Abstract
Motivation: Limited availability of data has hindered the development of algorithms that can identify functionally meaningful regulatory single nucleotide polymorphisms (rSNPs). Given the large number of common polymorphisms known to reside in the human genome, the identification of functional rSNPs via laboratory assays will be costly and time-consuming. Therefore appropriate bioinformatics strategies for predicting functional rSNPs are necessary. Recent data from the Encyclopedia of DNA Elements (ENCODE) Project has significantly expanded the amount of available functional information relevant to non-coding regions of the genome, and, importantly, led to the conclusion that many functional elements in the human genome are not conserved.
Results: In this article we describe how ENCODE data can be leveraged to probabilistically determine the functional and phenotypic significance of non-coding SNPs (ncSNPs). The method achieves excellent sensitivity (∼80%) and specificity (∼99%) based on a set of known phenotypically relevant and non-functional SNPs. In addition, we show that our method is not overtrained through the use of cross-validation analyses.
Availability: The software platforms used in our analyses are freely available (http://www.cs.waikato.ac.nz/ml/weka/). In addition, we provide the training dataset (Supplementary Table 3), and our predictions (Supplementary Table 6), in the Supplementary Material.
Contact: nschork@scripps.edu.
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Approximately 10 million common single nucleotide polymorphisms (or SNPs; with >1 % allele frequency) populate the human genome, the vast majority of which reside in non-coding regions (The International HapMap Consortium, 2003). Furthermore, it has been estimated that 50% of genes are associated with a common SNP that alters its expression (Buckland et al., 2006). The extent to which these polymorphisms underlie disease predisposition is unknown, but likely to be quite significant. Due to the incredibly large number of non-coding SNPs (ncSNPs), which may potentially be involved in disease by altering gene expression, identification of the specific polymorphisms altering gene expression is not feasible using current laboratory assays and technologies. In addition, most of these laboratory assays exploit reporter-based systems and as such are further complicated by variations in gene expression regulation from one cell type to the next.
Genome-wide association studies (GWAS) are currently routinely being used to identify common polymorphisms that underlie disease susceptibility in the population at large (Kraft and Cox, 2008). Initial results from these studies suggest that a small number of low penetrance polymorphisms—the majority of which have odds ratios for disease susceptibility often less than 1.5—contribute to genetic predisposition to common diseases, with the vast majority of the genetic component of these diseases yet to be characterized (Wray et al., 2007). These studies test only a small subset of SNPs in an attempt to find disease associated haplotypes, and thus do not necessarily lead to the identification of the individual causative SNPs. The genomic regions within which these susceptibility SNPs reside often have no obvious biological relationship to disease, raising questions about how best to determine this relationship. In fact, many of the SNPs found to be associated with diseases via GWAS analyses reside in non-coding and/or possibly uncharted regulatory regions of the genome (Damani and Topol, 2007; Matthew, 2008). There are number of factors, including but not limited to, power, small individual locus effect sizes, gene–environment interactions and multiple testing issues, that have more than likely hindered the identification of additional disease risk polymorphisms in GWAS settings (Cordell and Clayton, 2005; Eberle et al., 2007).
One approach to overcoming problems in the identification of disease susceptibility loci in GWAS and other association analysis settings is to computationally prioritize candidate SNPs for their likely impact on disease susceptibility. This can be pursued either before carrying out association studies by attaching weights to variations based on their known biological or disease-association effects, or after performing an association study by investigating the biological or disease-association properties of the genomic regions harboring the most strongly associated SNPs. A number of methods have been designed for this purpose, but are typically restricted to predictions involving the functional effects of SNPs within protein-coding regions, specifically non-synonymous SNPs (nsSNPs) which result in a change of the encoded amino acid (Mooney, 2005; Ng and Henikoff, 2006; Torkamani and Schork, 2007). The reasons for this restriction are the relative scarcity of training data for disease-associated SNPs falling outside protein-coding regions, and the relative ease of assigning predictive attributes, such as amino acid conservation and structural features of proteins, to protein-coding SNPs as compared to ncSNPs. Computational strategies focused on the identification and prediction of the functional effects of nucleic acid substitutions within transcription factor binding sites have also been developed, but many are restricted to either solely elucidating relevant binding site motifs and determining whether a SNP falls within these motifs (Andersen et al., 2008; Kel et al., 2003; Roth et al., 1998), or predicting the functional affects of substitutions within these motifs where an abundance of functional information is available (Michal et al., 2008). Other features beyond the existence of a transcription factor binding site, such as changes in the ‘openness’ of the DNA or the existence of epigenetic marks, may alter gene expression and, consequently, result in disease (Shames et al., 2007). A subset of these features were considered in a study similar to ours (Montgomery et al., 2007), however the methodology exploited in this study relied heavily upon prior knowledge of known transcriptional start sites.
Next-generation human genome annotation projects, specifically the Encyclopedia of DNA Elements (ENCODE) Project, whose goal is to identify and characterize functional elements within the human genome, have provided a wealth of information about the biological significance of human non-coding genomic regions, extending our knowledge of these regions far beyond the level of basic nucleotide sequence (ENCODE Project Consortium, 2007). This information is not limited to transcription factor binding site motifs, but rather extends to all non-coding regions, including 5′ -upstream, 3′ -downstream and untranslated genomic regions. In this article, we describe how ENCODE data can be leveraged to probabilistically determine the functional and phenotypic significance of ncSNPs. We take advantage of the currently characterized ENCODE genomic regions (which in total comprise ∼1% of the genome), and show that based on ENCODE-derived genomic parameters alone, we can predict with great confidence which SNPs are likely to be functional in these regions. Our strategy can be generalized to the genome as a whole as the availability of a complete functional annotation of the genome is developed.
2 METHODS
2.1 Training data
RefSeq annotated genes residing in the ENCODE regions were obtained from the UCSC Genome Browser (le Cessie and van Houwelingen, 1992). Known disease causing regulatory (rSNPs) were collected by querying the Human Gene Mutation Database (HGMD) with the gene symbols corresponding to all known genes within the ENCODE regions (Stenson et al., 2003). A total of 102 known disease-causing SNPs in 22 genes were identified. The majority (73%) of these deleterious rSNPs fell in 5′ -upstream regions. Precise UCSC Genome Browser human reference positions were determined for the disease SNPs by a BLAT search of the sequence adjacent to the disease SNP (Kent, 2002) using build hg18 of the human genome. All Single Nucleotide Polymorphism database (dbSNP) annotated SNPs residing within the ENCODE regions, and their positions within the UCSC human reference sequence, were obtained from the UCSC Genome Browser (120 063 SNPs). These SNPs were filtered for all SNPs residing within 5 kb of a gene using BioMart to query the Ensembl Database (Flicek et al., 2008). All SNPs residing within coding regions or introns were removed by their annotation in the UCSC Genome Browser (11 249 SNPs). Thus, ∼10% of SNPs residing within the ENCODE regions (or 0.1% of all SNPs in the human genome) were ultimately prioritized by our method. Presumably neutral SNPs were chosen from this dataset by selecting SNPs that have been validated as true SNPs and not sequencing errors, and have a minor allele frequency >40% based on the use of BioMart to query the HapMap Database (The International HapMap Consortium, 2003). A high minor allele frequency was chosen to enrich for SNPs that are more likely to be neutral, as it has been observed that nsSNPs that are more common are likely to be neutral, and this is also likely to be the case for ncSNPs as well, as it has been reported that the probability of an nsSNP being deleterious is inversely related to its minor allele frequency (Gorlov et al., 2008). The use of these location and allele frequency filters resulted in a total of 1049 presumably neutral SNPs.
2.2 Predictive attributes
All ENCODE attributes, except for the gene prediction attributes (391 total), were considered as ‘predictors’ of functionality and were initially assigned to each of the SNPs in the training dataset. Though it is known that the probability that a ncSNP is functional is associated with the nature of the actual nucleotide transition or transversion in question (13), given the small size of the known disease SNP dataset, nucleotide identities were withheld from the classifier to avoid potential biases resulting from a small training set. Finally, the 28-way conservation profile (multiZ28) (Miller et al., 2007) and conserved elements predictions (phastConsElements28wayPlacMammal) (Siepel et al., 2005) from the UCSC genome browser computed for each SNP were included as predictors. For each predictor, SNPs mapping within a chromosomal interval associated with a value, or signal strength, were assigned the value of that corresponding interval. The full list of initial predictors is presented in Supplementary Table 1.
Table 1.
Predictive attributes and their categorization
| Predictive attribute | X2 | Category |
|---|---|---|
| AffyChIpHl60PvalCtcfHr32 | 780 | RFBS |
| AffyChIpHl60SignalStrictp63_ActD | 115 | RFBS |
| StanfordChipK562Sp1 | 95 | RFBS |
| UppsalaChipHnf3b | 148 | RFBS |
| YaleChipRfbrDeserts | 109 | RFBS |
| YaleChIPSTAT1HeLaMaskLess50mer38bpPval | 260 | RFBS |
| YaleChIPSTAT1HeLaMaskLess50mer50bpPval | 93 | RFBS |
| SangerChipH3acHeLa | 488 | HM |
| SangerChipHitH3K4me3K562 | 710 | HM |
| UcsdNgHeLaDmH3K4_p30 | 121 | HM |
| UcsdNgHeLaH3K4me3_p0 | 159 | HM |
| StanfordPromotersAGS | 484 | PI |
| StanfordPromotersAverage | 495 | PI |
| StanfordPromotersCRL1690 | 585 | PI |
| StanfordPromotersMG63 | 495 | PI |
| StanfordPromotersPanc1 | 586 | PI |
| StanfordPromotersU87 | 481 | PI |
| NhgriDnaseHsChipPvalK562 | 319 | DHS |
| UWRegulomeBaseCaCo2 | 473 | DHS |
| UWRegulomeBaseEryAdult | 263 | DHS |
| UWRegulomeBaseEryFetal | 404 | DHS |
| UWRegulomeBaseHepG2 | 72 | DHS |
| UWRegulomeBaseHuh7 | 73 | DHS |
| UWRegulomeBaseK562 | 561 | DHS |
| UWRegulomeBaseP0041NC | 41 | DHS |
| YaleAffyNeutRNATransMap | 659 | TA |
| YaleAffyPlacRNATars | 86 | TA |
| YaleAffyPlacRNATransMap | 688 | TA |
Predictive power is represented in terms of the χ2-value.
To select the attributes with the highest individual predictive value while controlling for correlations and redundancy among the ENCODE data attributes, the training data was subjected to attribute selection using the CfsSubsetEval evaluator of Weka using the greedy search method known as ‘BestFirst’ (Witten and Frank, 2005). Examples of highly significant, non-trivial correlations (correlations not derived from ENCODE datasets generated at the same site), are presented in Supplementary Table 2 and visually in Supplementary Image 1. Ultimately, 28 predictive attributes were selected in this manner (Table 1). These attributes fall into five general categories: regulatory factor binding sites (RFBS), histone modifications (HM), promoter identification (PI) based on luciferase reporter assays, DNaseI hypersensitive sites (DHS) and transcriptional activity (TA). Interestingly, sequence conservation or identification of conserved elements, identified previously as informative markers for discriminating between functional and neutral rSNPs, were not among the remaining most significant predictors, suggesting a potential role for lineage-specific regulatory mechanisms that mediate human disease. The training data is given in Supplementary Table 3.
2.3 Cross-validation
Cross-validation was carried out by randomly splitting the training set into 3 (3-fold cross-validation) or 10 (10-fold cross-validation) groups, then performing predictions on each group using the remaining groups as the training data. Randomized introduction of new ‘disease-SNPs’ was performed by randomly selecting 102 SNPs from the 11 249 SNPs mapping within 5 kb of a gene, performing feature selection as above, then performing predictions using the new selected features.
3 ALGORITHM
3.1 Prediction scheme
Given the small size of the training data (102 disease SNPs and 1049 likely neutral disease SNPs), and the almost certain conditional independence violation of our predictive attributes, we surmised that either a naïve Bayes classifier (George et al., 1995) or a ridge logistic regression (le Cessie and van Houwelingen, 1992; Malo et al., 2008) would produce the most powerful and compelling predictive models or classifiers of disease associated SNPs based on ENCODE attributes. The naïve Bayes classifier, encoded in Weka, was implemented using a normal distribution estimator. The ridge logistic regression classifier, also encoded in Weka, was implemented with ridge estimator value of 1.0 × 10−8, and without limits on the number of iterations required for convergence. Classifiers were judged based upon their average F-measure to control for the limited availability of disease SNPs. Ultimately, the ridge logistic regression resulted in a higher F-measure based on 10-fold cross-validation and was thus chosen as the final classifier (average F-measure 0.92 versus 0.81 for naïve Bayes). Logistic regression also gave the highest average F-measure as compared to other classifiers, including support vector machine, decision trees and nearest neighbor classifiers (Supplementary Table 4). The threshold probability to call a SNP functional was set at the value that gave the highest average F-measure: 0.50. Accuracy was ultimately measured in a variety of ways including the area under the curve (AUC), determined empirically, receiver operator characteristic curves (ROC), and the Matthew's correlation coefficient (MCC).
4 IMPLEMENTATION
4.1 Accuracy
The logistic regression method accurately identified disease-causing SNPs with 83.3% accuracy and neutral SNPs with 99.5% accuracy (AUC=0.960 ± 0.003, MCC=0.877) [Fig. 1, Table 2 (wRF)]. Cross-validation analysis confirmed a high level of predictive power for this model, although predictions for disease-causing mutations suffered from the loss of training data in this analysis: 10-fold cross-validation resulted in 77.4% disease SNP accuracy and 99.0% neutral SNP accuracy (AUC=0.938 ± 0.005, MCC=0.809); 3-fold cross-validation resulted in 77.5% disease SNP accuracy and 98.6% neutral SNP accuracy (AUC=0.930 ± 0.005, MCC=0.789) (Fig. 1, Table 2). Ultimately, accuracy for predicting disease-causing SNPs is weakened when a larger number of SNPs in the training data are withheld for the cross-validation analyses, while neutral SNP accuracy is only slightly reduced.
Fig. 1.
ROC curves generated from training and testing the classifier based on the full dataset (black), 10-fold cross-validation (green) and 3-fold cross-validation (red). Note the modest differences between testing based on the full dataset and cross-validation. AUC is shown in Table 2.
Table 2.
Accuracy of predictions
| Test set | Area under the curve | Matthew's correlation coefficient | Balanced error rate | True positive(%) | True negative (%) | Correctly classified (%) |
|---|---|---|---|---|---|---|
| (wRF) Full training set | 0.960 ± 0.003 | 0.877 | 0.086 | 83.3 | 99.5 | 98.1 |
| (wRF) 10-Fold cross-validation | 0.938 ± 0.005 | 0.809 | 0.118 | 77.4 | 99.0 | 97.0 |
| (wRF) 3-Fold cross-validation | 0.930 ± 0.005 | 0.789 | 0.120 | 77.5 | 98.6 | 96.7 |
| (nRF) Full training-set | 0.948 ± 0.004 | 0.876 | 0.095 | 81.4 | 99.7 | 98.1 |
| (nRF) 10-Fold cross-validation | 0.927 ± 0.005 | 0.813 | 0.122 | 76.5 | 99.1 | 97.1 |
| (nRF) 3-Fold cross-validation | 0.927 ± 0.005 | 0.819 | 0.117 | 77.5 | 99.1 | 97.2 |
| Random disease SNPs | 0.678 ± 0.015 | 0.146 | 0.485 | 3.0 | 99.9 | 91.3 |
wRF = with regulatory factors, nRF = without regulatory factors.
To demonstrate that the predictive power of our method does not result from a random selection of ENCODE attributes which happen to differentiate our disease-causing and neutral SNPs, 10 random datasets were generated in which the neutral SNPs were kept the same as our original dataset, but 102 new ‘disease SNPs’ were randomly selected from all SNPs mapping within 5 kb of genes in the ENCODE regions. These datasets were then subjected to the same feature selection and prediction scheme as described earlier. Table 2 presents the average performance of the predictions across all 10 random datasets (random disease SNPs). Note the low MCC (0.146) and proportion of ‘disease SNPs’ (3.0%) identified correctly. This result confirms that our list of predictive attributes (Table 1), do, in fact, accurately distinguish disease-causing SNPs from neutral SNPs in a biologically meaningful manner.
4.2 Generalizability
To improve the generalizability of our predictions, regulatory factor binding site-related at-tributes were removed from the set of predictive attributes (nRF, Table 2), to eliminate any bias resulting from known disease SNPs which alter specific regulatory factor binding sites. This adjustment resulted in a small loss of predictive power when the full training set is used to make predictions (MCC=0.876 versus 0.877), but decreases the loss in accuracy during cross-validation (10-fold cross-validation MCC=0.813 versus 0.809, 3-fold cross-validation MCC 0.819 versus 0.789), suggesting removal of the regulatory factor binding site attributes will improve performance on rSNPs affecting genes not represented in the training data. This training data is given in Supplementary Table 5. This generalized predictor was ultimately applied to a large set of SNPs mapping within 5 kb of a gene in the Encode regions (see Methods Section 2; the results are presented in Supplementary Table 6).
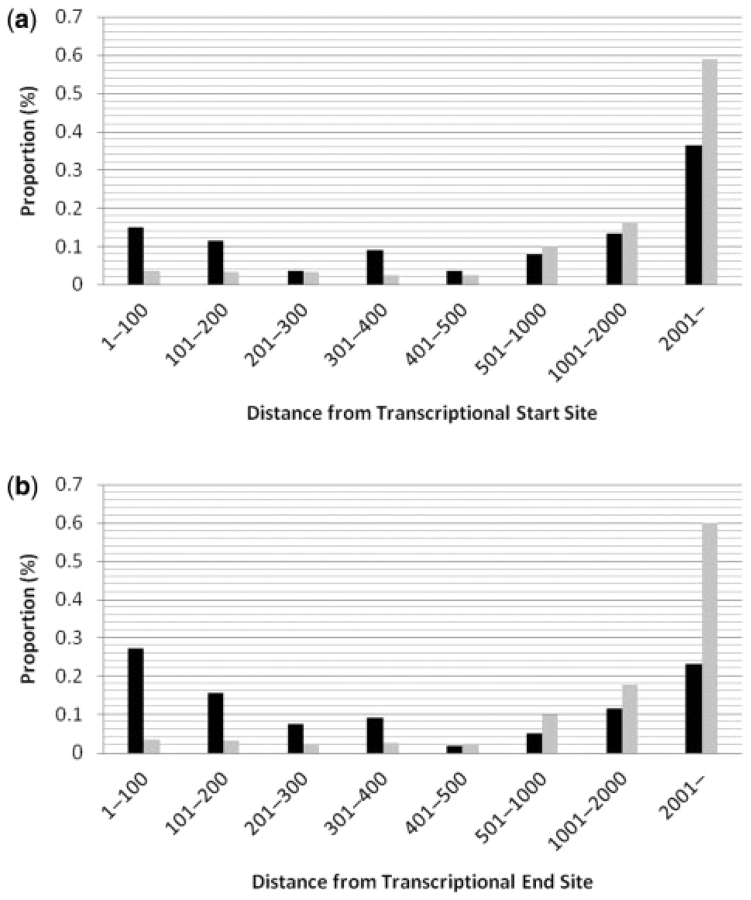
Within this set of 11 249 SNPs, 275 SNPs (2.4%) were predicted to affect gene expression. These SNPs, nearby genes and the gene disease associations are presented in Supplementary Table 7. Forty-two percent of predicted functional regulatory polymorphisms occurred within the proximal promoter (first 500 bp before the transcriptional start site). Thirty-five percent of these predicted functional regulatory polymorphisms within the proximal promoter occurred within the first 100 bp before the transcriptional start site, consistent with the bias in the distribution of functional polymorphisms observed by Buckland et al. (2005). In fact, the distribution of predicted functional polymorphisms exactly mirrors the distribution of confirmed functional polymorphisms observed by Buckland et al. (2005), including a slight excess of functional polymorphisms residing between 301 bp and 400 bp from the transcriptional start site (Fig. 2a). Furthermore, we predict a large number of functional polymorphisms in distant regulatory regions (more than 2 kb away from the transcriptional start site), suggesting that functional polymorphisms affecting long range regulatory elements are important mediators of gene expression. The distribution of predicted functional polymorphisms downstream of genes is similar to the upstream distribution, but with a much stronger bias for functional SNPs closer to the transcriptional end site (61% within the first 500 bp of the transcriptional end site) (Fig. 2b). Predicted neutral polymorphisms occur much more frequently at sites distal from the transcriptional start or end sites (Fig. 2a and 2b).
Fig. 2.
(a) The proportion of 5′-upstream predicted functional (black bars) and neutral (gray bars) SNPs are displayed relative to their distance from the nearest gene transcriptional start sites. The distribution within the proximal promoter (first 500 bp) mirrors the distribution of known functional polymorphisms as described by Buckland et al. (b) The proportion of 3′-downstream predicted functional (black bars) and neutral (gray bars) SNPs are displayed relative to their distance from the nearest gene transcriptional end sites.
Although it is expected that a greater proportion of functional SNPs will lie within the proximal promoter of genes (5.8% of SNPs within the proximal promoter are predicted to be functional compared to 1.4% of SNPs beyond the first 500 bp), there is the possibility that this bias will result in false positive predictions within the proximal promoter. To address this issue we collected a list of functionally characterized SNPs located within the proximal promoter but with no functional affect from a survey conducted by Buckland et al. (2005). Note that functional SNPs identified in the Buckland et al. survey and other surveys (Hoogendoorn et al., 2003; Ng and Henikoff, 2006; Rockman and Wray, 2002) corresponded to disease SNPs within our dataset and could not be used as a positive set for independent verification. Of 648 SNPs analyzed by Buckland et al., 14 neutral SNPs were located in the ENCODE regions. All 14 SNPs were predicted to be neutral by our method. While this represents a small verification dataset, it suggests that the high degree of specificity observed in our initial analyses is applicable to SNPs residing within the proximal promoter and is not an artifact resulting from the fact that the majority of SNPs reside outside of this region.
5 DISCUSSION
Due to the limited availability of data, development of algorithms to prioritize rSNPs has been difficult. In this article, we describe how ENCODE data can be used to probabilistically prioritize regulatory variations. This method may be useful in identifying common disease associated rSNPs or can be used to prioritize rare ncSNPs identified via resequencing studies. The expansion of ENCODE annotated regions, especially promoter identification, and the corresponding availability of a larger training set of confirmed functional variants, should significantly improve the generalizability of this approach. Although a portion of the ENCODE regions represent a random sample of the genome, it is possible that this sampling has led to ascertainment bias in comparison to the genome as a whole. With these restrictions on the available training data, we were still able to achieve excellent sensitivity (∼80%) and specificity (∼99%) with confidence that the method is not overtrained.
Surprisingly, conservation was not selected as a significant predictive attribute. Comparison of the χ2-value for conservation predictors (≈100) places conservation in the lower end of our selected attributes. Either the more powerful attributes render conservation redundant, or, lineage-specific regulatory elements render conservation less informative in terms of disease prediction.
The accuracy of our predictions was obtained by identifying important regulatory sites through their degree of ‘openness’, as in DNaseI hypersensitive sites, transcriptional activity and epigenetic marks identifying sites important for transcription. It is possible that some of these attributes correlate with previously described predictive attributes, such as GC content or distance from transcriptional start sites (Montgomery et al., 2007). The ENCODE predictors identify promoter regions in a variety of ways. Transcriptional activity identifies these sites in a relatively straightforward way, by determining whether or not a genomic region is able to drive the transcription of a reporter gene. The epigenetic marks more accurately pinpoint these promoter sites by determining histone H3 acylation and H3K4 methylation sites. These epigenetic marks have been shown to distinctly mark the 5′ regions of transcriptionally active genes and tend not to extend into the transcribed regions (Liang et al., 2004). Therefore, these predictors are likely able to identify promoter regions for both known and yet-to-be-characterized genes or non-coding RNAs. DNaseI hypersensitive sites are able to define many other types of regulatory elements, including insulators, enhancers and silencers (Burgess-Beusse et al., 2002; Felsenfeld, 1996; Gross and Garrard, 1988). Any single predictor defines regulatory regions in broad sections, and it is likely that only the combination of the above predictors is able to more accurately define critical regulatory regions. Still, it is more than likely that our method is capable of defining small portions of the genome which contain critical regulatory elements, rather than pinpointing specific nucleotides of importance.
With the above caveats in mind, the method described in this article should significantly improve the ability to identify ncSNPs relevant to disease and provides a starting point for the investigation of functional non-coding polymorphisms. The limitations in resolution and applicability to the whole genome should be relatively straightforward to overcome upon the expansion of the ENCODE regions, as well as the availability of a larger and more general training set.
Supplementary Material
ACKNOWLEDGEMENTS
A.T. is a Scripps Genomic Medicine Dickinson Scholar.
Funding: N.J.S. and his laboratory are supported in part by the following research grants: The National Heart Lung and Blood Institute Family Blood Pressure Program (FBPP; U01 HL064777-06); the National Institute on Aging Longevity Consortium (U19 AG023122-01); the National Institute of Mental Health Consortium on the Genetics of Schizophrenia (COGS; 5 R01 HLMH065571-02); the NIMH-funded Genetic Association Information Network Study of Bipolar Disorder National (1 R01 MH078151-01A1); National Institutes of Health grants: N01 MH22005, U01 DA024417-01, and P50 MH081755-01; Scripps Genomic Medicine and the Scripps Translational Science Institute.
Conflict of Interest: none declared.
REFERENCES
- Andersen MC, et al. In silico detection of sequence variations modifying transcriptional regulation. PLoS Comput. Biol. 2008;4:e5. doi: 10.1371/journal.pcbi.0040005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckland PR, et al. Strong bias in the location of functional promoter polymorphisms. Hum. Mutat. 2005;26:214–223. doi: 10.1002/humu.20207. [DOI] [PubMed] [Google Scholar]
- Buckland PR. The importance and identification of regulatory polymorphisms and their mechanisms of action. Biochim. Biophys. Acta. 2006;1762:17–28. doi: 10.1016/j.bbadis.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Burgess-Beusse B, et al. The insulation of genes from external enhancers and silencing chromatin. Proc. Natl Acad. Sci. USA. 2002;99:16433–16437. doi: 10.1073/pnas.162342499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordell HJ, Clayton DG. Genetic association studies. Lancet. 2005;366:1121–1131. doi: 10.1016/S0140-6736(05)67424-7. [DOI] [PubMed] [Google Scholar]
- Damani SB, Topol EJ. Future use of genomics in coronary artery disease. J. Am. Coll. Cardiol. 2007;50:1933–1940. doi: 10.1016/j.jacc.2007.07.062. [DOI] [PubMed] [Google Scholar]
- Eberle MA, et al. Power to detect risk alleles using genome-wide tag SNP panels. PLoS Genet. 2007;3:1827–1837. doi: 10.1371/journal.pgen.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin unfolds. Cell. 1996;86:13–19. doi: 10.1016/s0092-8674(00)80073-2. [DOI] [PubMed] [Google Scholar]
- Flicek P, et al. Ensembl 2008. Nucleic Acids Res. 2008;36:D707–D714. doi: 10.1093/nar/gkm988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George H, et al. Estimating continuous distributions in Bayesian classifiers. Proceedings of the Eleventh Conference on Uncertainty in Artificial Intelligence; 1995. pp. 338–345. [Google Scholar]
- Gorlov IP, et al. Shifting paradigm of association studies, value of rare singlenucleotide polymorphisms. Am. J. Hum. Genet. 2008;82:100–112. doi: 10.1016/j.ajhg.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross DS, Garrard WT. Nuclease hypersensitive sites in chromatin. Annu. Rev. Biochem. 1988;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- Hoogendoorn B, et al. Functional analysis of human promoter polymorphisms. Hum. Mol. Genet. 2003;12:2249–2254. doi: 10.1093/hmg/ddg246. [DOI] [PubMed] [Google Scholar]
- Karolchik D, et al. The UCSC Genome Browser Database, 2008 update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kel AE, et al. MATCH, a tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003;31:3576–3579. doi: 10.1093/nar/gkg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. BLAT–the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft P, Cox DG. Study designs for genome-wide association studies. Adv. Genet. 2008;60:465–504. doi: 10.1016/S0065-2660(07)00417-8. [DOI] [PubMed] [Google Scholar]
- Cessie S, van Houwelingen JC. Ridge estimators in logistic regression. Appl. Stat. 1992;41:191–201. [Google Scholar]
- Liang G, et al. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc. Natl Acad. Sci. USA. 2004;101:7357–7362. doi: 10.1073/pnas.0401866101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malo N, et al. Accommodating linkage disequilibrium in genetic-association analyses via ridge regression. Am. J. Hum. Genet. 2008;82:375–385. doi: 10.1016/j.ajhg.2007.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew CG. New links to the pathogenesis of Crohn disease provided by genome-wide association scans. Nat. Rev. Genet. 2008;9:9–14. doi: 10.1038/nrg2203. [DOI] [PubMed] [Google Scholar]
- Michal L, et al. Functional characterization of variations on regulatory motifs. PLoS Genet. 2008;4:e1000018. doi: 10.1371/journal.pgen.1000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller W, et al. 28-way vertebrate alignment and conservation track in the UCSC Genome Browser. Genome Res. 2007;17:1797–1808. doi: 10.1101/gr.6761107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery SB, et al. A survey of genomic properties for the detection of regulatory polymorphisms. PLoS Comput. Biol. 2007;3:e106. doi: 10.1371/journal.pcbi.0030106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney S. Bioinformatics approaches and resources for single nucleotide polymorphism functional analysis. Brief. Bioinform. 2005;6:44–56. doi: 10.1093/bib/6.1.44. [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. Predicting the effects of amino acid substitutions on protein function. Annu. Rev. Genomics Hum. Genet. 2006;7:61–80. doi: 10.1146/annurev.genom.7.080505.115630. [DOI] [PubMed] [Google Scholar]
- Rockman MV, Wray GA. Abundant raw material for cis-regulatory evolution in humans. Mol. Biol. Evol. 2002;19:1991–2004. doi: 10.1093/oxfordjournals.molbev.a004023. [DOI] [PubMed] [Google Scholar]
- Roth FP, et al. Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole-genome mRNA quantitation. Nat. Biotechnol. 1998;16:939–945. doi: 10.1038/nbt1098-939. [DOI] [PubMed] [Google Scholar]
- Shames DS, et al. DNA methylation in health, disease, and cancer. Curr. Mol. Med. 2007;7:85–102. doi: 10.2174/156652407779940413. [DOI] [PubMed] [Google Scholar]
- Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenson PD, et al. Human Gene Mutation Database (HGMD), 2003 update. Hum. Mutat. 2003;21:577–581. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- Torkamani A, Schork NJ. Accurate prediction of deleterious protein kinase polymorphisms. Bioinformatics. 2007;23:2918–2925. doi: 10.1093/bioinformatics/btm437. [DOI] [PubMed] [Google Scholar]
- Witten IH, Frank E. Data Mining, Practical Machine Learning Tools and Techniques. 2nd. San Francisco: Morgan Kaufmann; 2005. [Google Scholar]
- Wray NR, et al. Prediction of individual genetic risk to disease from genomewide association studies. Genome Res. 2007;17:1520–1528. doi: 10.1101/gr.6665407. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.