Abstract
Bacillus cereus Fnr is a member of the Crp/Fnr (cyclic AMP-binding protein/fumarate nitrate reduction regulatory protein) family of helix-turn-helix transcriptional regulators. It is essential for the expression of hbl and nhe enterotoxin genes independently of the oxygen tension in the environment. We studied aerobic Fnr binding to target sites in promoters regulating the expression of enterotoxin genes. B. cereus Fnr was overexpressed and purified as either a C-terminal His-tagged (FnrHis) fusion protein or an N-terminal fusion protein tagged with the Strep-tag (IBA BioTAGnology) (StrepFnr). Both recombinant Fnr proteins were produced as apoforms (clusterless) and occurred as mixtures of monomers and oligomers in solution. However, apoFnrHis was mainly monomeric, while apoStrepFnr was mainly oligomeric, suggesting that the His-tagged C-terminal extremity may interfere with oligomerization. The oligomeric state of apoStrepFnr was dithiothreitol sensitive, underlining the importance of a disulfide bridge for apoFnr oligomerization. Electrophoretic mobility shift assays showed that monomeric apoFnr, but not oligomeric apoFnr, bound to specific sequences located in the promoter regions of the enterotoxin regulators fnr, resDE, and plcR and the structural genes hbl and nhe. The question of whether apoFnr binding is regulated in vivo by redox-dependent oligomerization is discussed.
The facultative anaerobic, spore-forming Bacillus cereus has gained notoriety as an opportunistic human pathogen that can cause a wide range of diseases from periodontitis and endophthalmitis to meningitis in immunocompromised patients. However, most of the reported illnesses involving B. cereus are food-borne intoxications, classified as emetic and diarrheal syndromes (17, 30). Diarrheal syndrome may result from the production in the human host's small intestine of various extracellular factors including hemolysin BL (Hbl), nonhemolytic enterotoxin (Nhe), and cytotoxin CytK (17, 26). The genes encoding these potential virulence factors belong to the PlcR regulon (1, 7, 20, 31).
B. cereus will grow efficiently by anaerobic glucose fermentation in amino acid-rich media supplemented with glucose as the major source of carbon and energy (3, 21, 29, 33, 34). The ability of B. cereus to grow well under these conditions is controlled by both the two-component system ResDE (4) and the redox regulator Fnr (33, 34). Unlike ResDE, B. cereus Fnr has been shown to be essential for fermentative growth and for enterotoxin synthesis under both anaerobiosis and aerobiosis (33, 34). Fnr protein is a member of the large Crp/Fnr (cyclic AMP-binding protein/fumarate nitrate reduction regulatory protein) superfamily of transcription factors that coordinate physiological changes in response to a variety of metabolic and environmental stimuli (16). Members of the family are predicted to be structurally related to the catabolite gene activator protein of Escherichia coli, Crp (also known as the cyclic AMP receptor protein) (10). Like all the members of the Crp/Fnr family, B. cereus Fnr contains an N-terminal region made up of antiparallel β-strands able to accommodate a nucleotide, and a C-terminal helix-turn-helix structural motif. In addition, it contains a C-terminal extension with four cysteine residues considered, in Bacillus subtilis, to coordinate a [4Fe-4S]+2 center that serves as a redox sensor (27). The B. subtilis Fnr forms a stable dimer that is independent of both the oxygen tension in the environment and FeS cluster formation. However, the presence of an intact [4Fe-4S]+2 cluster is required for it to bind to a specific DNA-binding site and for subsequent transcriptional activation (27).
Structurally, the predicted Fnr of B. cereus resembles the B. subtilis Fnr (27). Therefore, the FeS cluster could also be a key component required for the DNA-binding activity of B. cereus Fnr under anaerobiosis. However, our previous results suggested that unlike B. subtilis Fnr, B. cereus Fnr may also exist in an active state under aerobiosis and thus conserve some site-specific DNA-binding properties. To address this specificity further and elucidate the mechanism by which Fnr regulates enterotoxin gene expression in aerobically growing B. cereus cells, we characterized the DNA-binding activities of purified aerobic Fnr. To this end, we overproduced full-length Fnr in Escherichia coli with two different tags. We showed that both recombinant Fnr proteins were produced in apo forms (devoid of FeS cluster) under oxic conditions. Recombinant Fnr containing a C-terminal polyhistidine-tagged sequence was shown to be mainly monomeric in solution, while N-terminally Strep-tagged Fnr occurred mainly as oligomers. Only the monomeric forms of both recombinant apoFnr proteins were found to bind to the promoter regions of fnr itself, the pleiotropic regulator genes resDE and plcR, and the structural enterotoxin genes hbl and nhe. Finally, our results pointed to some new unusual properties of Fnr that may have physiological relevance in the redox regulation of enterotoxin expression, enterotoxin expression being both directly and indirectly (via ResD and PlcR) regulated by apoFnr under aerobiosis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Escherichia coli strain TOP 10 (Invitrogen) [F− mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 araD139Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG] was used as the general cloning host, and strain BL21 CodonPlus(DE3)-RIL (Stratagene) [F− ompT hsdS(rB− mB−) dcm+ Tetr gal λ (DE3) endA Hte [argU ileY leuW Camr] was used to overexpress fnr. Both E. coli strains were routinely grown in Luria broth with vigorous agitation at 37°C. The wild-type B. cereus F4430/73 (32) and fnr mutant (34) were grown as previously described.
General molecular methods.
Restriction endonuclease and T4 DNA ligase were obtained from Promega and used in accordance with the manufacturer's instructions. Genomic DNA of B. cereus was purified by the method of Guinebretiere and Nguyen-The (11). Plasmid DNA was purified using anion-exchange columns (Promega). PCR amplification of DNA was carried out with Taq polymerase using the manufacturer's specifications (Roche Molecular Biochemicals) for reaction conditions. The 5′ end of the resDE mRNA was mapped from a 5′ rapid amplification of cDNA ends (5′ RACE) PCR product obtained with the 3′/5′ RACE kit (Roche Molecular Biochemicals). For this purpose, we used total RNA extracted from B. cereus F4430/73 cells harvested at μmax, i.e., the maximal expression of the resDE operon. Briefly, the first-strand cDNA was synthesized from total RNA with fnr-specific primer SP1 (5′-GCCTGGTAAAGATGGCATTG-3′), avian myeloblastosis virus reverse transcriptase, and the deoxynucleotide mixture of the 3′/5′ RACE kit as recommended by the manufacturer. After purification and dA tailing of the cDNA, a PCR with the dT anchor oligonucleotide primer and the specific fnr SP2 primer (5′-GATGATGAGGATCGTATTVGTCG-3′), followed by a nested PCR with SP3 primer (5′-GAGAGTGCGCAGCGGGTAGAG-3′), yielded a PCR product of 190 bp, as revealed by 2% agarose gel electrophoresis. This PCR product was purified and sequenced.
Cloning and overexpression of recombinant Fnr.
The coding sequence for B. cereus fnr was PCR amplified from B. cereus F4430/73 genomic DNA using either primers PET101F (5′-CACCGTGGCAAACAGTATGACATTATCT-3′) and PET101R (5′-ATCAATGCTACAAA CAGAAGC-3′) or primers PET52F (5′-CCCGGGATGACATTATCTCAAGATTTAAAAGAA-3′; SmaI restriction site in bold type) and PET52R (5′-GAGCTCCTAATCAATGCTACAAACAGAAGCA-3′; SacI restriction site in bold type). The amplicons were cloned as a blunt-end PCR product into pET101/D-TOPO (Invitrogen) and as a SmaI-SacI fragment into the corresponding sites of pET-52b(+) (Novagen), yielding pET101fnr and pET52fnr, respectively. B. cereus Fnr was produced as a C-terminal fusion with a His tag (FnrHis) using pET101fnr and as an N-terminal fusion with a Strep-tag (IBA BioTAGnology) (StrepFnr) using pET52fnr in E. coli BL21 CodonPlus(DE3)-RIL (Stratagene). Recombinant cells were grown at 37°C in Luria broth with 100 μg ml−1 ampicillin. When the optical density at 600 nm reached ∼1.0, protein production was triggered by adding isopropyl-β-d-thiogalactopyranoside (IPTG) with a final concentration of 0.2 mM (pET101fnr) or 0.4 mM (pET52fnr). Cells were grown for 16 h at 20°C.
Purification of FnrHis.
Cells from a 4.8-liter culture were harvested by centrifugation (10,000 × g, 15 min), resuspended in buffer A (50 mM sodium phosphate buffer [pH 7.0], 300 mM NaCl), and incubated with 0.5 mg/ml lysozyme for 30 min under gentle agitation. Cells were lysed by sonication for 3 min at 80% of maximum amplitude using a Vibra cell ultrasonifier (Fisher Bioblock Scientific). Cell debris was removed by centrifugation at 20,000 × g for 20 min. The supernatant was run through a 5-ml Co2+ immobilized metal ion affinity chromatographic column (Clontech) equilibrated with buffer A. The column was washed with 50 ml of buffer A and then with 25 ml of buffer A containing 10 mM imidazole, and the protein was eluted with 5 ml of buffer A containing 150 mM imidazole. The eluted fraction was desalted on a Sephadex G25 column (Amersham Pharmacia Biotech) and concentrated using Nanosep 30-kDa molecular-mass-cutoff devices (Omega disc membrane; Pall Filtron). Concentrated samples were run through a 104-ml Superdex SD200 column (Amersham Biosciences) equilibrated with buffer B (100 mM Tris-HCl [pH 8], 150 mM NaCl, 1 mM dithiothreitol [DTT]). Protein was stored as pellets in liquid nitrogen.
Purification of StrepFnr.
Cells from a 6-liter culture were harvested by centrifugation at 10,000 × g for 15 min, resuspended in 120 ml of buffer C (25 mM Tris-HCl [pH 8], 1 mM DTT), and incubated with 0.2 mg·ml−1 of lysozyme and 0.5 mM EDTA for 10 min at 30°C. Cells were lysed by sonication as described above for the purification of FnrHis. Cell debris was removed by centrifugation at 43,000 × g for 1 h, and the resulting supernatant was run through a 30-ml DEAE-cellulose column (DE52; Whatman) equilibrated with buffer C. The column was then washed with the same buffer. Nonretained fractions were adjusted to pH 7 with 1 M KH2PO4 and run through a 30-ml hydroxyapatite agarose column (HA Ultrogel; Pall Corporation) equilibrated with buffer D (50 mM KH2PO4 [pH 7], 1 mM DTT). The column was developed with a linear gradient from 50 to 200 mM KH2PO4 at a flow rate of 2 ml/min. Fractions containing recombinant Fnr were pooled and concentrated to 48 mg·ml−1 by ultrafiltration through an Omega disc membrane (30-kDa cutoff; diameter, 43 mm; Pall Filtron). A polishing step was then carried out with gel filtration on a 104-ml Superdex SD200 column (Amersham Biosciences) equilibrated with buffer D containing 150 mM NaCl. The purified protein was stored as pellets in liquid nitrogen.
Protein biochemical analyses.
Protein concentrations were determined by either a bicinchoninic acid assay according to the manufacturer's instructions (Interchim) or a biuret method insensitive to thiols (22). Bovine serum albumin was used as a standard. Overproduction of Fnr in induced cultures and its purification were monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The Laemmli method was used for SDS-PAGE (18). Proteins were stained with Coomassie brilliant blue. The reducing agent β-mercaptoethanol was omitted to analyze the disulfide form of apoFnr. The molecular mass of apoFnr was accurately measured with an Esquire 3000plus ion trap mass spectrometer equipped with a nanoelectrospray on-line ion source (Bruker Daltonics) essentially as described previously (6). Before mass measurement, purified apoFnr was desalted with a ZipTipC18 (Millipore) and diluted in 50% acetonitrile-1% formic acid (vol/vol).
DLS.
The quaternary structure of purified apoFnr in solution was measured by dynamic light scattering (DLS). Samples were centrifuged and run through a 24-ml Superdex 200 column (HR10/30) equilibrated and run at a flow rate of 0.5 ml/min with 50 mM Tris-HCl (pH 8.3) containing 120 mM NaCl and 0.05% NaN3 filtered at 0.1 μm. The column was operated with an Agilent 1100 series reverse-phase high-performance liquid chromatography system equipped with a G1322A degasser, G1311A quaternary pump, and G1313A autosampler. The elution profile was monitored with a G1315B diode array detector (Agilent), a miniDawn Tristar multiangle laser static light scattering detector (three angles, 45°, 90°, and 135°) coupled to a DynaPro Titan light scattering instrument (Wyatt Technology) placed at 90° and an Optilab rEX differential refractometer (Wyatt Technology). The 90° multiangle light scattering detector was calibrated with pure toluene, and bovine serum albumin was then used to normalize the other detector (45° and 135°) in the corresponding buffer.
Chemical cross-linking of FnrHis.
FnrHis in 10 mM 3-(N-morpholino)propanesulfonic acid (MOPS) buffer (pH 7.75) was treated with protein cross-linking agents, N-hydroxysulfosuccinimide (5 mM) and 1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride (EDC) (12.5 mM). The reaction mixture contained protein at a concentration of 5 μM in a total reaction mixture volume of 20 μl. The reaction was allowed to proceed for 30 min at room temperature and stopped by adding 25 mM β-mercaptoethanol. The products were analyzed by 12% nondenaturing SDS-PAGE and detected by Western blotting using an anti-His antibody.
ApoFnr antiserum preparation.
Polyclonal antibodies against apoFnr were generated in-house. Rabbits were immunized with a total of 2 mg of purified FnrHis, administered in four equal doses over a 90-day period, and bled on day 120. Antisera specificities were checked by Western blotting.
Western blot analysis.
B. cereus protein extracts were prepared as follows: cells were harvested by centrifugation, resuspended in buffer containing 8 M urea, 4% (wt/vol) CHAPS ([3-[(3-cholamidopropyl)-dimethylammonio]propanesulfonate]), and mechanically disrupted using a FastPrep instrument (FP120; Bio101, Thermo Electron Corporation). Cell debris was removed by centrifugation (3,500 × g, 10 min, 4°C). Proteins were then filtered and resolved by SDS-PAGE under nonreducing conditions (18). Resolved proteins were transferred to nitrocellulose membranes (Amersham Bioscience) in a Bio-Rad liquid/liquid transfer unit. As appropriate, apoFnr was detected with either anti-His antibodies (FnrHis) or anti-Strep antibodies (StrepFnr) or with a 1:2,000 dilution of polyclonal rabbit serum. The blotted membranes were developed with a 1:2,000 dilution of peroxidase-conjugated goat anti-rabbit immunoglobulin G (Sigma) and an enhanced chemiluminescence substrate (Immobilon Western; Millipore).
EMSA.
The 5′ untranslated regions (5′UTRs) of fnr, resDE, plcR, hbl, and nhe were PCR amplified with the following primer pairs: FnrF (5′-CGAACACTTCAGCAGGCATA-3′) and FnrR (5′-AATGTCATACTGTTTGCCAC-3′), ResDF (5′-TGGGATCCCAAAAGAGGTTTG-3′) and ResDR (5′-CGATCCTCATCATCTACAAT-3′), PlcRF (5′-TATGTTTGTGCAAGGCGAAC-3′) and PlcRR (5′-CCTAATTTTTCTGCGTGCAT-3′), Hbl1F (5′-GGTAAGCAAGTGGGTGAAGC-3′) and Hbl1R (5′-AATCGCAAATGCAGAGCACAA-3′), Hbl2F (5′-TTAACTTAATTCATATAACTT-3′) and Hbl2R (5′-TACGCATTAAAAATTTAAT-3′), and NheF (5′-TGTTATTACGACAGTTCCAT-3′) and NheR (5′-CTGTAACCAATAACCCTGTG-3′). The forward primers were 5′ end labeled with T4 polynucleotide kinase (Promega) and [γ-32P]ATP (Amersham Biosciences). The 5′-32P-labeled amplicons were purified using High Pure PCR product purification columns (Roche). Electrophoretic mobility shift assays (EMSAs) were performed by incubating labeled DNA fragments (1,000 cpm per reaction mixture) with the specified amount of purified Fnr in 50 mM Tris-HCl (pH 7.5) buffer containing 50 mM KCl, 0.1 mM EDTA, 10% glycerol, 4 mM DTT, 4 mM MgCl2, 0.5 μg of bovine serum albumin, and 1 μg of poly(dI-dC)/ml in a final volume of 10 μl. Binding reaction mixtures were incubated for 30 min at 37°C and then loaded onto a 4% or 6% nondenaturing polyacrylamide gel run with Tris-borate-EDTA buffer at 4°C and 200 V. Labeled products were quantified using a Molecular Dynamics PhosphorImager.
RESULTS
Overexpression and purification of two recombinant Fnr proteins.
From the sequence alignment of 13 B. cereus Fnr homologues (see Fig. S1 in the supplemental material), two possible alternative translation initiation starts could be identified for B. cereus F4430/73: GTG as previously defined (34) or ATG 12 nucleotides further on. Taking into account this information, two procedures were developed for the aerobic production of B. cereus F4430/73 Fnr in E. coli cells: (i) expression of a His-tagged fusion protein from pET101/D-TOPO to release a Fnr variant (FnrHis) that begins with the valine encoded by the predicted start codon of B. cereus (34) and contains 30 additional amino acids at the C-terminal end, and (ii) expression of a protein from pET-52b(+) fused to Strep-tag (IBA BioTAGnology) to release an Fnr variant (StrepFnr) that contains 22 additional amino acids at the N-terminal end. In the latter variant, the first amino acid of the native Fnr is the methionine located four amino acids after the valine encoded by the predicted start codon (see Fig. S1 in the supplemental material). The FnrHis protein was purified using cobalt affinity chromatography. Because preliminary tests indicated that StrepFnr bound very weakly to Strep-Tactin Sepharose (IBA BioTAGnology), the tagged protein was purified by means of three successive chromatography runs not based on the tag affinity. On SDS-polyacrylamide gels, purified FnrHis (29 kDa) and StrepFnr (28 kDa) exhibited the expected molecular masses (see Fig. S2 in the supplemental material). The exact average molecular mass of StrepFnr determined by mass spectrometry was 27,913 ± 2 Da. This value corresponds almost perfectly (75-ppm deviation) to the expected polypeptide sequence, except that the initial formyl-methionine is cleaved (theoretical value of 27,911 Da). This maturation probably also occurs with the FnrHis protein.
UV-visible absorption spectrum of both aerobic recombinant forms of Fnr showed a single peak at 280 nm (data not shown), suggesting that there is no absorbing prosthetic group (14). This indicates that both recombinant Fnr forms were purified as apoproteins under aerobiosis.
Oligomeric state of both recombinant apoFnr proteins.
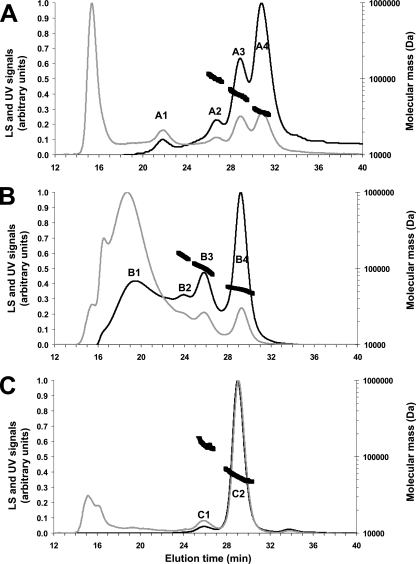
DLS was used to examine the oligomeric state of the two recombinant forms of apoFnr. DLS reveals the homogeneity and oligomeric state of proteins when resolved by gel filtration based on the scattering of visible light by particles (5). The oligomerization states of FnrHis and StrepFnr were analyzed using a DynaPro Titan DLS instrument and attendant software ASTRA. Figure 1 shows the elution profiles obtained for both proteins and the molecular mass estimates derived from the light scattering signal. Besides a peak of aggregates at 16 min, FnrHis was resolved into four elution peaks at 22.0 (A1), 26.6 (A2), 28.8 (A3), and 30.7 (A4) min elution time as detected on the UV trace (Fig. 1A). The molar mass across peak A1 could not be determined because of a polydisperse distribution (the molecular mass varied from 170 to 400 kDa). This strongly suggests that this peak contained aggregates that interacted with the column, but their proportion was low, as the DLS signal was very weak. In contrast, the distribution of molar masses across peaks A2, A3, and A4 was constant, indicating a monodisperse distribution (i.e., a homogeneous molecule) for each peak with molecular masses of 98 (A2), 60 (A3), and 30 (A4) kDa (±3%). This indicates that FnrHis occurs mainly as a mixture of trimers, dimers, and monomers in solution. Considering the relative mass ratio that can be estimated from the UV trace, the predominant form was the monomer (70%). The light scattering trace obtained with StrepFnr showed the presence of aggregates (peak B1) and three peaks with molecular masses of 157 (B2), 106 (B3), and 54 (B4) kDa (±3%) (Fig. 1B). These peaks unambiguously correspond to the hexameric, tetrameric, and dimeric forms of StrepFnr, respectively. In this case, the dimeric form (33%) formed the largest population. The same DLS experiment was repeated in reducing conditions with 10 mM DTT in the elution buffer. Complete disappearance of the hexameric form and almost complete disappearance of the tetrameric form (C1) were observed (Fig. 1C). The dimeric form (C2) was thus predominant (89%). Hence, the addition of reductant affected the oligomerization state of StrepFnr in solution. Intermolecular disulfide bridges are involved in the formation of the highest oligomeric forms. In addition, the absence of monomers in reducing conditions suggests that the dimers observed were either noncovalently linked structures or DTT-resistant, covalently linked structures.
FIG. 1.
Gel filtration and DLS chromatograms of purified Fnr proteins. FnrHis (A), StrepFnr (B) and reduced StrepFnr (C) were injected (∼300 μg in 100 μl) into a Superdex 200 column (HR 10/30) with 50 mM Tris-HCl (pH 8.3)-120 mM NaCl as the eluant at a flow rate of 0.5 ml/min. DTT (10 mM) was added to the elution buffer to determine the oligomeric state of reduced StrepFnr in panel C. The black and gray lines correspond to the light scattering (LS) signal and the UV signal recorded at 280 nm, respectively. These signals were normalized as a ratio from 0.0 to 1.0 for comparison (left y axis). The molecular mass estimates of the major peaks are also indicated by thick black broken lines (right y axis).
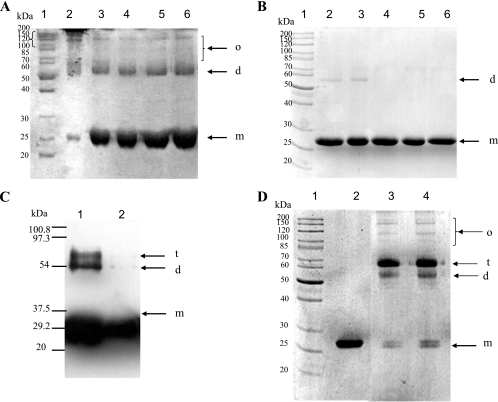
When purified StrepFnr underwent SDS-PAGE under nonreducing conditions (no DTT or β-mercaptoethanol), a multiple-band pattern was observed, revealing the presence of a mixed population of monomeric, dimeric, and higher oligomeric forms in relative ratios compatible with those found in a DLS experiment (Fig. 2A). Under reducing conditions (with DTT), the two major species were the monomeric and dimeric forms. Increasing the concentration of DTT from 10 mM to 200 mM caused the total reduction of dimeric species to monomeric forms. These data indicate that most of the protein was reticulated through disulfide bridges, but a significant amount of dimeric StrepFnr could either not be completely reduced by DTT or remained particularly stable in the electrophoresis conditions used. In contrast, only a very small fraction of FnrHis was found to remain dimeric after 10 mM DTT treatment (Fig. 2B). This suggests that the oligomeric FnrHis population detected by DLS contained mainly noncovalently linked structures. To investigate the ability of monomeric FnrHis to form covalently linked structures, a fraction of the purified protein was treated with either the chemical cross-linker EDC or with the divalent thiol-reactive agent diamide. The first cross-linker modifies an ionic interaction into a covalent link, while the latter mimics disulfide bridge formation. Figure 2 shows the reaction products analyzed by SDS-PAGE. The formation of oligomers from monomers could be evidenced using both EDC (Fig. 2C) and diamide (Fig. 2D). Homodimers and homotrimers were the major products. As expected when using cross-linkers, these entities migrated at relative molecular weights slightly lower than the exact weights because of their more rigid structures. Surprisingly, the band corresponding to apoFnr monomer appeared as a discrete doublet after treatment with diamide, reflecting a possible induced conformational change trapped by intrapolypeptide cross-links (Fig. 2D). In conclusion, FnrHis monomers were able to self-associate and form higher-order covalently linked structures in the presence of cross-linkers. This suggests that unlike StrepFnr, FnrHis does not tend to form covalently linked homodimers or, more specifically, intermolecular disulfide bridges.
FIG. 2.
SDS-PAGE analysis of the oligomeric nature of StrepFnr and FnrHis. (A and B) Effect of DTT on StrepFnr (A) and FnrHis (B) oligomerization. Purified proteins were incubated with 0, 10, 50, 100, or 200 mM DTT (lanes 2 to 6, respectively). Recombinant proteins were then subjected to nonreducing SDS-PAGE. The arrows show the positions of monomers (m), dimers (d), and higher oligomers (o). Lane 1 contains molecular mass standard proteins. (C) SDS-PAGE profile of FnrHis cross-linked with EDC. FnrHis (5 μM) was cross-linked with EDC. Products were visualized by immunoblotting with anti-His antibody. Lane 1, cross-linked FnrHis; lane 2, untreated FnrHis. (D) Nondenaturing SDS-PAGE profile of FnrHis cross-linked with diamide. Lane 1, molecular mass standard proteins; lane 2, untreated FnrHis; lanes 3 and 4, disulfide-linked FnrHis with 1 mM and 10 mM diamide, respectively. The arrows show the positions of monomers (m), dimers (d), trimers (t), and higher oligomers (o).
Detection of endogenous apoFnr in B. cereus F4430/73 cells.
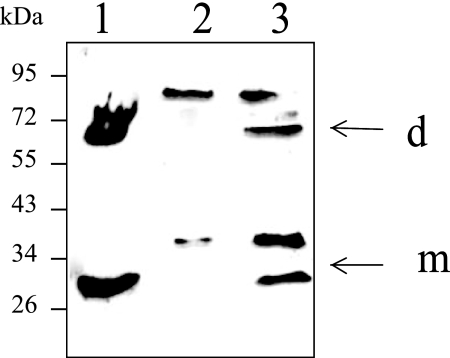
To determine whether the formation of disulfide-linked homodimers might be of physiological relevance, we tested the presence of various forms of endogenous apoFnr in aerobically grown B. cereus cells (4). Figure 3 shows the Western blot detection performed with apoFnr antiserum following SDS-PAGE under nonreducing conditions. The antiserum reacted with two bands of the sizes expected for the monomeric (∼30-kDa) and dimeric forms (∼60-kDa) of apoFnr in wild-type cells, but not in fnr mutant cells. Two other protein bands of 40 and 80 kDa cross-reacted with apoFnr antiserum in wild-type cells (Fig. 3, lane 3). As these bands were also observed in the fnr mutant cells (Fig. 3, lane 2), they were not related to Fnr. Finally, these results indicated that the apoFnr antiserum can be used efficiently for the detection of endogenous apoFnr in B. cereus F4430/73 cells and, more importantly, that some dimeric apoFnr could be disulfide linked in B. cereus.
FIG. 3.
Western blot detection of endogenous Fnr species from B. cereus cells. Lysates of wild-type B. cereus F4430/73 and fnr mutant were probed with polyclonal Fnr antiserum. Both strains were grown in regulated batch culture (pH 7.2) under aerobiosis (4). Proteins were separated by nonreducing SDS-PAGE. Lane 1, StrepFnr purified from E. coli; lane 2, fnr mutant; lane 3, wild-type strain. The putative identities shown on the right were determined for the wild-type strain on the basis of results obtained with both recombinant Fnr and fnr mutant strains. The arrows show the positions of monomer (m) and dimer (d) forms. The positions and masses (in kilodaltons) of molecular mass markers are given to the left of the gel.
Binding of apoFnr to the 5′UTRs of fnr, resDE, plcR, hbl, and nhe.
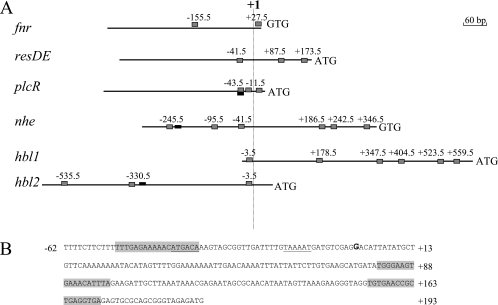
The amino acid residues forming the REX3R motif within the helix-turn-helix DNA-binding domain of Crp regulatory proteins are strictly conserved in the potential DNA-binding domain of B. cereus F4430/73 Fnr as in its homologues found in strains belonging to the B. cereus group (12). Accordingly, Fnr of B. cereus F4430/73 was assumed to bind to DNA motifs similar to the TGTGAN6TCACA consensus sequence defined in previous work (2, 16). Using the Virtual tool of the Prodoric database and the corresponding E. coli Crp position weight matrix, we scanned the 5′UTRs of regulatory and structural genes of B. cereus F4430/73 enterotoxins. Figure 4A shows the locations of predicted Fnr-binding boxes for fnr, resDE, plcR, nhe, hbl1, and hbl2 promoters and their positions relative to the transcriptional start point of each gene/operon. Except for resDE (Fig. 4B), the transcriptional start sites were identified in previous studies (1, 4). Three putative Fnr-binding sites were found in the 5′UTRs of the enterotoxin gene regulators fnr, resDE, and plcR. Eight potential Fnr-binding sites were found in the nhe promoter region: four were located upstream of the transcriptional start site and four were downstream. The hbl promoter region contained 11 potential Fnr-binding sites, 4 located upstream of the +1 site and 7 downstream.
FIG. 4.
Potential Fnr-binding sites in the 5′ untranslated regions of fnr, resDE, plcR, hbl, and nhe. All numbering is relative to the transcription start site at position +1. (A) Potential Fnr-binding sites are shown relative to the transcription start site as gray boxes. PlcR boxes are shown as black boxes. (B) Genetic organization of the resDE promoter region. The transcriptional start site (+1) determined by 5′ RACE PCR is in bold type. The putative −35 and −10 motifs are underlined. Putative Crp/Fnr boxes are indicated by a gray background.
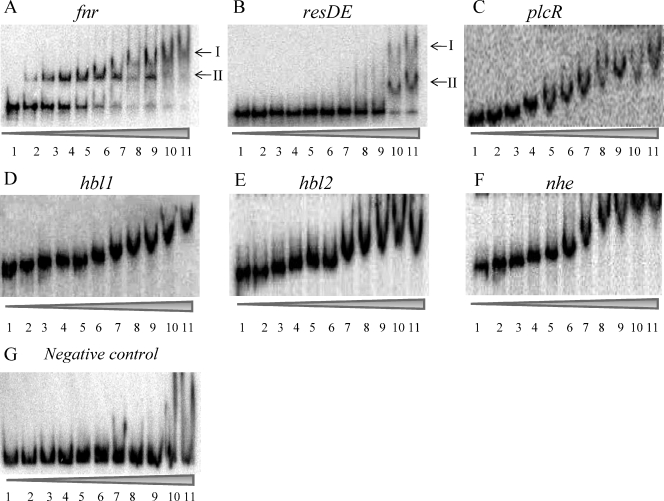
To test whether apoFnr bound to the Fnr boxes predicted from the nucleotide sequence analysis, EMSAs were performed with both StrepFnr and FnrHis and DNA fragments containing 5′UTRs of fnr, resDE, plcR, hbl, and nhe. In view of its size (1,157 bp), the 5′UTR of hbl was first divided into two overlapping fragments of 636 bp (hbl1) and 610 bp (hbl2) as defined in Fig. 4A. Figure 5 shows the EMSA results for the six fragments. FnrHis bound to all the regions tested, while no DNA-binding activity could be detected with StrepFnr. The specificity of the binding was evidenced from the disappearance of complexes in competition assays using 50-fold excess of homologous unlabeled promoter regions and by the absence of any competition when an unlabeled heterologous DNA was used (data not shown). EMSAs with the negative control (Fig. 5G) showed that a shift above 6 μM apoFnr should be considered the result of nonspecific binding. In addition, the behavior of apoFnr differed markedly in the gel-shift titration assay depending on the promoter regions. ApoFnr bound to fnr and resDE promoter regions in an ordered fashion giving two retarded species (complex I and II) below 6 μM. In contrast, an increasing amount of apoFnr resulted in a gradual decrease in the mobility of the protein-DNA complexes for plcR, hbl, and nhe promoter regions, which appeared to be stabilized at higher protein concentrations. This suggests that, as more protein was added, the protein complex bound to the DNA increased proportionally in size, with the added apoFnr being distributed evenly among all the complexes. The smearing of these species during EMSA also suggested that these high-molecular-weight complexes were not stable and dissociated during electrophoresis. The EMSA data also showed that the plcR, hbl, and nhe 5′UTRs were bound by apoFnr with a lower affinity (equilibrium dissociation constant [KD] of ≤0.4 μM) than the resDE and fnr promoter regions (KDs of 3 and 4.5 μM, respectively).
FIG. 5.
Binding of apoFnr to 5′UTRs of fnr, resDE, plcR, hbl, and nhe genes determined by EMSAs. DNAs corresponding to fnr (A), resDE (B), plcR (C), hbl1 (D), hbl2 (E), nhe (F), and a negative control (G) were bound with increasing concentrations of apoFnr as indicated by the height of the triangle below the gel. The results presented are representative examples of an experiment performed in triplicate with either purified FnrHis or with reduced StrepFnr (purified StrepFnr plus 200 mM DTT). Lanes 1 to 11 contain 0, 0.2, 0.4, 0.6, 0.8, 1, 2, 3, 4, 5, and 6 μM apoFnr protein, respectively.
To test whether the oligomeric state regulated the DNA-binding activity of both FnrHis and StrepFnr, the effect of the reducing agent DTT (200 mM) on the binding of StrepFnr and the effect of the oxidizing agent diamide (1 mM) on the binding of FnrHis to all promoter regions were investigated. Adding reductant resulted in the generation of StrepFnr-DNA complex patterns similar to those obtained with FnrHis (Fig. 5). The effect of DTT was reversible, while the addition of diamide (1 mM) abolished StrepFnr binding (data not shown). Likewise, FnrHis showed no DNA-binding activity in the presence of diamide (data not shown). Thus, the oligomeric state of apoFnr was found to critically affect its binding activity. The data also indicate that apoFnr was able to bind the fnr, resDE, plcR, hbl, and nhe 5′UTRs only when present predominantly as a monomer.
DISCUSSION
Our previous studies showed that aerobic enterotoxin expression was regulated by both the transcriptional regulator Fnr and oxygen availability (or redox state) under aerobiosis (4, 33). In the present work, we describe experimental evidence for redox regulation of enterotoxin gene expression mediated by Fnr through its DNA-binding properties.
B. cereus apoFnr was overexpressed in E. coli and purified as either a C-terminal His-tagged (FnrHis) or an N-terminal Strep-tagged (StrepFnr) fusion protein. Unlike FnrHis, StrepFnr was purified without an affinity chromatography step. The reason was the poor affinity of the Strep-tag peptide (IBA BioTAGnology) for streptavidin (Strep-Tactin) due to its fusion to the N terminus of Fnr (19). No such problem was encountered in the case of Strep-tagged B. subtilis Fnr (27). This different behavior may be explained by the marked difference in the two N-terminal polypeptide sequences. Both recombinant Fnr (FnrHis and StrepFnr) were produced in multiple oligomeric apoforms. The distribution of quaternary structures was shown to differ between the two tagged variants. Purified FnrHis was predominantly monomeric, while StrepFnr was predominantly oligomeric, and the oligomerization of StrepFnr appeared to be due to the formation of disulfide bridges. Data obtained from crystal structure analysis of a member of the Crp/Fnr family showed that dimerization involved the C-terminal domain (13). This suggests that extension of B. cereus Fnr at its C terminus may introduce steric hindrance that reduces flexibility and/or affects interdomain communication. In turn, this would result in a less permissive, locked conformation, rendering the thiol group less exposed for pairing to form the disulfide bond.
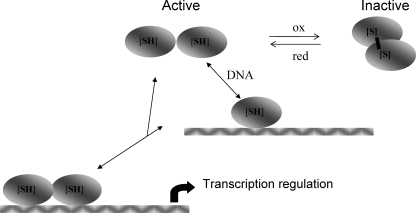
Our results showed that the active DNA-binding form of both recombinant apoFnrs was the monomer. Diamide treatment inactivated monomeric apoFnr in a DTT-reversible manner, suggesting that it was subject to redox regulation. In addition, we detected the presence of disulfide-linked endogenous dimers in B. cereus cells. Taken together, these findings suggest that formation of stabilized dimeric apoFnr by means of one or more SS bonds may be a regulatory mechanism that controls Fnr binding under exposure to oxidizing conditions. Figure 6 shows the scheme we propose for the reversible activation/inactivation of B. cereus apoFnr. It implies that this protein mediates a response to oxygen concentration and/or redox state causing the repression or activation of relevant genes. Such a thiol-based redox switch has been observed with Desulfitobacterium dehalogenans CrpK, a member of the Crp/Fnr family (24, 25). In this bacterium, the redox switch involves formation of an intermolecular disulfide bond that links two CprK subunits in an inactive dimer. Although it belongs to the same family, B. cereus Fnr contains three more cysteines than CprK does and should have the capacity to bind a FeS cluster like B. subtilis Fnr (27). For this reason, our findings are original. Additional work is now required to determine which of the seven cysteine residues are involved in this redox state sensing.
FIG. 6.
Proposal for the regulation of apoFnr activity by a thiol-disulfide redox switch. Brackets indicate that one or more disulfide bonds may be involved.
Many transcription factors bind DNA to form dimeric protein-DNA complexes. For these proteins, there are two limiting pathways that can describe the route of complex assembly. The protein can dimerize first and then associate with DNA (dimer pathway), or it can follow a pathway in which two monomers bind DNA sequentially and assemble their dimerization interface while bound to DNA (monomer pathway) (15). Many regulators bind DNA by the dimer pathway, and this is the case for Fnr of B. subtilis and E. coli under anaerobiosis (27). Under aerobiosis, apoFnr is produced as an inactive monomer in E. coli (28) and as an inactive dimer in B. subtilis (27). Because only the monomeric form of B. cereus apoFnr binds to DNA, we propose that Fnr binding in B. cereus occurs via the monomer pathway under aerobiosis (Fig. 6). Binding through the monomer pathway allows a dimeric transcription factor to respond rapidly to stimuli and to locate its target site quickly without becoming entrapped kinetically at a nonspecific site (23). Therefore, in addition to a faster assembly of apoFnr-DNA complexes in response to oxygen tension in the environment allowed by the monomer pathway, an efficient way to discriminate between specific and nonspecific target sites is also provided.
Since apoFnr bound to the promoter regions of fnr itself, the two-component system resDE genes, the virulence regulator plcR gene, and the enterotoxin genes hbl and nhe, we concluded that apoFnr directly controlled both its own expression and that of resDE, plcR, hbl, and nhe (34). The relatively low DNA-binding affinity observed for apoFnr suggests that other factors may be involved in DNA recognition as well as in protein-DNA complex stabilization (16). For example, it is conceivable that apoFnr operates with a specific oxidoreductase system or that for some other reason the cytoplasmic environment provided by B. cereus enhances its site-specific DNA-binding ability. In addition, interaction of apoFnr with one or more other regulatory proteins may facilitate its interaction with DNA. High-affinity binding to 5′UTRs of enterotoxin genes may require apoFnr-PlcR interaction insofar as PlcR (1) possesses binding sites close to the predicted Fnr-binding sites (Fig. 4A). Another possible interaction partner of apoFnr is the redox regulator ResD (4).
Transcriptional regulators such as members of the Crp/Fnr family interact with the α subunit of RNA polymerase (RNAP) (10). It has been shown that the protein-protein interaction increases the affinity of both partners to the promoter site (2). The contacts established between a Crp/Fnr protein and RNAP involve three patches of surface-exposed amino acids (called activating regions 1, 2, and 3) of Crp/Fnr protein. These contacts depend on the specific architecture of each promoter. The Crp/Fnr-dependent promoters can be grouped into three classes (labeled I, II, and III) based on the number and position of the Crp/Fnr-binding sites relative to the start of transcription and on the mechanism for transcription activation (2). The upstream DNA-binding site in class I promoters is centered either at position −61.5 (i.e., its axis of symmetry is between positions −61 and −62) or one to three helical turns further upstream (i.e., −71.5, −82.5, or −92.5). In class II promoters, the symmetry axis of the binding site is located at position −41.5 relative to the transcription start site, thus overlapping with the −35 region. Class III promoters comprise two or more DNA-binding sites for Crp/Fnr and have various architectures according to both the spacing between the DNA-binding sites and the distance between the Crp/Fnr-DNA-binding sites and RNAP-DNA-binding sites. In the case of B. cereus, the locations of predicted Crp/Fnr-binding sites upstream of the transcriptional start site suggest that the B. cereus fnr promoter region is a class I activating promoter, while resDE and plcR promoter regions are class II promoters. The nhe and hbl promoters are different and may be considered class III Crp/Fnr-dependent activated promoters. However, nhe, hbl, and to a lesser extent fnr, resDE, and plcR promoter regions, also contain predicted Crp/Fnr boxes located close to the −10 region and/or downstream of the transcriptional start site, i.e., at positions different from those found in classical Crp/Fnr-activated promoters. Comparable results were found for E. coli and B. subtilis Fnr (9, 27), where repression of transcription is mediated by Fnr binding to sites in different locations than in activating sites. Thus, we hypothesize that the regulation of enterotoxin gene expression involves an interplay of transcriptional activation and repression by Fnr. Repression may be mediated by occupancy of sites located downstream of the +1 site. In conclusion, the mechanism of Fnr-dependent regulation of enterotoxin in B. cereus is undoubtedly complex, and further extensive studies are required to examine the essential role of the downstream binding sites. Importantly, both hbl and nhe promoters have a long UTR (Fig. 4A), making it likely that mechanisms at the posttranscriptional level also control their expression. Such regulation could involve interaction between transcriptional regulators and ribosomal proteins (8). Finally, deciphering the complexities of this Fnr-dependent regulation is necessary to fully understand the mechanisms employed by B. cereus to ensure optimal virulence gene expression in response to changes in oxygen tension such as those encountered during infection in a human host.
In conclusion, this work shows that unlike its homologue in B. subtilis (12, 27), B. cereus Fnr is able to function as a transcriptional factor independently of the integrity of the FeS cluster. Thus, B. cereus Fnr illustrates the great versatility of the archetypal Crp/Fnr structure for transducing environmental signals to the transcriptional apparatus. More importantly, this study expands our knowledge of the molecular mechanisms used in B. cereus to modulate the transcriptional level of enterotoxin genes in response to redox variations.
Supplementary Material
Acknowledgments
J.E. held a fellowship from the Ministère de la Recherche et de l'Enseignement Supérieur.
We thank Christine Meyer (CEA-Grenoble) for her help and technical advice in protein purification, Bernard Fernandez (CEA-Marcoule) for conducting DLS experiments, Jean-Charles Gaillard (CEA-Marcoule) for mass spectrometry measurements, and Valérie Tanchou (CEA-Marcoule) for her kind help in the production of polyclonal antibodies.
Footnotes
Published ahead of print on 18 April 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Agaisse, H., M. Gominet, O. A. Okstad, A. B. Kolsto, and D. Lereclus. 1999. PlcR is a pleiotropic regulator of extracellular virulence factor gene expression in Bacillus thuringiensis. Mol. Microbiol. 321043-1053. [DOI] [PubMed] [Google Scholar]
- 2.Busby, S., and R. H. Ebright. 1999. Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 293199-213. [DOI] [PubMed] [Google Scholar]
- 3.Duport, C., S. Thomassin, G. Bourel, and P. Schmitt. 2004. Anaerobiosis and low specific growth rates enhance hemolysin BL production by Bacillus cereus F4430/73. Arch. Microbiol. 18290-95. [DOI] [PubMed] [Google Scholar]
- 4.Duport, C., A. Zigha, E. Rosenfeld, and P. Schmitt. 2006. Control of enterotoxin gene expression in Bacillus cereus F4430/73 involves the redox-sensitive ResDE signal transduction system. J. Bacteriol. 1886640-6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Folta-Stogniew, E. 2006. Oligomeric states of proteins determined by size-exclusion chromatography coupled with light scattering, absorbance, and refractive index detectors. Methods Mol. Biol. 32897-112. [DOI] [PubMed] [Google Scholar]
- 6.Gabant, G., J. Augier, and J. Armengaud. 2008. Assessment of solvent residues accessibility using three Sulfo-NHS-biotin reagents in parallel: application to footprint changes of a methyltransferase upon binding its substrate. J. Mass Spectrom. 43360-370. [DOI] [PubMed] [Google Scholar]
- 7.Gohar, M., O. A. Okstad, N. Gilois, V. Sanchis, A. B. Kolsto, and D. Lereclus. 2002. Two-dimensional electrophoresis analysis of the extracellular proteome of Bacillus cereus reveals the importance of the PlcR regulon. Proteomics 2784-791. [DOI] [PubMed] [Google Scholar]
- 8.Gray, J. P., J. W. Davis II, L. Gopinathan, T. L. Leas, C. A. Nugent, and J. P. Vanden Heuvel. 2006. The ribosomal protein rpL11 associates with and inhibits the transcriptional activity of peroxisome proliferator-activated receptor-alpha. Toxicol. Sci. 89535-546. [DOI] [PubMed] [Google Scholar]
- 9.Green, J., A. S. Irvine, W. Meng, and J. R. Guest. 1996. FNR-DNA interactions at natural and semi-synthetic promoters. Mol. Microbiol. 19125-137. [DOI] [PubMed] [Google Scholar]
- 10.Green, J., C. Scott, and J. R. Guest. 2001. Functional versatility in the CRP-FNR superfamily of transcription factors: FNR and FLP. Adv. Microb. Physiol. 441-34. [DOI] [PubMed] [Google Scholar]
- 11.Guinebretiere, M. H., and C. Nguyen-The. 2003. Sources of Bacillus cereus contamination in a pasteurized zucchini purée processing line, differentiated by two PCR-based methods. FEMS Microbiol. Ecol. 43207-215. [DOI] [PubMed] [Google Scholar]
- 12.Guinebretiere, M. H., F. L. Thompson, A. Sorokin, P. Normand, P. Dawyndt, M. Ehling-Schulz, B. Svensson, V. Sanchis, C. Nguyen-The, M. Heyndrickx, and P. De Vos. 2008. Ecological diversification in the Bacillus cereus group. Environ. Microbiol. 10851-865. [DOI] [PubMed] [Google Scholar]
- 13.Joyce, M. G., C. Levy, K. Gabor, S. M. Pop, B. D. Biehl, T. I. Doukov, J. M. Ryter, H. Mazon, H. Smidt, R. H. van den Heuvel, S. W. Ragsdale, J. van der Oost, and D. Leys. 2006. CprK crystal structures reveal mechanism for transcriptional control of halorespiration. J. Biol. Chem. 28128318-28325. [DOI] [PubMed] [Google Scholar]
- 14.Khoroshilova, N., H. Beinert, and P. J. Kiley. 1995. Association of a polynuclear iron-sulfur center with a mutant FNR protein enhances DNA binding. Proc. Natl. Acad. Sci. USA 922499-2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kohler, J. J., S. J. Metallo, T. L. Schneider, and A. Schepartz. 1999. DNA specificity enhanced by sequential binding of protein monomers. Proc. Natl. Acad. Sci. USA 9611735-11739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korner, H., H. J. Sofia, and W. G. Zumft. 2003. Phylogeny of the bacterial superfamily of Crp-Fnr transcription regulators: exploiting the metabolic spectrum by controlling alternative gene programs. FEMS Microbiol. Rev. 27559-592. [DOI] [PubMed] [Google Scholar]
- 17.Kotiranta, A., K. Lounatmaa, and M. Haapasalo. 2000. Epidemiology and pathogenesis of Bacillus cereus infections. Microbes Infect. 2189-198. [DOI] [PubMed] [Google Scholar]
- 18.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 19.Maier, T., N. Drapal, M. Thanbichler, and A. Bock. 1998. Strep-tag II affinity purification: an approach to study intermediates of metalloenzyme biosynthesis. Anal. Biochem. 25968-73. [DOI] [PubMed] [Google Scholar]
- 20.Okstad, O. A., M. Gominet, B. Purnelle, M. Rose, D. Lereclus, and A. B. Kolsto. 1999. Sequence analysis of three Bacillus cereus loci carrying PIcR-regulated genes encoding degradative enzymes and enterotoxin. Microbiology 1453129-3138. [DOI] [PubMed] [Google Scholar]
- 21.Ouhib, O., T. Clavel, and P. Schmitt. 2006. The production of Bacillus cereus enterotoxins is influenced by carbohydrate and growth rate. Curr. Microbiol. 53222-226. [DOI] [PubMed] [Google Scholar]
- 22.Pelley, J. W., C. W. Garner, and G. H. Little. 1978. A simple rapid biuret method for the estimation of protein in samples containing thiols. Anal. Biochem. 86341-343. [DOI] [PubMed] [Google Scholar]
- 23.Pomerantz, J. L., S. A. Wolfe, and C. O. Pabo. 1998. Structure-based design of a dimeric zinc finger protein. Biochemistry 37965-970. [DOI] [PubMed] [Google Scholar]
- 24.Pop, S. M., N. Gupta, A. S. Raza, and S. W. Ragsdale. 2006. Transcriptional activation of dehalorespiration. Identification of redox-active cysteines regulating dimerization and DNA binding. J. Biol. Chem. 28126382-26390. [DOI] [PubMed] [Google Scholar]
- 25.Pop, S. M., R. J. Kolarik, and S. W. Ragsdale. 2004. Regulation of anaerobic dehalorespiration by the transcriptional activator CprK. J. Biol. Chem. 27949910-49918. [DOI] [PubMed] [Google Scholar]
- 26.Ramarao, N., and D. Lereclus. 2006. Adhesion and cytotoxicity of Bacillus cereus and Bacillus thuringiensis to epithelial cells are FlhA and PlcR dependent, respectively. Microbes Infect. 81483-1491. [DOI] [PubMed] [Google Scholar]
- 27.Reents, H., I. Gruner, U. Harmening, L. H. Bottger, G. Layer, P. Heathcote, A. X. Trautwein, D. Jahn, and E. Hartig. 2006. Bacillus subtilis Fnr senses oxygen via a [4Fe-4S] cluster coordinated by three cysteine residues without change in the oligomeric state. Mol. Microbiol. 601432-1445. [DOI] [PubMed] [Google Scholar]
- 28.Reinhart, F., S. Achebach, T. Koch, and G. Unden. 2008. Reduced apo-fumarate nitrate reductase regulator (apoFNR) as the major form of FNR in aerobically growing Escherichia coli. J. Bacteriol. 190879-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosenfeld, E., C. Duport, A. Zigha, and P. Schmitt. 2005. Characterisation of aerobic and anaerobic vegetative growth of the food-borne pathogen Bacillus cereus. J. Can. Microbiol. 51149-158. [DOI] [PubMed] [Google Scholar]
- 30.Schoeni, J. L., and A. C. Wong. 2005. Bacillus cereus food poisoning and its toxins. J. Food Prot. 68636-648. [DOI] [PubMed] [Google Scholar]
- 31.Slamti, L., and D. Lereclus. 2002. A cell-cell signaling peptide activates the PlcR virulence regulon in bacteria of the Bacillus cereus group. EMBO J. 214550-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spira, W. M., and J. M. Goepfert. 1975. Biological characteristics of an enterotoxin produced by Bacillus cereus. Can. J. Microbiol. 211236-1246. [DOI] [PubMed] [Google Scholar]
- 33.Zigha, A., E. Rosenfeld, P. Schmitt, and C. Duport. 2006. Anaerobic cells of Bacillus cereus F4430/73 respond to low oxidoreduction potential by metabolic readjustments and activation of enterotoxin expression. Arch. Microbiol. 185222-233. [DOI] [PubMed] [Google Scholar]
- 34.Zigha, A., E. Rosenfeld, P. Schmitt, and C. Duport. 2007. The redox regulator Fnr is required for fermentative growth and enterotoxin synthesis in Bacillus cereus F4430/73. J. Bacteriol. 1892813-2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.