Abstract
Now that many genomes have been sequenced and the products of newly identified genes have been annotated, the next goal is to engineer the desired phenotypes in organisms of interest. For the phenotypic engineering of microorganisms, we have developed novel artificial transcription factors (ATFs) capable of reprogramming innate gene expression circuits in Escherichia coli. These ATFs are composed of zinc finger (ZF) DNA-binding proteins, with distinct specificities, fused to an E. coli cyclic AMP receptor protein (CRP). By randomly assembling 40 different types of ZFs, we have constructed more than 6.4 × 104 ATFs that consist of 3 ZF DNA-binding domains and a CRP effector domain. Using these ATFs, we induced various phenotypic changes in E. coli and selected for industrially important traits, such as resistance to heat shock, osmotic pressure and cold shock. Genes associated with the heat-shock resistance phenotype were then characterized. These results and the general applicability of this platform clearly indicate that novel ATFs are powerful tools for the phenotypic engineering of microorganisms and can facilitate microbial functional genomic studies.
INTRODUCTION
Historically, many unique cellular traits of microorganisms have been identified and modified, not only for the benefit of human life, but also for industrial applications. To improve the characteristics of useful microorganisms, several approaches have been explored to expedite the screening for new phenotypes of interest. Because of its simplicity and convenience, random mutagenesis has been widely used for the selection of desired phenotypes (1,2). However, mutagenesis approaches rely on labor-intensive work and luck. Therefore, more rational approaches have been employed for the manipulation of genes relevant to a specific function (3–5). Most rational approaches involve either the deletion or overexpression of a single gene or sequential multigenic modifications (4,5). However, the engineering of desired phenotypes in organisms often requires the complete reprogramming of innate gene expression circuits, a process that necessitates multigenic transcriptional coordination. Even with the most sophisticated computational methods for physiological pathway analysis, complete identification of the precise genes involved in achieving a desired phenotype is almost impossible. Therefore, to permit multiple simultaneous gene expression changes, researchers recently developed methods for engineering of the global transcription machinery (6,7). Although this methodology permits changes in the expression of many genes that allow organisms to access novel cellular phenotypes, its use has been restricted either to a specific genotype or to housekeeping genes that are generally expressed and thought to be involved in routine cellular metabolism (7). Here, we have developed novel artificial transcription factors (ATFs) to completely reprogram innate gene expression circuits in Escherichia coli and thus to elicit broad perturbations in the E. coli transcriptome.
Zinc fingers (ZFs) are well-characterized and highly specific DNA-binding domains found in a wide variety of transcriptional regulatory proteins (8,9). Because of their diversity and modular structure, ZF domains have provided an attractive framework with which to construct diverse, novel ATFs (10–17).
The cyclic AMP receptor protein (CRP) is a global transcription factor that regulates gene expression at >200 different promoters in E. coli (18–21). CRP activates transcription by interacting with RNA polymerase through CRP's functionally independent transcriptional activation domains. Because of the existence of these various domains (20), CRP could be engineered and used as a transcriptional effector domain to construct novel ATFs.
In this study, we selected 40 human ZFs with diverse DNA-binding specificities and combinatorially assembled them to construct a library of DNA-binding domains that consisted of 3 ZFs each. These DNA-binding domains were then fused to a CRP effector domain to create a fusion protein that either activated or repressed transcription regardless of the presence of endogenous transcription factors. Introduction of these novel ATFs into E. coli induced various phenotypic changes such as resistance to heat shock, osmotic pressure and cold shock. The bacterial strains were selected that displayed the newly acquired industrially important traits and the genes associated with the selected phenotypes were identified and characterized.
MATERIALS AND METHODS
Strains and plasmids
E. coli XL1-Blue (Stratagene, La Jolla, CA) and E. coli K-12 MG1655 were used for this experiment. To test an EGFP-based reporter system, MG1655ΔaraBAD was constructed by λ-Red-mediated markerless deletion (22). For complementation assays, the genes related to the thermotolerance phenotype, cpxP, ompW and the marRAB operon were deleted from the MG1655 E. coli genome by the above method, producing MG1655ΔcpxP, MG1655ΔompW and MG1655ΔmarRAB, respectively. The T7lac promoter of the pETDuet-1 vector (Novagen, San Diego, CA) was replaced by either a tac promoter or an arabinose-inducible promoter, generating pETtac and pETara, respectively. The pBR322 origin of the high-copy plasmid pETtac was replaced by P15A origin, generating low-copy plasmid pACYCtac. pETtac was used for the expression of the ATF libraries. pACYCtac was used for the expression of the T2 ATF to assess effects of T2 ATF copy number on the thermotolerance phenotype. In addition, for the complementary assays, the genes related to the thermotolerance phenotype, cpxP and ompW, were amplified with the polymerase chain reaction (PCR) from the MG1655 genomic DNA, and the amplified cpxP and ompW genes were cloned into pETtac individually or together, producing pETtac-cpxP, pETtac-ompW and pETtac-cpxP/ompW, respectively. pETara was used for the expression of the 3-ZF-CRP fusion proteins, which consisted of 3 ZFs fused to various CRP derivatives, to find the most effective CRP effector domain for this study (Figure 1A and Supplementary Table 2).
Figure 1.
Engineering a CRP-based effector domain for novel ATFs. (A) A schematic representation of CRP that shows the regions of the protein used as effector domains in relation to known structural features. The numbers indicate positions of the amino acid residues (Supplementary Table 1). (B) Activation of the EGFP reporter gene by 3-ZF-CRP fusion proteins in an EGFP-based reporter system. 3-ZF recognizes the target DNA sequence (5′-GCG GCG GGG-3′) upstream of the tac promoter of the reporter gene (−67 to −50), and 3-ZF-CRP fusion proteins activate transcription of the EGFP reporter gene upon induction with arabinose. E. coli cells that harbor both the reporter plasmid and the 3-ZF-CRP encoding plasmid were grown in LB medium with 0.2% arabinose (to induce 3-ZF-CRP fusion expression) and without IPTG (to repress EGFP expression) at 37°C for 10 h and then analyzed. (C) Repression of the EGFP reporter gene by 3-ZF-CRP fusion proteins in an EGFP-based reporter system. 3-ZF recognizes the target DNA sequence (5′-GCG GCG GGG-3′) downstream of the tac promoter of the reporter gene (+24 to +41), and 3-ZF-CRP fusion proteins repress transcription of the EGFP reporter gene upon induction with arabinose. E. coli cells that harbored both the reporter plasmid and 3-ZF-CRP encoding plasmid were grown under the same conditions described above, except that 0.5 mM IPTG was added at inoculation (to induce EGFP expression). Expression of the EGFP reporter gene regulated by a 3-ZF-CRP fusion protein is given as relative fluorescence intensity values. All fluorescence intensities were represented as a relative value compared with that of cells that contained the reporter plasmid and a control plasmid that did not encode a 3-ZF-CRP fusion protein and measured in triplicate. ‘+1’ indicates the transcription initiation site. Error bars represent SD.
Selection of a potent CRP effector domain with an EGFP-based reporter system
Reporter plasmids (pEGFP-A and pEGFP-R) were constructed by inserting a tac promoter sequence containing the target DNA-binding site for the 3-ZF-CRP fusion proteins and the gene encoding enhanced green fluorescent protein (EGFP) into pACYC184 (New England Biolabs, Beverly, MA). pEGFP-A was constructed by inserting 2 copies of the target DNA sequence (5′-GCG GCG GGG-3′) upstream of the tac promoter (between positions −67 to −50) of pACYC184, followed by insertion of the EGFP gene downstream of the promoter. pEGFP-R was constructed by inserting 2 copies of the target DNA sequence (5′-GCG GCG GGG-3′) downstream of the tac promoter (between positions +24 to +41) of pACYC184, followed by insertion of the EGFP gene downstream of the promoter. To investigate the effector activity of various CRP derivatives, MG1655ΔaraBAD was co-transformed with either pEGFP-A or pEGFP-R and a plasmid encoding a 3-ZF-CRP fusion protein. Activity of the 3-ZF-CRP fusion proteins was analyzed by assessing their ability to activate or repress transcription of the EGFP gene in pEGFP-A and pEGFP-R, respectively.
To examine the effector activity of CRP derivatives to activate transcription of the EGFP gene, transformants that harbored both a plasmid that encoded a 3-ZF-CRP fusion protein and pEGFP-A were grown in LB medium with 0.2% arabinose (to induce 3-ZF-CRP fusion protein expression) and without IPTG (to repress EGFP expression) at 37°C for 10 h. To measure the effector activity of CRP derivatives to repress transcription of the EGFP gene, transformants harboring both a plasmid that encoded a 3-ZF-CRP fusion protein and pEGFP-R were grown under the same conditions described above, but 0.5 mM IPTG was added at the time of inoculation (to induce EGFP expression). After harvesting cells by centrifugation, cell pellets were washed once with TE buffer, and the cell densities of the various preparations were made to be equivalent by the addition of varying amounts of the TE buffer. Fluorescence of these cell preparations were measured in an SLM-AMINCO 8100 spectrofluorometer (Spectronic Instruments, Rochester, NY) at an excitation wavelength of 488 nm and an emission wavelength of 507 nm. All fluorescence intensities were represented as relative values compared with that of a preparation of bacterial cells that contained the reporter plasmid and a control plasmid that did not encode a 3-ZF-CRP fusion protein.
Construction of an ATF library
An ATF library was constructed with 40 ZFs and CRP D1 (Supplementary Table 2). To construct a DNA fragment that encodes a ZF, two 66-nt oligonucleotides (oligos), where 19-nt sequences at each 3′ end are complementary to each other, are synthesized. These oligos are hybridized and filled in by T4 DNA polymerase, producing a 113-bp duplex DNA segment. The resulting 113-bp DNA segment has a sequence encoding a ZF in the middle, preceded by an 18-bp linker sequence containing an XmaI site (bold) at the 5′ end (5′-CCCGGGGAAAAACCGTAC-3′), which is frequently found at the junction between ZF-encoding DNA segments (at their 5′ ends) in naturally occurring ZF proteins. An AgeI site, an in-frame stop codon (TAA), and an EcoRI site (5′-ACCGGTTAAGAATTC-3′) were also incorporated into the 3′ region of the 113-bp ZF-encoding DNA segment. The 113-bp products of this fill-in reaction were then digested with XmaI and EcoRI, and subsequently cloned into pUC19 (New England Biolabs) digested with the same enzymes.
To construct an ATF library, a DNA segment that encoded CRP D1 was PCR-amplified from MG1655 genomic DNA using the following forward (CRP-F) and reverse (CRP-R) primers, respectively: CRP-F, 5′-CATGCCATGGTGCTTGGCAAACCGCAAACA-3′ [which contained an NcoI site (bold)]; and CRP-R, 5′-CCGGAATTCACCGGTAGAACAGCCGACAATCTGAC-3′ [which contained an AgeI site and an EcoRI site (bold)]. The PCR-amplified CRP D1 DNA, after digestion with NcoI and EcoRI, was cloned into pETtac that had been digested with the same enzymes, producing pETtac-CRP D1. Equal amounts of each single-finger plasmid were mixed together to form a pool. The single-finger plasmid pool was digested with XmaI and EcoRI and then ligated into the AgeI/EcoRI-digested pETtac-CRP D1 to generate a series of pETtac-CRP D1-1ZF library vectors, each encoding CRP D1 and one ZF. After transformation of E. coli XL1-Blue with these plasmids, the pETtac-CRP D1-1ZF library vectors were purified from the transformants.
Subsequently, the pETtac-CRP D1-1ZF library vectors that encoded CRP D1 and one ZF were digested with AgeI and EcoRI and then ligated to the pool of the XmaI/EcoRI-digested single-finger. After transformation of E. coli with the products of these ligation reactions, plasmids were purified, which now encoded CRP D1 and two ZFs (the pETtac-CRP D1-2ZF library vectors).
To the AgeI/EcoRI-digested pETtac-CRP D1-2ZF library vectors, the pool of XmaI/EcoRI-digested single-finger was ligated to generate the final ATF library used in the experiments herein, which encoded CRP D1 and three ZFs (the pETtac-CRP D1-3ZF library vectors) (14).
Screening for desirable phenotypes
E. coli MG1655 was electrotransformed with 10 ng of ATF library vector described above, with a transformation efficiency of 1 × 108 cfu/μg DNA. The resulting 106 transformants which exceeded the complexities of the ATF libraries were analyzed for thermotolerance, osmotolerance and cold-tolerance upon ATF expression.
To screen for the thermotolerant strains, transformants were cultured in LB liquid medium with 0.5 mM IPTG for 3 h at 37°C to induce ATF expression. Cells were then incubated for 2 h at 55°C. After heat treatment, cells were plated on LB agar plates and incubated overnight at 37°C. The surviving cells were screened again for the thermotolerant phenotype and cell growth was assessed. The growth experiments were performed in 100 ml LB medium containing 0.5 mM IPTG at both 37 and 50°C. Cell growth was monitored by measuring the optical density (O.D.) at 600 nm.
For the screening of the osmotolerant phenotype, transformants were cultured in LB liquid medium with 0.5 mM IPTG for 3 h at 37°C. Cells were then plated on minimal A agar plates with 0.6 M NaCl and 0.5 mM IPTG. After 2 days of incubation at 37°C, the growing colonies were selected and individually analyzed for their osmotolerance. For the screening of the cold-tolerance phenotype, transformants were cultured in LB liquid medium with 0.5 mM IPTG for 3 h at 37°C. Cells were then plated on LB agar plates with 0.5 mM IPTG. After 2 days of incubation at 15°C, the growing colonies were selected and individually analyzed for their cold-tolerance.
Plasmids that encoded ATFs were rescued from the selected cells that showed desirable phenotypes, and the ATF sequences of the rescued plasmid were confirmed by DNA sequencing. The plasmids that contained the ATFs were retransformed back into E. coli to confirm the phenotypic changes.
DNA microarray analysis of the thermotolerant cells
E. coli cells transformed with either a plasmid encoding the T2 ATF or the pETtac vector were cultured in LB liquid medium with 0.5 mM IPTG at 50°C. Total RNA was isolated from exponential-phase cultures with an RNeasy mini kit (QIAGEN, Valencia, CA) and RNAprotect Bacteria Reagent (QIAGEN), and the RNA preparations were subjected to reverse transcription. cDNA labelled with either Cy3-dUTP (C, wild-type E. coli transformed with a control plasmid) or Cy5-dUTP (LT2 and HT2, cells expressing the T2 ATF on low- and high-copy plasmids, respectively) was synthesized from each preparation of total RNA by random priming. Labelled cDNA probes were purified and hybridized to a DNA microarray slide (TwinChipTM E. coli chip, Digital Genomic, Seoul, Korea) that contained the complete E. coli genome. The slides were scanned by Scanarray lite (Packard Bioscience, Boston, MA) and analyzed by the GenePix Pro 3.0 software (Axon Instrument, Union City, CA).
RESULTS
Engineering of CRP as an effector domain
A potent effector domain is needed to construct ATFs capable of carrying out transcriptional activation or repression of a gene(s). Therefore, we examined the transcriptional activities of wild-type CRP and several CRP derivatives in order to identify effective effector domains for the generation of ATFs (Supplementary Table 1). Wild-type CRP (amino acid residues 1–209), CRP D1 (residues 1–180), CRP D2 (residues 137–190), CRP D3 (residues 137–180) and CRP D4 (residues 151–168) were selected as candidate effector domains for our novel ATFs (Figure 1A). These selected CRP effector domains were then each fused to a DNA-binding domain that consisted of 3 ZFs (Z23, Z18 and Z18, ordered in the N- to C- terminal direction) and binds specifically to the 9-base pair (bp) DNA sequence 5′-GCG GCG GGG-3′ (Supplementary Table 2).
We used a reporter plasmid in which the EGFP gene was used as a reporter to select the most potent effector domains. Two different reporter plasmids were constructed by inserting the target DNA sequences for the 3-ZF domain into two different regions of the reporter gene. Specifically, the resulting plasmids were pEGFP-A, which contained the 3-ZF target sequence upstream of the reporter promoter (−67 to −50), and pEGFP-R, which contained the 3-ZF target sequence downstream of the promoter (+24 to +41) (Figure 1).
The ATFs that were constructed by combining the 3-ZF domain with one of various CRP derivatives were shown to efficiently regulate transcription of the reporter gene. Among CRP derivatives, CRP D1 showed both strong activation and repression, depending on the position of the 3-ZF target DNA sequence (Figure 1B and C). Therefore, CRP D1 was chosen as an effector domain for the remainder of this study. An ATF that consisted of the 3-ZF domain alone (no effector domain) very poorly either activated or repressed transcription, depending on the position of the 3-ZF target DNA sequences.
Construction of ATF libraries
ZFs have exquisite sequence specificity and modularity. Amino acid residues at positions −1, 2, 3 and 6 in the α-helix of a ZF confer its specificity, recognizing a unique 3-bp of DNA sequence (8,9). We compared the amino acid residues at critical base-contacting positions of ZFs encoded by sequences in the human genome and selected a total of 40 ZFs that specify distinct DNA-binding specificities (Supplementary Table 2).
A library of DNA-binding domains consisting of 3 ZFs each was constructed by combinatorial assembly of the selected 40 ZFs (13,14). This process allowed the preparation of 6.4 × 104 (= 40 × 40 × 40) distinct, 3-ZF DNA-binding domains that targeted selected 9-bp target DNA sequences. These DNA-binding domains were then linked to a potent transcriptional effector domain (CRP D1) to create ATFs (14).
Screening for desirable phenotypic changes
We introduced the ATF libraries constructed with high-copy plasmid into E. coli and allowed ATFs to be constitutively expressed. We then screened the transformed bacteria for specific phenotypes, such as resistance to heat shock, osmotic pressure and cold shock.
The phenotype that we screened for first was that of improved thermotolerance (Figure 2A). After heat treatment of a pool of 106 ATF transformants at 55°C for 2 h, a condition that severely inhibits the growth of wild-type E. coli (17,23), 50 ATF transformants survived. The selected thermotolerant cells not only survived but grew well under the condition of heat treatment (at 55°C for 2 h), whereas growth of wild-type cells was completely inhibited under the same condition. In contrast, both wild-type and the selected thermotolerant cells showed similar growth patterns when grown at the optimal growth temperature, 37°C, without heat treatment.
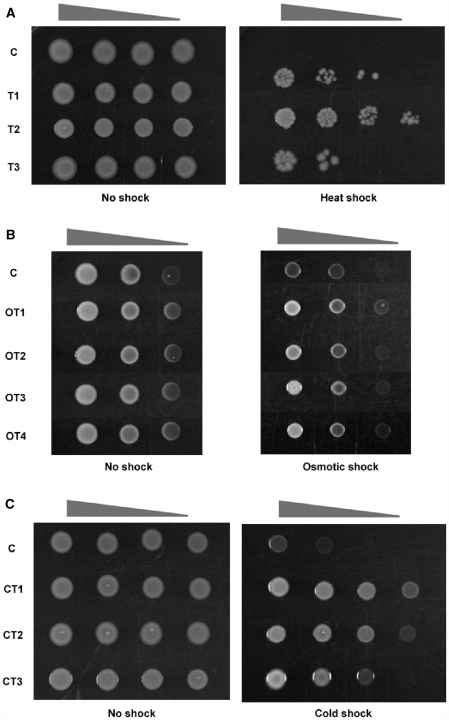
Figure 2.
Various phenotypic changes induced by ATFs. (A) A thermotolerant phenotype induced by ATFs. The growth of thermotolerant cells (T1, 2 and 3) and control cells (C) was monitored after incubation for 2 h at 37°C (No shock) or for 2 h at 55°C (Heat shock). The triangles above each panel indicate tenfold serial dilutions (1 : 1 to 1 : 1000, left to right) of spotted cells. (B) An osmotolerant phenotype induced by ATFs. The growth of cells was monitored on minimal A plates containing 0.6 M NaCl. The triangles above each panel indicate fivefold serial dilutions (1 : 1 to 1 : 25, left to right) of spotted cells. OT1-OT4, osmotolerant cell isolates 1–4, respectively. (C) A cold-tolerant phenotype induced by ATFs. Cells were cultured at 15°C, and cell growth was monitored. The triangles above each panel indicate fivefold serial dilutions (1 : 1 to 1 : 125, left to right) of spotted cells. CT1 and CT2, cold-tolerant cell isolates 1 and 2, respectively. For all panels, C indicates the wild-type E. coli transformed with a control plasmid without any previous selection. After the first round of phenotypic screening, all of the phenotypic changes were confirmed by plasmid rescue, sequence analysis and retransformation.
Second, to screen for cells with improved osmotolerance, ATF transformants were grown on a minimal A medium containing 0.6 M NaCl, which almost completely inhibits the growth of wild-type cells (24,25). Of 106 ATF transformants screened, 150 ATF transformants survived on a minimal A medium containing 0.6 M NaCl. The surviving cells were then subjected to further screening (Figure 2B). The selected ATF transformants showed an improvement in osmotolerance at the concentration of 0.6 M NaCl. In the absence of osmotic pressure (as encountered in 0.6 M NaCl), both wild-type and the selected osmotolerant cells exhibited similar growth patterns.
Finally, we screened for ATFs that induced a cold-tolerance phenotype (26–28) in E. coli (Figure 2C). To screen for cold-tolerant cells, the pool of 106 ATF transformants was grown at 15°C in LB medium, wherein most wild-type E. coli cells stop their growth. Thirteen of 106 ATF transformants survived at 15°C. Only the selected cold-tolerant cells grew well at 15°C, even though both the selected cold-tolerant and wild-type cells showed similar growth patterns when grown at the optimal growth temperature, 37°C.
To confirm that the various phenotypic improvements were indeed caused by the ATFs, the plasmids that encoded the ATFs were rescued from the selected cells and then retransformed into wild-type E. coli. Of 106 wild-type E. coli cells retransformed with each rescued ATF plasmid, 50 transformants were randomly selected and analyzed for their respective phenotypic changes (thermotolerance, osmotolerance and cold-tolerance). The phenotypic changes in all the selected retransformants were the same as those in the primary transformants. The selected ATFs induced only their respective phenotypes and random ATFs did not confer the above phenotypes upon E. coli cells. The identities of the selected ATFs along with their respective phenotypic changes are presented in Supplementary Table 3.
Of all the best-growing thermotolerant mutant cells, the T2 thermotolerant cell expressing an ATF on high-copy plasmid (HT2) exhibited the most improved cellular viability under the stressful condition (heat treatment at 55°C for 2 h). Therefore, the remainder of this study focuses on this particular mutant cell, which expressed the T2 ATF. The T2 ATF was composed of CRP D1 and 3 ZFs (Z13, Z2 and Z25, ordered in the N- to C- terminal direction) (Supplementary Table 3). The putative binding site for the T2 ATF was inferred to be 5′-NGA TTC (G/A)CT-3′ from the published literature (8,13,29,30).
In addition, the T2 ATF generated the different degree of thermotolerance, when it was expressed at various levels with different expression vectors. The T2 transformants expressing T2 ATF on low-copy plasmid (LT2) were also more heat resistant than wild-type cells. However their heat resistance was less than that of T2 ATF on high-copy plasmid (HT2) under the given heat-stress condition (at 55°C for 2 h). This observation suggests that various levels of phenotypic changes can be obtained by controlling the level of ATF expression using different copy number plasmids.
Identification of genes associated with thermotolerance
Using cDNA prepared from RNA isolated from the E. coli transformant containing the T2 ATF on low- and high-copy plasmids, we performed genome-scale gene expression profiling analyses of the T2 transformant with E. coli DNA microarrays. Transcription profiling of the T2 thermotolerant cells revealed that the T2 ATF caused differential expression of hundreds of genes relative to the wild-type cells containing a control plasmid without the ATF. Specifically, a total of 202 genes in the transformant with HT2 were differentially expressed by more than 4-fold compared to the control (133 up-regulated and 69 down-regulated) (Supplementary Tables 4 and 5). Similarly, 107 genes were altered in the transformant with LT2 (61 up-regulated and 46 down-regulated) (Supplementary Tables 6 and 7). Among these genes, 45 genes were altered in a similar fashion in both HT2 and LT2 (25 up-regulated and 20 down-regulated). These results suggest that the T2 ATF is affecting a subset of important genes responsible for thermotolerance, implying that thermotolerance is a phenotype controlled by those genes.
Although the transcriptional reprogramming that occurred in the T2 thermotolerant cells was quite broad, there was some clustering of genes that encoded chaperons and proteins involved in membrane functions and protection against environmental stresses (23,31,32) (Supplementary Tables 4–7). Genes with no known function were also found to be differentially expressed in T2 thermotolerant cells relative to the control.
We noticed that the genes that showed the largest change in gene expression in T2 thermotolerant cells (both HT2 and LT2), relative to the control, were those involved in the protection of bacteria against various forms of environmental stresses (Table 1). To determine whether these genes are direct targets of the T2 ATF, we analyzed the regulatory sequences to search for sequence elements that matched the putative binding site for the T2 ATF. Indeed, many of the genes affected by the T2 ATF housed binding sites for this transcription factor in their regulatory regions. Among those genes, cpxP, ompW and marRAB exhibited the greatest change in their expression. The T2 ATF was found to bind to the putative T2 ATF-binding sites present in the regulatory regions of cpxP, ompW and marRAB by a DNA band-shift assay (data not shown), implying that they are direct targets of the T2 ATF. Therefore, cpxP, ompW and marRAB were selected for further study (Table 1).
Table 1.
Genes associated with thermotolerance
| Gene | Functiona | log2 fold-changeb (high/low)c |
|---|---|---|
| Increased expression | ||
| ompW | Outer membrane protein | 6.1/5.2 |
| cpxP | Chaperone involved in resistance to extracytoplasmic stress | 5.3/2.8 |
| yliH | Conserved protein | 4.9/4.1 |
| ybaS | Predicted glutaminase | 4.3/1.8 |
| ybaT | Predicted transporter | 4.2/1.4 |
| Decreased expression | ||
| marR | MarR transcriptional repressor | −3.5/−1.3 |
| marA | MarA transcriptional activator | −3.2/−1.2 |
| marB | Multiple antibiotic resistance protein | −2.7/−1.3 |
aFrom the EcoCyc database (http://ecocyc.org).
blog2 fold-change in gene expression between the T2 thermotolerant mutant and the wild-type E. coli containing a control plasmid that lacked an ATF, both grown at 50°C (average of duplicate experiments).
chigh and low, log2 fold-change of T2 thermotolerant mutant cell expressing the T2 ATF on high- and low-copy plasmids, respectively.
cpxP and ompW, which are known to be involved in resistance to environmental stresses (33–35), were the most highly activated by the T2 ATF.
In addition, a number of genes were repressed by the T2 ATF. The largest decrease in gene expression was seen for the marRAB operon that is known to encode proteins that have a role in multiple antibiotic resistance. The MarRAB protein, for example, is a transcription factor that regulates the expression of several E. coli genes involved in responses to multiple antibiotics and oxidative stress. Transcription profiling revealed that genes regulated by the marRAB operon exhibited the great change in expression levels in the T2 thermotolerant mutant cell, relative to the control. For example, the hdeA and slp genes, which are known to be repressed by MarRAB in wild-type cells, were strongly activated 4- to 16-fold in the T2 cells (Supplementary Table 4). These results imply that the T2 ATF inhibits the expression of MarRAB, which subsequently results in either the activation or repression of genes that are differentially regulated by this versatile transcriptional regulatory protein.
To examine the functional relevance of these target genes to the thermotolerance phenotype in E. coli, we either overexpressed or knocked out the cpxP and ompW genes. We also knocked out the marRAB operon and examined the response of the mutant strain to heat treatment (Figure 3). When the cpxP or ompW gene was individually overexpressed in wild-type cells, the resulting cells were more resistant to the lethal effects of heat shock than wild-type cells. But they were less heat resistant than the T2 thermotolerant cells (HT2 and LT2) under heat-stress conditions. In addition, co-expression of cpxP and ompW in wild-type cells led to more heat tolerance than the wild-type cells overexpressing cpxP or ompW individually. The cpxP and ompW co-overexpressing cells were nearly as resistant as the LT2 thermotolerant cell, but much less resistant than the HT2 thermotolerant cell. Despite the increase in heat tolerance of the cpxP- or/and ompW-overexpressing cells, the basal growth rate of these strains in the absence of heat stress was similar to that of wild-type cells. Furthemore, the cpxP-deleted mutant exhibited a wild-type phenotype, whereas the ompW-deleted mutant was less heat resistant than wild-type cells.
Figure 3.
Identification of target genes associated with thermotolerance. (A) Analysis of thermotolerance by overexpression or knockout of the cpxP and/or ompW genes and knockout (Δ) of the marRAB operon. Growth of cells was monitored after incubation for 2 h at 37°C (No shock) or for 2 h at 55°C (Heat shock). The triangles above each panel indicate tenfold serial dilutions (1 : 1 to 1 : 1000, left to right) of spotted cells. (B) Growth profiles in LB medium (100 ml) at 37 and 50°C, respectively. Inset graph shows growth profiles of LT2 cells expressing T2 ATF on low-copy plasmid in LB medium (100 ml) at 50°C. The O.D.s were measured with a spectrophotometer at 600 nm at different time intervals. The growth experiments were performed in triplicate. Error bars represent SD. C, the wild-type E. coli transformed with a control plasmid without any previous selection. cpxP and ompW, cells overexpressing cpxP and ompW, respectively. cpxP + ompW, cells co-expressing cpxP and ompW. ΔcpxP, cpxP knockout mutant. ΔompW, ompW knockout mutant. ΔmarRAB, marRAB knockout mutant.
Concerning the marRAB, the marRAB knockout mutant did not show any thermotolerance even though the marRAB was highly repressed in the T2 thermotolerant cell (HT2). This observation indicates that this operon itself is insufficient to constitute the thermotolerance phenotype. Thus, MarRAB may not participate directly in the development of thermotolerace of E. coli.
Taken together, the individual effect of cpxP, ompW and marRAB on the thermotolerance was not strong enough to confer a HT2-like thermotolerance, even though their expression level was significantly influenced by the T2 ATF in the HT2 thermotolerant cell. Detailed mechanism of these genes involved in the thermotolerance should be further studied.
DISCUSSION
The intricate interaction of multiple genes throughout the entire gene expression network controls the phenotypic variations in cells and organisms. Herein, we have demonstrated the applicability of ATF libraries to induce genetic perturbations (through transcriptional activation or repression) and thus to induce phenotypic variations in E. coli.
Transcription factors have modular structures that consist of a DNA-binding domain and an effector (activation or repression) domain. Libraries of proteins that contain only a DNA-binding domain have been used to repress transcription (12,17). However, DNA-binding domains that do not contain an effector domain cannot activate the expression of genes, because activation of transcription depends upon the activity of an effector domain. Using CRP as an effector domain, our ATFs can activate the expression of genes. Furthermore, prokaryotic transcription factors can have dual activity as either an activator or a repressor (36). Activation or repression mediated by transcription factors is linked to the distance of the regulator-binding sites relative to promoters (36). The preferred sites for activators are located between nucleotide positions −80 and −30. Repressor-binding sites are generally located downstream from nucleotide position −30. We have also shown that our ATFs can function as both an activator and a repressor as described above (Figure 1B and C).
We induced various phenotypic changes in E. coli with our ATFs and the genes associated with the heat-resistance phenotype were identified. We found that the expression of over two hundred genes was significantly affected by the T2 ATF that induces a thermotolerance phenotype. RNA for the transcriptomic analysis was isolated from exponential-phase cultures to see the biggest difference in gene expression between the wild-type and the thermotolerant cells. Therefore, the result of the transcriptomic analysis presented does not address the difference in the general expression profile, but the gene expression difference under a specific condition (4 h after inoculation at 50°C) because the wild-type and the thermotolerant cells have very different growth rates at 50°C. The genes that show the largest increase in gene expression under our experimental condition, including cpxP and ompW, are highly involved in the protection of bacteria against various forms of environmental stresses. CpxP is a chaperone involved in resistance to extracytoplasmic stress (34,35). This result is consistent with the previous report that many chaperones are heat shock proteins, which are expressed in response to elevated temperatures or other cellular stresses. It is well known that proper protein folding is disrupted when cells are subjected to heat treatment, and our findings suggest that CpxP may be involved in protein folding under heat shock conditions. OmpW is an outer membrane protein that forms an eight-stranded beta-barrel with a hydrophobic channel (33). The function of OmpW is unknown, but recent data suggest that it may be involved in the protection of bacteria against various forms of environmental stresses, such as temperature extremes, salinity and availability of nutrients or oxygen. We also reason that the overexpression of OmpW might provide structural stability to the cell. It is known that high temperatures lead to an increase in the fluidity of the cellular membranes (37,38). Excessive membrane fluidity gives rise to leaky membranes, and, thus a corresponding loss of function of membrane proteins. Therefore, the changes in the E. coli's plasma membrane through the overexpression of OmpW may be associated with the mutated cell's ability to withstand elevated temperatures.
Overexpression of a single or double gene(s) among the top-candidate genes identified in the microarray analysis (e.g. cpxP and/or ompW) did not produce a thermotolerance phenotype as robust as the phenotype induced by the T2 ATF. It seems that the ability of the ATF to elicit simultaneous modification to the expression levels of many genes is critically important for obtaining highly improved thermotolerance phenotypes. For instance, transcriptional profiling revealed that T2 ATF represses the marRAB operon, which in turn leads to increased expression of genes such as hdeA and slp that are repressed by products of marRAB (39). HdeA is a chaperon and Slp is an outer membrane lipoprotein and both are involved in resistance to stress conditions (i.e. at low pH) (40) and so are likely to be among the many proteins contributing to the thermotolerance phenotype.
The ATF approach that we describe can cause nuanced changes in gene expression. In cases where the expression level of a particular gene is critical to obtaining the desired phenotypes, subtle changes in the gene expression might be necessary to allow cells to achieve them. For instance, the knockout of the marRAB operon failed to impart an improved thermotolerance phenotype, which indicates that the altered level of marRAB expression induced by T2 ATF rather than the complete elimination of these genes is necessary to achieve this phenotype. Additional levels of control over cellular phenotype are also possible by regulating the expression of the ATF. We have observed that our ATF can generate various levels of phenotypic improvements, such as the different degree of thermotolerance, when they were expressed at various levels with different expression vectors (Figure 3B). In addition, a conditional induction of a desired phenotype is possible with ATFs under an inducible promoter. We have observed that ATF expressed under an inducible promoter (a tac promoter) can conditionally generate desired phenotypes (data not shown). Moreover, a phenotypic transfer from one strain to another strain of the same species is straightforward with our approach. The same phenotypic changes were observed when the selected ATFs were expressed in other strains such as E. coli BL21 and XL1-Blue (data not shown). Finally, we have observed that our ATF can generate desired phenotypes more efficiently than random mutation. Random mutagenesis approach could generate osmotolerant and cold-tolerant cells, but not to the extent that we have obtained with our ATFs. Indeed, 50/106 thermotolerant cells were obtained under the heat treatment condition (at 55°C for 2 h), but cells that are tolerant to heat at 55°C could not be obtained by random mutation under our experimental condition. Also, ATFs could generate osmotolerance and cold-tolerance phenotypes (150/106 and 13/106 cells, respectively) more efficiently than random mutation (approximately 1/107 cells for each phenotype) under our experimental conditions.
Our results and the general applicability of this platform demonstrate that ATFs can be used as a powerful tool for phenotypic engineering and functional genomic analysis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was supported in part by grants from 21C Frontier Program of Microbial Genomics and Applications (MG05-0204-1-0), the Molecular & Cellular BioDiscovery Research Program (M1-0106-00-0200) from the Ministry of Science and Technology of Korea, the Korea Science & Engineering Foundation Grant (R01-2005-000-11010-0), and the Basic Research Program of the Korea Research Foundation Grant (KRF-2004-042-D00072). The Open Access publication charges were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Serina S, Nozza F, Nicastro G, Faggioni F, Mottl H, Dehò G, Polissi A. Scanning the Escherichia coli chromosome by random transposon mutagenesis and multiple phenotypic screening. Res. Microbiol. 2004;155:692–701. doi: 10.1016/j.resmic.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 2.Mormann S, Lömker A, Rückert C, Gaigalat L, Tauch A, Pühler A, Kalinowski J. Random mutagenesis in Corynebacterium glutamicum ATCC 13032 using an IS6100-based transposon vector identified the last unknown gene in the histidine biosynthesis pathway. BMC Genomics. 2006;7:205. doi: 10.1186/1471-2164-7-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alper H, Jin YS, Moxley JF, Stephanopoulos G. Identifying gene targets for the metabolic engineering of lycopene biosynthesis in Escherichia coli. Metab. Eng. 2005;7:155–164. doi: 10.1016/j.ymben.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Alper H, Miyaoku K, Stephanopoulos G. Construction of lycopene-overproducing E. coli strains by combining systematic and combinatorial gene knockout targets. Nat. Biotechnol. 2005;23:612–616. doi: 10.1038/nbt1083. [DOI] [PubMed] [Google Scholar]
- 5.Ro DK, Paradise EM, Ouellet M, Fisher KJ, Newman KL, Ndungu JM, Ho KA, Eachus RA, Ham TS, Kirby J. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440:940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- 6.Alper H, Moxley J, Nevoigt E, Fink GR, Stephanopoulos G. Engineering yeast transcription machinery for improved ethanol tolerance and production. Science. 2006;314:1565–1568. doi: 10.1126/science.1131969. [DOI] [PubMed] [Google Scholar]
- 7.Alper H, Stephanopoulos G. Global transcription machinery engineering: A new approach for improving cellular phenotype. Metab. Eng. 2007;9:258–267. doi: 10.1016/j.ymben.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 8.Wolfe SA, Nekludova L, Pabo CO. DNA recognition by Cys2His2 zinc finger proteins. Annu. Rev. Biophys. Biomol. Struct. 2000;29:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- 9.Greisman HA, Pabo CO. A general strategy for selecting high-affinity zinc finger proteins for diverse DNA target sites. Science. 1997;275:657–661. doi: 10.1126/science.275.5300.657. [DOI] [PubMed] [Google Scholar]
- 10.Blancafort P, Magnenat L, Barbas C.F., III Scanning the human genome with combinatorial transcription factor libraries. Nat. Biotechnol. 2003;21:269–274. doi: 10.1038/nbt794. [DOI] [PubMed] [Google Scholar]
- 11.Pabo CO, Peisach E, Grant RA. Design and selection of novel Cys2His2 zinc finger proteins. Annu. Rev. Biochem. 2001;70:313–340. doi: 10.1146/annurev.biochem.70.1.313. [DOI] [PubMed] [Google Scholar]
- 12.Kim JS, Pabo CO. Transcriptional repression by zinc finger peptides: exploring the potential for applications in gene therapy. J. Biol. Chem. 1997;272:29795–29800. doi: 10.1074/jbc.272.47.29795. [DOI] [PubMed] [Google Scholar]
- 13.Bae KH, Kwon YD, Shin HC, Hwang MS, Ryu EH, Park KS, Yang HY, Lee DK, Lee Y, Park J, et al. Human zinc fingers as building blocks in the construction of artificial transcription factors. Nat. Biotechnol. 2003;21:275–280. doi: 10.1038/nbt796. [DOI] [PubMed] [Google Scholar]
- 14.Park KS, Lee DK, Lee H, Lee Y, Jang YS, Kim YH, Yang HY, Lee SI, Seol W, Kim JS. Phenotypic alteration of eukaryotic cells using randomized libraries of artificial transcription factors. Nat. Biotechnol. 2003;21:1208–1214. doi: 10.1038/nbt868. [DOI] [PubMed] [Google Scholar]
- 15.Jamieson AC, Miller JC, Pabo CO. Drug discovery with engineered zinc-finger proteins. Nat. Rev. Drug. Discov. 2003;2:361–368. doi: 10.1038/nrd1087. [DOI] [PubMed] [Google Scholar]
- 16.Snowden AW, Zhang L, Urnov F, Dent C, Jouvenot Y, Zhong X, Rebar EJ, Jamieson AC, Zhang HS, Tan S, et al. Repression of vascular endothelial growth factor A in glioblastoma cells using engineered zinc finger transcription factors. Cancer. Res. 2003;63:8968–8976. [PubMed] [Google Scholar]
- 17.Park KS, Jang YS, Lee HR, Kim JS. Phenotypic alteration and target gene identification using combinatorial libraries of zinc finger proteins in prokaryotic cells. J. Bacteriol. 2005;187:5496–5499. doi: 10.1128/JB.187.15.5496-5499.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rhodius VA, West DM, Webster CL, Busby SJ, Savery NJ. Transcription activation at class II CRP-dependent promoters: the role of different activating regions. Nucleic Acids Res. 1997;25:326–332. doi: 10.1093/nar/25.2.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rhodius VA, Busby SJ. Transcription activation by the Escherichia coli cyclic AMP receptor protein: determinants within activating region 3. J. Mol. Biol. 2000;299:295–310. doi: 10.1006/jmbi.2000.3736. [DOI] [PubMed] [Google Scholar]
- 20.Harman JG. Allosteric regulation of the cAMP receptor protein. Biochem. Biophys. Acta. 2001;1547:1–17. doi: 10.1016/s0167-4838(01)00187-x. [DOI] [PubMed] [Google Scholar]
- 21.Busby S, Ebright RH. Transcription activation by catabolite activator protein (CAP) J. Mol. Biol. 1999;293:199–213. doi: 10.1006/jmbi.1999.3161. [DOI] [PubMed] [Google Scholar]
- 22.Sung BH, Lee CH, Yu BJ, Lee JH, Lee JY, Kim MS, Blattner FR, Kim SC. Development of a biofilm production-deficient Escherichia coli strain as a host for biotechnological applications. Appl. Environ. Microbiol. 2006;72:3336–3342. doi: 10.1128/AEM.72.5.3336-3342.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friedman SM, Hossain M, Hasson TH, Kawamura A. Gene expression profiling of intrinsic thermotolerance in Escherichia coli. Curr. Microbiol. 2006;52:50–54. doi: 10.1007/s00284-005-4578-6. [DOI] [PubMed] [Google Scholar]
- 24.von Weymarn N, Nyyssola A, Reinikainen T, Leisola M, Ojamo H. Improved osmotolerance of recombinant Escherichia coli by de novo glycine betaine biosynthesis. Appl. Microbiol. Biotechnol. 2001;55:214–218. doi: 10.1007/s002530000515. [DOI] [PubMed] [Google Scholar]
- 25.Gowrishankar J, Jayashree P, Rajkumari K. Molecular cloning of an osmoregulatory locus in Escherichia coli: increased proU gene dosage results in enhanced osmotolerance. J. Bacteriol. 1986;168:1197–1204. doi: 10.1128/jb.168.3.1197-1204.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shirano Y, Shibata D. Low temperature cultivation of Escherichia coli carrying a rice lipoxygenase L-2 cDNA produces a soluble and active enzyme at a high level. FEBS Lett. 1990;271:128–130. doi: 10.1016/0014-5793(90)80388-y. [DOI] [PubMed] [Google Scholar]
- 27.Xia B, Etchegaray JP, Inouye M. Nonsense mutations in cspA cause ribosome trapping leading to complete growth inhibition and cell death at low temperature in Escherichia coli. J. Biol. Chem. 2001;276:35581–35588. doi: 10.1074/jbc.M103871200. [DOI] [PubMed] [Google Scholar]
- 28.Qing G, Ma LC, Khorchid A, Swapna GV, Mal TK, Takayama MM, Xia B, Phadtare S, Ke H, Acton T. Cold-shock induced high-yield protein production in Escherichia coli. Nat. Biotechnol. 2004;22:877–882. doi: 10.1038/nbt984. [DOI] [PubMed] [Google Scholar]
- 29.Blancafort P, Segal DJ, Barbas C.F., III Designing transcription factor architectures for drug discovery. Mol. Pharmacol. 2004;66:1361–1371. doi: 10.1124/mol.104.002758. [DOI] [PubMed] [Google Scholar]
- 30.Sera T, Uranga C. Rational design of artificial zinc-finger proteins using a nondegenerate recognition code table. Biochemistry. 2002;41:7042–7081. doi: 10.1021/bi020095c. [DOI] [PubMed] [Google Scholar]
- 31.Riehle MM, Bennett AF, Lenski RE, Long AD. Evolutionary changes in heat-inducible gene expression in lines of Escherichia coli adapted to high temperature. Physiol. Genomics. 2003;14:47–58. doi: 10.1152/physiolgenomics.00034.2002. [DOI] [PubMed] [Google Scholar]
- 32.Harcum SW, Haddadin FT. Global transcriptome response of recombinant Escherichia coli to heat-shock and dual heat-shock recombinant protein induction. J. Ind. Microbiol. Biotechnol. 2006;33:801–814. doi: 10.1007/s10295-006-0122-3. [DOI] [PubMed] [Google Scholar]
- 33.Hong H, Patel DR, Tamm LK, van den Berg B. The outer membrane protein OmpW forms an eight-stranded beta-barrel with a hydrophobic channel. J. Biol. Chem. 2006;281:7568–7577. doi: 10.1074/jbc.M512365200. [DOI] [PubMed] [Google Scholar]
- 34.Danese PN, Silhavy TJ. CpxP, a stress-combative member of the Cpx regulon. J. Bacteriol. 1998;180:831–839. doi: 10.1128/jb.180.4.831-839.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DiGiuseppe PA, Silhavy TJ. Signal detection and target gene induction by the CpxRA two-component system. J. Bacteriol. 2003;185:2432–2440. doi: 10.1128/JB.185.8.2432-2440.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wagner R. Oxford, England: Oxford University Press; 2000. Transcription regulation in prokaryotes. pp. 199–207 and pp. 211–217. [Google Scholar]
- 37.Sinensky M. Homeoviscous adaptation: a homeostatic process that regulates the viscosity of membrane lipids in Escherichia coli. Proc. Natl Acad. Sci. USA. 1974;71:522–525. doi: 10.1073/pnas.71.2.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Los DA, Murata N. Membrane fluidity and its roles in the perception of environmental signals. Biochim. Biophys. Acta. 2004;1666:142–157. doi: 10.1016/j.bbamem.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 39.Seoane AS, Levy SB. Identification of new genes regulated by the marRAB operon in Escherichia coli. J. Bacteriol. 1995;177:530–535. doi: 10.1128/jb.177.3.530-535.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hong W, Jiao W, Hu J, Zhang J, Liu C, Fu X, Shen D, Xia B, Chang Z. Periplasmic protein HdeA exhibits chaperone-like activity exclusively within stomach pH range by transforming into disordered conformation. J. Biol. Chem. 2005;280:27029–27034. doi: 10.1074/jbc.M503934200. [DOI] [PubMed] [Google Scholar]
- 41.Weber IT, Steitz TA. Structure of a complex of catabolite gene activator protein and cyclic AMP refined at 2.5Å resolution. J. Mol. Biol. 1987;198:311–326. doi: 10.1016/0022-2836(87)90315-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.