The structure of the complex of the hexanucleotide duplex d(CGCGCA)·d(TGCGCG) with hexammineruthenium(III) ion shows a tautomeric shift in the adenine base and a consequent disruption of the A·T base pair.
Keywords: hexammineruthenium(III) ion, Z-DNA, tautomeric shift
Abstract
The hexamer duplex d(CGCGCA)·d(TGCGCG) was crystallized with hexammineruthenium(III) ions in an orthorhombic space group; the crystals diffracted to 1.54 Å resolution. Strong ion interactions with the adenine base induce a tautomeric shift from the amino to the imino form. Consequently, the A·T base pairing is disrupted. This structural study may be relevant to metal toxicity.
1. Introduction
The interactions of metals and metal complexes with DNA have been widely studied, particularly in antitumour chemistry (Hud & Polak, 2001 ▶; Egli, 2002 ▶; Reedijk, 1999 ▶). For example, one of the most successful anticancer drugs is cis-[PtCl2(NH3)2]. It is referred to as cisplatin (Reedijk, 2003 ▶) and forms an adduct with guanine bases in the DNA helix. Among nonplatinum complexes, ruthenium(III) complexes are widely used as antimetastatic agents (Gallori et al., 2000 ▶; Brabec, 2002 ▶; Alessio et al., 2004 ▶). Ruthenium(III) pentammine helps prevent carcinogenesis (Rubin et al., 1983 ▶). Experimental studies have shown that these ruthenium compounds interact with DNA (Gallori et al., 2000 ▶) and modify its chemistry and activity (Brabec, 2002 ▶). A study showed that this ligand binds to DNA through the guanine bases (Ho et al., 1987 ▶). The hexacoordinated form of this ion, hexammineruthenium(III) chloride, has been shown to be a strong inducer of Z-DNA (Thomas & Messner, 1988 ▶; Gueron et al., 2000 ▶), like its analogue cobalt hexammine chloride. Crystal structures of the complexes of ruthenium and cobalt hexammine with DNA show that these ions interact with guanine bases (Ho et al., 1987 ▶; Gessner et al., 1985 ▶; Brennan et al., 1986 ▶; Harper et al., 1998 ▶; Thiyagarajan et al., 2004 ▶). In the crystal structure of the complex of d(CGCGCG)2 with [Co(NH3)6]3+ (Gessner et al., 1985 ▶), the ion interacts with guanine and phosphate acceptor sites and stabilizes the Z-DNA form. Ho et al. (1987 ▶) showed a similar type of interaction of [Ru(NH3)6]3+ with the same hexamer sequence, although the ruthenium ion is not as effective as the cobalt ion in stabilizing Z-DNA.
The interaction of metal complexes with Z-DNA has also been studied in the crystal structures of sequences containing A·T base pairs. Brennan et al. (1986 ▶) showed that the decadeoxyoligonucleotide d(CGTACGTACG) is stabilized in the Z-form by cobalt hexammine binding to the guanine bases, although this structure is disordered and two backbone conformations were observed. The structure of the hexamer d(TGCGCA)2 determined by Harper et al. (1998 ▶) showed two ordered cobalt hexammine ions binding to guanine and phosphate acceptor sites. This binding played a role in stabilizing both the Z-DNA conformation and the crystal packing. In addition, the phosphate backbone exists in two equally populated discrete conformations at one nucleotide step around phosphate 11. Recently, the crystal structures of the hexamer sequence d(CGCGCA)·d(TGCGCG) complexed with [Co(NH3)6]3+ in two different space groups were reported from our laboratory (Thiyagarajan et al., 2004 ▶, 2005 ▶). In both crystals the cobalt hexammine ions make specific interactions with the adenine base. In one of them, the orthorhombic crystal, the ion interaction induces a tautomeric shift in the adenine and a consequent A·T wobble base pairing. The ion binding also mediates the inter-helical interactions and stabilizes the crystal packing.
Here, the structure of the hexamer sequence d(CGCGCA)·d(TGCGCG) complexed with hexammineruthenium(III) chloride, i.e. [Ru(NH3)6]3+, is presented. The details of the interactions of the ion with the DNA structure are discussed. Like the cobalt ion, Ru3+ also makes strong interactions with adenine and guanine bases, leading to destabilization of the base pairs but not of the DNA helix.
2. Materials and methods
2.1. Crystallization and data collection
The single strands of the complementary hexamer duplex were purchased from M/s Microsynth, Switzerland and annealed to form the duplex. Hexammineruthenium(III) chloride was purchased from Sigma–Aldrich Chemical Co. (St Louis, MO, USA). Crystals were grown at room temperature (293 K) by the hanging-drop vapour-diffusion method. We obtained two crystal forms at two slightly different ion concentrations. For both crystals, the crystallization drop contained 1 mM DNA in sodium cacodylate buffer pH 6.9 and 0.05 mM spermine. The hexammineruthenium(III) concentration was 0.8 and 0.4 mM for the two crystal forms, respectively. The drop was equilibrated against 50% methylpentanediol in the reservoir. Crystals of dimensions 0.2 × 0.1 × 0.1 mm and 0.15 × 0.1 × 0.1 mm, respectively, were used for data collection on a MAR Research imaging-plate system at the GNR Laboratory for Structural Biology, Central Leather Research Institute, Chennai, India. The data set from the crystal grown at the higher concentration of hexammine was indexed in the orthorhombic space group P212121. The other data set was indexed in the hexagonal space group P65. The orthorhombic crystal diffracted to 1.54 Å resolution, while the hexagonal crystal diffracted to the far lower resolution of 2.60 Å.
Structure solution using the hexagonal data set yielded a disordered duplex. The final R factor after refinement for this data set is 25.7%, with a free R value of 36.3%. Intensity analysis of the data set indicated the possibility of merohedral twinning (French & Wilson, 1978 ▶). However, the data could only be partially detwinned (Yeates, 1997 ▶). Furthermore, calculations using the partially detwinned data did not improve the solution. Because of the twinning, the disorder and the low resolution, the final model is not free of errors. Thus, in this paper we only discuss the structure in the orthorhombic system. Table 1 ▶ gives the data-collection and refinement details for this system.
Table 1. Data-collection and refinement details of the P212121 structure.
Values in parentheses are for the last shell.
| Resolution (Å) | 1.54 |
| Completeness (%) | 90.8 (94.2) |
| Rmerge (%) | 5.3 (35.78) |
| Unit-cell parameters (Å) | a = 17.95, b = 30.84, c = 44.60 |
| No. of reflections | 3000 |
| Multiplicity | 5.04 |
| Mean I/σ(I) | 4.7 |
| R factor (%) | 18.9 |
| Rfree (%) | 22.3 |
2.2. Structure solution and refinement
The structure was solved by molecular replacement using AMoRe (Navaza, 1994 ▶) from the CCP4 suite (Collaborative Computational Project, Number 4, 1994 ▶). As this crystal is isomorphous to the cobalt complex, the coordinates of d(CGCGCA)·d(TGCGCG) in the cobalt hexammine complex (Thiyagarajan et al., 2005 ▶) were used as the starting model.
8% of the data in the resolution range 25.0–1.54 Å were flagged for cross-validation and the remainder were used to refine the solution using REFMAC5 (Murshudov et al., 1997 ▶) from the CCP4 suite (Collaborative Computational Project, Number 4, 1994 ▶). The structure fitted well into the 2F o − F c electron-density map. 51 water molecules were located, some with partial occupancy. In the DNA helix, the phosphate groups at C3, C5, C9 and C11 have two alternate conformations. We set the occupancy factors such that the B factors of these atoms were almost equal. One strong blob of density that appeared alone in the electron-density map even at the 6σ level was identified as a hexammineruthenium(III) ion. Four of the amine groups could be clearly located around the Ru atom. The other amine groups were placed by geometrical considerations. The average Ru—NH3 bond length is 2.09 Å after refinement, confirming that these are indeed amine groups (Engelhardt et al., 1995 ▶) and not water molecules. (This point is discussed in greater detail later.) The final R factor is 21.6% (R free = 25.5%). Since the difference Fourier map showed negative density at the ion position, we lowered the occupancy factors for these atoms to 0.7. This move alone reduced the R factor by 2% and R free by 3%. The final R factor is 18.9% (R free = 22.3%) and the final difference Fourier map is free of gross unexplained features. Any further uncertainties about the final model, for example about the correct placement of the A·T base pair, were eliminated by attempting refinement of other models and calculating omit maps. In none of these cases could we reasonably interpret the maps in any way other than that reported here. The temperature factors of all the atoms of the structure, the water molecules and the hexammineruthenium(III) ion fall within acceptable ranges. The average thermal parameter is 16.4 Å2 for all atoms.
Conformational and helical parameter calculations were carried out using the programs FREEHELIX (Dickerson, 1998 ▶), CURVES (Lavery & Sklenar, 1988 ▶) and X3DNA (Lu & Olson, 2003 ▶).
3. Results and discussion
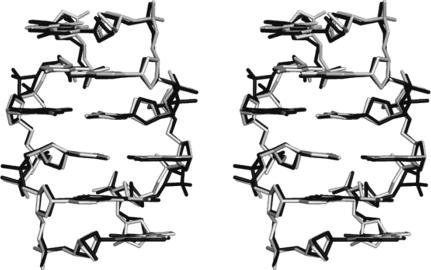
The sequence crystallizes as a left-handed Z-type helix that is very similar in conformation and crystal packing to that in the complex with cobalt hexammine (Thiyagarajan et al., 2005 ▶). Fig. 1 ▶ shows a superposition of the helix with the analogous helix in the cobalt complex. The root-mean-square deviation in atomic positions after least-squares superposition is 0.5 Å. Thus, while the overall helical structure in the present report conforms not only to the canonical Z-type helical structure but also to the structure in the cobalt complex, there are variations and differences, especially in the backbone conformations, which may be related to metal-ion interactions.
Figure 1.
Least-squares superposition of the hexamer in the ruthenium complex (black) with the respective hexamer in the cobalt complex (grey).
3.1. Helix structure
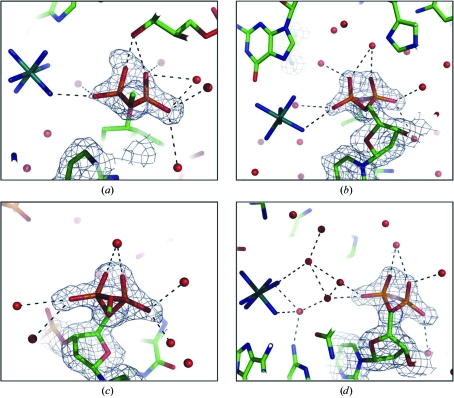
One noticeable feature of the present orthorhombic structure is in the backbone at residues C3, C5, C9 and C11, where the phosphate groups show two alternate conformations, termed ZI and ZII (Wang et al., 1979 ▶). In the ZI helix the backbone torsion angles ζ and α assume gauche − and trans conformations, respectively, while in ZII they are gauche + and trans, respectively. Harper et al. (1998 ▶) showed that the hexamer sequence d(TGCGCA)2 had both ZI and ZII conformations at phosphate position 11. In the cobalt complex of the present sequence (Thiyagarajan et al., 2005 ▶), C5 and C9 were identified as being in the ZII conformation, possibly as a result of ion binding. In the present structure, the four phosphate groups at C3, C5, C9 and C11 showed a mixture of ZI and ZII conformations at each site (Fig. 2 ▶). The phosphate groups in the ZII conformation at residues C3 and C5 interact directly with the ruthenium ion, while the phosphate group at C11 is bound to the ion through two water molecules. The backbone atoms at residue C9 do not bind to the ion, but form hydrogen bonds to water molecules. A comparison of Z-DNA hexamer crystal structures (Harper et al., 1998 ▶) showed that the ZI and ZII conformations are not specific to sequence. In a recent review, Subirana & Soler-Lopez (2003 ▶) showed that the phosphate groups in Z-DNA crystal structures are often disordered and may occupy alternate positions when cations are present. Thus, the alternate conformations at four phosphate sites of the present structure may be related to the ion interaction.
Figure 2.
The conformation of the phosphate groups at residues C3 (a), C5 (b), C9 (c) and C11 (d); the respective 2F obs − F calc electron-density maps are contoured at the 1.0σ level. The dotted lines represent hydrogen bonds.
3.2. Ion interactions
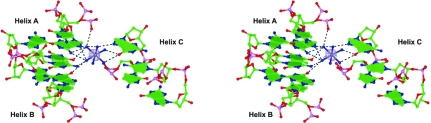
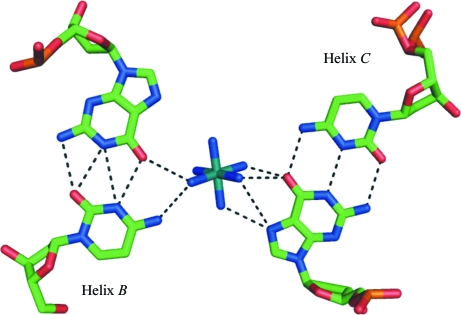
One hexammineruthenium(III) ion was clearly located from the Fourier map (Fig. 3 ▶). Since the f′′ value for ruthenium of 3.3 e is sufficiently large to be detected with a laboratory X-ray source, its position was confirmed by calculating an anomalous difference map using the CCP4 suite (Collaborative Computational Project, Number 4, 1994 ▶). Such a map was calculated with the experimental amplitudes (F + obs − F − obs) corresponding to Bijvoet pairs of reflections and the phases of the refined model without the Ru atom. The electron density calculated in this way is also shown in Fig. 3 ▶. It is clear that the entire hexammineruthenium(III) moiety makes strong interactions with A6 of one helix and simultaneously with the G12·C1 base pair and G4 of a symmetry-related duplex. The interaction distances are given in Table 2 ▶.
Figure 3.
Stereoview of two base pairs from each of the three symmetry-related helices (A, x, y, z; B, −x + ½, −y, z + ½; C, x + ½, −y + ½, −z) interacting with the ion. The electron density shown for the ruthenium ion corresponds to the anomalous difference map.
Table 2. Details of ion–DNA interaction geometries.
| Atom 1 | Atom 2 | Distance (Å) | Remarks |
|---|---|---|---|
| [Ru(NH3)6]3+ (N1) | A6 (N7) | 2.92 | |
| [Ru(NH3)6]3+ (N6) | A6 (N7) | 2.91 | |
| [Ru(NH3)6]3+ (N4) | A6 (N6) | 2.97 | |
| [Ru(NH3)6]3+ (N6) | A6 (N6) | 2.95 | |
| [Ru(NH3)6]3+ (N6) | G12 (O6) | 3.20 | |
| [Ru(NH3)6]3+ (N6) | C1 (N4) | 3.14 | |
| [Ru(NH3)6]3+ (N3) | G4 (O6) | 2.90 | |
| [Ru(NH3)6]3+ (N5) | G4 (O6) | 2.76 | |
| [Ru(NH3)6]3+ (N1) | G4 (N7) | 2.90 | |
| G12 (O6) | A6 (N6) | 3.28 | Cross-strand hydrogen bond |
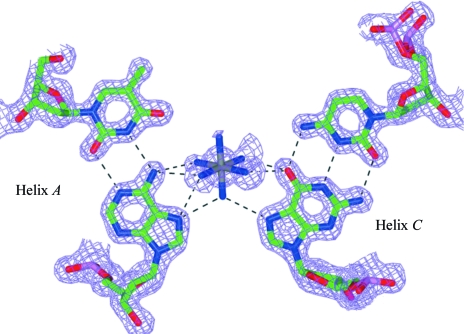
The interaction of the ion with the adenine base is the strongest interaction. The amine groups of the hexammineruthenium(III) ion make possible hydrogen bonds with N7 of adenine (Fig. 4 ▶). Previous studies on metals such as Co, Mo, Hg, Ru and Pt have shown that metal binding to N7 of the adenine base leads to protonation of N1 and destabilization of the base pair (Tajmir-Riahi, 1991 ▶; Day et al., 1994 ▶; Zamora et al., 1997 ▶; Burda et al., 2000 ▶; Velders et al., 2001 ▶; Sponer et al., 2002 ▶). Thus, metal binding stabilizes the imino tautomeric form of adenine by reducing the energy required for the tautomeric shift (Burda et al., 2000 ▶; Velders et al., 2001 ▶; Sponer et al., 2002 ▶). Protonation of N1 could be induced by metal binding to N7 of adenine either directly (Zamora et al., 1997 ▶; Burda et al., 2000 ▶) or indirectly via a ligand (Tajmir-Riahi, 1991 ▶). The present structure shows indirect binding of the Ru ion to N7 of adenine through the NH3 ligand, leading to protonation of N1 and consequent destabilization of the A·T base pair. The base-pairing scheme resembles the G·T wobble base pair (Ho et al., 1985 ▶; Kennard, 1985 ▶) frequently observed in RNA structures stabilized by metal hexammines (Cate & Doudna, 1996 ▶).
Figure 4.
Ruthenium-ion interactions with helix A and its symmetry-related helix C. The electron-density map was calculated with (2F obs − F calc) as coefficients and contoured at the 1.0σ level.
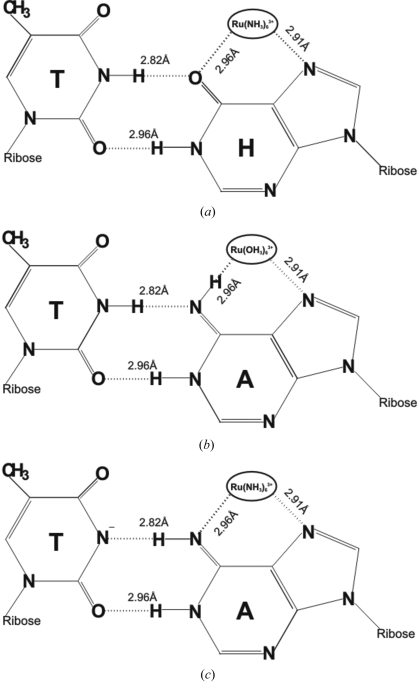
The ion also interacts with N6 of the adenine base. There are clearly some inconsistencies here. If the N6 of adenine is in the imino form, it could act as the proton acceptor for the ‘wobble’ hydrogen bond with N3 of thymine. In that case, it could not accept another hydrogen bond from NH3 of the ion. The nature of the ion–N6 interaction and furthermore the A–T interaction is therefore unclear. There are three possible resolutions to this conundrum. The first possibility is that the adenine base is oxidized to hypoxanthine and N6 is in fact O6. Such oxidation has been reported previously (Ponnamperuma et al., 1961 ▶; Karran & Lindahl, 1980 ▶; Wink et al., 1991 ▶; Nguyen et al., 1992 ▶; Grosjean et al., 1996 ▶) and would neatly explain all the aspects of the adenine–ion interaction as well as the A–T interaction in the present case, as shown in Fig. 5 ▶(a). However, we are constrained to reject this explanation for the following reasons. Firstly, the conditions required for the oxidation of adenine to hypoxanthine are the presence of oxidizing agent (Wink et al., 1991 ▶; Nguyen et al., 1992 ▶), ionizing radiation (Ponnamperuma et al., 1961 ▶), high temperature (Karran & Lindahl, 1980 ▶) or enzymatic conversion (Grosjean et al., 1996 ▶) and these do not exist in the present case. There is no oxidizing agent present either in the stock DNA solution or in the crystallization drop. Also, ruthenium(II) or ruthenium(III) ions are not oxidizing agents and cannot convert adenine to hypoxanthine (Velders et al., 2001 ▶; Hotze et al., 2002 ▶). Secondly, the rate of such oxidation at room temperature, even when oxidizing agents are present, is too low (Shapiro, 1995 ▶) to be a credible possibility in the present case. Furthermore, it is unlikely that radiation damage during data collection has lead to the conversion (Ponnamperuma et al., 1961 ▶), since this would imply rapid loss of crystal integrity. We did not observe any such damage during the data-collection process. One may further consider the possibility that hypoxanthine has inadvertently replaced adenine during the synthesis of the sequence. This possibility can again be eliminated because the sample we obtained from M/s Microsynth was not only purified by HPLC but also had its sequence checked by mass spectrometry prior to shipping. Moreover, when used in other experiments, as for example in the hexagonal structure, the same sample clearly showed the normal A·T base pairs, leading us to conclude that the base is indeed adenine and not hypoxanthine.
Figure 5.
Schematic sketch of three possible explanations of ruthenium-ion interactions with the A6·T7 base pair. (a) Adenine is modified to hypoxanthine (H). (b) The amine groups of the ruthenium ion are exchanged with water. (c) The N3 imino proton of thymine is transferred to the environment.
The second possible resolution of the problem postulates that the NH3 groups surrounding the Ru3+ ion have undergone exchange with water (leading to the interactions illustrated in Fig. 5 ▶ b). Again, this has been shown to be possible (Shriver & Atkins, 1999 ▶), but with the very low rate of about 10−5 s−1. Martin & Swaddle (1974 ▶) suggested that the radiolytic decomposition of cobalt(III) hexammine leads to the exchange of amine groups. A similar effect may also be possible in the present ruthenium ion. However, as described in §2, the geometry of the ion and surrounding ligands make it explicit that Ru3+ is hexacoordinated to NH3 and not to water.
The third possibility is the one that we believe describes the actual situation in the present structure. It is known that exchange of DNA imino protons at N3 of thymine with solvent protons occurs via a base-pair disruption reaction that brings the imino proton into an open state. In this state, the imino proton can be transferred to proton acceptors (Englander & Kallenbach, 1984 ▶; Gueron & Leroy, 1995 ▶). The acceptors can be N atoms of other bases within the same DNA molecule or proton acceptors present in solution (e.g. OH− and water; Gueron & Leroy, 1995 ▶). The thymine (T) then becomes T(−) and N3 can be a hydrogen-bond acceptor. Consequently, the tautomer of adenine stabilized by hydrogen bonding to the amine group of ruthenium ion forms a wobble base pair with thymine. Fig. 5 ▶(c) gives a schematic sketch of these possible interactions.
In an attempt to further clarify the situation, we have used extended Hückel theory (EHT) to calculate the possible energetic preference for each of the three possible interactions using the software package ArgusLab 4.0.1 (Thompson, 2004 ▶). The atomic coordinates were set to the three possible conformations shown in Fig. 5 ▶. The EHT calculations showed that there were only very small differences in the three energy values. The energies are −160.88, −163.40 and −160.18 au for the first, second and third conformations, respectively. Thus, the energy calculations do not rule out any of the three possibilities, but neither do they strongly support any one of them over the others.
The other ion interactions involve the G4·C9 and G12·C1 base pairs of a neighbouring symmetry-related helix (Fig. 6 ▶). The O6 of G4 makes a pair of bifurcated hydrogen bonds with the two amine groups of the ion. Similarly, N7 of the same base makes another pair of bifurcated hydrogen bonds with the ion.
Figure 6.
Geometry of the interaction of the ruthenium ion with helix B and helix C. The dotted lines represent hydrogen bonds.
The ion interactions with the G12·C1 base pair involve binding to O6 of G12 and N4 of C1. This disrupts the base pairing (Fig. 6 ▶). The value of the shear at the terminal base pair is −1.59 Å, in contrast to −1.14 Å in the cobalt structure and −0.22 Å in the undisturbed Z helix (Wang et al., 1979 ▶). The difference compared with the cobalt structure might be related to the larger ionic radius of Ru3+ (0.82 Å) compared with Co3+ (0.68 Å).
To summarize, we suggest that the hexammineruthenium(III) ion has induced a tautomeric shift of the adenine base from the amino to the imino form. In addition, the thymine is deprotonated, leading to the wobble A·T base pair. Both hydrogen bonds in the base pair have proton donors on the adenine base and proton acceptors on the thymine base. The ion also disrupts the G·C base pair. Finally, the overall helix conformation and the ion interactions in the present case are very similar to the those of the cobalt structure.
Supplementary Material
PDB reference: hexammineruthenium(III) ion interactions with Z-DNA, 2 hto, r2ftosf
PDB reference: 2htt, r2httsf
Acknowledgments
We thank the Department of Biotechnology, Government of India for financial support under grant No. BT/PR5015/BRB/10/365/2004. We also thank the Universities Grants Commission and the Department of Science and Technology, Government of India for support under the CAS programme and the FIST programme, respectively. The X-ray data-collection facility at the GNR Centre for Structural Biology, Central Leather Research Institute, Chennai is acknowledged.
References
- Alessio, E., Mestroni, G., Bergamo, A. & Sava, G. (2004). Curr. Top. Med. Chem.4, 1525–1535. [DOI] [PubMed]
- Brabec, V. (2002). Prog. Nucleic Acid Res. Mol. Biol.71, 1–68. [DOI] [PubMed]
- Brennan, R. G., Westhof, E. & Sundaralingam, M. (1986). J. Biomol. Struct. Dyn.3, 649–665. [DOI] [PubMed]
- Burda, J. V., Sponer, J. & Leszczynski, J. (2000). J. Biol. Inorg. Chem.5, 178–188. [DOI] [PubMed]
- Cate, J. H. & Doudna, J. A. (1996). Structure, 4, 1221–1229. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Day, E. F., Crawford, C. A., Felting, K., Dunbar, K. R. & Christou, G. (1994). J. Am. Chem. Soc.116, 9339–9340.
- Dickerson, R. E. (1998). Nucleic Acids Res.26, 1906–1926. [DOI] [PMC free article] [PubMed]
- Egli, M. (2002). Chem. Biol.9, 277–286. [DOI] [PubMed]
- Engelhardt, L. M., Reynolds, P. A. & Sobolev, A. N. (1995). Acta Cryst. C51, 1045–1047.
- Englander, S. W. & Kallenbach, N. R. (1984). Q. Rev. Biophys.16, 521–655. [DOI] [PubMed]
- French, S. & Wilson, K. (1978). Acta Cryst. A34, 517–525.
- Gallori, E., Vettori, C., Alessio, E., Vilchez, F. G., Vilaplana, R., Orioli, P., Casini, A. & Messori, L. (2000). Arch. Biochem. Biophys.376, 156–162. [DOI] [PubMed]
- Gessner, R. V., Quigley, G. J., Wang, A. H.-J., van der Marel, G. A., van Boom, J. H. & Rich, A. (1985). Biochemistry, 24, 237–240. [DOI] [PubMed]
- Grosjean, H., Auxilien, S., Constantinesco, F., Simon, C., Corda, Y., Becker, H. F., Foiret, D., Morin, A., Jin, Y. X., Fournier, M. & Fourrey, J. L. (1996). Biochimie, 78, 488–501. [DOI] [PubMed]
- Gueron, M., Demaret, J. P. & Filoche, M. (2000). Biophys. J.78, 1070–1083. [DOI] [PMC free article] [PubMed]
- Gueron, M. & Leroy, J. L. (1995). Methods Enzymol.261, 383–413. [DOI] [PubMed]
- Harper, A., Brannigan, J. A., Buck, M., Hewitt, L., Lewis, R. J., Moore, M. H. & Schneider, B. (1998). Acta Cryst. D54, 1273–1284. [DOI] [PubMed]
- Ho, P. S., Frederick, C. A., Quigley, G. J., van der Marel, G. A., van Boom, J. H., Wang, A. H.-J. & Rich, A. (1985). EMBO J.4, 3617–3623. [DOI] [PMC free article] [PubMed]
- Ho, P. S., Frederick, C. A., Saal, D., Wang, A. H.-J. & Rich, A. (1987). J. Biomol. Struct. Dyn.4, 521–534. [DOI] [PubMed]
- Hotze, A. C. G., Broekhuisen, M. E. T., Velders, A. H., van der Schilden, K., Haasnoot, J. G. & Reedijk, J. (2002). Eur. J. Inorg. Chem.2002, 369–376.
- Hud, N. V. & Polak, M. (2001). Curr. Opin. Struct. Biol.11, 293–301. [DOI] [PubMed]
- Karran, P. & Lindahl, T. (1980). Biochemistry19, 6005–6011. [DOI] [PubMed]
- Kennard, O. (1985). J. Biomol. Struct. Dyn.3, 205–226. [DOI] [PubMed]
- Lavery, R. & Sklenar, H. (1988). J. Biomol. Struct. Dyn.6, 63–91. [DOI] [PubMed]
- Lu, X.-J. & Olson, W. K. (2003). Nucleic Acids Res.31, 5108–5121. [DOI] [PMC free article] [PubMed]
- Martin, A. & Swaddle, T. W. (1974). Can. J. Chem.52, 2751–2759.
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed]
- Navaza, J. (1994). Acta Cryst. A50, 157–163.
- Nguyen, T., Brunson, D., Crespi, C. L., Penman, B. W., Wishnok, J. S. & Tannenbaum, S. R. (1992). Proc. Natl Acad. Sci. USA, 89, 3030–3034. [DOI] [PMC free article] [PubMed]
- Ponnamperuma, C., Lemmon, R. M., Bennett, E. L. & Calvin, M. (1961). Science, 134, 113. [DOI] [PubMed]
- Reedijk, J. (1999). Curr. Opin. Chem. Biol.3, 236–40. [DOI] [PubMed]
- Reedijk, J. (2003). Proc. Natl Acad. Sci. USA, 100, 3611–3616. [DOI] [PMC free article] [PubMed]
- Rubin, J. R., Sabat, M. & Sundaralingam, M. (1983). Nucleic Acids Res.11, 6571–6586. [DOI] [PMC free article] [PubMed]
- Shapiro, R. (1995). Orig. Life Evol. Biosph.25, 83–98. [DOI] [PubMed]
- Shriver, D. F. & Atkins, P. W. (1999). Inorganic Chemistry. Oxford University Press.
- Sponer, J., Leszczynski, J. & Hobza, P. (2002). Biopolymers, 61, 3–31. [DOI] [PubMed]
- Subirana, J. A. & Soler-Lopez, M. (2003). Annu. Rev. Biophys. Biomol. Struct.32, 27–45. [DOI] [PubMed]
- Tajmir-Riahi, H. A. (1991). J. Biomol. Struct. Dyn.8, 1169–1186. [DOI] [PubMed]
- Thiyagarajan, S., Rajan, S. S. & Gautham, N. (2004). Nucleic Acids Res.32, 5945–5953. [DOI] [PMC free article] [PubMed]
- Thiyagarajan, S., Rajan, S. S. & Gautham, N. (2005). Acta Cryst. D61, 1125–1131. [DOI] [PubMed]
- Thomas, T. J. & Messner, R. P. (1988). Biochimie.70, 221–226. [DOI] [PubMed]
- Thompson, M. A. (2004). ArgusLab 4.0.1. WA Planaria Software LLC, Seattle, USA.
- Velders, A. H., van der Geest, B., Kooijman, H., Spek, A. L., Haasnoot, J. G. & Reedijk, J. (2001). Eur. J. Inorg. Chem.2001, 369–372.
- Wang, A. H.-J., Quigley, G. J., Kolpak, F. J., Crawford, J. L., van Boom, J. H., van der Marel, G. A. & Rich, A. (1979). Nature (London), 282, 680–686. [DOI] [PubMed]
- Wink, D. A., Kasprzak, K. S., Maragos, C. M., Elespuru, R. K., Misra, M., Dunams, T., Cebula, T. A., Koch, W. H., Andrews, A. W., Allen, J. S. & Keefer, L. K. (1991). Science, 254, 1001–1003. [DOI] [PubMed]
- Yeates, T. O. (1997). Methods Enzymol.276, 344–358. [PubMed]
- Zamora, F., Kunsman, M., Sabat, M. & Lippert, B. (1997). Inorg. Chem.36, 1583–1587. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: hexammineruthenium(III) ion interactions with Z-DNA, 2 hto, r2ftosf
PDB reference: 2htt, r2httsf