Abstract
DivIB, also known as FtsQ in gram-negative organisms, is a division protein that is conserved in most eubacteria. DivIB is localized at the division site and forms a complex with two other division proteins, FtsL and DivIC/FtsB. The precise function of these three bitopic membrane proteins, which are central to the division process, remains unknown. We report here the characterization of a divIB deletion mutant of Streptococcus pneumoniae, which is a coccus that divides with parallel planes. Unlike its homologue FtsQ in Escherichia coli, pneumococcal DivIB is not required for growth in rich medium, but the ΔdivIB mutant forms chains of diplococci and a small fraction of enlarged cells with defective septa. However, the deletion mutant does not grow in a chemically defined medium. In the absence of DivIB and protein synthesis, the partner FtsL is rapidly degraded, whereas other division proteins are not affected, pointing to a role of DivIB in stabilizing FtsL. This is further supported by the finding that an additional copy of ftsL restores growth of the ΔdivIB mutant in defined medium. Functional mapping of the three distinct α, β, and γ domains of the extracellular region of DivIB revealed that a complete β domain is required to fully rescue the deletion mutant. DivIB with a truncated β domain reverts only the chaining phenotype, indicating that DivIB has distinct roles early and late in the division process. Most importantly, the deletion of divIB increases the susceptibility to β-lactams, more evidently in a resistant strain, suggesting a function in cell wall synthesis.
Cell division is a vital process; indeed, it may define life itself. Understanding cell division in microorganisms is also important in order to design innovative therapeutic strategies. In this work, we investigated an aspect of bacterial cell division in the pathogen Streptococcus pneumoniae, which is estimated by the WHO to cause more than 1 million deaths per year worldwide. The pneumococcus has the shape of an elongated ellipsoid surrounded by a circular outgrowth of cell wall that marks the site of the next division, perpendicular to the long axis. The term “ovococci” was recently proposed to distinguish such cells (streptococci, enterococci, and lactococci) from truly spherical cocci (e. g., Staphylococcus aureus) (53). Pneumococcal cells are generally found as diplococci or short chains. The mechanisms of division, and more generally morphogenesis, have been studied mostly in the model organisms Escherichia coli and Bacillus subtilis (18, 20, 23). These numerous studies have uncovered several components of the divisome, which can be defined, if not as a complex, at least as the functional ensemble of proteins localized at the division site and participating in the process. Eight conserved, mostly essential proteins constitute the core of the divisome: FtsZ, FtsA, FtsK, FtsQ/DivIB, FtsL, FtsB/DivIC, FtsW, and FtsI. This core set of proteins is found in both gram-positive and gram-negative organisms. Additional, less conserved division proteins are found in various phylogenetic lineages. The core division proteins are listed here in the conditional order of their recruitment at the division site of E. coli (for a review, see reference 23). Progress has been made on several aspects of cell division, and some functions can be attributed to several division proteins (20, 23). FtsZ forms polymers with a circular distribution on the cytoplasmic side of the membrane at the site of division and governs the recruitment of the other proteins. The dimerization or polymerization of FtsA stabilizes the FtsZ ring, and FtsA may mediate the interaction between FtsZ and the membrane (31, 47). FtsK is involved in chromosome segregation and membrane fusion. FtsI, which is a septal penicillin-binding protein, and probably FtsW participate in cell wall synthesis. However no precise function is known for the three proteins FtsQ/DivIB, FtsL, and FtsB/DivIC.
Despite some differences in the conditional order of recruitment between E. coli and B. subtilis (20), it appears that recruitment of FtsQ/DivIB, FtsL, and FtsB/DivIC occurs in the middle of the process. In E. coli, the localization of FtsQ at the division site is a prerequisite for the recruitment of FtsL and FtsB, and the presence of FtsL and FtsB at the division site is mutually dependent (7, 22). In B. subtilis, the presence of FtsL and DivIC at mid-cell also depends on the presence of DivIB and reciprocally, at least at the temperature at which DivIB is essential (14, 30). These results suggested strongly the existence of a complex of these three proteins. Through their extracellular region predicted to form a coiled coil, a direct interaction between FtsL and FtsB/DivIC was expected. The formation of the ternary complex was confirmed by yeast and bacterial triple-hybrid experiments (16, 28). It was isolated by coimmunoprecipitation in E. coli (6) and reconstituted in vitro with recombinant soluble forms of the pneumococcal proteins (37).
DivIB is a bitopic membrane protein with a variable N-terminal cytoplasmic region, a single transmembrane segment, and a periplasmic or extracellular domain containing a conserved region of about 200 amino acids adjacent to the transmembrane segment. The extracellular domain is important for the function and is necessary and sufficient for the localization of FtsQ/DivIB, provided that it is anchored to the membrane (5, 9, 12, 27, 29), although the transmembrane segment also contributes to the septal localization of FtsQ/DivIB (46, 51). FtsQ is a low-abundance protein, with an estimated abundance of about 50 molecules per cell in E. coli (9). DivIB in B. subtilis and S. pneumoniae was estimated to be somewhat more abundant, with 5,000 and 200 molecules per cell, respectively (37, 44).
Differences also exist between various species in regard to the essentiality of ftsQ/divIB under laboratory conditions. The gene ftsQ is essential in E. coli (5, 9, 12, 27). In Streptomyces coelicolor, divIB is dispensable for mycelial growth but is required for cell division (34). In B. subtilis, divIB is essential only at high temperature (2). This essentiality of divIB could be attributed to its role in stabilizing at high temperature the essential FtsL. FtsL has a high turnover in B. subtilis and disappears rapidly after its expression is stopped (14). At high temperature in a thermosensitive divIB mutant, FtsL also disappears rapidly, indicating the stabilizing role of DivIB. Note that this effect could explain the absence of FtsL at the division site in the divIB thermosensitive mutant (14). In addition, overexpression of FtsL alleviates the essentiality of divIB (14). In B. subtilis, FtsL is degraded by the membrane protease YluC (3). It is not known if the removal of FtsL itself is important or if degradation products are also effectors of the regulation of cell division. In E. coli, FtsL is also regulated at the transcriptional level by DnaA to stop cell division in case of DNA damage (25). In contrast to its effect on FtsL, DivIB regulates negatively the amount of DivIC in B. subtilis. That is, the amount of DivIC decreases in the absence of FtsL when DivIB is present but remains constant in the absence of DivIB (14, 16).
The study of morphologically distinct organisms can bring important new insights, as exemplified by recent studies on the division of Caulobacter crescentus (4, 19), Staphylococcus aureus (39), or S. coelicolor (1, 32). With this aim of providing new data on the function of DivIB, we report here the characterization of a divIB deletion mutant of S. pneumoniae. Whereas both B. subtilis and E. coli are rod-shaped bacteria, S. pneumoniae is an elongated coccus, or “ovococcus,” with parallel planes of division (53). Like B. subtilis, the pneumococcus is gram positive, while E. coli is gram negative. In E. coli, the dcw cluster forms essentially one operon and ftsQ is adjacent to ftsA and ftsZ, whereas in B. subtilis, the divIB gene is separated from ftsA and ftsZ by intervening genes. In S. pneumoniae, the dcw genes are distributed in three distinct regions and divIB is associated with mur genes that are involved in cell wall synthesis but is separated from ftsA and ftsZ (33).
The periplasmic region of FtsQ/DivIB is organized in three domains termed α, β, and γ (Fig. 1), following a structural study carried out with recombinant DivIB from Geobacillus stearothermophilus (43) and recent localization experiments in B. subtilis (51). The crystal structures of the periplasmic domains of FtsQ from E. coli and Yersinia enterocolitica have been reported recently (49). The α domain is proximal to the cytoplasmic membrane and coincident with the polypeptide transport-associated domain, which was proposed previously to function as a molecular chaperone (43, 45, 49). The α domain in recombinant soluble form of the extracellular part of DivIB from S. pneumoniae, like that of G. stearothermophilus, is readily digested by trypsin and was found to be largely unfolded by nuclear magnetic resonance spectroscopy (S. Masson and A. Zapun, unpublished data). The β domain is compact and resistant to tryptic digestion (covering residues 220 to 361, as determined by N-terminal sequencing (VKEYDIVA) and electrospray mass spectrometry (1,5841 Da) (I. Petit, unpublished data). The structure of the β domain from G. stearothermophilus was solved by nuclear magnetic resonance spectroscopy (43). The α and β domains constitute the 200-residue-long conserved region of FtsQ/DivIB. The C-terminal γ domain either is highly variable or is absent is some species (e.g., Haemophilus influenzae and Legionella pneumophila). The sequence of the C-terminal tail suggests that it is unfolded, and it is digested by trypsin in recombinant DivIB proteins. The 15 C-terminal residues of E. coli FtsQ that could be attributed to a γ domain, on the basis of sequence analysis, were found to be disordered in the crystal structure (49). Some uncertainties regarding the limit of the β and γ domains are discussed and addressed in this work by complementation experiments with pneumococcus.
FIG. 1.
Sequence alignment of the DivIB extracytoplasmic region. The alignment was generated with ClustalW, using in addition to the sequences shown those of Yersinia pestis, Vibrio cholerae, C. crescentus, H. influenzae, Mycobacterium tuberculosis, S. coelicolor, Bacillus bacilliformis, Corynebacterium glutamicum, Rhizobium meliloti, Listeria monocytogenes, Enterococcus hirae, S. aureus, L. pneumophila, Neisseria meningitidis, Borrelia burgdorferi, Streptococcus pyogenes, and Geobacillus kaustophilus. The domains were defined by limited tryptic proteolysis of recombinant pneumococcal DivIB. Residue 336 corresponds to the C terminus of the β domain defined by the tryptic digestion of DivIB from G. stearothermophilus (43).
Lastly, several clues point to a role of FtsQ/DivIB in the regulation of cell wall synthesis at a late stage of division (11, 35). No gene encoding an FtsQ/DivIB orthologue was identified in the genomes of bacteria without cell walls (32). Moreover, one E. coli mutant isolated following culture over many generations in the presence of penicillin and sucrose grows as a protoplast L-form type. Such mutants divide with a much reduced amount of cell wall. In one such strain the ftsQ gene codes for a truncated protein in the periplasmic domain after residue 131 (48). In E. coli, the overexpression of FtsQ exacerbates the dominant negative effect of PBP3 mutants (38). Also, when divIB is deleted in B. subtilis, sporulation efficiency drops dramatically (41), but the polar septa that form nevertheless are thicker, supporting a role of DivIB in the regulation of the synthesis of the peptidoglycan during sporulation (41). Lastly, two sib mutants (for “suppressor of divIB”) were isolated by spreading the divIB null mutant of B. subtilis on nutritive agar at 49°C. These suppressor mutations result in single amino acid substitutions in PBP2B (the orthologue of FtsI), which participate in the synthesis of the septal peptidoglycan during division. Both amino acid changes are in the noncatalytic N-terminal domain of the extracytoplasmic region of PBP2B and are sufficient to circumvent the need for DivIB (16). Finally, the coordinated expression of divIB and murB is important for growth and sporulation in B. subtilis (42). Increased expression of divIB reduces the requirement for MurB, an enzyme that participate in the synthesis of the peptidoglycan precursor. Here, we report another observation that supports a role for DivIB in peptidoglycan synthesis, as the deletion of divIB increases specifically the sensitivity to β-lactam antibiotics known to target cell wall synthesis.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and transformation.
The bacterial strains used in this work are listed in Table 1. Cells were grown at 37°C in an atmosphere of 95% air and 5% CO2 in 40 ml of Bacto Todd-Hewitt (TH) medium (Difco) or in a chemically defined (CD) medium (50) supplemented with 0.5 g liter−1 choline but without l-asparagine, l-cystine, hydroxy-l-proline, and l-tyrosine. Strains harboring resistance determinants were routinely grown in the presence of kanamycin (500 mg liter−1) or chloramphenicol (4.5 mg liter−1). Antibiotics were omitted for the determination of growth curves. For growth in CD broth, the inocula were washed three times with CD medium prior to inoculation. Alternatively, 4.5-ml cultures were grown in triplicates in 12-well cell culture plates (Cellstar; Greiner Bio-One) sealed with a transparent film (Manco Crystal Clear sealing tape). Plates were incubated at 37°C, and the turbidity was monitored at 595 nm in a Fluostar reader (BMG optima). Competent cells were generated and transformations were carried out as described previously (13).
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Genotype | Description | Source or reference |
|---|---|---|---|
| Strains | |||
| R6 | Wild type | Noncapsulated D39 derivative | Laboratory stock |
| R61 | R6 ami::PM-divIB, Kanr | R6 transformed with a ligation mixture of NcoI/BamHI-digested pCEP and divIB PCR fragment, Kanr | This study |
| R62 | R6 ami::PM-divIC, Kanr | R6 transformed with a ligation mixture of NcoI/BamHI-digested pCEP and divIC PCR fragment, Kanr | This study |
| R63 | R6 ami::PM-ftsL, Kanr | R6 transformed with a ligation mixture of NcoI/BamHI-digested pCEP and ftsL PCR fragment, Kanr | This study |
| R62X5204 | R6 pbp2x5204 | R6 transformed with pbp2x of strain 5204 | 8 |
| B2 | R6 divIB::cat | Camr | This study |
| B21 | R6 divIB::cat ami::PM-divIB | B2 transformed with a ligation mixture of NcoI/BamHI-digested pCEP and divIB PCR fragment, Camr Kanr | This study |
| B22 | R6 divIB::cat ami::PM-divIC | B2 transformed with a ligation mixture of NcoI/BamHI-digested pCEP and divIC PCR fragment, Camr Kanr | This study |
| B23 | R6 divIB::cat ami::PM-ftsL | B2 transformed with a ligation mixture of NcoI/BamHI-digested pCEP and ftsL PCR fragment, Camr Kanr | This study |
| B24 | R6 divIB::cat bgaA::PfcsK-divIB | B2 transformed with pLIM101, Camr Kanr | This study |
| B25 | R6 divIB::cat bgaA::PfcsK-divIBΔγ | B2 transformed with pLIM102, Camr Kanr | This study |
| B26 | R6 divIB::cat bgaA::PfcsK-divIBE222A | B2 transformed with pLIM103, Camr Kanr | This study |
| B27 | R6 divIB::cat bgaA::PfcsK-divIBΔβγ | B2 transformed with pLIM104, Camr Kanr | This study |
| B28 | R6 divIB::cat bgaA::PfcsK-divIBS337* | B2 transformed with pLIM105, Camr Kanr | This study |
| B29 | R6 divIB::cat bgaA::PfcsK-divIBΔec | B2 transformed with pLIM106, Camr Kanr | This study |
| B22X5204 | R6 divIB::catpbp2x5204 | R62X5204 transformed with divIB::cat PCR product, Camr | This study |
| Plasmids | |||
| pCEP | Expression platform at ami locus with maltose-induced promoter | 26 | |
| pLIM100 | Expression platform at bgaA locus with fucose-induced promoter | L. Roux and A. Zapun, unpublished data | |
| pLIM101 | Modification of pLIM100 for expression of DivIB | This study | |
| pLIM102 | Modification of pLIM100 for expression of DivIBΔγ | This study | |
| pLIM103 | Modification of pLIM100 for expression of DivIBE222A | This study | |
| pLIM104 | Modification of pLIM100 for expression of DivIBΔβγ | This study | |
| pLIM105 | Modification of pLIM100 for expression of DivIBS337* | This study | |
| pLIM106 | Modification of pLIM100 for expression of DivIBΔec | This study |
Plasmids and DNA constructions.
The plasmids and primers used in this work are listed in Tables 1 and 2. To construct divIB, divIC, and ftsL null strains, competent cells of the R6 strain were transformed with PCR products consisting of a chloramphenicol resistance (cat) cassette flanked by fragments, about 500 bp long, homologous to the regions adjacent to each target genes, according to the method of Fadda et al. (21), and plated onto chloramphenicol-containing Columbia blood agar. Transformants were further isolated twice on chloramphenicol plates. Insertion in the divIB gene was checked by the length of a PCR product obtained with the external primers divIB-F0 and divIB-R7. The absence of DivIB was checked by immunoblotting.
TABLE 2.
Primers used in this study
| Function and primer name | Sequence (5′ to 3′)a | Restriction site |
|---|---|---|
| Deletion of divIB | ||
| divIB-F1 | GCTCTTGGCGATAGCAAAAT | |
| divIB-R2 | TCAAACAAATTTTCATCAAGCTTCTTTAGACTCTGGTTCTTCTTT | |
| divIB-F3 | AAAGAAGAACCAGAGTCTAAAGAAGCTTGATGAAAATTTGTTTGA | |
| divIB-R4 | GTCAATCGAATTAAATCGGATGTTCTAGAACTAGTGGATCCCCCGG | |
| divIB-F5 | CCGGGGGATCCACTAGTTCTAGAACATCCGATTTAATTCGATTGAC | |
| divIB-R6 | CCCCAAAAATTCACCATCTG | |
| Deletion of divIC | ||
| divIC-F1 | AGACCGTTTTGGAGAATACCC | |
| divIC-R2 | TCAAACAAATTTTCATCAAGCTTGTATTCATTTTGAATAAAAGAATT | |
| divIC-F3 | AATTCTTTTATTCAAAATGAATACAAGCTTGATGAAAATTTGTTTGA | |
| divIC-R4 | TTGATACTGAGTTTGCAAGTCTGTCTAGAACTAGTGGATCCCCCGG | |
| divIC-F5 | CCGGGGGATCCACTAGTTCTAGACAGACTTGCAAACTCAGTATCAA | |
| divIC-R6 | TCATTCGCATAAACAGGAACC | |
| Deletion of ftsL | ||
| ftsL-F1 | AAGTGAAGCCGATTGAGACAA | |
| ftsL-R2 | TCAAACAAATTTTCATCAAGCTTGTTTAAGTTGCATCTGTAGTAT | |
| ftsL-F3 | ATACTACAGATGCAACTTAAACAAGCTTGATGAAAATTTGTTTGA | |
| ftsL-R4 | CTTGCTTGGCATCGTCCAATTTCTAGAACTAGTGGATCCCCCGG | |
| ftsL-F5 | CCGGGGGATCCACTAGTTCTAGAAATTGGACGATGCCAAGCAAG | |
| ftsL-R6 | TTGTCCGTTTGGGTAACTACG | |
| Gene amplification for ligation into pCEP | ||
| divIB-D1 | CTGCCATGGCGTCAAAAGATAAGAAAAATGAGG | NcoI |
| divIB-R | CGCCCTCGAGGATCCACTAGTCTAGCGACGCGATGAACGC | BamHI |
| divIC-D1 | CTGCCATGGCGTCTAAAAATATTGTACAATTGAATAATTC | NcoI |
| divIC-R | CGCCCTCGAGGATCCACTAGTTCACCTTTGAAGCAAGTCAGG | BamHI |
| ftsL-D1 | CTGCCATGGCGGCAGAAAAAATGGAAAAAACAGG | NcoI |
| ftsL-R | CGCCCTCGAGGATCCACTAGTTTACTCCGCTATTCTAATATTTTCATTG | BamHI |
| Gene amplification for ligation into pET-30b(+) | ||
| divIB-F | GGTGCCATATGTCGACGTCAAAAGATAAGAAAAATGAGGACAAAGAAACC | NdeI |
| External primer | ||
| divIB-F0 | GAAAGAACTAACAGAGCGCTAC | |
| divIB-R7 | GATACCTGTTGCAAGTCCAGC | |
| amiF-F1 | CCGAATTAGCACGTTATCAAAAAGG | |
| treR-R | CGCCAAATCGCTGAGCAG | |
| Mutagenesis of divIB in pET30b-divIB | ||
| divIB E222A-F | GGTCAAGGCATATGATATTGTGGCC | |
| divIB E222A-R | GGCCACAATATCATATGCCTTGACC | |
| divIB Δγ-F | GGAAAAAGCCTAGCTAGCGGCCAAGG | NheI |
| divIB Δγ-R | CCTTGGCCGCTAGCTAGGCTTTTTCC | NheI |
| divIB S337*-F | GCCACAATTGTCAGAACCGTGAGTGGTCG | |
| divIB S337*-R | CGACCACTCACGGTTCTGACAATTGTGGC | |
| divIB Δβγ-F | CCAACTAAGTTCACTATCTAGATCAAGG | XbaI |
| divIB Δβγ-R | CCTTGATCTAGATAGTGAACTTAGTTGG | XbaI |
| divIB Δec-F | CAGTCCTTAATCGATCATGAAAGATATTCG | ClaI |
| divIB Δec-R | CGAATATCTTTCATGATCGATTAAGGACTG | ClaI |
Restriction sites are underlined.
To construct DivIB-, DivIC-, and FtsL-overexpressing strains, the respective genes were inserted in the chromosomes of the R6 and B2 strains downstream of the ami operon, under the control of a maltose-induced promoter (PM) and with a kanamycin resistance gene, by transformation with ligation mixtures of NcoI/BamHI-digested PCR products and pCEP plasmid (26). The insertions were checked by PCR with the primers amiF-F1 and treR-R, and the inserted genes were sequenced.
Alternatively, divIB, or alleles for truncated variants (divIBΔγ, divIBE222A, divIBS337*, divIBΔβγ, and divIBΔec) were introduced at the bgaA locus under the control of a fucose-regulated promoter (Pfcsk) with a kanamycin resistance gene, using the pLIM100 plasmid (L. Roux and A Zapun, unpublished data). The divIB PCR product (primers divIB-F and divIB-R) was introduced as a NdeI and BamHI fragment into pET30b (Novagen). The alleles coding for truncated proteins were obtained by the introduction of stop codons using the QuikChange site-directed mutagenesis kit (Stratagene) and the primers described in Table 2. The variant genes obtained were subcloned into plasmid pLIM100 with NdeI and BamHI restriction enzymes. All plasmids were checked for the desired mutations by DNA sequencing.
Evaluation of protein stability in the divIB-deleted strain.
Protein expression was inhibited in exponentially growing cultures in TH medium at an optical density at 600 nm of 0.3 by the addition of erythromycin (40 mg liter−1). Aliquots were withdrawn after various time intervals, cells were immediately harvested, and pellets were resuspended in 100 μl of lysis buffer (Cell Lytic [Sigma] supplemented with 1 mM MgCl2, 190 U liter −1 DNase, 10 mg liter −1 lysozyme, and Complete protease inhibitor cocktail [Roche]) and incubated 15 min at room temperature prior to the addition of 40 μl of reducing XT sample buffer (Bio-Rad) and boiling for 3 min. Samples (adjusted according to the optical density of the culture) were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on 4 to 12% gradient Criterion gels (Bio-Rad) and immunoblotting.
Optical microscopy.
For morphological observations using phase-contrast microscopy, 50 μl of exponentially growing culture was deposited on a slide and briefly heated over a flame before the overlaying of 50 μl of phosphate-buffered saline (PBS) containing 0.2 mg liter−1 of DAPI (4′,6′-diamidino-2-phenylindole) and mounting. Samples were examined with an Olympus BX61 microscope equipped with a UPFLN 100× O-2PH/1.3 objective and a QImaging Retiga-SRV 1394 cooled charge-coupled device camera. Image processing was performed using the Volocity software package. Cell measurements and analyses were performed with NIH ImageJ software version 1.36b (40).
Electron microscopy.
For whole-cell transmission electron microscopy (TEM), bacteria were negatively stained with 2% (wt/vol) uranyl acetate using the mica floatation technique. Briefly, 4 μl of bacteria at an optical density of 0.3, washed twice in PBS, was applied to the edge of a 5-mm2 piece of carbon-coated mica and allowed to infiltrate between the carbon and mica. After approximately 30 s, the carbon-mica was introduced into a well containing 500 μl of uranyl acetate and the carbon sheet was allowed to float on the stain for 30 s. A copper grid was then placed on the carbon, and the specimen was picked up with tweezers. The excess liquid was removed with blotting paper, and the specimen was air dried.
For TEM of ultrathin sections, exponentially growing cells were centrifuged at 7,000 × g for 3 min. The pellets were fixed for 1 h at room temperature in a solution of 2% glutaraldehyde in TEM buffer (100 mM sodium cacodylate, pH 6.8). Fixed cells were washed twice in TEM buffer and postfixed for 1 h in 1% osmium tetroxide in TEM buffer. The dehydration was performed using ethanol, and the cell pellets were embedded in Epon before sectioning. Thin sections were double stained with uranyl acetate and lead citrate. Specimens were examined using a Philips CM12 electron microscope equipped with a LaB6 filament operating at 100 kV.
For scanning electron microscopy, cultures were rapidly chilled, washed in 10 mM phosphate (pH 7.0), and fixed in 1% paraformaldehyde and 1.25% glutaraldehyde in 0.15 M sodium cacodylate buffer for 4 h at room temperature. Fixed cells were washed with PBS, transferred onto a circular cover glass, and dehydrated using increasing concentrations of acetone; this was followed by critical-point drying using CO2. Samples were then coated with platinum using an Emitech 575 turbo sputtering apparatus and examined with an FE Hitachi S4000 scanning electron microscope operating at 15 to 20 kV.
Susceptibility to antimicrobials.
The MICs of penicillin G, cefotaxime, ofloxacin, and gentamicin were determined using the E-test method. Liquid overnight cultures were plated at a 0.5 McFarland standard on Columbia 4% blood agar, E-test strips (AB Biodisk) were laid on the plates, and the MICs were read after 18 to 24 h of incubation.
The effect of penicillin G, cefotaxime, and sodium azide on liquid cultures in TH broth at 37°C was monitored by the optical density at 600 nm. The antimicrobials were added once the cultures had reached a density of 0.3. The β-lactams were added at fourfold their MICs in liquid broth (0.68 mg liter−1 and 0.34 mg liter−1 for R6 and B2, respectively), and NaN3 was at 0.2%.
RESULTS
Pneumococcal divIB is not essential for growth in rich medium.
To test whether divIB, divIC, and ftsL are required for cell division in S. pneumoniae, we tried to inactivate the three genes individually. The R6 laboratory strain was transformed with a PCR product consisting of a chloramphenicol resistance (cat) cassette flanked by extensions that are homologous to the regions 3′ and 5′ of each target gene. We were able to obtain insertion-deletion mutations in the divIB gene (about 50 colonies per transformation) but not in either ftsL or divIC, despite numerous attempts. Therefore, FtsL and DivIC appear to be essential in S. pneumoniae, but DivIB does not. The correct insertion of the cat cassette at the divIB locus was confirmed by PCR with external primers. In divIB-deleted strains, codons 100 through 297 of divIB are replaced by a stop codon and the cat cassette. The first 100 codons of divIB encode part of the N-terminal cytoplasmic region with a sequence unlikely to adopt a folded conformation. In the absence of the transmembrane segment, this N-terminal fragment of DivIB, if expressed, is unlikely to be functional. Five clones were examined for their growth rate and morphology by phase-contrast microscopy. One representative clone, termed B2, was chosen for further characterization.
Immunoblotting confirmed the absence of wild-type DivIB protein in B2 (not shown). The growth rate of strain B2 in TH medium, monitored by the culture turbidity, was comparable to that of the parent strain R6 (mass doubling time of 45 min measured in 12-well plates) during the exponential phase, and the maximum densities were similar. However, strain B2 lysed sooner than R6 following the stationary phase (not shown). B. subtilis deleted of divIB does not divide at high temperature, and we examined the growth of pneumococcus at 40°C, the maximum temperature that supports physiological growth of S. pneumoniae without morphological changes (D. Fadda and O. Massidda, unpublished data). The growth rates of R6 and B2 were similar and were higher at 40°C than at 37°C.
The absence of DivIB affects the morphology of pneumococcus.
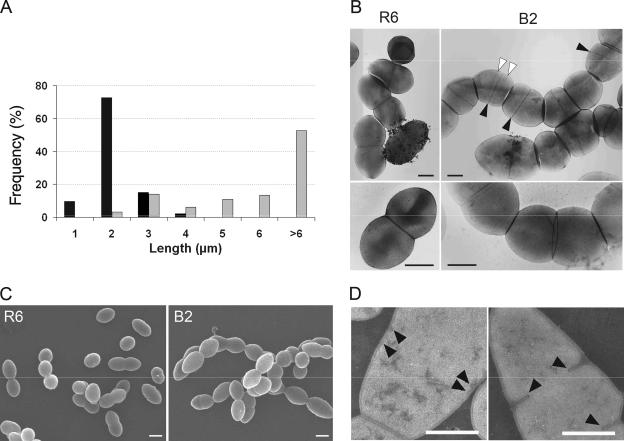
Examination by phase-contrast microscopy revealed that strain B2 formed longer and more abundant chains than the parental R6 strain. To evaluate this phenotype quantitatively, the chain length distribution was determined (Fig. 2A). While the average chain length of the wild-type R6 was 1.6 μm (corresponding mostly to diplococci) at an optical density at 600 nm of 0.45, most chains of B2 were longer than 6 μm in the same conditions. The average chain length seemed to drop at the inoculation of a fresh culture and increase with cell density. The increasing chain length would explain why the number of CFU increased more slowly that the total cell mass monitored by turbidity (not shown).
FIG. 2.
Phenotypes of the ΔdivIB mutant. Bacteria were grown in TH broth at 37°C under 5% CO2 to an optical density at 600 nm of 0.45 and prepared for microscopy. (A) Chain length distribution of strains R6 (black) and B2 (gray) (165 measurements each). (B) Transmission electron micrographs of negatively stained pneumococci (images were taken at nominal magnifications of ×10,000 or ×22,000 [top and bottom, respectively]). White arrowheads indicate equatorial rings, and black arrowheads point to septa. (C) Scanning electron micrographs. (D) Transmission electron micrographs of negatively stained thin sections of B2. Arrowheads point to nascent septa. Bars, 0.5 μm.
The morphology of most cells in B2 chains appeared normal as observed by phase-contrast light microscopy, scanning electron microscopy, and negative-stain TEM (Fig. 2B and C). Chains consist of linked diplococci and thus result likely from a defect in the last steps of cell separation. The transmission electron micrographs in Fig. 2B require some comments, as they permit the unusual visualization of the cell surface in transparency. The equatorial annular outgrowths are visible on some cells, as well as rings marking the outer edge of septa. The position of septa can be distinguished from cell-cell interfaces that appear as dark discs due to the trapping of dye between cells.
Besides morphologically normal cells, aberrantly shaped cells were also found in B2 cultures. They probably represented fewer than 5% of all cells, but they were difficult to quantify as there is a continuum of forms between normal cells and very clearly enlarged cells. Electron micrographs of negatively stained thin sections of such larger and deformed cells revealed the presence of multiple septa (Fig. 2D).
Pneumococcal divIB is essential in CD medium, and growth is rescued by an additional copy of ftsL.
In CD medium at 37°C, the R6 strain grew with a mass doubling time comparable to that in rich medium (45 min) after a long lag phase of about 10 hours following inoculation. In contrast, strain B2 did not grow in CD medium whereas it grew in TH medium, showing that divIB is essential under these growth conditions (Table 3). Addition of l-asparagine or l-tyrosine (20 mg liter−1) did not restore growth, nor did the addition of 1% NaCl. Confirming that the washing steps with CD medium did not kill B2, growth resumed following inoculation of TH medium. Complementation of B2 by insertion of divIB at the ami or bgaA locus restored growth in CD medium and the wild-type morphology, that is, a chain length shorter than 2 μm corresponding mostly to diplococci (Table 3; see Fig. 4). When inserted downstream of the ami operon, divIB was under the control of a maltose-inducible promoter (26). In the absence of the MalR repressor on a multicopy plasmid, this promoter is known to cause expression even without inducer (26). DivIB was slightly overexpressed in R6, with or without added maltose, as estimated by immunoblotting (not shown). In B2 complemented at the ami locus, the amount of DivIB produced in the absence of maltose was somewhat lower than in the wild-type R6 strain but was sufficient to restore wild-type morphology and growth in defined medium. With the aim of obtaining a lower basal expression level of DivIB, divIB was inserted at the dispensable bgaA locus (52) under the control of a fucose-inducible promoter (10, 36). The expression of DivIB in the B2 strain in the presence of fucose was over 10-fold greater than the wild-type level, as judged by immunoblotting. In the absence of fucose the amount of DivIB was comparable to that in wild-type R6, and the wild-type phenotypes were also restored. Addition of sucrose or glucose did not significantly reduce the basal levels of expression of DivIB with either promoter. It was therefore impossible to reduce the expression of DivIB enough to observe the fate of cells following depletion of DivIB.
TABLE 3.
Introduction of divIB, divIC, and ftsL at the ami locus in strains R6 and B2
| Parental strain | Genotype | Growtha in:
|
Average chain length (μm)b in:
|
||
|---|---|---|---|---|---|
| CD medium | TH medium | CD medium | TH medium | ||
| R6 | Wild type | + | + | 1.5 | 1.6 |
| ami::PM-divIB | + | + | 1.6 | 1.9 | |
| ami::PM-divIC | + | + | 1.4 | 1.9 | |
| ami::PM-ftsL | + | + | 1.4 | 3.3 | |
| B2 | divIB::cat | − | + | − | 6.6 |
| divIB::cat ami::PfcsK-divIB | + | + | 1.4 | 1.8 | |
| divIB::cat ami::PfcsK-divIC | − | + | − | 4.2 | |
| divIB::cat ami::PfcsK-ftsL | + | + | 3.4 | 6.5 | |
Growth was monitored at 590 nm in 12-well plates. +, doubling time of about 45 min; −, absence of growth for 35 h.
Strains were cultivated to an optical density at 600 nm of between 0.3 and 0.5, and 200 particles were measured for each strain.
FIG. 4.
Functional characterization of the DivIB domains. The cytoplasmic N-terminal domain is in white. The transmembrane segment is hatched. The three domains of the extracellular region are in various shades of gray. Alleles encoding DivIB variants were introduced in B2 at the bgaA locus. Exponentially growing cultures were analyzed by phase-contrast microscopy. For each strain, 200 particles were measured. Growth in CD medium was monitored in 12-well plates at 37°C. +, mass doubling of about 45 min. −, absence of growth at 35 h following the inoculation.
DivIB forms a complex with DivIC and FtsL (6, 16, 37). To test whether overexpression of DivIC or FtsL could compensate for the absence of DivIB, the divIC and ftsL genes were introduced downstream of the ami locus under the control of the maltose-induced promoter in B2 and in R6 for a control. The additional copy of ftsL rescued the growth of B2 in CD broth. However, the wild-type morphology was not restored by the additional ftsL copy in either TH or CD medium (Table 3). Surprisingly, the presence of an additional copy of ftsL caused a moderated chaining of R6 in TH but not in CD medium. In contrast to ftsL, the additional copy of divIC did not rescue B2 growth in CD medium, nor did it affect the phenotype of R6 (Table 3). Despite the additional gene, no measurable modification of the amount of FtsL or DivIC was observed by immunoblotting in either strain, in the absence or the presence of maltose.
Destabilization of FtsL in the absence of DivIB.
The results above highlight a functional link between FtsL and DivIB. Moreover, in B. subtilis, the proteolysis of FtsL in vivo is influenced by DivIB (14). These observations prompted us to look for effects of the absence of DivIB on the degradation of its partners FtsL and DivIC and other proteins involved in cell division or cell wall synthesis. Following the addition of the translation inhibitor erythromycin at the experimentally determined MIC where no lysis occurred, the amounts of various proteins in R6 and B2 were evaluated by immunoblotting (Fig. 3). In the absence of protein synthesis, the amount of FtsL in B2 decreased drastically after 60 min and it was not detected after 120 min, whereas it remained constant in R6 for 150 min (Fig. 3G). No important effect of the absence of DivIB on the stability in vivo of FtsK, PBP2x (FtsI), PBP1a, DivIVA, FtsZ, or DivIC was observed. The levels of these proteins remained nearly constant for 150 min following inhibition of protein synthesis, with the possible exception of PBP2x, which may show a slight decrease.
FIG. 3.
Stability in vivo of some division proteins in the absence of DivIB. Strains R6 and B2 were grown in TH broth to an optical density at 600 nm of 0.3 prior to addition of erythromycin (40 μg/ml) to inhibit protein synthesis. Aliquots withdrawn after the time intervals indicated were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting. (A to G) Immunoblots with sera against the proteins indicated on the right. (H) Corresponding Coomassie blue-stained gel of total cell lysates.
Functional analysis of the three extracellular domains of DivIB.
To investigate the functions of the various domains of DivIB in pneumococcus, we took advantage of the divIB null mutant and the expression platform at the bgaA locus to conduct complementation experiments. Various forms of DivIB truncated from the C terminus were thus expressed, and the growth and morphological phenotypes were examined. As stated above, expression of full-length DivIB restored both wild-type morphology and growth in CD medium. The empty expression platform, which consists of the fucose-induced promoter and a kanamycin resistance cassette, did not affect either growth or morphology of either strain (Fig. 4). Expression of the diverse truncated forms of DivIB did not affect the R6 strain, which also expresses the endogenous full-length DivIB.
To probe the function of the γ and β domains, three truncated forms, DivIBΔγ, DivIBS337*, and DivIBΔβγ, were created. In DivIBΔγ, the γ domain of DivIB was truncated in order to leave a β domain in accordance with our results from trypsin proteolysis (S. Masson and A. Zapun, unpublished data), sequence analysis using alignments, and hydrophobic cluster analysis (Fig. 1) (37, 49). In DivIBS337*, the protein DivIB was truncated further in order to match the C terminus of the β domain, as defined for the protein from Geobacillus stearothermophilus (43). DivIBΔβγ retains the α domain, whereas the whole extracellular domain is absent from DivIBΔec. The point mutant DivIBE222A was created to probe the role of this highly conserved residue at the junction of the α and β domains. With the exception of DivIBΔec, since antibodies were raised against the extracellular soluble part of DivIB, the expression of all the truncated forms was checked by immunoblot.
Results of the complementation experiments are given in Fig. 4. The nonconserved γ tail (residues 362 to 396) appears to be dispensable. However, residues 337 to 361, which form the C terminus of the β domain, are necessary to restore growth in CD medium but are not required to prevent chain formation in rich medium. Further truncations did not rescue either the growth in CD medium or the morphology phenotypes.
Replacement of the conserved glutamate 222 by an alanine permitted growth in defined medium but only slightly reduced the chain length in rich medium.
Cells with DivIB depleted are more susceptible to β-lactam antibiotics.
To test whether DivIB might be involved in cell wall synthesis, we measured the impact of the inactivation of divIB on the susceptibility to β-lactam antibiotics, which target the penicillin-binding proteins that assemble the peptidoglycan. B2 was more susceptible than the parental R6 strain to penicillin G and cefotaxime by a factor of 2 (Table 4). As the wild-type R6 strain is itself susceptible to β-lactams, these results concern minute concentrations of antibiotics. We therefore looked at the consequence of disrupting divIB in a derivative of the R6 strain with a reduced susceptibility to β-lactams due to the expression of a variant of PBP2x of clinical origin (strain 5204) that has a low affinity for β-lactams (R62X5204). In the resulting strain (termed B22X5204), the susceptibility to penicillin G and cefotaxime was increased fourfold (Table 4). The disruption of divIB had no effect on the susceptibility to ofloxacin, a fluoroquinolone that interferes with DNA replication and transcription, and to gentamicin, an aminoglycoside that inhibits protein synthesis.
TABLE 4.
Effect of divIB deletion on antimicrobial resistance
| Strain | MIC (mg liter−1)
|
|||
|---|---|---|---|---|
| Cefotaxime | Penicillin G | Ofloxacin | Gentamicin | |
| R6 | 0.032 | 0.032 | 1.5 | 24 |
| B2 | 0.015 | 0.015 | 1.5 | 24 |
| R62X5204 | 0.38 | 0.094 | 1.5 | 24 |
| B22X5204 | 0.094 | 0.023 | 1.5 | 24 |
Penicillin G and cefotaxime have lytic and nonlytic actions on pneumococcus, respectively. To test whether the deletion of divIB had a general effect on the susceptibility to lysis, we examined the fate of cultures following the addition of these β-lactams at fourfold their MICs during the exponential growth phase. Although the lysis induced by penicillin G was faster for B2 than for R6, cefotaxime did not cause lysis of either strain (not shown). The electron transport chain inhibitor azide (NaN3) at 0.2% caused the rapid growth arrest of both the R6 and B2 strains without causing lysis (not shown). These results indicate that the greater susceptibility of B2 to β-lactams does not result from a gross defect of the peptidoglycan causing a severe fragility of the cell wall.
DISCUSSION
Essentiality of DivIB and stabilization of FtsL.
The gene divIB/ftsQ is required for correct cell division in the rod-shaped bacteria E. coli and B. subtilis, but these bacteria differ regarding the essentiality of this gene. Unlike ftsQ in E. coli, divIB in S. pneumoniae is dispensable under some conditions, like in B. subtilis. In contrast, ftsL and divIC/ftsB are essential in the three organisms. Although division of pneumococcus can occur without divIB in the TH rich medium, longer chains of cells are formed, and a small fraction of cells are malformed with multiple septa (Fig. 2). Thus, pneumococcal divIB appears to influence a late stage of cell division, chain dispersion, but is also involved at an early stage of the division for proper septum formation. The defect in septation is possibly responsible for the failure to grow in defined medium.
Growth in defined medium was restored by an additional copy of ftsL (Table 3), although this second gene did not increase the amount of FtsL in exponentially growing cells (in TH medium). The amount of FtsL is not affected by the presence or absence of divIB, with or without an additional copy of ftsL. However, when protein synthesis is inhibited, the amount of FtsL drops dramatically in the absence of DivIB, whereas it remains constant in its presence (Fig. 3). These observations suggest that DivIB protects FtsL from degradation, as the absence of DivIB increases the turnover of FtsL. Indeed, as a normal amount of FtsL is maintained in the culture even in the absence of DivIB when translation can proceed, it is likely that the synthesis of FtsL is increased to compensate for its faster degradation.
When the defined medium is inoculated with cells grown previously in TH medium, growth resumes only after a lag phase of several hours. This lag probably allows for the readjustment of the metabolic pathways to the new conditions. It is possible that while cells undergo this adaptation, the level of FtsL drops below a critical threshold in the mutant, because its rate of synthesis may be insufficient to compensate for its faster degradation in the absence of DivIB. The additional copy of ftsL may generate enough FtsL synthesis to overcome this possible critical step during the lag phase due to the changing conditions.
The hypothesis discussed above implies that the essentiality of DivIB is mediated by that of FtsL. The relationship between DivIB and FtsL in S. pneumoniae resembles that in B. subtilis, where the essentiality of divIB at high temperature could be attributed to the reduced amount of FtsL (14, 15). In E. coli, in contrast, the essentiality of ftsQ appears to result not from protecting FtsL but rather from stabilizing the interaction between FtsL and FtsB, which do not associate and are not localized at the division site in the absence of FtsQ (6).
The reason underlying the defect in chain dispersion, in contrast, does not appear to involve FtsL. Indeed, a second copy of ftsL does not restore the normal cell separation. Chain dispersion depends on the action of peptidoglycan hydrolases, so that a defect in cell separation may result from a modified peptidoglycan that is less prone to degradation or from a deficit in the recruitment or activation of one or more hydrolases at the division site. Interestingly, the expression of a truncated form of DivIB that did not restore growth in defined medium, and is probably unable to interact with FtsL (see below), could restore normal chain length.
Domains of DivIB.
The extracytoplasmic part of DivIB consists of three domains (Fig. 1). We took advantage of the possibility of complementing the deleted strain to investigate the relative roles of these domains in S. pneumoniae (Fig. 4). Truncations were designed to produce C termini corresponding to domains resulting from proteolytic cleavage. The sequence of the C-terminal tail of DivIB or FtsQ (i. e., residues further than about 200 to 215 amino acids from the transmembrane segment) is not conserved, is of variable length (sometimes completely absent), and consists mostly of charged and polar residues. The C-terminal tail is therefore likely unimportant. This γ domain of pneumococcal DivIB is unlikely to be structured and was found to be dispensable for the interaction in vitro with the heterodimer formed by FtsL and DivIC (37). As divIBΔγ restores growth in CD medium and a normal chain length distribution, we conclude that the γ domain is dispensable for the function of DivIB. In contrast, the conserved region of the extracellular domain (i. e., the 200 to 215 residues following the transmembrane segment) is required for normal function, as complementation with a gene encoding DivIB deprived of its complete extracellular domain (divIBΔec) did not modify the phenotype of the deletion strain (Fig. 4). However, we could not check whether this truncated protein was expressed and stable. The conserved region of DivIB can itself be divided in two domains based on the susceptibility to tryptic digestion: an α domain that was sensitive to degradation and a β domain that was resistant. The existence of these domains is supported by the recently determined crystal structure of FtsQ (49). We found that the divIBΔβγ gene, which codes for a protein with an extracellular region containing only the α domain, did not restore wild-type phenotypes (Fig. 4), indicating that the β domain is required, as was found in B. subtilis (51).
An uncertainty exists regarding the C-terminal limit of the β domain. Limited proteolysis of recombinant pneumococcal DivIB produced a β domain reaching to residue 361, which includes the complete C-terminal part of the conserved, folded region, as determined by examination of sequence alignments and hydrophobic cluster analysis. Tryptic digestion of DivIB from Geobacillus stearothermophilus resulted in a shorter fragment extending to the corresponding residue 336 in the pneumococcal protein (43). In B. subtilis, residues following position 229 were found to include an epitope participating in the septal localization (51). Interestingly, a gene encoding DivIB truncated after residue 336 (divIB337*) failed to restore growth in defined medium but corrected the chaining in TH medium, showing that the two defects can be separated (Fig. 4). In E. coli, one ftsQ mutant was identified, corresponding to a similar truncation of the β domain (the last 29 residues), that is unable to recruit FtsL to the division site (11). Other truncations pointed to residues 250 to 256 of FtsQ for the recruitment of FtsL and FtsB in E. coli (24). Also in E. coli, amino acids 237 and 252 in the C-terminal part of the β domain were shown to be important for the recruitment of FtsL and FtsB (24, 49). These observations indicate that the C-terminal part of the β domain is necessary for the interaction with FtsL. The failure of divIB337* to restore growth in defined medium would then be consistent with the idea that the essentiality of DivIB results from a functional relationship with FtsL.
The most conserved residue in DivIB and FtsQ is a glutamic acid at the junction of the α and β domains. This glutamate is not required for the septal localization of DivIB in B. subtilis (51). To probe its function, the deletion strain was complemented with a gene coding for a protein with the point mutation E222A. The divIBE222A gene did restore growth in CD medium but did not prevent chaining in TH medium (Fig. 4), in contrast to divIB337* but like an additional copy of ftsL. Thus, the conserved junction of the α and β domains seems to be involved in a function of DivIB that influences a very late stage of the division process, cell dispersion, but is not involved in the essential function of DivIB.
A possible role of DivIB in cell wall synthesis.
In chains of the divIB deletion mutant, diplococcal cells appear to be attached by the tip of their poles, as if only cell dispersal was impaired, much like in a lytA or lytB mutant. LytA and LytB are peptidoglycan hydrolases participating in or required for cell separation, and a lack of these enzymes results in chaining (17). The formation of chains in the absence of DivIB could be due to a modification of the peptidoglycan composition, making the cell wall less prone to cleavage by hydrolases. However, no important modification in the amounts of the different muropeptides was detected in the divIB null mutant (W. Vollmer, personal communication), so if changes are real, they are small or localized.
If the small decrease in the amount of PBP2x observed in the absence of DivIB and protein synthesis (Fig. 3C) turns out to be real, this observation could indicate an interaction between DivIB and PBP2x and support previous data from two-hybrid experiments with the proteins from B. subtilis (14, 16). Another observation that could be consistent with a speculative function of DivIB in peptidoglycan synthesis is the increased susceptibility to β-lactam antibiotics of the divIB deletion mutant (Table 4). The lytic and nonlytic actions of penicillin G and cefotaxime were retained in the divIB deletion mutant, implying that the increased susceptibility to β-lactams was probably not the result of a generally increased susceptibility to lysis. No effect of the deletion of divIB on the susceptibility to drugs that target DNA or protein metabolism was found. Also, rapid growth arrest caused by azide (a poison of the electron transport chain) did not cause lysis of either the mutant the parental strain. In a strain that exhibits some resistance to β-lactams due to the presence of a low-affinity PBP2x, the absence of DivIB restores susceptibility to cefotaxime and penicillin G. This finding indicates that targeting the function of DivIB provides a way to restore the efficacy of the most tried and tested class of antibacterial drugs, the β-lactams.
Acknowledgments
This work was partly supported by European grant LSMH-CT-EUR-INTAFAR 2004-512138 to T. Vernet's laboratory and by a Ministry of Research (“Allocation de Recherche”) grant to A.L.G.
We thank Waldemar Vollmer for the peptidoglycan analysis, as well as Alessandro Riva, Anne Marie Di Guilmi, and Daphna Fenel for the electronic microscopy experiments and Isabelle Petit for the partial proteolysis of DivIB.
Footnotes
Published ahead of print on 25 April 2008.
REFERENCES
- 1.Angert, E. R. 2005. Alternatives to binary fission in bacteria. Nat. Rev. Microbiol. 3214-224. [DOI] [PubMed] [Google Scholar]
- 2.Beall, B., and J. Lutkenhaus. 1989. Nucleotide sequence and insertional inactivation of a Bacillus subtilis gene that affects cell division, sporulation, and temperature sensitivity. J. Bacteriol. 1716821-6834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bramkamp, M., L. Weston, R. A. Daniel, and J. Errington. 2006. Regulated intramembrane proteolysis of FtsL protein and the control of cell division in Bacillus subtilis. Mol. Microbiol. 62580-591. [DOI] [PubMed] [Google Scholar]
- 4.Briegel, A., D. P. Dias, Z. Li, R. B. Jensen, A. S. Frangakis, and G. J. Jensen. 2006. Multiple large filament bundles observed in Caulobacter crescentus by electron cryotomography. Mol. Microbiol. 625-14. [DOI] [PubMed] [Google Scholar]
- 5.Buddelmeijer, N., M. E. Aarsman, A. H. Kolk, M. Vicente, and N. Nanninga. 1998. Localization of cell division protein FtsQ by immunofluorescence microscopy in dividing and nondividing cells of Escherichia coli. J. Bacteriol. 1806107-6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buddelmeijer, N., and J. Beckwith. 2004. A complex of the Escherichia coli cell division proteins FtsL, FtsB and FtsQ forms independently of its localization to the septal region. Mol. Microbiol. 521315-1327. [DOI] [PubMed] [Google Scholar]
- 7.Buddelmeijer, N., N. Judson, D. Boyd, J. J. Mekalanos, and J. Beckwith. 2002. YgbQ, a cell division protein in Escherichia coli and Vibrio cholerae, localizes in codependent fashion with FtsL to the division site. Proc. Natl. Acad. Sci. USA 996316-6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carapito, R., L. Chesnel, T. Vernet, and A. Zapun. 2006. Pneumococcal beta-lactam resistance due to a conformational change in penicillin-binding protein 2x. J. Biol. Chem. 2811771-1777. [DOI] [PubMed] [Google Scholar]
- 9.Carson, M. J., J. Barondess, and J. Beckwith. 1991. The FtsQ protein of Escherichia coli: membrane topology, abundance, and cell division phenotypes due to overproduction and insertion mutations. J. Bacteriol. 1732187-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan, P. F., K. M. O'Dwyer, L. M. Palmer, J. D. Ambrad, K. A. Ingraham, C. So, M. A. Lonetto, S. Biswas, M. Rosenberg, D. J. Holmes, and M. Zalacain. 2003. Characterization of a novel fucose-regulated promoter (PfcsK) suitable for gene essentiality and antibacterial mode-of-action studies in Streptococcus pneumoniae. J. Bacteriol. 1852051-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, J. C., M. Minev, and J. Beckwith. 2002. Analysis of ftsQ mutant alleles in Escherichia coli: complementation, septal localization, and recruitment of downstream cell division proteins. J. Bacteriol. 184695-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, J. C., D. S. Weiss, J. M. Ghigo, and J. Beckwith. 1999. Septal localization of FtsQ, an essential cell division protein in Escherichia coli. J. Bacteriol. 181521-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chesnel, L., L. Pernot, D. Lemaire, D. Champelovier, J. Croize, O. Dideberg, T. Vernet, and A. Zapun. 2003. The structural modifications induced by the M339F substitution in PBP2x from Streptococcus pneumoniae further decreases the susceptibility to beta-lactams of resistant strains. J. Biol. Chem. 27844448-44456. [DOI] [PubMed] [Google Scholar]
- 14.Daniel, R. A., and J. Errington. 2000. Intrinsic instability of the essential cell division protein FtsL of Bacillus subtilis and a role for DivIB protein in FtsL turnover. Mol. Microbiol. 36278-289. [DOI] [PubMed] [Google Scholar]
- 15.Daniel, R. A., E. J. Harry, V. L. Katis, R. G. Wake, and J. Errington. 1998. Characterization of the essential cell division gene ftsL(yIID) of Bacillus subtilis and its role in the assembly of the division apparatus. Mol. Microbiol. 29593-604. [DOI] [PubMed] [Google Scholar]
- 16.Daniel, R. A., M. F. Noirot-Gros, P. Noirot, and J. Errington. 2006. Multiple interactions between the transmembrane division proteins of Bacillus subtilis and the role of FtsL instability in divisome assembly. J. Bacteriol. 1887396-7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Las Rivas, B., J. L. Garcia, R. Lopez, and P. Garcia. 2002. Purification and polar localization of pneumococcal LytB, a putative endo-beta-N-acetylglucosaminidase: the chain-dispersing murein hydrolase. J. Bacteriol. 1844988-5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.den Blaauwen, T., M. A. de Pedro, M. Nguyen-Disteche, and J. A. Ayala. 2008. Morphogenesis of rod-shaped sacculi. FEMS Microbiol. Rev. 32321-344. [DOI] [PubMed] [Google Scholar]
- 19.England, J. C., and J. W. Gober. 2001. Cell cycle control of cell morphogenesis in Caulobacter. Curr. Opin. Microbiol. 4674-680. [DOI] [PubMed] [Google Scholar]
- 20.Errington, J., R. A. Daniel, and D. J. Scheffers. 2003. Cytokinesis in bacteria. Microbiol. Mol. Biol. Rev. 6752-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fadda, D., C. Pischedda, F. Caldara, M. B. Whalen, D. Anderluzzi, E. Domenici, and O. Massidda. 2003. Characterization of divIVA and other genes located in the chromosomal region downstream of the dcw cluster in Streptococcus pneumoniae. J. Bacteriol. 1856209-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghigo, J. M., D. S. Weiss, J. C. Chen, J. C. Yarrow, and J. Beckwith. 1999. Localization of FtsL to the Escherichia coli septal ring. Mol. Microbiol. 31725-737. [DOI] [PubMed] [Google Scholar]
- 23.Goehring, N. W., and J. Beckwith. 2005. Diverse paths to midcell: assembly of the bacterial cell division machinery. Curr. Biol. 15R514-R526. [DOI] [PubMed] [Google Scholar]
- 24.Goehring, N. W., I. Petrovska, D. Boyd, and J. Beckwith. 2007. Mutants, suppressors, and wrinkled colonies: mutant alleles of the cell division gene ftsQ point to functional domains in FtsQ and a role for domain 1C of FtsA in divisome assembly. J. Bacteriol. 189633-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goranov, A. I., L. Katz, A. M. Breier, C. B. Burge, and A. D. Grossman. 2005. A transcriptional response to replication status mediated by the conserved bacterial replication protein DnaA. Proc. Natl. Acad. Sci. USA 10212932-12937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guiral, S., V. Henard, M. H. Laaberki, C. Granadel, M. Prudhomme, B. Martin, and J. P. Claverys. 2006. Construction and evaluation of a chromosomal expression platform (CEP) for ectopic, maltose-driven gene expression in Streptococcus pneumoniae. Microbiology 152343-349. [DOI] [PubMed] [Google Scholar]
- 27.Guzman, L. M., D. S. Weiss, and J. Beckwith. 1997. Domain-swapping analysis of FtsI, FtsL, and FtsQ, bitopic membrane proteins essential for cell division in Escherichia coli. J. Bacteriol. 1795094-5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karimova, G., N. Dautin, and D. Ladant. 2005. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J. Bacteriol. 1872233-2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katis, V. L., and R. G. Wake. 1999. Membrane-bound division proteins DivIB and DivIC of Bacillus subtilis function solely through their external domains in both vegetative and sporulation division. J. Bacteriol. 1812710-2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katis, V. L., R. G. Wake, and E. J. Harry. 2000. Septal localization of the membrane-bound division proteins of Bacillus subtilis DivIB and DivIC is codependent only at high temperatures and requires FtsZ. J. Bacteriol. 1823607-3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lara, B., A. I. Rico, S. Petruzzelli, A. Santona, J. Dumas, J. Biton, M. Vicente, J. Mingorance, and O. Massidda. 2005. Cell division in cocci: localization and properties of the Streptococcus pneumoniae FtsA protein. Mol. Microbiol. 55699-711. [DOI] [PubMed] [Google Scholar]
- 32.Margolin, W. 2000. Themes and variations in prokaryotic cell division. FEMS Microbiol. Rev. 24531-548. [DOI] [PubMed] [Google Scholar]
- 33.Massidda, O., D. Anderluzzi, L. Friedli, and G. Feger. 1998. Unconventional organization of the division and cell wall gene cluster of Streptococcus pneumoniae. Microbiology 1443069-3078. [DOI] [PubMed] [Google Scholar]
- 34.McCormick, J. R., and R. Losick. 1996. Cell division gene ftsQ is required for efficient sporulation but not growth and viability in Streptomyces coelicolor A3(2). J. Bacteriol. 1785295-5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nanninga, N. 1998. Morphogenesis of Escherichia coli. Microbiol. Mol. Biol. Rev. 62110-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ng, W. L., G. T. Robertson, K. M. Kazmierczak, J. Zhao, R. Gilmour, and M. E. Winkler. 2003. Constitutive expression of PcsB suppresses the requirement for the essential VicR (YycF) response regulator in Streptococcus pneumoniae R6. Mol. Microbiol. 501647-1663. [DOI] [PubMed] [Google Scholar]
- 37.Noirclerc-Savoye, M., A. Le Gouellec, C. Morlot, O. Dideberg, T. Vernet, and A. Zapun. 2005. In vitro reconstitution of a trimeric complex of DivIB, DivIC and FtsL, and their transient co-localization at the division site in Streptococcus pneumoniae. Mol. Microbiol. 55413-424. [DOI] [PubMed] [Google Scholar]
- 38.Piette, A., C. Fraipont, T. Den Blaauwen, M. E. Aarsman, S. Pastoret, and M. Nguyen-Disteche. 2004. Structural determinants required to target penicillin-binding protein 3 to the septum of Escherichia coli. J. Bacteriol. 1866110-6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pinho, M. G., and J. Errington. 2003. Dispersed mode of Staphylococcus aureus cell wall synthesis in the absence of the division machinery. Mol. Microbiol. 50871-881. [DOI] [PubMed] [Google Scholar]
- 40.Rasband, W. S. 1997-;-2007. ImageJ. National Institutes of Health, Bethesda, MD. http://rsb.info.nih.gov/ij/.
- 41.Real, G., S. Autret, E. J. Harry, J. Errington, and A. O. Henriques. 2005. Cell division protein DivIB influences the Spo0J/Soj system of chromosome segregation in Bacillus subtilis. Mol. Microbiol. 55349-367. [DOI] [PubMed] [Google Scholar]
- 42.Real, G., and A. O. Henriques. 2006. Localization of the Bacillus subtilis murB gene within the dcw cluster is important for growth and sporulation. J. Bacteriol. 1881721-1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robson, S. A., and G. F. King. 2006. Domain architecture and structure of the bacterial cell division protein DivIB. Proc. Natl. Acad. Sci. USA 1036700-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rowland, S. L., V. L. Katis, S. R. Partridge, and R. G. Wake. 1997. DivIB, FtsZ and cell division in Bacillus subtilis. Mol. Microbiol. 23295-302. [DOI] [PubMed] [Google Scholar]
- 45.Sanchez-Pulido, L., D. Devos, S. Genevrois, M. Vicente, and A. Valencia. 2003. POTRA: a conserved domain in the FtsQ family and a class of beta-barrel outer membrane proteins. Trends Biochem. Sci. 28523-526. [DOI] [PubMed] [Google Scholar]
- 46.Scheffers, D. J., C. Robichon, G. J. Haan, T. den Blaauwen, G. Koningstein, E. van Bloois, J. Beckwith, and J. Luirink. 2007. Contribution of the FtsQ transmembrane segment to localization to the cell division site. J. Bacteriol. 1897273-7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shiomi, D., and W. Margolin. 2007. Dimerization or oligomerization of the actin-like FtsA protein enhances the integrity of the cytokinetic Z ring. Mol. Microbiol. 661396-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Siddiqui, R. A., C. Hoischen, O. Holst, I. Heinze, B. Schlott, J. Gumpert, S. Diekmann, F. Grosse, and M. Platzer. 2006. The analysis of cell division and cell wall synthesis genes reveals mutationally inactivated ftsQ and mraY in a protoplast-type L-form of Escherichia coli. FEMS Microbiol. Lett. 258305-311. [DOI] [PubMed] [Google Scholar]
- 49.van den Ent, F., T. M. Vinkenvleugel, A. Ind, P. West, D. Veprintsev, N. Nanninga, T. den Blaauwen, and J. Lowe. 2008. Structural and mutational analysis of the cell division protein FtsQ. Mol. Microbiol. 68110-123. [DOI] [PubMed] [Google Scholar]
- 50.van de Rijn, I., and R. E. Kessler. 1980. Growth characteristics of group A streptococci in a new chemically defined medium. Infect. Immun. 27444-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wadsworth, K. D., S. L. Rowland, E. J. Harry, and G. F. King. 2008. The divisomal protein DivIB contains multiple epitopes that mediate its recruitment to incipient division sites. Mol. Microbiol. 671143-1155. [DOI] [PubMed] [Google Scholar]
- 52.Zahner, D., and R. Hakenbeck. 2000. The Streptococcus pneumoniae beta-galactosidase is a surface protein. J. Bacteriol. 1825919-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zapun, A., T. Vernet, and M. G. Pinho. 2008. The different shapes of cocci. FEMS Microbiol. Rev. 32345-360. [DOI] [PubMed] [Google Scholar]