Abstract
The Aeromonas hydrophila wb*O34 gene cluster of strain AH-3 (serotype O34) was cloned and sequenced. This cluster contains genes necessary for the production of O34-antigen lipopolysaccharide (LPS) in A. hydrophila. We determined, using either mutation or sequence homology, roles for the majority of genes in the cluster by using the chemical O34-antigen LPS structure obtained for strain AH-3. The O34-antigen LPS export system has been shown to be a Wzy-dependent pathway typical of heteropolysaccharide pathways. Furthermore, the production of A. hydrophila O34-antigen LPS in Escherichia coli K-12 strains is dependent on incorporation of the Gne enzyme (UDP-N-acetylgalactosamine 4-epimerase) necessary for the formation of UDP-galactosamine in these strains. By using rapid amplification of cDNA ends we were able to identify a transcription start site upstream of the terminal wzz gene, which showed differential transcription depending on the growth temperature of the strain. The Wzz protein is able to regulate the O34-antigen LPS chain length. The differential expression of this protein at different temperatures, which was substantially greater at 20°C than at 37°C, explains the previously observed differential production of O34-antigen LPS and its correlation with the virulence of A. hydrophila serotype O34 strains.
In gram-negative bacteria lipopolysaccharide (LPS) is one of the major structural and immunodominant molecules of the outer membrane. LPS consists of three moieties: lipid A, core oligosaccharide, and O-specific antigen or O side chain. The O antigen is the external component of LPS, and it consists of a polymer of oligosaccharide repeating units. Another interesting feature is the high chemical variability shown by the O-antigen LPS, which leads to similar genetic variation in the genes involved in LPS biosynthesis, the so-called wb* cluster (for a review, see reference 45). The genetics of O-antigen biosynthesis have been intensively studied in the Enterobacteriaceae, and it has been shown that the wb* clusters usually contain genes involved in biosynthesis of activated sugars, glycosyl transferases, and O-antigen polymerases and in O-antigen export. Despite heterogeneity in the structures of the O antigens, only three pathways for assembly of O antigens have been recognized: the Wzy-dependent pathway, the ABC transporter-dependent pathway, and the synthase-dependent pathway (45).
Mesophilic motile Aeromonas species are opportunistic and primary pathogens of a variety of aquatic and terrestrial animals, including humans; the clinical manifestations range from gastroenteritis to soft tissue infections, including septicemia and meningitis (5, 25). Mesophilic Aeromonas sp. serotype O34 strains have been recovered from moribund fish (33) and from clinical specimens (23); serotype O34 is the single most common Aeromonas serotype (24), accounting for 26.4% of all infections. Previous investigations have documented that serotype O34 strains are important causes of infections in humans (23, 24). The varied clinical picture of Aeromonas infections and gastroenteric illness, in particular, suggests that the pathogenic mechanisms of this bacterium are complex. The role of O34 antigen in adhesion, colonization, and serum resistance has been established previously (35, 38), as have the effects of growth temperature and osmolarity on the outer membrane components (especially the O-antigen LPS) and virulence of the A. hydrophila serotype O34 strains (1, 34, 36).
The A. hydrophila O-antigen cluster of strain PPD134/91 (serotype O18) has been sequenced, and seven biosynthetic genes for the synthesis of TDP-rhamnose and GDP-mannose, six putative transferase genes, one O-unit flippase gene, and one O-antigen chain length determinant gene were identified (56). Nevertheless, no mutants of this strain devoid of the O-antigen LPS were obtained, and the chemical structure of this strain is unknown.
In this paper we report the cloning, sequencing, and temperature regulation of the A. hydrophila O34-antigen LPS. We previously characterized chemically the structure of the O34 antigen and the LPS core of strain AH-3 belonging to serotype O34 (27, 28) (Fig. 1. To our knowledge, this is the first O-antigen LPS from this bacterium that has been completely studied genetically and chemically and also the O-antigen LPS found most frequently in several kinds of infections with this microorganism (24).
FIG. 1.
Chemical structures of the O34-antigen LPS (A) and the LPS core (B) from A. hydrophila strain AH-3 (27, 28). The O34-antigen LPS is linked to the Gal residue (indicated by italics) of the LPS core (28).
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. The Aeromonas serotype O18 strains used were Aer211 (California Department of Health Services), JCM3980 (Japan Collection of Microorganisms), PPD134/91 (Agri-Food and Veterinary Authorities, Singapore) (55), 613-90 (National Institute of Health, Japan), and three isolates from our laboratory. The Aeromonas serotype O34 strains used were Aer184 (California Department of Health Services), JCM3996 (Japan Collection of Microorganisms), PPD64/90(Agri-Food and Veterinary Authorities, Singapore), Ba5 (University of Montreal) (55), 519-92, 619-91, 614-91, 1051-88 (National Institute of Health, Japan), and six isolates from our laboratory. Aeromonas strains were routinely grown in tryptic soy broth (TSB) or on tryptic soy agar at 30°C unless otherwise stated, while Escherichia coli strains were grown in Luria-Bertani (LB) Miller broth and on LB Miller agar at 37°C. Kanamycin (50 μg ml−1), ampicillin (100 μg ml−1), tetracycline (20 μg ml−1), rifampin (100 μg ml−1), and chloramphenicol (20 μg ml−1) were added to different media.
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| E. coli strains | ||
| DH5α | F−endA hsdR17 (rK− mK+) supE44 thi-1 recA1 gyrA96 φ80lacZM15 | 19 |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac (F−proAB lacIqZΔM15 Tn10) | Stratagene |
| CLM4 | lacZ trp (sbcB-rfb) upp rel rpsL recA | 30 |
| S17-1 | hsdR pro recA RP4-2 in chromosome Km::Tn7 (Tc::Mu) | 9 |
| MC1061 | thi thr-1 leu-6 proA2 his-4 argE2 lacY1 galK2 ara-14 xyl-5 supE44 λpir | 10 |
| A. hydrophila strains | ||
| AH-3 | O34, wild type | 33 |
| AH-405 | AH-3, spontaneous Rifr | 1 |
| AH-901 | AH-3 rmlC::mini-Tn5Km-1 Rifr Kmr | This study |
| AH-902 | AH-3 manC::mini-Tn5Km-1 Rifr Kmr | This study |
| AH-405ΔwecA | AH-3 wecA mutant in frame with pDM4, Kmr | This study |
| AH-405Δwzy | AH-3 wzy mutant in frame with pDM4, Kmr | This study |
| AH-405Δwzz | AH-3 wzz mutant in frame with pDM4, Kmr | This study |
| PPD134/91 | O18, wild type | 56 |
| Plasmids | ||
| pRK2073 | Helper plasmid, Spcr | 9 |
| pLA2917 | Cosmid vector, Kmr Tcr | 28 |
| COS-O34 | pLA2917 cosmid with the complete wb*O34 region, Tcr | This study |
| pGEM-T | PCR cloning vector, Ampr | Promega |
| pGEMT-Gne | pGEM-T vector with complete gne gene of AH-3 | 11 |
| pBCSK | Cloning vector with lacZ gene, Cmr | Stratagene |
| pDM4 | pir dependent with sacAB genes, oriR6K Cmr | 10 |
| pDM4ΔwecA | pDM4 with AH-3::wecA::Km, Cmr Kmr | This study |
| pDM4Δwzy | pDM4 with AH-3::wzy::Km, Cmr Kmr | This study |
| pDM4Δwzz | pDM4 with AH-3::wzz::Km, Cmr Kmr | This study |
| pBAD33 | Arabinose-inducible expression vector, Cmr | ATCC |
| pBAD33-wecA | PBAD33 with complete wecA gene of AH-3, Cmr | This study |
| pBAD33-wzy | PBAD33 with complete wzy gene of AH-3, Cmr | This study |
| pBAD33-wzz | PBAD33 with complete wzz gene of AH-3, Cmr | This study |
| pRS550 | bla-kan-Tl4-BamHI-SmaI-EcoRI-lacZ+, Ampr Kmr | 51 |
| pRS-PWZZA | pRS550 with wzzA3 promoter-lacZ fusion, Ampr Kmr | This study |
| pRS-PWZZP | pRS550 with wzzPPD134/91 promoter-lacZ fusion, Ampr Kmr | This study |
| pACYC184 | Plasmid vector, Cmr Tcr | 47 |
| pACYC-PWZZA | pACYC184 with 6.3-kb StuI fragment of pRS-PWZZA, Cmr | This study |
| pACYC-PWZZP | pACYC184 with 6.3-kb StuI fragment of pRS-PWZZP, Cmr | This study |
| pACYC-lacZ | pACYC184 with 6.1-kb StuI fragment of pRS550, Cmr | This study |
Mini-Tn5Km-1 mutagenesis.
Conjugal transfer of transposition element mini-Tn5Km-1 from E. coli S17-1λpirKm-1 to A. hydrophila AH-405 (rifampin-resistant AH-3) was carried out by using a conjugal drop incubated for 6 h at 30°C with a ratio of S17-1λpirKm-1 to AH-405 to HB101/pRK2073 (helper plasmid) of 1:5:1 (by culture volume) (10). Serial dilutions of the mating mixture were plated on tryptic soy agar supplemented with rifampin and kanamycin in order to select mutants.
General DNA methods.
General DNA manipulations were done essentially as described previously (47). DNA restriction endonucleases, T4 DNA ligase, E. coli DNA polymerase (Klenow fragment), and alkaline phosphatase were used as recommended by the suppliers.
Cloning of DNA flanking mini-Tn5Km-1 insertions.
Chromosomal DNA of mini-Tn5Km-1 mutants was digested with EcoRI, PstI, and EcoRV, purified, ligated into the vector pBCSK (Stratagene), and introduced into E. coli XL1-Blue. Recombinant plasmids containing the transposon with flanking insertions were selected in LB medium plates supplemented with kanamycin and chloramphenicol. The mini-Tn5Km-1 flanking sequences were obtained by using primers specific for the I and O ends of mini-Tn5Km-1 (5′-AGATCTGATCAAGAGACAG-3′ and 5′-ACTTGTGTATAAGAGTCAG-3′, respectively), as well as primers M13for and T3.
DNA sequencing and computer analysis of sequence data.
Double-stranded DNA sequencing was performed by using the dideoxy chain termination method (49) with an ABI Prism dye terminator cycle sequencing kit (Perkin Elmer). Oligonucleotides used for genomic DNA amplification experiments and for DNA sequencing were purchased from Pharmacia LKB Biotechnology. The DNA sequence in all six frames was translated, and all open reading frame (ORFs) larger than 100 bp were inspected. Deduced amino acid sequences were inspected using the GenBank, EMBL, and Swiss-Prot databases and the BLASTP network service at the National Center for Biotechnology Information (3, 8). A protein family profile was obtained using the Pfam protein family database at the Sanger Center (8). Clustal W was used for multiple-sequence alignment.
Southern and dot blot hybridizations.
Southern blotting was performed by capillary transfer. For dot blot hybridization, the DNA was denatured by boiling it for 5 min, chilled on ice for another 5 min, and spotted onto a Hybond N1 (Amersham) nylon membrane. Probe labeling, hybridization, and detection were carried out using the enhanced chemiluminescence labeling and detection system (Amersham) according to the manufacturer's instructions.
LPS.
Cultures used for analysis of LPS were grown in TSB at 20 or 37°C. LPS was purified by the method of Galanos et al. (15), which resulted in a yield of 2.3% (dry cells). For screening purposes LPS was obtained after proteinase K digestion of whole cells (14). LPS samples were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) or SDS-Tricine-PAGE and were visualized by silver staining as previously described (14, 22). For chemical analysis, purified LPS samples were hydrolyzed with 1 N trifluoroacetic acid for 4 h at 100°C. Alditol acetates and methyl glycoside acetates were analyzed using an Agilent Technologies 5973N mass spectrometer equipped with a 6850A gas chromatography column and an RTX-5 capillary column (30 m by 0.25 [inside diameter]; flow rate of He carrier gas, 1 ml min−1; Restek) as previously described (10).
Mutant construction.
Chromosomal in-frame wecA, wzy, and wzz deletion mutants were constructed by allelic exchange as described by Milton et al. (11, 40). To obtain the AH-405ΔwecA mutant, DNA regions flanking the wecA gene were amplified using AH-3 chromosomal DNA and primers PA (5′-CGCGGATCCTGAACTTCCCCAAAGATTTACC-3′), PB (5′-CCCATCCACTAAACTTAAACAAAAATACTCCCAAAATCCCCAC-3′), PC (5′-TGTTTAAGTTTAGTGGATGGGGTGTTCAAAACTATTCGCACC-3′), and PD (5′-CGCGGATCCTCAGCAACGACACCAAGAGG-3′) in two sets of asymmetric PCRs. Two of these primers included BamHI sites (underlined in primers PA and PD). DNA fragments that were 530 bp (primers PA and PB) and 623 bp (primers PC and PD) long were obtained. The DNA fragment obtained with primers PA and PB contained the region from nucleotide 13579 in manB to nucleotide 14078, corresponding to the third base of the 20th codon of wecA. The DNA fragment obtained with primers PC and PD contained the region from nucleotide 15165, corresponding to the first base of the 383rd codon of wecA, to nucleotide 15757 in the rmlD gene. The DNA fragments obtained with primers PA and PB and with primers PC and PD were annealed at the overlapping region (underlined in primers PB and PC) and amplified by PCR as a single 1,132-bp fragment, using primers PA and PD. The fusion product was purified, BamHI digested, ligated into BglII-digested and phosphatase-treated vector pDM4, electroporated into E. coli MC1061 (λpir), and plated on LB agar plates containing chloramphenicol at 30°C to obtain plasmid pDM4ΔwecA.
The AH-405Δwzy mutant was constructed using AH-3 chromosomal DNA and primers YA (5′-ACGCGTCGACTATTTGGTACGCTGGTGGA-3′), YB (5′-CCCATCCACTAAACTTAAACAACCAGCGGAGAATGATGAT-3′), YC (5′-TGTTTAAGTTTAGTGGATGGGGGCGTGGGTTATTTCAGTC-3′), and YD (5′-ACGCGTCGACCATCAGATCGTCTGCCGTA-3′). Two of these primers included SalI sites (underlined in primers YA and YD). DNA fragments that were 627 bp (primers YA and YB) and 570 bp (primers YC and YD) long were obtained. The DNA fragment obtained with primers YA and YB contained the region from nucleotide 6646 in wbxD to nucleotide 7241, corresponding to the third base of the 36th codon of wzy. The DNA fragment obtained with primers YC and YD contained the region from nucleotide 8274, including the last base of the 36th wzy codon, to nucleotide 8812 in the wbxE gene. The DNA fragments obtained with primers YA and YB and with primers YC and YD were annealed at the overlapping region (underlined in primers YB and YC) and amplified by PCR as a single 1,176-bp fragment using primers YA and YD. The fusion product was purified, SalI digested, ligated into the SalI-digested and phosphatase-treated vector pDM4, electroporated into E. coli MC1061 (λpir), and plated on LB agar plates containing chloramphenicol at 30°C to obtain plasmid pDM4Δwzy. Similarly, primers ZA (5′-ACGCGTCGACTTCTGAGGTGAGTTTGGCC-3′), ZB (5′-CCCATCCACTAAACTTAAACACTGCGGCAACATCTTATCC-3′), ZC (5′-TGTTTAAGTTTAGTGGATGGGACTACCTTGCTTGGTGGCA-3′), and ZD (5′-ACGCGTCGACAAACAGCAGACCGGCAAAC-3′) were used to construct plasmid pDM4Δwzz to obtain the AH-405Δwzz mutant (for meaning of underlining, see above). Plasmid pDM4Δwzz included the region from nucleotide 16143 to the third base of the 15th codon of wzz (nucleotide 16676), the in-frame 21-bp linker, and the region from nucleotide 17637 in wzz to nucleotide 18129, including the last base of the 26th wzz codon.
Each mutated gene was transferred to the chromosome by homologous recombination using the λpir-dependent suicide plasmid pDM4 containing the counterselectable marker sacB. Triparental mating with mobilizing strain HB101/pRK2073 was used to transfer the plasmids containing the engineered in-frame deletions (pDM4ΔwecA, pDM4Δwzy, and pDM4Δwzz) into the A. hydrophila AH-405 rifampin-resistant (Rifr) strain. Transconjugants were selected on plates containing chloramphenicol and rifampin at 30°C. PCR analysis confirmed that the vector had integrated correctly into the chromosomal DNA. To complete the allelic exchange, the integrated suicide plasmid was forced to recombine out of the chromosome by growing organisms on agar plates containing 15% sucrose. Mutants were selected based on survival on plates containing 15% sucrose and loss of the chloramphenicol resistance marker of vector pDM4. The mutations were confirmed by sequencing of the whole constructs in amplified PCR products.
Plasmid construction and mutant complementation studies.
For complementation studies, the wecA, wzy, and wzz genes from A. hydrophila strain AH-3 were PCR amplified by using specific primer pairs and ligated to the plasmid vector pBAD33 (ATCC). The wecA gene was PCR amplified from A. hydrophila AH-3 chromosomal DNA with primers PF (5′-AAAAGTACTGAGTTATCAACGCCGTCAC-3′) and PR (5′-AAAACTGCACATCAACTGCCGTATAGGC-3′) as a 1,497-bp fragment. The PCR product was digested with ScaI and PstI (sites underlined) and ligated to the SmaI and PstI sites in previously digested pBAD33 to obtain plasmid pBAD33-wecA. Plasmid pBAD33-wzy was constructed by PCR amplification of a 1,596-bp DNA fragment from AH-3 chromosomal DNA using primers YF (5′-AAAAGTACTGCATCAAGAAGGTCGTTAG-3′) and YR (5′-AAAACTGCAGCCGCTCCAAGAAACGACTA-3′). The fragment was digested with ScaI and PstI (sites underlined) and ligated to the SmaI and PstI sites in previously digested pBAD33. For pBAD-wzz construction a 1,234-bp DNA fragment was PCR amplified from AH-3 chromosomal DNA with primers ZF (5′-AAAAGTACTACACTAGC GATCTTGAGGATAA-3′) and ZR (5′-ACGCGTCGACAGAGATGCTCACCCTTTTC-3′). The fragment was digested with ScaI and SalI (sites underlined) and ligated to the SmaI and SalI sites in previously digested pBAD33.
The plasmid constructs were transformed into E. coli LMG194 by electroporation, plated on LB agar plates containing chloramphenicol, and incubated at 30°C. Since the pBAD33 plasmid was derived from the pACYC184 plasmid, which was derived from the p15A plasmid (12, 20), plasmids pBAD33-wecA, pBAD33-wzy, and pBAD33-wzz were independently transferred into AH-405ΔwecA, AH-405Δwzy, and AH-405Δwzz, respectively, by triparental mating using mobilizing strain HB101/pRK2073. Transconjugants were selected on plates containing chloramphenicol and rifampin and confirmed by PCR. Each gene was expressed from the arabinose-inducible and glucose-repressible pBAD33 promoter. Repression from the araC promoter was achieved by growth in medium containing 0.2% (wt/vol) d-glucose, and induction was obtained by adding l-arabinose to a final concentration of 0.2% (wt/vol). The cultures were grown for 18 h at 30°C in TSB supplemented with chloramphenicol and 0.2% glucose. These cultures were diluted 1:100 in fresh medium (without glucose) and grown until the optical density at 600 nm (OD600) was about 0.2. l-Arabinose was then added, and the cultures were grown for another 2 h. Repressed controls were maintained in glucose-containing medium.
RT-PCR.
Total RNA was isolated from A. hydrophila AH-3 grown at 20 and 37°C in liquid medium (TSB) by using Trizol reagent (Invitrogen). To ensure that RNA was devoid of contaminating DNA, the preparation was treated with RNase-free DNase I (amplification grade; Invitrogen). First-strand cDNA was obtained using 1 μg of DNase-digested total RNA, random primers, and the Thermoscript reverse transcription (RT)-PCR system (Invitrogen) according to the manufacturer's instructions. A PCR without reverse transcriptase was also performed to confirm the absence of contaminating DNA in the RNA samples. The RT-PCR and PCR products were analyzed by agarose gel electrophoresis. Semiquantitative PCR provides image estimates of the amounts of amplified bands before the plateau is reached. For semiquantitative PCR, first-strand cDNA was obtained as described previously, and PCR amplification was performed using specific primers A3-B1 (5′-AATCCGGTATCCGCAAGAC-3′) and A3-CF2 (5′-TAACGCGCATCCTGATAG-3′) to amplify a 1,126-bp DNA fragment from the rmlB-rmlC region and primers GSP2-O34 (5′-GTCGAAAGAGCTGAAAACGG-3′) and GSP3-O34 (5′-GCTGGCTACATCAGATAATGAG-3′) to amplify a 253-bp DNA fragment from the wzz internal region. PCRs were performed using 100-μl mixtures containing 1× PCR buffer (Ecogen), 2 mM MgCl2, 1 μl of cDNA as the template, 1 μM of each primer, 200 μM of each deoxynucleoside triphosphate, and 2 U of Taq DNA polymerase (Ecogen). The following PCR program was used: preincubation at 94°C for 1 min and then 35 cycles of denaturation at 94°C for 45 s, annealing at 56°C for 30 s, and extending at 72°C for 1 min. To analyze the amount of cDNA template, 15-μl aliquots were removed for each PCR sample every five cycles starting at cycle 15 and ending at cycle 35. Amplicons at each time point were compared by using agarose gel electrophoresis with ethidium bromide staining. A. hydrophila ribosomal protein 16S rRNA primers were used as a control for the amount of cDNA template.
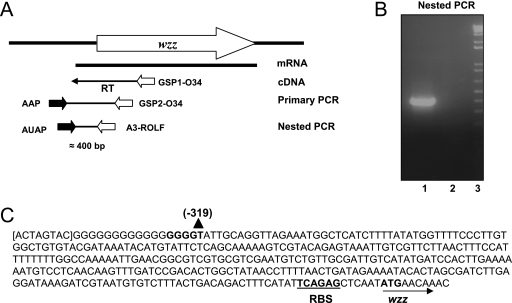
Mapping the transcription start site for the A. hydrophila AH-3 wzz gene by 5′ RACE PCR.
The A. hydrophila AH-3 wzz cDNA 5′ end was amplified using the 5′ random amplification of cDNA ends (RACE) system (version 2.0; Invitrogen). Total RNA was isolated from A. hydrophila AH-3, and 5 μg was pretreated with RNase-free DNase I (amplification grade; Invitrogen) used according to the manufacturer's protocol. The absence of contaminating DNA in the RNA samples was confirmed by PCR. First-strand cDNA was synthesized using the entire volume of DNase-digested total RNA (5 μg), the wzz internal primer GSP1-O34 (5′-TTATCTTCGGTTATACCAGATGC-3′), and the Thermoscript RT-PCR system (Invitrogen) at 57°C for 45 min. Reverse transcriptase was deactivated at 85°C for 5 min, and 1 μl of RNase H was then added and incubated at 37°C for 20 min. Purification of cDNA with S.N.A.P. columns and tailing of purified cDNA using terminal deoxynucleotidyl transferase and dCTP were done according to the 5′ RACE system (version 2.0) instructions. cDNA was confirmed after each step by PCR using nested primers GSP2-O34 and GSP3-O34.
Tailed cDNA was amplified by primary PCR using 10 μM of each primer, the 5′ RACE abridged anchor primer (primer AAP) and GSP2-O34. The PCR program used was 94°C for 1 min, followed by 35 cycles of 94°C for 45 s, 58°C for 30 s, and 72°C for 2 min and then by extension at 72°C for 5 min. Nested PCR was performed using the primary PCR product diluted 1:100 as the template and 10 mM of each nested primer abridged with the universal amplification primer AUAP and A3-ROLF (5′-TGCGGCAACATCTTATCC-3′). The PCR program used was 94°C for 1 min, followed by 35 cycles of 94°C for 45 s, 55°C for 30 s, and 72°C for 2 min and then by extension at 72°C for 5 min. PCR products were analyzed by agarose gel electrophoresis, and amplified bands were excised from the gel, purified, and sequenced with the A3-ROLF primer.
Construction of lacZ fusions to analyze promoter function.
Transcriptional fusions were constructed by inserting the DNA fragments into plasmid pRS550 (51). This plasmid carries a cryptic lacZ operon and genes that confer resistance to both kanamycin and ampicillin. To construct the wzzAH-3 promoter-lacZ fusion, a 220-bp DNA fragment, located 262 bp upstream of the wzz start codon, was amplified by PCR from genomic A. hydrophila AH-3 DNA using primers P34wzzF1A (5′-CGCGGATCCAGGTGAGTTTGGCCCTAAAA-3′) and P34wzzR1A (5′-CCGGAATTCGTACACAGCAAGGGAAA-3′). To construct the wzzPPD134/91 promoter-lacZ fusion, a 175-bp DNA fragment, located 19 bp upstream of the wzz start codon, was amplified by PCR from genomic A. hydrophila PPD134/91 (34) DNA using primers PPwzzF (5′-CGCGGATCCTCTTGATGCAATCAGAATCAGC-3′) and PPwzzR (5′-CCGGAATTCCCAATAACGACATGCCACAC-3′). PCR products were digested with BamHI and EcoRI (sites underlined) and ligated into pRS550 digested with both endonucleases for correct insert orientation. Transcriptional fusions were electroporated into E. coli DH5α cells and plated on LB agar containing ampicillin and kanamycin to obtain plasmids pRS-PWZZA and pRS-PWZZP, respectively. Recombinant colonies were confirmed by PCR using specific primers from the DNA insertion fragments.
Plasmids pACYC-PWZZA and pACYC-PWZZP were constructed by digestion of plasmids pRS-PWZZA and pRS-PWZZP, respectively, with StuI. StuI digestion generated three bands, and the 6.3-kb band was purified and ligated to HindIII-SalI-digested pACYC184, blunt ended, and dephosphorylated. Transcriptional fusions were electroporated into E. coli DH5α cells and selected on LB medium plates containing chloramphenicol. Recombinant colonies selected by using resistance to chloramphenicol and sensitivity to tetracycline were confirmed by PCR using specific primers from the DNA insertion fragments and then transferred to the A. hydrophila AH-405 rifampin-resistant strain by triparental mating.
β-Galactosidase assay.
Bacteria were grown overnight in LB medium, subcultured (1:200) in fresh medium, and incubated at 20 or 37°C until the OD600 was 0.4. β-Galactosidase activity (in Miller units) was calculated as follows (21, 39): A420/OD600 × 1,000/(assay time [in min] × volume [in ml]).
Nucleotide sequence accession number.
The complete nucleotide sequence of the A. hydrophila AH-3 wb*O34 gene cluster described here has been deposited in the GenBank database under accession number EU274663.
RESULTS
Kanamycin- and rifampin-resistant transconjugants resulting from mating between A. hydrophila AH-405 and E. coli S17-1λpir(pUTminiTn5Km1) were initially immunoscreened by colony blotting with antibodies against O34-antigen LPS obtained previously (36). Colonies that were negative in this colony blot analysis were also tested for sensitivity to bacteriophage PM1, a bacteriophage specific for this antigen (37). Among the approximately 1,500 transconjugants screened, two mutants (AH-901 and AH-902) were unable to react with specific antiserum against Aeromonas O34-antigen LPS, were resistant to bacteriophage PM1, and were characterized further. LPS gel (Fig. 2A and B) and Western blot analyses using the same antiserum showed that these mutants lacked the O34-antigen LPS. Southern blot analysis using a probe specific for the transposon demonstrated that each mutant had a single copy of the mini-transposon in its genome. The DNA flanking the transposon insertion of each mutant was isolated and cloned into pBCSK (see Materials and Methods). Kanamycin- and chloramphenicol-resistant colonies obtained from AH-901 EcoRI restriction and AH-902 EcoRV restriction had 2.1- and 2.2-kb insertion fragments, respectively. Sequence analysis of the fragment from AH-901 revealed an ORF that shared homology with different dTDP-4-dehydrorhamnose 3,5-epimerases (RmlC proteins) involved in dTDP-l-rhamnose or 6-deoxy-l-talose (l-6dTal) biosynthesis, while the same analysis for AH-902 revealed an ORF that shared homology with different mannose-1-phosphate guanylyltransferases (ManC proteins) involved in the biosynthesis of GDP-d-mannose. DNA probes that were specific for the rmlC- and manC-like genes were used to screen a previously constructed genomic library of A. hydrophila AH-3 (43). A single cosmid (COS-O34) containing intact copies of both genes was isolated.
FIG. 2.
(A) LPS from A. hydrophila AH-3 (wild type) (lane 1), mutants AH-901, AH-902, AH-405ΔwecA, and AH-405Δwzy (lanes 2, 3, 4, and 5, respectively), mutants AH-901 and AH-902 complemented with plasmids COS-O34 and AH-405ΔwecA, respectively (lanes 6 and 7, respectively), and mutant AH-405Δwzy complemented with pBAD33-wecA and pBAD33-wzy (lanes 8 and 9, respectively). LPS was extracted and analyzed by 12% SDS-PAGE as described by Darveau and Hancock (14). (B) SDS-Tricine-PAGE gels for LPS core of mesophilic Aeromonas strains. The lanes contained the same strains as the lanes in panel A. (C) LPS from A. hydrophila AH-3 (wild type) (lane 1), E. coli K-12 strain CLM4 (lane 2), strain CLM4 with the COS-O34 plasmid (lane 3), and strain CLM4 with the COS-O34 and pGEMT-Gne plasmids (lane 4). All the strains were grown at 20°C, and strains with pBAD33 plasmids were grown under induced conditions.
COS-O34 plasmid DNA conferred O34-antigen LPS production to mutants devoid of the O34-antigen LPS (AH-901 and AH-902 [Fig. 2A and B]) but not to E. coli strains DH5α (19) and CLM4 (30), which do not have the epimerase that converts UDP-GlcNAc to UDP-GalNAc. However, when the COS-O34 plasmid was coexpressed in the two E. coli strains with plasmid pGEMT-Gne (carrying the complete A. hydrophila gne gene [9]), the E. coli strains were able to produce O34-antigen LPS (Fig. 2C). No such O34-antigen LPS production in E. coli strains was obtained with the plasmid vector(s) alone.
Sequence analysis of the A. hydrophila AH-3 wb* gene cluster.
The nucleotide sequence of the plasmid COS-O34 insert was determined in order to identify the A. hydrophila AH-3 genes conferring O34-antigen production. An 18,156-bp sequence was determined in both directions by using oligonucleotides complementary to cosmid pLA2917 (43) sequences flanking the COS-O34 DNA insert. Other sequence-derived oligonucleotides were purchased (Amersham-Pharmacia Biotech) and used to complete the nucleotide sequence. Analysis of the sequenced region revealed 17 ORFs (Fig. 3A) transcribed in the same direction. Upstream or downstream of the last ORF no other ORFs involved in LPS biosynthesis were identified. These results suggest that this sequence corresponds to the O34-antigen wb* gene cluster. In all cases, putative Shine-Dalgarno sequences were found upstream of all ORF start codons. Computer analysis of the wb*O34 gene cluster sequence revealed a conserved JUMPstart sequence with the 8-bp ops (operon polarity suppressor) sequence (GGCGGTAG) 120 bp upstream from ORF1 required. The ops sequence is recognized by the bacterial antiterminator RfaH, which can be recruited by the transcription elongation complex to reduce pausing and termination at intergenic sites of polycistronic operons, allowing RNA polymerase to conclude transcription of the distal genes in large operons (4, 6). Also, a putative promoterlike sequence between ORF16 (rmlD) and ORF17 (wzz) was observed. Two rho-independent possible transcriptional terminator sequences downstream of ORF16 and ORF17 were identified (Fig. 3A). RT-PCR using specific primers and total RNA from A. hydrophila AH-3 grown in TSB at 20 and 37°C demonstrated that there was amplification between ORF1 and ORF16 (rmlB to rmlD) in both conditions. However, no amplification was obtained with oligonucleotide pairs from ORF16 (rmlD) to ORF17 (wzz), confirming that wzz is transcribed independently (Fig. 3A).
FIG. 3.
(A) Genetic organization of the A. hydrophila AH-3 wb*O34 region from the COS-O34 plasmid. The transcription direction and stops (lollipop symbols) are indicated. The positions of mini-Tn5 (AH-901 and AH-902) mutations are indicated. (B) Comparison of A. hydrophila PPD134/91 (wb*O18 cluster) with A. hydrophila AH-3 (wb*O34 cluster) from ORF1 to ORF17. aa, amino acids.
Analysis of the ORF deduced amino acid sequence.
The DNA sequence was translated in all six frames, and all ORFs were inspected. Computer database searching was carried out to tentatively identify the sequenced genes. Proteins similar to each ORF gene product were analyzed to determine the levels of similarity and identity (Table 2). This analysis revealed three sugar synthesis pathways, pathways for l-rhamnose, l-6dTal, and d-mannose. ORF1-, ORF2-, ORF3-, and ORF16- encoded proteins were homologous to RmlB (2), RmlA (57), RmlC, and RmlD (16, 17), respectively (Table 2). The products of rmlA through rmlD are responsible for the biosynthesis of dTDP-l-rhamnose from glucose-1-phosphate, and these four genes usually form a group in the wb* gene clusters (48). In this case, the order of the rml gene region was quite different because the rmlD gene was alone and completely separated from the other rml genes. The rmlC mutant (AH-901) is devoid of the O34-antigen LPS. The similarity searches for ORF4 showed that the protein encoded by this gene was similar to several enzymes, all of which had dTDP-6-deoxy-l-lyxo-hexulose reductase activity responsible for biosynthesis of the dTDP-6dTal from dTDP-4-keto-6-deoxy-l-mannose (Table 2). The dTDP-6-deoxy-l-lyxo-hexulose reductase (Tll) and the dTDP-l-rhamnose synthase (RmlD) catalyze the reduction of the same substrate for synthesis of dTDP-l-6dTal and dTDP-l-rhamnose, respectively. In Actinobacillus actinomycetemcomitans, the pathway for dTDP-l-6dTal biosynthesis has been characterized and involves the products of rmlA, rmlB, rmlC, and tll (29). This result suggested that ORF4 could correspond to the A. hydrophila tll gene. ORF13- and ORF14-encoded proteins were homologous to mannose-1-phosphate guanylyltransferase (ManC) and phosphomannomutase (ManB), respectively (26, 54) (Table 2). These two proteins work together to synthesize GDP-mannose from mannose-6-phosphate (48), a hexose sugar that is frequently found in O antigens of E. coli, Salmonella enterica, and other gram-negative bacteria. The manC mutant (AH-902) is devoid of the O34-antigen LPS.
TABLE 2.
Characteristics of the A. hydrophila AH-3 wb*O34 region
| ORF | Product | Nucleotide position | Protein size (amino acids) | Conserved domain(s) | Homologous protein (accession no.) | % Identity/ % similarity |
|---|---|---|---|---|---|---|
| ORF1 | RmlB | 322-1407 | 361 | NAD-dependent epimerase/dehydratase family (PF01370) | dTDP-glucose 4,6-dehydratase, A. hydrophila PPD134/91 (AAM74474) | 92/95 |
| dTDP-glucose 4,6-dehydratase, E. coli ATCC 8739 (ACA77260) | 82/89 | |||||
| ORF2 | RmlA | 1407-2270 | 287 | Nucleotidyl transferase (PF00483) | Glucose-1-phosphate thymidylyltransferase, A. hydrophila PPD134/91 (AAM74475) | 95/97 |
| Glucose-1-phosphate thymidylyltransferase, Yersinia pestis KIM (AMM83956) | 75/86 | |||||
| ORF3 | RmlC | 2283-2834 | 183 | dTDP-4-dehydrorhamnose 3,5-epimerase (PF00908) | dTDP-4-dehydrorhamnose 3,5-epimerase, A. hydrophila PPD134/91 (AAM74476) | 93/96 |
| dTDP-4-dehydrorhamnose 3,5-epimerase, E. coli S88 (CAN87671) | 60/75 | |||||
| ORF4 | Tll | 2837-3664 | 275 | NAD-dependent epimerase/dehydratase family (PF01370) | dTDP-6-deoxy-l-lyxo-hexulose reductase, E. coli (AAZ20758) | 56/72 |
| ORF5 | Wzx | 3664-5151 | 495 | Polysaccharide biosynthesis protein (PF01943) | O-unit flippase, A. hydrophila PPD134/91 (AAM74478) | 37/59 |
| O-antigen flippase, Salmonella enterica (AAK83016) | 26/45 | |||||
| ORF6 | WbxB | 5155-5661 | 168 | Bacterial transferase hexapeptide (PF00132) | O-Acetyltransferase, E. coli (AAZ20764) | 33/54 |
| ORF7 | WbxC | 5649-6245 | 198 | Bacterial transferase hexapeptide (PF00132) | O-Acetyltransferase, A. hydrophila PPD134/91 (AMM74480) | 35/55 |
| Acetyltransferase, Enterobacter sp. strain 638 (ABP61321) | 62/77 | |||||
| ORF8 | WbxD | 6230-7132 | 300 | Glycosyl transferase family 2 (PF00535) | Rhamnosyl transferase, A. hydrophila PPD134/91 (AAF45033) | 87/93 |
| Glycosyl transferase family 2, Enterobacter sp. strain 638 (ABP61315) | 44/63 | |||||
| ORF9 | Wzy | 7134-8381 | 415 | Integral membrane protein, A. hydrophila PPD134/91 (AAM74481) | 69/83 | |
| ORF10 | WbxE | 8378-9415 | 345 | Glycosyl transferase group 1 (PF00534) | Mannosyltransferase, A. hydrophila PPD134/91 (AAF45034) | 84/91 |
| Mannosyltransferase-like, Yersinia pestis KIM (NP668407) | 53/69 | |||||
| ORF11 | WbxF | 9419-10018 | 199 | Bacterial transferase hexapeptide (PF00132) | O-Acetyltransferase, A. hydrophila PPD134/91 (AAM74482) | 83/90 |
| Acyltransferase, E. coli W3110 (BAA15875) | 44/61 | |||||
| ORF12 | WbxG | 10008-11006 | 332 | Glycosyltransferase, E. coli (AAZ20765) | 27/50 | |
| ORF13 | ManC | 11008-12459 | 483 | Nucleotidyl transferase (PF00483) | Mannose-1-phosphate guanylyltransferase, A. hydrophila PPD134/91 (AAM74484) | 88/94 |
| Mannose-1-phosphate guanylyltransferase/mannose-6-phosphate isomerase, E. coli ATCC 8739 (ACA77251) | 63/82 | |||||
| ORF14 | ManB | 12452-13882 | 476 | Phosphoglucomutase/ phosphomannomutase (PF02878) | Phosphomannomutase, A. hydrophila PPD134/91 (AAF67004) | 82/90 |
| Phosphomannomutase, Salmonella enterica (Q00330) | 60/73 | |||||
| ORF15 | WecA | 14019-15212 | 397 | Bacterial sugar transferase (PF02397) | Undecaprenyl-galactosyl transferase, Leptospira borgpetersenii L550 (YP-797655) | 51/69 |
| ORF16 | RmlD | 15219-16121 | 300 | RmlD substrate binding domain (PF04321) | dTDP-4-dehydrorhamnose reductase, A. hydrophila PPD134/91 (AAM74486) | 73/81 |
| dTDP-4-dehydrorhamnose reductase, Salmonella enterica ATCC 9150 (YP150075) | 41/60 | |||||
| ORF17 | Wzz | 16632-17714 | 360 | Chain length determinant protein (PF02706) | Chain length determinant, A. hydrophila PPD134/91 (AAM74487) | 80/90 |
| Chain length determinant, E. coli K-12 (NP415119) | 30/47 |
Seven ORFs were identified as ORFs that encode different transferases. ORF6, ORF7, and ORF11 encoded proteins similar to O-acetyltransferases and were designated wbxB, wbxC, and wbxF, respectively (Table 2). These three transferases have a repeat structure composed of tandem repeats of an (LIV)GX4 hexapeptide that was found in different transferase protein families (44, 52). WbxB and WbxF each contain three hexapeptide tandem repeats (positions 66 to 83, 105 to 122, and 123 to 140 and positions 88 to 105, 132 to 149, and 150 to 167, respectively), and WbxC contains five hexapeptide tandem repeats (positions 48 to 65, 73 to 90, 93 to 110, 134 to 151, and 152 to 169). Furthermore, WbxB, WbxC, and WbxF have conserved domains for acetyltransferases (GenBank accession number COG0110) and acyltransferases (GenBank accession number PRK09677). These three proteins may play a role in the addition of acetyl groups at different positions of dTDP-l-6dTal, since A. hydrophila O34 antigen is acetylated by three acetyl groups. 6dTalI is O acetylated stoichiometrically at position 2, and 6dTalII has two O-acetyl groups (25). On the basis of the A. hydrophila O34-antigen structure, we expected three sugar transferase genes. ORF8, ORF10, and ORF12 encoded proteins that showed homology to different rhamnosyl transferases, mannosyl transferases, and glycosyl transferases, respectively (Table 2). These proteins may function in transfer of the synthesized nucleotide sugar monomers to the growing O unit, and these three genes were designated wbxD, wbxE, and wbxG, respectively. WbxD has a glycosyl transferase family 2 domain, WbxE has a glycosyltransferase family 1 domain (7), and WbxG does not have conserved domains. The ORF15 product showed similarities with various UDP-galactose-lipid carrier transferases and galactosyltransferases, which catalyze the addition of galactose to an oligosaccharide precursor or a lipid intermediate and have a bacterial sugar transferase domain (Pfam accession number PF02397) (Table 2). Hydrophobicity analysis and identification of four putative transmembrane domains (amino acid residues 24 to 46, 53 to 75, 85 to 102, and 214 to 235) suggest that this protein is an integral membrane protein and could be a putative transferase (WecA) involved in the initiation of the assembly of the O units.
This cluster also contains three saccharide-processing genes which encode proteins that have potential transmembrane domains. ORF5 was designated wzx because of the levels of similarity and identity of its product to O-antigen flippases involved in O-antigen translocation across the membrane, as well as because of the presence of a polysaccharide biosynthesis protein domain (Pfam accession number PF01943) (Table 2). Furthermore, hydrophobicity analysis and identification of 14 putative transmembrane domains of the Wzx protein (amino acid residues 12 to 34, 44 to 66, 87 to 107, 127 to 149, 162 to 179, 184 to 206, 227 to 249, 259 to 281, 303 to 325, 345 to 367, 380 to 397, 402 to 424, 436 to 455, and 460 to 482) suggest that this protein is indeed an integral membrane protein (31). The ORF9-encoded protein showed homology to different Wzy proteins involved in polymerization of the O-antigen polysaccharide. Hydrophobicity analysis and identification of 10 transmembrane domains (amino acid residues 7 to 29, 33 to 55, 68 to 85, 100 to 122, 129 to 148, 163 to 185, 194 to 216, 226 to 244, 331 to 353, and 377 to 399) with a high periplasmic loop and the carboxy-terminal region located on the cytoplasmic face of the membrane suggest that this protein is also an integral membrane protein. Furthermore, an RX2L motif (R99 to L102) at the end of periplasmic loop 2 and an HX11G motif (H308 to G320) at periplasmic loop 4 were found, similar to other O polymerases (13, 50). These results suggest that the role of the Wzy protein is as an O34-antigen polymerase in a Wzy-dependent biosynthetic pathway. The protein encoded by the last gene in the cluster showed similarity to some chain length determinants (Wzz) from different bacteria and had a chain length determinant protein domain (Pfam accession number PF02706) between amino acids 16 and 155 characteristic of the Wzz family. This protein has a chain length determinant protein domain characteristic of the Wzz family between amino acids 16 and 155, two potential transmembrane domains (amino acid residues 41 to 63 and 332 to 354) in the N- and C-terminal regions, and a large hydrophilic central domain in the periplasmic face of the plasma membrane with different putative coiled-coil regions, like proteins that belong to the polysaccharide copolymerase superfamily (53). The similarity and topology suggested the possible role of this wzz gene in regulation of the A. hydrophila O34-antigen chain length.
Characterization of LPS from wild-type and mutants cells grown at 20 and 37°C.
We isolated total LPS from wild-type strain AH-3 and derived mutants grown at 20 and 37°C in TSB. We used a relationship between Glc (a unique sugar residue in the LPS core [Fig. 1]) and Man (a unique sugar residue in the O-antigen repeating unit [Fig. 1]) to establish the ratio of LPS core to O34 repeating units. No Man was observed in the LPS of polar mutants (AH-901 and AH-902) and AH-405ΔwecA grown at either 20 or 37°C, which was characteristic of a complete lack of O34-antigen LPS (Fig. 2A and B). A ratio of 1/1 was observed for the LPS of AH-405Δwzy grown at either 20 or 37°C, which was characteristic of wzy mutants with a single repetition of the O34-antigen LPS (Fig. 2A and B). The LPS of the AH-405Δwzz strain grown at either 20 or 37°C showed a ratio of 1/4, indicating an average of four O34-antigen units for each LPS core molecule with the distribution shown in Fig. 4. Finally, the LPS of AH-3 (wild type) grown at 20°C showed a ratio of approximately 1/20, which indicated that on average there were approximately 20 O34-antigen units per LPS core molecule with the distribution shown in Fig. 4. However, the LPS of the same strain grown at 37°C showed a ratio of approximately 1/2, with the distribution shown in Fig. 4.
FIG. 4.
Electrophoresis of LPS from A. hydrophila AH-3 (wild type) (lanes 1 and 4), mutant AH-405Δwzz (lanes 2 and 5), and mutant AH-405Δwzz complemented with plasmid pBAD33-wzz (lanes 3 and 6). The strains in lanes 1, 2, and 3 were grown at 20°C, while the strains in lanes 4, 5, and 6 were grown at 37°C. The arrow in lane 4 indicates the small amount of large O34-antigen LPS repeating units observed in the wild-type strain grown at 37°C. The strain with the pBAD33-wzz plasmid was grown under induced conditions.
The subpopulations of LPS molecules as they appeared in gels for the wild-type strain grown at 20 and 37°C are completely different. R-form LPS contains lipid A and core (marked by Glc in A. hydrophila AH-3, as previously stated), and S-form LPS contains the R-form plus the O-antigen repeating units (marked by Man in A. hydrophila AH-3, as previously mentioned). At 20°C there is a subpopulation of the S-form of LPS with a different amount of O34-antigen repeating units (the average number was 20, but this included LPS molecules with 1 to >40 repetitions). However, at 37°C the LPS molecule subpopulation is formed mainly by R-form LPS (LPS core), occasionally with some molecules with S-form LPS with a very high number of O34-antigen repeating units, resulting in an average of two repeating units that are not distributed in a uniform way (Fig. 4). This fact could be correlated with the downregulation of the wzz gene observed at 37°C (see below) and the role of Wzz from A. hydrophila wb*O34 in the distribution of the chain repetitions. It is important to point out that the AH-405Δwzz mutant had an LPS molecule subpopulation of S-forms with from 1 to approximately 10 O34-antigen repeating units, whether it was grown at 20 or 37°C (Fig. 4).
Most of the mutants (AH-901, AH-902, AH-405ΔwecA, and AH-405Δwzy) exhibited the A. hydrophila AH-3 LPS phenotype when they were complemented with the corresponding genes in the pBAD33 plasmid, when they were grown at either 20 or 37°C. However, this was not the case for AH-405Δwzz mutant, as shown in Fig. 4. Mutant AH-405Δwzz complemented with pBAD33-wzz produced the same LPS profile whether it was grown at 20 or 37°C, which corresponded to the LPS profile of A. hydrophila AH-3 grown at 20°C, with a ratio of Glc to Man of approximately 1/20 and the distribution of the subpopulation of LPS molecules shown in Fig. 4. It is important to note that in this case the wzz gene is under the control of the promoter from the plasmid vector.
A. hydrophila AH-3 wb*O34 regulation by temperature.
To analyze whether the transcription of the wb*O34 gene cluster is temperature regulated, a semiquantitative RT-PCR analysis of the rmlB-rmlC region and the wzz gene for cultures grown at 20 and 37°C was carried out. The results showed that the transcription of the rmlB-rmlC region at 20°C was similar to that at 37°C, in contrast to wzz transcription, which was significantly higher at 20°C than at 37°C (Fig. 5A), suggesting that at 37°C transcription of wzz was downregulated.
FIG. 5.
(A) RT-PCR DNA fragments obtained from the rmlB-rmlC region and wzz internal region of A. hydrophila AH-3 genomic DNA isolated when the strain was grown at 20 and 37°C. (B) β-Galactosidase activities of E. coli DH5α with the pRS-PWZZA (wzzAH-3 promoter-lacZ fusion) and pRS-PWZZP (wzzPPD134/91 promoter-lacZ fusion) plasmids and of A. hydrophila AH-405 with the pACYC-PWZZA (wzzAH-3 promoter-lacZ fusion) and pACYC-PWZZP (wzzPPD134/91 promoter-lacZ fusion) plasmids were determined for cultures grown at 20°C (open bars) and 37°C (shaded bars) (19, 36). As a control we also determined the activities of E. coli DH5α with the pRS550 plasmid vector and A. hydrophila AH-405 with pACYC-lacZ at both growth temperatures.
RACE was used to determine the exact location of the A. hydrophila AH-3 wzz promoter (Fig. 6). Primary PCR of tailed cDNA using primers AAP (abridged anchor primer) and GSP2-O34 failed to give the expected amplification bands, but nested PCR using primers AUAP (abridged universal amplification primer) and A3-ROLF resulted in one unique DNA band, at approximately 400 bp. The DNA sequence of the amplified band indicated that it was tailed with G residues and that the wzz transcription start site was located at nucleotide −319 upstream of the wzz translation start site. However, due to the C tailing, it is not possible to rule out the possibility that the start site is located at the G residue at nucleotide −320, −321, −322, or −323 upstream of the wzz gene.
FIG. 6.
Amplification of the A. hydrophila AH-3 wzz cDNA 5′ end performed using the 5′ RACE system (version 2.0; Invitrogen). (A) RACE scheme showing the primers and products described in Materials and Methods. (B) Amplicon obtained by nested PCR using primers AUAP and A3-ROLF. Lane 1, primary PCR template; lane 2, PCR negative control; lane 3, molecular weight standard (Invitrogen). (C) Location of the transcriptional start site (GGGGT) for A. hydrophila AH-3 wzz (indicated by bold type). RBS, ribosome binding site.
The β-galactosidase activities at 20 and 37°C of the pRS- PWZZA transcriptional fusion, containing the A. hydrophila AH-3 wzz 5′ RACE predicted promoter, and the pRS- PWZZP transcriptional fusion, carrying the A. hydrophila PPD134/91 intergenic region between rmlD and wzz, were monitored and compared with that of the E. coli DH5α host strain with the pRS550 plasmid vector alone. As shown in Fig. 5B, the β-galactosidase activity of pRS-PWZZA was about four times greater at 20°C than at 37°C, in contrast to pRS-PWZZP, which showed similar β-galactosidase activities at the two temperatures. The background levels for E. coli DH5α with the pRS550 vector alone were not significant and were similar at the two temperatures. Furthermore, the β-galactosidase activities at 20 and 37°C of the pACYC-PWZZA and pACYC-PWZZP transcriptional fusions in A. hydrophila AH-405 were monitored and compared with those of pACYC-lacZ in the same A. hydrophila strain. Fusion pACYC-PWZZA in A. hydrophila AH-405 exhibited six- to sevenfold greater β-galactosidase activity at 20°C than at 37°C. Fusion pACYC-PWZZP exhibited similar levels of β-galactosidase activity at the two temperatures. A. hydrophila AH-405 with pACYC-lacZ did not show significant β-galactosidase activity. These results confirm that there is a DNA sequence in the upstream region of the A. hydrophila AH-3 wzz gene that is able to function as a temperature-regulated promoter.
Several Aeromonas O18 strains tested (n = 6) showed identical LPS profiles in gels when they were grown at 20 or 37°C with full expression of O-antigen LPS. All these strains were unable to hybridize with the wzzAH-3 promoter. All the A. hydrophila O34 strains tested (n = 14) showed LPS profiles similar to those of strain AH-3 at 20 and 37°C, and all of them showed hybridization with the wzzAH-3 promoter.
DISCUSSION
The O34-antigen wb* gene cluster had a G+C content of 44.3%, which is lower than the value expected for species belonging to the genus Aeromonas (57 to 65%) and is characteristic of some wb* clusters that usually have G+C contents at least 10% less than the species average. The encoded proteins are consistent with the chemical structure of the O34-antigen LPS (Fig. 1A); there are biosynthetic genes for the production of the monosaccharide residues mannose (d-Man) and l-6dTal corresponding to ORF13 and ORF14 and to ORF1 to ORF4, respectively, while genes encoding the WbxE (ORF10) and WbxD (ORF8) transferases are also present. There are also three putative acetyltransferases (WbxB, WbxC and WbxF, encoded by ORF6, ORF7, and ORF11, respectively), which is in agreement with the three different acyl residues found in the O34-antigen LPS chemical structure. Two main known pathways for O-antigen export have been established (45), the Wzy-dependent pathway for heteropolysaccharide structures and the ABC2 transporter-dependent pathway mainly for homopolysaccharides. The presence of ORF5 (Wzx) and ORF9 (Wzy) showed that O34-antigen LPS belongs to the first pathway. Furthermore, the findings are in agreement with the fact that a Wzy nonpolar mutant has only a single O34-antigen LPS repetition. To synthesize O antigens, monomers are assembled on a lipid carrier (undecaprenol phosphate) by enzymes encoded in the wb* gene cluster before their incorporation into the LPS molecule; the ORF15 product is an undecaprenol-sugar-P-transferase (WecA) able to initiate assembly of the O34-antigen units, as the nonpolar mutant is unable to produce this antigen. Although the O34 repeat unit presumably begins with N-acetylgalactosamine (d-GalNAc) transfer, the A. hydrophila WecA protein has a topology similar to that of WbaP: an N-terminal region containing three transmembrane helices, a large central periplasmic loop, and a C-terminal domain containing the last transmembrane helix and a large cytoplasmic tail, which have sugar phosphate transferase activity (46). Nevertheless, the d-GalNAc residue should be incorporated by a transferase into the O34 repeating unit, and WbxG (ORF12) is the most likely candidate. ORF16 (RmlD) is necessary for the biosynthesis of rhamnose (not present in O34-antigen LPS) but not for the biosynthesis of l-6dTal (42), which is in agreement with the finding that a mutation in this gene does not alter the O34-antigen LPS (data not shown). The biosynthesis of l-rhamnose is characterized by the presence of four genes (rmlA to rmlD) that are always adjacent, but in this case rmlD seems to be replaced by tll (for l-6dTal biosynthesis [42]) and rmlD is displaced to the end of the O34 wb* cluster. The ORF17 product is a Wzz protein that is able to regulate the O34-antigen chain length, as described for other bacteria (41). The periplasmic face of this protein had different putative coiled-coil regions which in other bacteria are essential for its function, because it interacts at least with Wzy and the O antigen, changing the Wzz conformation (18). Recently, Marolda et al. suggested that the region predicted to be coiled coils is important for Wzz function because the coiled coils maintain the native conformation of the protein (32). The Wzz protein becomes the protein regulated by temperature in O34-antigen LPS formation.
Genes of the A. hydrophila wb*O34 cluster are distributed in two independent transcripts: ORF1 to ORF16 and ORF17. Semiquantiative RT-PCR, transcriptional fusion with lacZ, and the β-galactosidase activities when a strain was grown at 20 and 37°C demonstrated that the transcription levels of ORF1 to ORF16 were similar at the two temperatures and that there was differential transcription of ORF17 (more transcription at 20°C than at 37°C). The complete DNA sequence for Aeromonas O-antigen LPS from strain PPD134/91 (serotype O18) was described previously (56) and is very similar to the A. hydrophila AH-3 wb*O34 sequence. Comparison of these two O-antigen clusters showed that most ORFs exhibit levels of amino acid and nucleotide identity greater than 80 and 74%, respectively, although some ORFs (ORF9, ORF14, ORF15, 1 ORF6, and ORF17) show slightly reduced levels of identity. ORF5, ORF6, and ORF7, which encode an O-antigen flippase and two O-acetyltransferases, respectively, exhibit lower levels of amino acid and nucleotide identity (Fig. 3B). However, the chemical O-antigen LPS structure of strain PPD134/91 is unknown, and no mutants of this strain were isolated and characterized. Nevertheless, the O-antigen LPS chemical structure of this strain should be similar to that of strain AH-3 due to the high levels of similarity of many of the genes. Also, the cross-reactivity between serotype O18 and O34 Aeromonas strains is well known. The PPD134/91 strain showed identical LPS profiles when it was grown at 20 and at 37°C, with clear O-antigen LPS repetitive bands. When we analyzed the DNA sequence of strain PPD134/91, the distance between ORF16 (RmlD) and ORF17 (Wzz) is less than the distance between these ORFs in strain AH-3. Transcriptional fusions carrying the A. hydrophila PPD134/91 intergenic region between rmlD and wzz showed similar β-galactosidase activities when the bacterium was grown at 20°C and at 37°C.
Furthermore, the wzz transcription start site in strain AH-3 (nucleotide −319) located by RACE is in an area which is not present in strain PPD134/91 due to its smaller size. This fact explains the differences in production of the O-antigen LPSs at different growth temperatures in these strains; and while the region with the wzz transcription start site in strain AH-3 located by RACE was found in several Aeromonas serotype O34 strains by DNA hybridization, negative results were obtained with two different Aeromonas serotype O18 strains tested (data not shown). The subpopulation of LPS molecules shown in gels for wild-type strain AH-3 grown at 20 and 37°C (Fig. 4) could be correlated with the downregulation of the wzz gene observed at 37°C and the role of Wzz from A. hydrophila wb*O34 in the distribution of the chain repetitions. The different amounts and distributions of the O34-antigen LPS repeating units are closely related to the susceptibility to nonimmune serum. Thus, AH-3::rmlC and AH-3::manC mutants were sensitive to nonimmune serum, the AH-405Δwzy mutant with a single repetition of the O34 antigen was partially sensitive to nonimmune serum, and the AH-405Δwzz mutant was always more resistant to serum than AH-405Δwzy. The sensitivity to serum and the infectivity values for the AH-405Δwzz mutant grown at both temperatures are similar to those observed for AH-3 at 37°C. The O34-antigen LPS biosynthesis is also regulated by osmolarity (1); strain AH-3 is rough at 37°C with low osmolarity but smooth at 37°C with high osmolarity (1), and the virulence changes in relation to these environmental conditions and the presence of O34-antigen LPS.
Acknowledgments
This work was supported by Plan Nacional de I + D and FIS grants (Ministerio de Educación, Ciencia y Deporte and Ministerio de Sanidad, Spain) and by Generalitat de Catalunya. N.J., S.V., and R.C. received predoctoral fellowships from Generalitat de Catalunya and Universidad de Barcelona.
We thank Maite Polo for her technical assistance.
Footnotes
Published ahead of print on 11 April 2008.
REFERENCES
- 1.Aguilar, A., S. Merino, X. Rubires, and J. M. Tomás. 1997. The influence of osmolarity on lipopolysaccharide and virulence of Aeromonas hydrophila serotype O:34 strains grown at 37°C. Infect. Immun. 651245-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allard, S. T. M., M.-F. Giraud, C. Whitfield, M. Graninger, P. Messner, and J. H. Naismith. 2001. The crystal structure of dTDP-d-glucose 4,6-dehydratase (RmlB) from Salmonella enterica serovar Typhimurium, the second enzyme in the dTDP-l-rhamnose pathway. J. Mol. Biol. 307283-295. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Artsimovitch, I., and R. Landick. 2002. The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell 109193-203. [DOI] [PubMed] [Google Scholar]
- 5.Austin, B., and C. Adams. 1996. Fish pathogens, p. 197-243. In B. Austin, M. Altwegg, P. J. Gosling, and S. W. Joseph (ed.), The genus Aeromonas. John Wiley and Sons, New York, NY.
- 6.Bailey, M. J., C. Hughes, and V. Koronakis. 1997. RfaH and the ops element, components of a novel system controlling bacterial transcription elongation. Mol. Microbiol. 26845-851. [DOI] [PubMed] [Google Scholar]
- 7.Campbell, J. A., G. J. Davies, V. Bulone, and B. Henrissat. 1997. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem. J. 326929-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bateman, A., E. Birney, L. Cerruti, R. Durbin, L. Etwiller, S. R. Eddy, S. Griffiths-Jones, K. L. Howe, M. Marshall, and E. L. Sonnhammer. 2002. The Pfam protein families database. Nucleic Acids Res. 30276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Canals, R., N. Jiménez, S. Vilches, M. Regué, S. Merino, and J. M. Tomás. 2006. A gene (uridine diphosphate N-acetylgalactosamine 4-epimerase) is essential for mesophilic Aeromonas serotype O:34 virulence. Infect. Immun. 74537-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Canals, R., N. Jiménez, S. Vilches, M. Regué, S. Merino, and J. M. Tomás. 2007. The role of Gne and GalE in the virulence of Aeromonas hydrophila serotype O34. J. Bacteriol. 189540-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Canals, R., S. Ramirez, S. Vilches, G. Horsburgh, J. G. Shaw, J. M. Tomás, and S. Merino. 2006. Polar flagellum biogenesis in Aeromonas hydrophila. J. Bacteriol. 188542-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang, A. C. Y., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 1341141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daniels, C., C. Vindurampulle, and R. Morona. 1998. Overexpression and topology of the Shigella flexneri O-antigen polymerase (Rfc/Wzy). Mol. Microbiol. 281211-1222. [DOI] [PubMed] [Google Scholar]
- 14.Darveau, R. P., and R. E. Hancock. 1983. Procedure for isolation of bacterial lipopolysaccharides from both smooth and rough Pseudomonas aeruginosa and Salmonella typhimurium strains. J. Bacteriol. 155831-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galanos, C., O. Lüderitz, and O. Westphal. 1969. A new method for the extraction of R lipopolysaccharides. Eur. J. Biochem. 9245-249. [DOI] [PubMed] [Google Scholar]
- 16.Giraud, M.-F., F. M. Gordon, C. Whitfield, P. Messner, S. A. McMahon, and J. H. Naismith. 1999. Purification, crystallization and preliminary structural studies of dTDP-6-deoxy-d-xylo-4-hexulose 3,5-epimerase (RmlC), the third enzyme of the dTDP-l-rhamnose synthesis pathway, from Salmonella enterica serovar Typhimurium. Acta Crystallogr. Sect. D 55706-708. [DOI] [PubMed] [Google Scholar]
- 17.Giraud, M.-F., H. J. McMiken, G. A. Leonard, P. Messner, C. Whitfield, and J. H. Naismith. 1999. Overexpression, purification, crystallization and preliminary structural study of dTDP-6-deoxy-l-lyxo-4-hexulose reductase (RmlD), the fourth enzyme of the dTDP-l-rhamnose synthesis pathway, from Salmonella enterica serovar Typhimurium. Acta Crystallogr. Sect. D 552043-2046. [DOI] [PubMed] [Google Scholar]
- 18.Guo, H., K. Lokko, Y. Zhang, W. Yi, Z. Wu, and P. G. Wang. 2006. Overexpression and characterization of Wzz of Escherichia coli O86:H2. Protein Expr. Purif. 4849-55. [DOI] [PubMed] [Google Scholar]
- 19.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166557-580. [DOI] [PubMed] [Google Scholar]
- 20.Heeb, S., Y. Itoh, T. Nishijyo, U. Schnider, C. Keel, J. Wade, U. Walsh, F. O'Gara, and D. Haas. 2000. Small, stable shuttle vectors based on the minimalpVS1 replicon for use in Gram-negative plant-associated bacteria. Mol. Plant-Microbe Interact. 13232-237. [DOI] [PubMed] [Google Scholar]
- 21.Hernandez, V. J., and H. Bremer. 1990. Guanosine tetraphosphate (ppGpp) dependence of the growth rate control of rrnB P1 promoter activity in Escherichia coli. J. Biol. Chem. 26511605-11614. [PubMed] [Google Scholar]
- 22.Hitchcock, P. J., and T. M. Brown. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154269-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janda, J. M., L. S. Guthertz, R. P. Kokka, and T. Shimada. 1994. Aeromonas species in septicemia: laboratory characteristics and clinical observations. Clin. Infect. Dis. 1977-83. [DOI] [PubMed] [Google Scholar]
- 24.Janda, J. M., S. L. Abbott, S. Khashe, G. H. Kellogg, and T. Shimada. 1996. Further studies on biochemical characteristics and serologic properties of the genus Aeromonas. J. Clin. Microbiol. 341930-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janda, J. M., and S. L. Abbott. 1998. Evolving concepts regarding the genus Aeromonas: an expanding panorama of species, disease presentations, and unanswered questions. Clin. Infect. Dis. 27332-344. [DOI] [PubMed] [Google Scholar]
- 26.Jensen, S. O., and P. R. Reeves. 2001. Molecular evolution of the GDP-mannose pathway genes (manB and manC) in Salmonella enterica. Microbiology 147599-610. [DOI] [PubMed] [Google Scholar]
- 27.Knirel, Y. A., A. S. Shaskov, S. N. Senchenkova, S. Merino, and J. M. Tomás. 2002. Structure of the O-polysaccharide of Aeromonas hydrophila O34: a case of random O-acetylation of 6-deoxy-l-talose. Carbohydr. Res. 3371381-1386 [DOI] [PubMed] [Google Scholar]
- 28.Knirel, Y. A., E. Vinogradov, N. Jimenez, S. Merino, and J. M. Tomás. 2004. Structural studies on the R-type lipopolysaccharide of Aeromonas hydrophila. Carbohydr. Res. 339787-793. [DOI] [PubMed] [Google Scholar]
- 29.Mäki, M., and R. Renkonen.2004. Biosynthesis of 6-deoxyhexose glycans in bacteria. Glycobiology 141R-15R. [DOI] [PubMed] [Google Scholar]
- 30.Marolda, C. L., and M. A. Valvano. 1993. Identification, expression, and DNA sequence of the GDP-mannose biosynthesis region encoded by the O7 rfb gene cluster of strain VW187 (Escherichia coli O7:K1). J. Bacteriol. 175148-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marolda, C. L., J. Vicarioli, and M. A. Valvano. 2004. Wzx proteins involved in biosynthesis of O antigen function in association with the first sugar of the O-specific lipopolysaccharide subunit. Microbiology 1504095-4105. [DOI] [PubMed] [Google Scholar]
- 32.Marolda, C. L., E. R. Haggerty, M. Lung, and M. A. Valvano. 2008. Functional analysis of predicted coiled-coil regions in the Escherichia coli K-12 O-antigen polysaccharide chain length determinant Wzz. J. Bacteriol. 1902128-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merino, S., V. J. Benedí, and J. M. Tomás. 1989. Aeromonas hydrophila strains with moderate virulence. Microbios 59165-173. [PubMed] [Google Scholar]
- 34.Merino, S., A. Aguilar, X. Rubires, and J. M. Tomás. 1998. Mesophilic Aeromonas sp. strains from different serotypes: the influence of growth temperature and osmolarity on lipopolysaccharide and virulence. Res. Microbiol. 149407-416. [DOI] [PubMed] [Google Scholar]
- 35.Merino, S., S. Camprubí, and J. Tomás. 1991. The role of the lipopolysaccharide in complement killing of Aeromomas hydrophila strains from serotype O:34. J. Gen. Microbiol. 1371583-1590. [DOI] [PubMed] [Google Scholar]
- 36.Merino, S., S. Camprubí, and J. M. Tomás. 1992. Effect of growth temperature on outer-membrane components and virulence of Aeromonas hydrophila strains of serotype O:34. Infect. Immun. 604343-4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Merino, S., S. Camprubí, and J. M. Tomás. 1992. Characterization of an O-antigen bacteriophage from Aeromonas hydrophila. Can. J. Microbiol. 38235-240. [DOI] [PubMed] [Google Scholar]
- 38.Merino, S., X. Rubires, A. Aguilar, J. F. Guillot, and J. M. Tomás. 1996. The role of the O-antigen polysaccharide on the colonization in vivo of the germfree chicken gut by Aeromonas hydrophila serogroup O:34. Microb. Pathog. 20325-333. [DOI] [PubMed] [Google Scholar]
- 39.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 40.Milton, D. L., R. O'Toole, P. Horstedt, and H. Wolf-Watz. 1996. Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 1781310-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murray, G. L., S. R. Attridge, and R. Morona. 2006. Altering the length of the lipopolysaccharide O antigen has an impact on the interaction of Salmonella enterica serovar Typhimurium with macrophages and complement. J. Bacteriol. 1882735-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakano, Y., N. Suzuki, Y. Yoshida, T. Nezu, Y. Yamashita, and T. Koga. 2000. Thymidine diphosphate-6-deoxy-l-lyxo-4-hexulose reductase synthesizing dTDP-6-deoxy-l-talose from Actinobacillus actinomycetemcomitans. J. Biol. Chem. 2756806-6812. [DOI] [PubMed] [Google Scholar]
- 43.Nogueras, M. M., S. Merino, A. Aguilar, V. J. Benedí, and J. M. Tomás. 2000. Cloning, sequencing, and role in serum susceptibility of porin II from mesophilic Aeromonas sp. Infect. Immun. 681849-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raetz, C. R., and S. L. Roderick. 1995. A left-handed parallel beta helix in the structure of UDP-N-acetylglucosamine acyltransferase. Science 270997-1000. [DOI] [PubMed] [Google Scholar]
- 45.Raetz, C. R. H., and C. Whitfield. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71635-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saldías, M. S., K. Patel, C. L. Marolda, M. Bittner, I. Contreras, and M. A. Valvano. 2008. Distinct functional domains of the Salmonella enterica WbaP transferase that is involved in the initiation reaction for synthesis of the O antigen subunit. Microbiology 154440-453. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 48.Samuel, G., and P. Reeves. 2003. Biosynthesis of O-antigens: genes and pathways involved in nucleotide sugar precursor synthesis and O-antigen assembly. Carbohydr. Res. 3382503-2519. [DOI] [PubMed] [Google Scholar]
- 49.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 745463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schild, S., A.-K. Lamprecht, and J. Reidl. 2005. Molecular and functional characterization of O antigen transfer in Vibrio cholera. J. Biol. Chem. 28025936-25947. [DOI] [PubMed] [Google Scholar]
- 51.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 5385-96. [DOI] [PubMed] [Google Scholar]
- 52.Wang, X. G., L. R. Olsen, and S. L. Roderick. 2002. Structure of the lac operon galactoside acetyltransferase. Structure 10581-588. [DOI] [PubMed] [Google Scholar]
- 53.Whitfield, C., and K. Larue. 2008. Stop and go: regulation of chain length in the biosynthesis of bacterial polysaccharides. Nat. Struct. Mol. Biol. 15121-123. [DOI] [PubMed] [Google Scholar]
- 54.Yang, Y.-H., Y.-B. Kangb, K.-W. Lee, T.-H. Lee, S.-S. Park, B.-Y. Hwang, and B.-G. Kima. 2005. Characterization of GDP-mannose pyrophosphorylase from Escherichia coli O157:H7 EDL933 and its broad substrate specificity. J. Mol. Catal. B Enzym. 371-8. [Google Scholar]
- 55.Yu, H. B., P. S. S. Rao, H. C. Lee, S. Vilches, S. Merino, J. M. Tomas, and K. Y. Leung. 2004. A type III secretion system is required for Aeromonas hydrophila AH-1 pathogenesis. Infect. Immun. 721248-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang, Y. L., E. Arakawa, and K. Y. Leung. 2002. Novel Aeromonas hydrophila PPD134/91 genes involved in O-antigen and capsule biosynthesis. Infect. Immun. 702326-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zuccotti, S., D. Zanardi, C. Rosano, L. Sturla, M. Tonetti, and M. Bolognesi. 2001. Kinetic and crystallographic analyses support a sequential-ordered bi bi catalytic mechanism for Escherichia coli glucose-1-phosphate thymidylyltransferase. J. Mol. Biol. 313831-843. [DOI] [PubMed] [Google Scholar]