Abstract
Protein aggregation is the phenomenon of protein self-association potentially leading to detrimental effects on physiology, which is closely related to numerous human diseases such as Alzheimer's and Parkinson's disease. Despite progress in understanding the mechanism of protein aggregation, how natural selection against protein aggregation acts on subunits of protein complexes and on proteins with different contributions to organism fitness remains largely unknown. Here, we perform a proteome-wide analysis by using an experimentally validated algorithm TANGO and utilizing sequence, interactomic and phenotype-based functional genomic data from yeast, fly, and nematode. We find that proteins that are capable of forming homooligomeric complex have lower aggregation propensity compared with proteins that do not function as homooligomer. Further, proteins that are essential to the fitness of an organism have lower aggregation propensity compared with nonessential ones. Our finding suggests that the selection force against protein aggregation acts across different hierarchies of biological system.
Keywords: natural selection, protein aggregation, functional genomics, proteome, Drosophila melanogaster, Caenorhabditis elegans
Proteins are essential working machineries in living organisms. To be functionally active, a protein needs to fold into a unique 3-dimensional structure. Cells possess a wide variety of protective mechanisms to facilitate efficient protein folding in a crowded environment and degrade those proteins when the folding is unsuccessful. It is estimated that more than 30% of the newly synthesized proteins are degraded by proteasome due to translation errors or improper folding (Schubert et al. 2000). Misfolded proteins that escape the quality control mechanisms can form aggregates and thereby lead to malfunctioning of associated biological processes (Chiti and Dobson 2006). Protein aggregation has been associated with more than 30 diseases (Chiti and Dobson 2006).
In addition to the production of misfolded proteins during protein synthesis, there are other occasions when proteins may form aggregates: mounting evidence shows that partial folding or unfolding may be frequent in various cellular functions, such as cell signaling (Dixon et al. 2004; Sawada et al. 2006), transcription (Radhakrishnan et al. 1997), and trafficking/translocation (Daughdrill et al. 1997). As a result, the exposure of certain protein regions that are buried in the native structure may cause inappropriate interactions with other identical molecules, leading to the formation of aggregates.
Given the compromising effects that protein aggregation has on normal cellular functions, it remains a key question how natural selection acts against protein aggregation to reduce its negative effects. Recent studies have revealed selection pressures at both sequence and structure level to prevent aggregation (Otzen et al. 2000; Richardson JS and Richardson DC 2002; Steward et al. 2002; Parrini et al. 2005; Monsellier and Chiti 2007). For example, at sequence level, Broome and Hecht (2000) showed that alternating polar and nonpolar amino acids are disfavored by evolutionary selection in natural protein to avoid aggregation. Rousseau et al. (2006) illustrated that the regions flanking the sequences with high aggregation propensity are often enriched by proline or charged residues to inhibit aggregation. Monsellier et al. (2007) found that the clustering of residues with high aggregation propensity in primary sequence is negatively selected. Structurally, the aggregation-prone sequences are usually buried in the native state and therefore are protected from forming aggregates when the native state is stable (Linding et al. 2004). The peripheral strands in β-sheet proteins, which can potentially form intermolecular interactions, are protected by various sequence and structure features such as inward-pointing charged residues, proline, and loop coverage (Richardson JS and Richardson DC 2002; Wang and Hecht 2002).
In addition, several proteome-wide analyses have offered significant insight into the relationship between selection against aggregation and genomic context or organism complexity (Bastolla et al. 2004; Tartaglia et al. 2005). Here, to shed light on how selection against protein aggregation acts on subunits of protein complexes and on proteins with different contributions to organism fitness, we analyze the proteomes of 3 organisms: Saccharomyces cerevisiae, Drosophila melanogaster, and Caenorhabditis elegans, by combining sequence, interactomic and phenotype-based functional genomic data.
Some proteins form homooligomers to perform cellular functions. As the formation of functional homooligomers and usually nonfunctional aggregates, both result from protein self-association, these 2 processes essentially compete with each other (fig. 1) (Ding et al. 2002). Therefore, to function effectively, it is plausible that proteins, which form homooligomeric complexes, are subject to higher selection pressure against forming aggregates as compared with other proteins that do not form oligomers.
FIG. 1.—
There is a competition between the formation of physiological oligomers and “aberrant” oligomers that are on the pathway of forming aggregates.
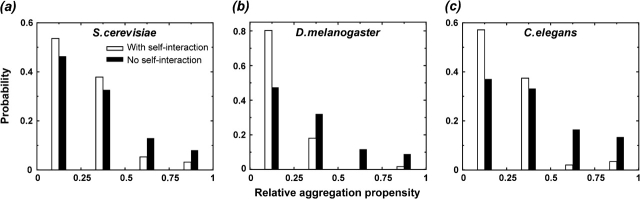
To test this hypothesis, we compare the aggregation propensity of proteins that have experimental evidence of self-interactions with those that lack such evidence. The aggregation propensity of individual proteins is calculated by an experimentally validated algorithm TANGO (see Methods). To measure the relative aggregation propensity of each protein in a given organism, we use the ratio between its TANGO score and the maximal TANGO score in the corresponding organism. The experimental evidence for protein self-interaction is collected from the data sets of protein–protein interactions from S.cerevisiae, D.melanogaster, and C.elegans (see Methods). We find that the former class of proteins has significantly lower aggregation propensities (0.25 ± 0.19S.cerevisiae, 0.14 ± 0.14D.melanogaster, and 0.25 ± 0.18C.elegans) than the latter (0.32 ± 0.25S.cerevisiae, 0.31 ± 0.25D.melanogaster, and 0.37 ± 0.27C.elegans), which was supported by a histogram comparison (fig. 2) and Kolmogorov–Smirnov test (PS.cerevisiae < 4 × 10−13, PD.melanogaster < 4 × 10−37, PC.elegans < 3 × 10−10), suggesting a higher selection pressure against protein aggregation for self-interacting proteins. In addition, we show that this observation is not due to the confounding factors such as difference in the size (length) distribution of these 2 protein classes or differential enrichment of natively unfolded proteins (Supplementary Material online).
FIG. 2.—
The histograms of relative aggregation propensity of proteins that have experimental evidence of self-interactions (open) and those that lack such evidence (filled) are plotted for (a) Saccharomyces cerevisiae, (b) Drosophila melanogaster, and (c) Caenorhabditis elegans with the bin size of 0.25.
Cellular function impairment caused by protein aggregate formation may ultimately lead to the decrease of individual fitness. Therefore, it is conceivable that the natural selection against aggregation will be evident in the light of fitness contribution of individual proteins to an organism. To assess this reasoning, we further study how proteins that have distinct contributions to organism fitness differ in their inherent aggregation propensities.
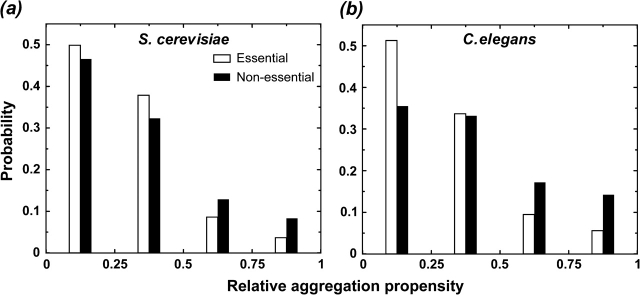
With the advent of functional genomic technologies, the relative contribution of individual genes (proteins) to overall organism fitness has been evaluated at a genome-wide scale for both single and multicellular organisms including S.cerevisiae and C.elegans. The evaluation was performed by characterizing the phenotypes (e.g., growth rate, viability, and embryo morphology) of the organism when a gene is either completely deleted from the genome or its transcript is depleted by RNA interference (RNAi) knockdown. We compare the aggregation propensity of proteins that are essential to organism fitness (see Methods) with nonessential ones. We find that the former category of proteins has significantly lower aggregation propensities than the latter (fig. 3, PS.cerevisiae < 5 × 10−8, PC.elegans < 3 × 10−105, Kolmogorov–Smirnov test), suggesting that the sequences of essential proteins are subject to a stronger selection against aggregation than those of nonessential ones. We also show that this observation is not attributed to the confounding factors such as the size (length) of proteins, self-interacting, or differential enrichment of natively unfolded proteins in the data set (Supplementary Material online).
FIG. 3.—
The histograms of relative aggregation propensity of proteins that are essential to organismal fitness (open) and nonessential ones (filled) are plotted for (a) Saccharomyces cerevisiae and (b) Caenorhabditis elegans with the bin size of 0.25.
In summary, our study reveals stronger selections against protein aggregation on proteins functioning through self-assembly and essential proteins compared with non–self-interacting and nonessential ones, respectively, which suggests that selection force against protein aggregation acts across different hierarchies.
Methods
The protein sequences from S.cerevisiae were obtained from the Saccharomyces genome database (ftp://genome-ftp.stanford.edu/, 6 October 2006). The protein sequences of D.melanogaster were obtained from the Ensembl genome database (ftp://ftp.ensembl.org/, version Drosophila_melanogaster.BDGP4.3.41). The protein sequences of C.elegans were obtained from the WormBase (available at: ftp://ftp.wormbase.org/, version wormpep176); (Chen et al. 2005).
The aggregation propensity of each protein was calculated by TANGO, which estimates how thermodynamically probable a segment from a protein/peptide is in a cross-β-aggregate conformation in comparison with other conformations such as random coils, α-helix, and β-turn (Fernandez-Escamilla et al. 2004). The TANGO algorithm has an accuracy of more than 90% in identifying aggregation-prone segments against a set of 176 experimentally validated peptides (Fernandez-Escamilla et al. 2004).
For S.cerevisiae, we use protein–protein interaction data sets from Database of Interacting Proteins (DIPs) (Xenarios et al. 2002), Munich Information Center for Protein Sequences (Mewes et al. 1998), 2 high-throughput yeast 2-hybrid (Y2H) experiments (Uetz et al. 2000; Ito et al. 2001), and 2 mass spectrometry analyses for protein complex (Gavin et al. 2002; Ho et al. 2002). For D.melanogaster, we use data sets from DIP and 2 high-throughput Y2H experiments (Giot et al. 2003; Stanyon et al. 2004). For C.elegans, we use the data set from a high-throughput Y2H experiment (Li et al. 2004).
Most of the essential genes of S.cerevisiae were identified from a comprehensive data set (http://chemogenomics.stanford.edu/supplements/01yfh/files/orfgenedata.txt) of single-gene deletion experiment (Deutschbauer et al. 2005) as those genes, which are required for the viability of S.cerevisiae. We also find other essential genes from the function annotations in Saccharomyces Genome Database (ftp://genome-ftp.stanford.edu/). Most of the essential genes of C.elegans were identified from several large-scale RNAi screens (Supplementary Material online) where the phenotype as a result of single-gene knockdown was directly observed. We refer to those genes, the knockdown of which led to lethality as essential genes. We also find other essential genes that are annotated as “lethal” from the function annotations in the WormBase (Chen et al. 2005).
Supplementary Material
Supplementary methods, table S1, and figures S1 and S2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We thank Julie Ahringer for her help with the RNAi data sets of C. elegans and Raymond Lee and Igor Antoshechkin for their help with the phenotype data set from the WormBase. We also thank Shantanu Sharma for reading and providing helpful suggestions on the manuscript. This work was supported in part by the American Heart Association grant no. 0665361U and the National Institutes of Health grant R01GM080742.
References
- Bastolla U, Moya A, Viguera E, van Ham RC. Genomic determinants of protein folding thermodynamics in prokaryotic organisms. J Mol Biol. 2004;343:1451–1466. doi: 10.1016/j.jmb.2004.08.086. [DOI] [PubMed] [Google Scholar]
- Broome BM, Hecht MH. Nature disfavors sequences of alternating polar and non-polar amino acids: implications for amyloidogenesis. J Mol Biol. 2000;296:961–968. doi: 10.1006/jmbi.2000.3514. [DOI] [PubMed] [Google Scholar]
- Chen N, Harris TW, Antoshechkin I, et al. (31 co-authors) WormBase: a comprehensive data resource for Caenorhabditis biology and genomics. Nucleic Acids Res. 2005;33:D383–D389. doi: 10.1093/nar/gki066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev of Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- Daughdrill GW, Chadsey MS, Karlinsey JE, Hughes KT, Dahlquist FW. The C-terminal half of the anti-sigma factor, FlgM, becomes structured when bound to its target, sigma 28. Nat Struct Biol. 1997;4:285–291. doi: 10.1038/nsb0497-285. [DOI] [PubMed] [Google Scholar]
- Deutschbauer AM, Jaramillo DF, Proctor M, Kumm J, Hillenmeyer ME, Davis RW, Nislow C, Giaever G. Mechanisms of haploinsufficiency revealed by genome-wide profiling in yeast. Genetics. 2005;169:1915–1925. doi: 10.1534/genetics.104.036871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F, Dokholyan NV, Buldyrev SV, Stanley HE, Shakhnovich EI. Molecular dynamics simulation of the SH3 domain aggregation suggests a generic amyloidogenesis mechanism. J Mol Biol. 2002;324:851–857. doi: 10.1016/s0022-2836(02)01112-9. [DOI] [PubMed] [Google Scholar]
- Dixon RD, Chen Y, Ding F, Khare SD, Prutzman KC, Schaller MD, Campbell SL, Dokholyan NV. New insights into FAK signaling and localization based on detection of a FAT domain folding intermediate. Structure. 2004;12:2161–2171. doi: 10.1016/j.str.2004.09.011. [DOI] [PubMed] [Google Scholar]
- Fernandez-Escamilla AM, Rousseau F, Schymkowitz J, Serrano L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat Biotechnol. 2004;22:1302–1306. doi: 10.1038/nbt1012. [DOI] [PubMed] [Google Scholar]
- Gavin AC, Bosche M, Krause R, et al. (38 co-authors) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Giot L, Bader JS, Brouwer C, et al. (49 co-authors) A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- Ho Y, Gruhler A, Heilbut A, et al. (46 co-authors) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SM, Armstrong CM, Bertin N, et al. (48 co-authors) A map of the interactome network of the metazoan C-elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linding R, Schymkowitz J, Rousseau F, Diella F, Serrano L. A comparative study of the relationship between protein structure and beta-aggregation in globular and intrinsically disordered proteins. J Mol Biol. 2004;342:345–353. doi: 10.1016/j.jmb.2004.06.088. [DOI] [PubMed] [Google Scholar]
- Mewes HW, Hani J, Pfeiffer F, Frishman D. MIPS: a database for protein sequences and complete genomes. Nucleic Acids Res. 1998;26:33–37. doi: 10.1093/nar/26.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsellier E, Chiti F. Prevention of amyloid-like aggregation as a driving force of protein evolution. EMBO Rep. 2007;8:737–742. doi: 10.1038/sj.embor.7401034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsellier E, Ramazzotti M, de Laureto PP, Tartaglia GG, Taddei N, Fontana A, Vendruscolo M, Chiti F. The distribution of residues in a polypeptide sequence is a determinant of aggregation optimized by evolution. Biophys J. 2007;93:4382–4391. doi: 10.1529/biophysj.107.111336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otzen DE, Kristensen O, Oliveberg M. Designed protein tetramer zipped together with a hydrophobic Alzheimer homology: a structural clue to amyloid assembly. Proc Natl Acad Sci USA. 2000;97:9907–9912. doi: 10.1073/pnas.160086297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrini C, Taddei N, Ramazzotti M, Degl'Innocenti D, Ramponi G, Dobson CM, Chiti F. Glycine residues appear to be evolutionarily conserved for their ability to inhibit aggregation. Structure. 2005;13:1143–1151. doi: 10.1016/j.str.2005.04.022. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan I, Perez-Alvarado GC, Parker D, Dyson HJ, Montminy MR, Wright PE. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator:coactivator interactions. Cell. 1997;91:741–752. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- Richardson JS, Richardson DC. Natural beta-sheet proteins use negative design to avoid edge-to-edge aggregation. Proc Natl Acad Sci USA. 2002;99:2754–2759. doi: 10.1073/pnas.052706099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau F, Serrano L, Schymkowitz JW. How evolutionary pressure against protein aggregation shaped chaperone specificity. J Mol Biol. 2006;355:1037–1047. doi: 10.1016/j.jmb.2005.11.035. [DOI] [PubMed] [Google Scholar]
- Sawada Y, Tamada M, Dubin-Thaler BJ, Cherniavskaya O, Sakai R, Tanaka S, Sheetz MP. Force sensing by mechanical extension of the Src family kinase substrate p130Cas. Cell. 2006;127:1015–1026. doi: 10.1016/j.cell.2006.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert U, Anton LC, Gibbs J, Norbury CC, Yewdell JW, Bennink JR. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–774. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- Stanyon CA, Liu GZ, Mangiola BA, Patel N, Giot L, Kuang B, Zhang HM, Zhong JH, Finley RL. A Drosophila protein-interaction map centered on cell-cycle regulators. Genome Biol. 2004;5:R96. doi: 10.1186/gb-2004-5-12-r96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward A, Adhya S, Clarke J. Sequence conservation in Ig-like domains: the role of highly conserved proline residues in the fibronectin type III superfamily. J Mol Biol. 2002;318:935–940. doi: 10.1016/S0022-2836(02)00184-5. [DOI] [PubMed] [Google Scholar]
- Tartaglia GG, Pellarin R, Cavalli A, Caflisch A. Organism complexity anti-correlates with proteomic beta-aggregation propensity. Protein Sci. 2005;14:2735–2740. doi: 10.1110/ps.051473805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, et al. (20 co-authors) A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Wang W, Hecht MH. Rationally designed mutations convert de novo amyloid-like fibrils into monomeric beta-sheet proteins. Proc Natl Acad Sci USA. 2002;99:2760–2765. doi: 10.1073/pnas.052706199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xenarios I, Salwinski L, Duan XJ, Higney P, Kim SM, Eisenberg D. DIP, the database of interacting proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res. 2002;30:303–305. doi: 10.1093/nar/30.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.