhood. J Pediatr. 2007;150:531–534. doi: 10.1016/j.jpeds.2007.01.044. [DOI] [PubMed] [Google Scholar]
- Sohal RS. Oxidative stress hypothesis of aging. Free Radic Biol Med. 2002;33:573–574. doi: 10.1016/s0891-5849(02)00885-7. [DOI] [PubMed] [Google Scholar]
- Sorensen L, Ekstrand M, Silva JP, Lindqvist E, Xu B, Rustin P, Olson L, Larsson NG. Late-onset corticohippocampal neurodepletion attributable to catastrophic failure of oxidative phosphorylation in MILON mice. J Neurosci. 2001;21:8082–8090. doi: 10.1523/JNEUROSCI.21-20-08082.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spelbrink JN, Toivonen JM, Hakkaart GAJ, et al. In vivo functional analysis of the human mitochondrial DNA polymerase POLG expressed in cultured human cells. J Biol Chem. 2000;275:24818–24828. doi: 10.1074/jbc.M000559200. [DOI] [PubMed] [Google Scholar]
- Spinazzola A, Viscomi C, Fernandez-Vizarra E, et al. MPV17 encodes an inner mitochondrial membrane protein and is mutated in infantile hepatic mitochondrial DNA depletion. Nat Genet. 2006;38:570–575. doi: 10.1038/ng1765. [DOI] [PubMed] [Google Scholar]
- Spradling AC, Rubin GM. Transposition of cloned P elements into Drosophila germ line chromosomes. Science. 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- Spradling AC, Stern DM, Kiss I, Roote J, Laverty T, Rubin GM. Gene disruptions using P transposable elements: an integral component of the Drosophila genome project. Proc Natl Acad Sci USA. 1995;92:10824–10830. doi: 10.1073/pnas.92.24.10824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D, Hancock JM, Webb DA, Tautz C, Dover GA. Complete sequences of the rRNA genes of Drosophila melanogaster. Mol Biol Evol. 1988;5:366–376. doi: 10.1093/oxfordjournals.molbev.a040500. [DOI] [PubMed] [Google Scholar]
- Van Goethem G, Dermaut B, Lofgren A, Martin JJ, Van Broeckhoven C. Mutation of POLG is associated with progressive external ophthalmoplegia characterized by mtDNA deletions. Nat Genet. 2001;28:211–212. doi: 10.1038/90034. [DOI] [PubMed] [Google Scholar]
- Waddel WJ, Hill C. A simple ultraviolet spectrophotometric method for determination of protein. J Lab Clin Med. 1956;48:311–314. [PubMed] [Google Scholar]
- Wang Y, Kaguni LS. Baculovirus expression reconstitutes Drosophila mitochondrial DNA polymerase. J Biol Chem. 1999;274:28972–28977. doi: 10.1074/jbc.274.41.28972. [DOI] [PubMed] [Google Scholar]
- Wang Y, Farr CL, Kaguni LS. Accessory subunit of mitochondrial DNA polymerase from Drosophila embryos. Cloning, molecular analysis, and association in the native enzyme. J Biol Chem. 1997;272:13640–13646. doi: 10.1074/jbc.272.21.13640. [DOI] [PubMed] [Google Scholar]
- Wolkow CA. Life span: getting the signal from the nervous system. Trends Neurosci. 2002;25:212–216. doi: 10.1016/s0166-2236(02)02133-1. [DOI] [PubMed] [Google Scholar]
- Yu SY, Yoo SJ, Yang L, Zapata C, Srinivasan A, Hay BA, Baker NE. A pathway of signals regulating effector and initiator caspases in the developing Drosophila eye. Development. 2002;129:3269–3278. doi: 10.1242/dev.129.13.3269. [DOI] [PubMed] [Google Scholar]
- Zheng J, Edelman SW, Tharmarajah G, Walker DW, Pletcher SD, Seroude L. Differential patterns of apoptosis in response to aging in Drosophila. Proc Natl Acad Sci USA. 2005;102:12083–12088. doi: 10.1073/pnas.0503374102. [DOI] [PMC free article] [PubMed] [Google Scholar]