Abstract
Lipid A isolated from several bacteria (Escherichia coli, Pseudomonas aeruginosa, Salmonella enterica, and various strains of Yersinia) showed abundant formation of pyrophosphate anions upon ion dissociation. Pyrophosphate [H3P2O7]− and/or [HP2O6]− anions were observed as dominant fragments from diphosphorylated lipid A anions regardless of the ionization mode (matrix-assisted laser desorption ionization or electrospray ionization), excitation mode (collisional activation or infrared photoexcitation), or mass analyzer (time-of-flight/time-of-flight, tandem quadrupole, Fourier transform–ion cyclotron resonance mass spectrometry). Dissociations of anions from model lipid phosphate, pyrophosphate, and hexose diphosphates confirmed that pyrophosphate fragments were formed abundantly only in the presence of an intact pyrophosphate group in the analyte molecule and were not due to intramolecular rearrangement upon ionization, ion-molecule reactions, or rearrangement following activation. This indicated that pyrophosphate groups are present in diphosphorylated lipid A from a variety of Gram-negative bacteria.
Keywords: pyrophosphorylation, tandem mass spectrometry, Yersinia pestis
Lipopolysaccharide (LPS) is the primary constituent of the outer leaflet of the outer membrane of Gram-negative bacteria (1). In addition to being the major surface molecule in Gram-negative bacteria, LPS is also considered a major pathogenic factor. Lipid A, also referred to as endotoxin, is the hydrophobic membrane anchor of LPS and is known to be a potent inducer of the host innate immune system (1, 2). Structurally, lipid A is characterized as a phosphoglycolipid defined by a conserved diglucosamine disaccharide with structural variations occurring by fatty acid position and identity, phosphorylation, and additional monosaccharide modification. Alteration of lipid A structure (i.e., changes in acylation, phosphorylation, and glycosylation) greatly affects the bacterium's virulence and can occur via a variety of environmental stimuli including divalent ion concentration, temperature, and other growth conditions (1, 3–6).
The phosphorylation pattern of lipid A has been shown to be important for its biological activity. For example, removal of a phosphate group has been shown to substantially reduce lipid A toxicity (7, 8) and interleukin-1 induction capacity (9). By contrast, masking of lipid A phosphate groups (e.g., addition of aminoarabinose) has been shown to affect bacterial resistance to host cationic antimicrobial peptides (10). The biochemical effects of phosphate groups in lipid A have been attributed to their negative charge that affects recognition by the Toll-like receptor 4 and further LPS-induced signaling in the host immune response to bacterial infection (11). Furthermore, monosaccharide modification to lipid A is thought to occur via an ester linkage with the phosphate substituents.
The biosynthesis of lipid A, as characterized in Escherichia coli, involves LPS intermediates that have a 1-position pyrophosphate and a 4′-position monophosphate (12, 13). An inner membrane enzyme (LpxT) has been recently identified that transfers a phosphate group to the lipid A moiety to form the 1-position pyrophosphate structure (12). However, lipid A is commonly described as bisphosphorylated with monophosphate attachment at the 1- and 4′-positions of the glucosamine dimer backbone or monophosphorylated with phosphate attachment at either the 1- or 4′-position. We intend to demonstrate that several Gram-negative bacteria produce diphosphorylated lipid A that retains the pyrophosphate substituent. This finding is important for further studies of biochemical modifications of LPS that involve yet unknown mechanisms (12, 13).
Our particular emphasis is centered on Yersinia pestis (Yp), the causative agent of the plague, which is a highly invasive and often lethal Gram-negative pathogen that is transmitted to the mammalian host by either fleabite or inhalation of an infectious droplet (14). Because of its potential use as a bioterrorism agent, Yp has been classified by the Center for Disease Control and Prevention as a Class A select agent. Transmission from the flea vector to the mammalian host induces temperature-dependent structural modification of Yp lipid A (6, 15–17). At the mammalian host temperature of 37°C, the primary structure of lipid A consists of a β-1,6-linked diglucosamine disaccharide with two phosphate moieties at the 1- and 4′-positions, amide-linked 3-hydroxymyristic acids at the 2- and 2′-positions, and ester-linked 3-hydroxymyristic acids at the 3- and 3′-positions (Fig. 1Inset) (6, 15–18). However, at a temperature typical of the flea vector (21°C), the primary lipid A structure is hexa-acylated. The additional fatty acids, palmitoleic acid (C16:1) and lauric acid (C12), are thought to be ester-linked via the 3-hydroxy position of the 3 and 3′ fatty acids, respectively, to form acyloxyacyl groups (6, 15–19). Furthermore, Yp lipid A from both temperature variants has been reported to contain modifications of aminoarabinose and phosphoethanolamine (6, 15, 16, 18, 19). Despite extensive research, ambiguity remains to date as to the exact location of the acyloxyacyl fatty acids, phosphate moieties, and other modifications.
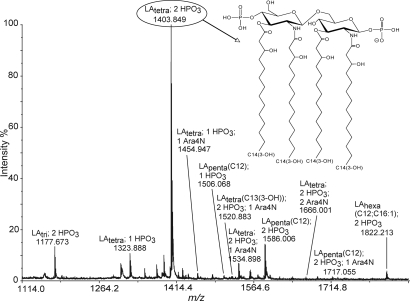
Fig. 1.
Negative ion mode MALDI-TOF mass spectrum of lipid A from Yp grown at 37°C. (Inset) Structure representing a previously proposed structure for m/z 1,404 (6, 15–18). Shorthand notation is as follows: LA, lipid A; subscript, number of acyl chains and, unless otherwise noted, acyl chains are 3-hydroxymyristic acid with the exception of C12 (lauric acid) and C16:1 (palmitoleic acid); HPO3, phosphate group; Ara4N, aminoarabinose.
Diphosphorylated¶ lipid A is generally presumed to be phosphorylated at the 1- and 4′-positions. Our initial analysis of Yp lipid A grown at 37°C and 21°C suggested that several diphosphorylated lipid A structures from these two growth conditions were pyrophosphorylated rather than bisphosphorylated.‖ In the present study, we used a multifaceted mass spectrometric approach to investigate the presence of pyrophosphate in diphosphorylated lipid A. Specifically, tetra-acylated diphosphorylated lipid A extracted from Yp grown at 37°C is used as the benchmark diphosphorylated lipid A sample. Additional mass spectrometric analysis was performed on numerous diphosphorylated lipid A structures from Yp grown at 37°C and 21°C, Escherichia coli, Pseudomonas aeruginosa, and Salmonella enterica. We report that all lipid A structures isolated from Yp strain KIM6+ (20), whose (M–H)− anion contained two phosphate groups, were mixtures of bisphosphate and pyrophosphate structures. Most importantly, we suggest that the presence of pyrophosphate in diphosphorylated lipid A is not exclusively reserved for Yp but is a general phenomenon among Gram-negative bacterial lipid A structures.
Structural elucidation of lipid A has been achieved by a variety of chemical methods coupled to analytical techniques (21–24). Of particular interest, the use of mass spectrometry (MS) has proven to be an effective technique for determination of lipid A structure (25–37). Negative-ion mode matrix-assisted laser desorption ionization (MALDI) time-of-flight (TOF) MS has proven to be very useful in initial characterization of lipid A structures (30–32, 38, 39). The benefit of MALDI TOF/TOF MS to ionize, isolate, and fragment select components in lipid A mixtures provides a ready means for comprehensive structural characterization. Electrospray ionization (ESI) linear ion trap (LIT)–Fourier transform (FT)–ion cyclotron resonance (ICR) MS has recently been used to reveal great diversity in Francisella tularensis subspecies novicida lipid A structures available between insect and mammalian host growth conditions (37). This hybrid mass spectrometer has proven beneficial for structure analysis because the FT-ICR mass analyzer provides accurate mass measurements of the deprotonated precursor molecules and tandem MS fragments, and the LIT can offer multiple stages of collision-induced dissociation (CID) by MSn. This combined effort of dual hybrid mass analyzers permits increased confidence in structure assignment.
Results
MALDI TOF MS of Lipid A Extracted from Yp Grown at 37°C.
Lipid A isolated from Yp grown at 37°C was analyzed in negative ion mode by MALDI TOF MS (Fig. 1). The resulting mass spectrum was similar to previous reports (6, 15). The most abundant anion giving the base peak at m/z 1,404 corresponded to a singly deprotonated lipid A with four 3-hydroxymyristic acid residues and two phosphate groups attached the glucosamine backbone dimer. The additional ions in the mass spectrum were identified based on molecular mass and comparison with literature [supporting information (SI) Table S1]. In particular, we focused on characterizing the dominant m/z 1,404 ion by implementing several mass spectrometric techniques.
MALDI TOF/TOF MS of the Yp Lipid A Anion at m/z 1,404.
To further investigate the structure of lipid A extracted from Yp grown at 37°C, the base peak in the MALDI TOF mass spectrum, m/z 1,404, was isolated and fragmented in the TOF/TOF mass spectrometer (Fig. 2A and Scheme 1). Analysis of the tandem mass spectrum showed product ions grouped in three distinct m/z regions (Table S2). Neutral loss of 3-hydroxymyristic acid, phosphoric acid, and combinations of these two characterized the high-abundant product ions observed in the high m/z region (m/z 800–1,404). The low-abundant product ions located in the middle m/z region (m/z 200–800) were a result of glycosidic cleavages with and without neutral losses of 3-hydroxymyristic acid and phosphoric acid. The third region of low m/z (m/z 50–200) was dominated by high-abundant pyrophosphate and phosphate product ions. We note that although the fragment ion elemental compositions have been unequivocally established from accurate mass measurements, the ion structures shown in Scheme 1 are only tentative, thereby acknowledging that multiple isomers could be present.
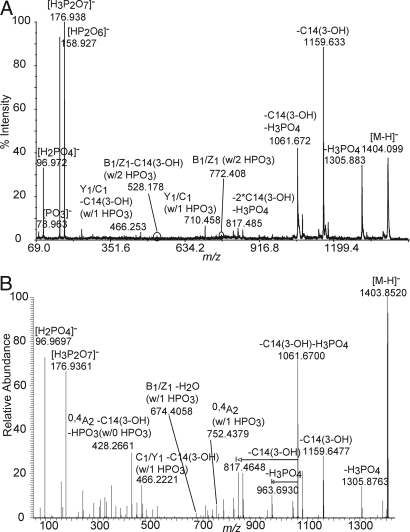
Fig. 2.
Negative ion mode MALDI-TOF/TOF MS2 spectrum of m/z 1,404 of lipid A from Yp grown at 37°C (A) and negative ion mode ESI LTQ-FT IRMPD MS2 spectrum of m/z 1,404 of lipid A from Yp grown at 37°C (B).
Scheme 1.
Dissociation of m/z 1,404 rationalized by a pyrophosphorylated lipid A structure. Note that although the pyrophosphate moiety is shown at the 1-position, it could have been placed at the 4′-position. The structure of m/z 1,404 is a heterogeneous mixture of bisphosphate and pyrophosphate, and therefore the structure in this scheme is not the only conceivable phosphorylation arrangement.
The two most abundant product ions, m/z 177 and 159, were assigned the molecular formulas [H3P2O7]− and [HP2O6]−, respectively, based on accurate mass measurements (Table S2). These are pyrophosphate-related ions that indicate the presence of a P-O-P linkage in lipid A anions. Furthermore, ions at m/z 528 and 772, although of low abundance, corresponded to glycosidic bond cleavages where the resulting anion retained not one but two phosphate groups. The product ion at m/z 772 was identified as a B1/Z1 ion, and the product ion at m/z 528 was identified as a B1/Z1 minus 3-hydroxymyristic acid, using the nomenclature described by Costello (40). It should be noted that because of the symmetric nature of the lipid A structure at m/z 1,404, B/Z and C/Y ions cannot be distinguished. Of particular note, more abundant product ions from glycosidic bond cleavages where the anion contained only one phosphate moiety were observed, and these ions were located at m/z 710 (C1/Y1 ion) and 466 (C1/Y1 ion minus 3-hydroxymyristic acid) and at m/z 692 (B1/Z1 ion) and 448 (B1/Z1 ion minus 3-hydroxymyristic acid).
Pyrophosphate Precursor Ions from Yp Lipid A.
To establish whether the presence of pyrophosphate anions could be an artifact of the MALDI process and/or some postsource event (i.e., by rearrangement or ion-molecule reaction) in the TOF/TOF mass spectrometer, we analyzed lipid A from Yp grown at 37°C in negative ion mode with an ESI tandem quadrupole mass spectrometer, using both product ion and precursor ion scans. Electrospray ionization is well established as a very gentle ionization method that preserves analyte structures from solution to the gas phase. The product ion scan of m/z 1,404 (Fig. S1) resulted in a tandem mass spectrum that was very similar to that of the tandem mass spectrum from the MALDI TOF/TOF mass spectrometer. Most importantly, the middle m/z region displayed glycosidic and cross-ring fragments, and the low m/z region was represented by very abundant phosphate and pyrophosphate product ions. Additional evidence was obtained by performing a precursor ion scan on the pyrophosphate m/z 159 anion (Fig. S2). The resulting tandem mass spectrum revealed that m/z 1,404 was one of the precursor ions for the product ion at m/z 159. In addition, there were two other pyrophosphate precursors in the spectrum (m/z 1,535 and 1,666) that corresponded to lipid A anions preserving one or two aminoarabinose moieties, as discussed later.
ESI LTQ-FT MS of Lipid A from Yp Grown at 37°C.
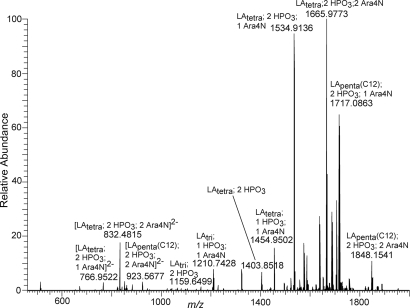
Lipid A extracted from Yp grown at 37°C was analyzed in negative ion mode with an ESI LTQ-FT MS (Fig. 3 and Table S3). Comparison of the MALDI TOF/TOF mass spectrum with the ESI LTQ-FT mass spectrum revealed several notable differences. The base peak, m/z 1,404, in the MALDI TOF mass spectrum is actually of low relative abundance in the ESI LTQ-FT mass spectrum. This observation was readily explained by the “softer” ionization technique of ESI that allowed for retention of the labile aminoarabinose modification. As evident from the ESI mass spectrum, the labile aminoarabinose modification was retained and abundant ions at m/z 1,535 and 1,666 corresponded to diphosphoryl tetra-acylated lipid A plus one aminoarabinose (1,404 + 131 = 1,535) and diphosphoryl tetra-acylated lipid A plus two aminoarabinose (1,404 + 131 + 131 = 1,666). Another obvious difference between the MALDI TOF mass spectrum and the ESI LTQ-FT mass spectrum was the presence of doubly deprotonated species in the ESI LTQ-FT mass spectrum due to the tendency of ESI to produce more multiply charged ions than MALDI.
Fig. 3.
Negative ion mode ESI LTQ-FT mass spectrum of lipid A from Yp grown at 37°C. Refer to Fig. 1 for shorthand notation. All ions are singly deprotonated unless otherwise noted.
Pyrophosphate Ion Detection upon CID and Infrared Multiphoton Dissociation (IRMPD).
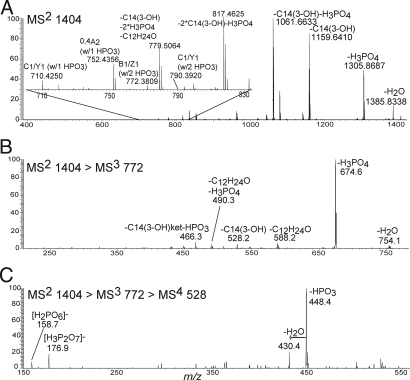
The capabilities of LIT-FTMS were further used to investigate the structure of lipid A at m/z 1,404 in a series of negative ion mode CID MSn experiments. The tandem mass spectrum of the ion at m/z 1,404 (Fig. 4A and Table S4) resembled the tandem mass spectra of m/z 1,404 from the other instruments and ionization methods. The most abundant product ions were a result of neutral losses of 3-hydroxymyristic acid and phosphoric acid. Additionally, there were several low abundant product ions resulting from glycosidic and cross-ring fragmentation. Of particular note, the m/z 772 ion corresponded to a B1/Z1 fragment with two phosphate moieties. The presence of pyrophosphate ions was established through a multistep (MS4) dissociation sequence, as follows. The glycosidic product ion B1/Z1 at m/z 772 with two phosphates (Fig. 4B) was produced, isolated, and dissociated to m/z 528 which retained two phosphate groups. The m/z 528 fragment ion was isolated and dissociated in the MS4 mass spectrum to produce pyrophosphate fragment ions at m/z 159 and 177 (Fig. 4c).
Fig. 4.
Negative ion mode ESI LTQ-FT CID MS2 spectrum of m/z 1,404 of lipid A from Yp grown at 37°C (A), ESI LTQ CID MS3 of m/z 772 derived from m/z 1,404 (B), and ESI LTQ CID MS4 of m/z 528 derived from m/z 772 and 1,404 (C). Structures for m/z 1,404, 772, and 528 ions can be found in Scheme 1.
The high mass resolution capabilities of FT MS were further used to investigate the structure of the lipid A ion at m/z 1,404 using IRMPD (Fig. 2B and Table S5). The added benefit of IRMPD is that adjusting the IR laser duration and power allows for multiple fragmentation pathways and thereby providing a more informative tandem mass spectrum. The number of product ions observed in the IRMPD tandem mass spectrum was significantly increased in comparison to the CID tandem mass spectra generated by the TOF/TOF, tandem quadrupole, and the LTQ-FT mass spectrometers (cf. Fig. 2 A and B). In particular, abundant pyrophosphate (m/z 177) and phosphate (m/z 97) product ions were clearly observed that were unambiguously identified by accurate mass measurements (Table S5). It should be pointed out that all product ions identified in the IRMPD tandem mass spectrum were also identified in the CID MSn spectra.
Other Diphosphoryl Lipid A Structures from Yp Grown at 37°C and 21°C.
The formation of pyrophosphate fragment ions, as discussed in detail for the m/z 1,404 precursors, were unambiguously observed for all lipid A anions from Yp grown at 37°C that contained a total of two phosphate groups. The same observation was made for lipid A from Yp grown at 21°C, where all ions whose lipid A structure had two phosphate moieties displayed high-abundant pyrophosphate product ions in their tandem mass spectrum as well (data not shown). Thus, we concluded that this behavior was common to different lipid A ions regardless of the presence of aminoarabinose or mode of bacterial growth.
Diphosphorylated Lipid A from E. coli F583 (Rd mutant) and Other Gram-negative Bacteria.
Diphosphorylated lipid A from E. coli F583 (Rd mutant) represents a readily available purified lipid A species whose structure is presumed to contain two phosphate groups located at the 1- and 4′-positions of the diglucosamine backbone (ref. 41 and references therein). Two ions were identified in the MALDI TOF mass spectrum whose m/z value corresponded to lipid A structures with two phosphate substituents (Fig. S3a). Subsequent MALDI TOF/TOF tandem mass spectra yielded pyrophosphate anions as the most abundant product ions (Fig. S3 b and c).
Several other diphosphorylated lipid A samples from common Gram-negative bacteria were analyzed by negative ion mode MALDI TOF/TOF MS (Table 1). In summary, all tandem mass spectra where the precursor ion corresponded to a lipid A structure with two phosphate moieties contained very abundant pyrophosphate product ions at m/z 159 and 177.
Table 1.
List of selected Gram-negative bacteria whose diphosphorylated lipid A structure resulted in pyrophosphate product ions.
| Gram-negative bacterium | Strain | Diphosphoryl lipid A structure | Pyrophosphate productions |
|---|---|---|---|
| Escherichia coli | F583 (Rd mutant) | Yes | Yes |
| Pseudomonas aeruginosa | PAK | Yes | Yes |
| Salmonella enterica serovar typhimurium | 14028 | Yes | Yes |
| Yersinia enterocolitica | Clinical isolate | Yes | Yes |
| Yersinia pestis grown at 21°C | KIM6+ | Yes | Yes |
| Yersinia pestis grown at 37°C | KIM6+ | Yes | Yes |
| Yersinia pseudotuberculosis | PB1 0:1b | Yes | Yes |
MS/MS Analysis of Monophosphate, Bisphosphate, and Pyrophosphate Diacylglycerol and Glycan Standards.
With the consistent detection of pyrophosphate fragment ions, a question arose as to whether these may be due to chemical reactions during ionization or rearrangement of gas-phase ions upon excitation. We addressed this question with pyrophosphate and phosphate anions from 1,2-dioleoyl-sn-glycerol-3-phosphate (18:1 DGMP) and dioleoylglycerol pyrophosphate (18:1 DGPP). As expected, the product ion scan for the deprotonated 18:1 DGMP precursor ion displayed product ions at m/z 79 and 97, which correspond to phosphate anions (Fig. S4). However, no pyrophosphate product ions were observed in this mass spectrum. In contrast, the tandem mass spectrum of the deprotonated 18:1 DGPP precursor ion displayed very abundant product anions at m/z 159 (pyrophosphate) and m/z 79 (phosphate) (Fig. S5). Interestingly, ions at m/z 177 and 97 were also present but were in very low abundance.
Another model we investigated involved fructose-1,6-bisphosphate and glucose-1,6-bisphosphate, which represent simple bisphosphorylated glycan standards to determine if pyrophosphate ions could be produced by condensation of two phosphate groups in the dissociating ion. The deprotonated fructose-1,6-bisphosphate anion was isolated and fragmented (Fig. S6). The most abundant product ions in the tandem mass spectrum were at m/z 79 and 97 corresponding to phosphate anions. In addition, particularly relevant, weak pyrophosphate product ions at m/z 159 and 177 were observed in the tandem mass spectrum. MALDI TOF/TOF MS analysis of glucose-1,6-bisphosphate resulted in a very similar tandem mass spectrum where high-abundant phosphate anions were observed in conjunction with weak pyrophosphate anions. Notably, the low relative abundance of the pyrophosphate fragment ions from the hexose bisphosphates was strikingly different from those observed for lipid A ions.
To further exclude the possibility of pyrophosphate ion formation from lipid A ionization, we separated the monophosphorylated and diphosphorylated species from Yp grown at 37°C by on-line LC (Fig. S7a) and obtained high-resolution tandem mass spectra with IRMPD. The tandem mass spectrum of the diphosphorylated fraction at m/z 1,404 showed abundant pyrophosphate anions at m/z 159 and 177 (Fig. S7b). In contrast, the tandem mass spectrum of the monophosphorylated fraction at m/z 1,324 showed only the monophosphate anion at m/z 97 (Fig. S7c).
Discussion
The above-presented mass spectrometric analysis provided strong evidence for the presence of pyrophosphate in diphosphorylated forms of Yp lipid A. Moreover, this unexpected feature of lipid A structure was not particular to specific growth temperatures, species, or even genus, but rather a general phenomenon for many Gram-negative bacteria. We note that indications of pyrophosphate groups based on low-resolution mass spectrometry were previously reported for lipid A from Pseudoalteromonas haloplanktis (42) and Salmonella typhimurium (30, 43).
Pyrophosphate moieties are known to be present in triphosphorylated lipid A structures. For example, E. coli K-12 has a lipid A structure with a 1-position pyrophosphate and a 4′-position monophosphate (44, 45). A recent report linked pyrophosphate forming periplasmic phosphorylation of lipid A by LpxT to the presence of undecaprenyl pyrophosphate as the phosphate donor (12). Our finding of pyrophosphorylated structures present in diphosphorylated lipid A raises new questions of the biochemical pathways that lead to the formation of the pyrophosphate group by phosphate transfer or its preservation upon dephosphorylation of triphosphorylated lipid A. We believe that the unequivocal detection of pyrophosphate structures using the combined mass spectrometric approach described here will be useful in further biological studies of lipid A from pathogenic bacteria.
Conclusions
Diphosphorylated lipid A from several Gram-negative bacteria produce characteristic high-abundance pyrophosphate ions under a variety of mass spectrometric conditions. The multifaceted mass spectrometric approach confirmed the presence of a pyrophosphate moiety in several diphosphorylated lipid A structures. Of particular interest, pyrophosphate product ions were not only observed for Yp but were also identified in several other common Gram-negative bacteria. We conclude that diphosphorylated lipid A are heterogeneous mixtures of pyrophosphate and bisphosphate structures. This finding may have important implications in microbiology, in particular, with regard to the formation of pyrophosphorylated variants of lipid A and their toxicity when produced from pathogenic bacteria.
Experimental Procedures
Bacterial Strains and Growth Conditions.
Yp KIM6+ (20) was grown in Luria broth (LB) (pH 7.4) at 37°C and 21°C with aeration and harvested in the late exponential phase.
LPS Purification and Lipid A Isolation.
Yp LPS was extracted by using a hot phenol/water extraction method (46). Further treatment of LPS with RNase A, DNase Ι, and proteinase K ensured removal of contaminating nucleic acids and proteins (47). Hydrolysis of LPS to isolate lipid A was accomplished with 1% SDS at pH 4.5 as described in ref. 48.
Materials.
Dioleoylglycerol pyrophosphate (18:1 DGPP) and 1,2-dioleoyl-sn-glycerol-3-phosphate (18:1 DGMP) were purchased from Avanti Polar Lipids. d-Fructose-1,6-bisphosphate and α-d-glucose-1,6-bisphosphate were purchased from Sigma–Aldrich. Diphosphoryl lipid A from Escherichia coli F583 (Rd mutant) was purchased from Sigma–Aldrich. Lipid A samples from the following organisms were obtained from the Ernst laboratory (University of Washington): Pseudomonas aeruginosa (strain PAK), Salmonella enterica serovar typhimurium (strain 14028), Yersinia enterocolitica (clinical isolate), and Yersinia pseudotuberculosis (strain PB1 0:1b).
Matrix-Assisted Laser Desorption Ionization Time-of-Flight/Time-of-Flight Mass Spectrometry.
Lipid A was analyzed by MALDI in the negative ion mode on a 4700 Proteomics Analyzer (Applied Biosystems). Samples were dissolved in 10 μl of a mixture of 5-chloro-2-mercaptobenzothiazole (20 mg/ml) in chloroform/methanol/water 4:4:1 (vol/vol/vol), and 0.5 μl of sample was analyzed by MALDI-TOF MS and TOF/TOF MS. Both MS and MS/MS data were acquired in reflectron mode with a Nd:YAG laser with a 200-Hz repetition rate, and up to 3,750 shots were accumulated for each spectrum. The precursor isolation window was set to ±5 Da. MS/MS spectra were acquired with collision energies of 1 keV, and air was used as the collision gas. Instrument calibration and all other tuning parameters were optimized by using HP Calmix (Sigma). Data were acquired and processed by using Data Explorer (Applied Biosystems).
Electrospray Ionization Linear Ion Trap Fourier Transform Ion Cyclotron Resonance Mass Spectrometry.
Lipid A was analyzed by ESI in the negative ion mode on an LTQ-FT linear ion trap Fourier transform ion cyclotron resonance mass spectrometer (Thermo Fisher). Samples were diluted to ≈0.3–1.0 mg/ml in chloroform/methanol (1:1) and infused at a rate of 0.5–1.0 μl/min via a fused silica capillary (75 μm i.d./360 μm o.d.) with an ≈15-μm spray tip (New Objective). Instrument calibration and tuning parameters were optimized by using a solution of Ultramark 1621 (Lancaster Pharmaceuticals). For experiments acquired in the ICR cell, resolution was set to 100 K and ion populations were held constant by automatic gain control at 1.0 × 106 and 5.0 × 105 for MS and MS/MS, respectively. For tandem mass spectra, the precursor ion selection window was set to 4–8 Da and the collision energy was set to 30% on the instrument scale. The CID MSn analysis in the linear ion trap was acquired with an ion population of 1.0 × 104 and maximum fill time of 200 ms. The subsequent MS3 and MS4 events had an isolation window of 2 Da with a collision energy of 25%. All spectra were acquired over a time period of 1–2 min and averaged. Typically, MS and MS2 events were mass-analyzed in the ICR cell, and MS3 and MS4 were mass analyzed in the LTQ. Infrared multiphoton dissociation (IRMPD) MS2 events were acquired in the ICR cell by using similar detection parameters to those described above. Precursor ions were irradiated by IR photons produced by a CO2 laser [Synrad firestar Series V20, Model FSV20SFB; 75 W (10.2–10.8 μm)] with pulse durations of 20–100 ms and pulse power of 20–80%. Data were acquired and processed with Xcalibur (version 1.4; Thermo Fisher), using seven-point Gaussian smoothing. On-line liquid chromatography ESI tandem MS experiments were preformed by interfacing a custom-fabricated microcolumn (fused-silica capillary) packed with silica to the LTQ-FT ESI source.
Electrospray Ionization Tandem Quadrupole Mass Spectrometry.
Lipid A was analyzed by ESI in the negative ion mode on a Sciex API III tandem quadrupole mass spectrometer (PerkinElmer). Samples were diluted to ≈0.3–1.0 mg/ml in chloroform/methanol (1:1) and infused at a rate of 0.5–1.0 μl/min through a fused silica capillary (i.d. 100 μm) by using a syringe pump (Harvard Apparatus Model 11). The instrument was operated with the following settings: needle voltage, −4,300 V; counter electrode, −650 V; nebulizer gas pressure, 20 psi; curtain gas pressure, 10 psi; declustering potential, −35 V; collision cell entrance potential, −10 V; collision cell exit potential, −15 V; and collision gas, argon. Tandem MS data were acquired in both product ion and precursor ion scan modes.
Supplementary Material
Acknowledgments.
We thank Drs. Alexander Scherl (Department of Medicinal Chemistry, University of Washington), Martin Sadilek (Department of Chemistry, University of Washington), and Priska D. von Haller (Proteomics Resource, University of Washington). We also thank the Murdock foundation for funds to purchase the MALDI TOF/TOF MS machine, as well as the National Institute of Allergy and Infectious Diseases (Grant 1U54 AI57141-01) and the National Institute on Environmental Health Sciences (Grant 5P30 ES007033-10) for generous funding and support.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. A.S. is a guest editor invited by the Editorial Board.
Diphosphorylated lipid A refers to lipid A structures containing two phosphate substituents irrespective of their location.
This article contains supporting information online at www.pnas.org/cgi/content/full/0800445105/DCSupplemental.
Bisphosphorylated lipid A refers to lipid A structures containing both a 1-position monophosphate and a 4 -position monophosphate.
References
- 1.Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trent MS. Biosynthesis, transport, and modification of lipid A. Biochem Cell Biol. 2004;82:71–86. doi: 10.1139/o03-070. [DOI] [PubMed] [Google Scholar]
- 3.Ernst RK, Guina T, Miller SI. How intracellular bacteria survive: Surface modifications that promote resistance to host innate immune responses. J Infect Dis. 1999;179(Suppl 2):S326–S330. doi: 10.1086/513850. [DOI] [PubMed] [Google Scholar]
- 4.Guo L, et al. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell. 1998;95:189–198. doi: 10.1016/s0092-8674(00)81750-x. [DOI] [PubMed] [Google Scholar]
- 5.Miller SI, Ernst RK, Bader MW. LPS, TLR4, and infectious disease diversity. Nat Rev Microbiol. 2005;3:36–46. doi: 10.1038/nrmicro1068. [DOI] [PubMed] [Google Scholar]
- 6.Rebeil R, Ernst RK, Gowen BB, Miller SI, Hinnebusch BJ. Variation in lipid A structure in the pathogenic Yersiniae. Mol Microbiol. 2004;52:1363–1373. doi: 10.1111/j.1365-2958.2004.04059.x. [DOI] [PubMed] [Google Scholar]
- 7.Rietschel ET, et al. Bacterial endotoxin: Molecular relationship of structure to activity and function. FASEB J. 1994;8:217–225. doi: 10.1096/fasebj.8.2.8119492. [DOI] [PubMed] [Google Scholar]
- 8.Baldridge JR, et al. Taking a toll on human disease: Toll-like receptor 4 agonists as vaccine adjuvants and monotherapeutic agents. Exp Opin Biol Ther. 2004;4:1129–1138. doi: 10.1517/14712598.4.7.1129. [DOI] [PubMed] [Google Scholar]
- 9.Loppnow H, et al. IL-1 induction-capacity of defined lipopolysaccharide partial structures. J Immunol. 1989;142:3229–3238. [PubMed] [Google Scholar]
- 10.Trent MS, Stead CM, Tran AX, Hanskins JV. Diversity of endotoxin and its impact on pathogenesis. J Endotox Res. 2006;12:205–223. doi: 10.1179/096805106X118825. [DOI] [PubMed] [Google Scholar]
- 11.Gangloff M, Gay NJ. MS-2: The Toll gatekeeper in endotoxin signalling. Trends Biochem Sci. 2004;29:294–300. doi: 10.1016/j.tibs.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 12.Touze T, Tran AX, Hankins JV, Mengin-Lecreulx, Trent MS. Periplasmic phosphorylation of lipid A is linked to the synthesis of undecaprenyl phosphate. Mol Microbiol. 2008;67:264–277. doi: 10.1111/j.1365-2958.2007.06044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou Z, Lin S, Cotter RJ, Raetz CR. Lipid A modifications characteristic of Salmonella typhimurium are induced by NH4VO3 in Escherichia coli K12. Detection of 4-amino-4-deoxy-L-arabinose, phosphoethanolamine and palmitate. J Biol Chem. 1999;274:18503–18514. doi: 10.1074/jbc.274.26.18503. [DOI] [PubMed] [Google Scholar]
- 14.Perry RD, Fetherston JD. Yersinia pestis—Etiologic agent of plague. Clin Microbiol Rev. 1997;10:35–66. doi: 10.1128/cmr.10.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawahara K, Tsukano H, Watanabe H, Lindner B, Matsuura M. Modification of the structure and activity of lipid A in Yersinia pestis lipopolysaccharide by growth temperature. Infect Immun. 2002;70:4092–4098. doi: 10.1128/IAI.70.8.4092-4098.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knirel YA, et al. Temperature-dependent variations and intraspecies diversity of the structure of the lipopolysaccharide of Yersinia pestis. Biochemistry. 2005;44:1731–1743. doi: 10.1021/bi048430f. [DOI] [PubMed] [Google Scholar]
- 17.Rebeil R, et al. Characterization of late acyltransferase genes of Yersinia pestis and their role in temperature-dependent lipid A variation. J Bacteriol. 2006;188:1381–1388. doi: 10.1128/JB.188.4.1381-1388.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aussel L, et al. Novel variation of lipid A structures in strains of different Yersinia species. FEBS Lett. 2000;465:87–92. doi: 10.1016/s0014-5793(99)01722-6. [DOI] [PubMed] [Google Scholar]
- 19.Dalla Venezia N, Minka S, Bruneteau M, Mayer H, Michel G. Lipopolysaccharides from Yersinia pestis: Studies on lipid A of lipopolysaccharides Ι and ΙΙ. Eur J Biochem. 1985;151:399–404. doi: 10.1111/j.1432-1033.1985.tb09115.x. [DOI] [PubMed] [Google Scholar]
- 20.Une T, Brubaker RR. In vivo comparison of avirulent Vwa- and Pgm- or Pstr phenotypes of Yersiniae. Infect Immun. 1984;43:895–900. doi: 10.1128/iai.43.3.895-900.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brandenburg K, Richter W, Koch MH, Meyer HW, Seydel U. Characterization of the nonlamellar cubic and HII structures of lipid A from Salmonella enterica serovar Minnesota by x-ray diffraction and freeze-fracture electron microscopy. Chem Phys Lipids. 1998;91:53–69. doi: 10.1016/s0009-3084(97)00093-5. [DOI] [PubMed] [Google Scholar]
- 22.Fukuoka S, et al. Physico-chemical analysis of lipid A fractions of lipopolysaccharide from Erwinia carotovora in relation to bioactivity. Biochim Biophys Acta. 2001;1510:185–197. doi: 10.1016/s0005-2736(00)00347-3. [DOI] [PubMed] [Google Scholar]
- 23.Brandenburg K, et al. Biophysical characterization of triacyl monosaccharide lipid A partial structures in relation to bioactivity. Biophys J. 2002;83:322–333. doi: 10.1016/S0006-3495(02)75172-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brecker L. Nuclear magnetic resonance of lipid A—The influence of solvents on spin relaxation and spectral quality. Chem Phys Lipids. 2003;125:27–39. doi: 10.1016/s0009-3084(03)00055-0. [DOI] [PubMed] [Google Scholar]
- 25.Hashimoto M, et al. Chemical structure and immunobiological activity of lipid A from Prevotella intermedia ATCC 25611 lipopolysaccharide. FEBS Lett. 2003;543:98–102. doi: 10.1016/s0014-5793(03)00414-9. [DOI] [PubMed] [Google Scholar]
- 26.Qureshi N, Takayama K, Heller D, Fenselau C. Position of ester groups in the lipid A backbone of lipopolysaccharides obtained from Salmonella typhimurium. J Biol Chem. 1983;258:12947–12951. [PubMed] [Google Scholar]
- 27.Qureshi N, Takayama K, Ribi E. Purification and structural determination of nontoxic lipid A obtained from the lipopolysaccharide of Salmonella typhimurium. J Biol Chem. 1982;257:11808–11815. [PubMed] [Google Scholar]
- 28.Seid RC, Jr, Bone WM, Phillips LR. Identification of ester-linked fatty acids of bacterial endotoxins by negative ion fast atom bombardment mass spectrometry. Anal Biochem. 1986;155:168–176. doi: 10.1016/0003-2697(86)90242-3. [DOI] [PubMed] [Google Scholar]
- 29.Takayama K, Qureshi N, Ribi E, Cantrell JL. Separation and characterization of toxic and nontoxic forms of lipid A. Rev Infect Dis. 1984;6:439–443. doi: 10.1093/clinids/6.4.439. [DOI] [PubMed] [Google Scholar]
- 30.Guo L, et al. Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP-phoQ. Science. 1997;276:250–253. doi: 10.1126/science.276.5310.250. [DOI] [PubMed] [Google Scholar]
- 31.Kaltashov IA, Doroshenko V, Cotter RJ, Takayama K, Qureshi N. Confirmation of the structure of Lipid A Derived from the Lipopolysaccharide of Rhodobacter sphaeroides by a combination of MALDI, LS IMS, and tandem mass spectrometry. Anal Chem. 1997;69:2317–2322. doi: 10.1021/ac9612943. [DOI] [PubMed] [Google Scholar]
- 32.Lindner B. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry of lipopolysaccharides. Methods Mol Biol. 2000;145:311–325. doi: 10.1385/1-59259-052-7:311. [DOI] [PubMed] [Google Scholar]
- 33.Boue SM, Cole RB. Confirmation of the structure of lipid A from Enterobacter agglomerans by electrospray ionization tandem mass spectrometry. J Mass Spectrom. 2000;35:361–368. doi: 10.1002/(SICI)1096-9888(200003)35:3<361::AID-JMS943>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 34.Chan S, Reinhold VN. Detailed structural characterization of lipid A: Electrospray ionization coupled with tandem mass spectrometry. Anal Biochem. 1994;218:63–73. doi: 10.1006/abio.1994.1141. [DOI] [PubMed] [Google Scholar]
- 35.Kussak A, Weintraub A. Quadrupole ion-trap mass spectrometry to locate fatty acids on lipid A from gram-negative bacteria. Anal Biochem. 2002;307:131–137. doi: 10.1016/s0003-2697(02)00004-0. [DOI] [PubMed] [Google Scholar]
- 36.Sforza S, et al. Determination of fatty acid positions in native lipid A by positive and negative electrospray ionization mass spectrometry. J Mass Spectrom. 2004;39:378–383. doi: 10.1002/jms.598. [DOI] [PubMed] [Google Scholar]
- 37.Shaffer SA, Harvey MD, Goodlett DR, Ernst RK. Structural heterogeneity and environmentally regulated remodeling of Francisella tularensis subspecies novicida lipid A characterized by tandem mass spectrometry. J Am Soc Mass Spectrom. 2007;18:1080–1092. doi: 10.1016/j.jasms.2007.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Worrall TA, Lin S, Cotter RJ, Woods AS. On-probe sample purification of lipids for matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J Mass Spectrom. 2000;35:647–650. doi: 10.1002/(SICI)1096-9888(200005)35:5<647::AID-JMS973>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 39.Therisod H, Labas V, Caroff M. Direct microextraction and analysis of rough-type lipopolysaccharides by combined thin-layer chromatography and MALDI mass spectrometry. Anal Chem. 2001;73:3804–3807. doi: 10.1021/ac010313s. [DOI] [PubMed] [Google Scholar]
- 40.Costello CE, Vath JE. Tandem mass spectrometry of glycolipids. Methods Enzymol. 1990;193:738–768. doi: 10.1016/0076-6879(90)93448-t. [DOI] [PubMed] [Google Scholar]
- 41.Raetz CRH. Biochemistry of endotoxins. Annu Rev Biochem. 1990;59:129–170. doi: 10.1146/annurev.bi.59.070190.001021. [DOI] [PubMed] [Google Scholar]
- 42.Ummarino S, Corsaro MM, Lanzetta R, Parrilli M, Peter-Katalinić J. Determination of phosphorylation sites in lipooligosaccharides from Pseudoalteromonas haloplanktis TAC 125 grown at 15°C and 25°C by nano-electrospray ionization quadrupole time-of-flight tandem mass spectrometry. Rapid Commun Mass Spectrom. 2003;17:2226–2232. doi: 10.1002/rcm.1179. [DOI] [PubMed] [Google Scholar]
- 43.Rietschel ET, Brade L, Lindner B, Zähringer U. In: Bacterial Endotoxic Lipopolysaccharides. Morrison DC, Ryan JL, editors. Vol 1. Boca Raton, FL: CRC Press; 1992. pp. 3–41. Molecular Biochemistry and Cellular Biology. [Google Scholar]
- 44.Rosner MR, Tang J, Barzilay I, Khorana HG. Structure of the lipopolysaccharide from an Escherichia coli heptose-less mutant. I. Chemical degradations and identification of products. J Biol Chem. 1979;254:5906–5917. [PubMed] [Google Scholar]
- 45.Zhou Z, White KA, Polissi A, Georgopoulos C, Raetz CRH. Function of Escherichia coli MsbA, an essential ABC family transporter, in lipid A and phospholipid biosynthesis. J Biol Chem. 1998;273:12466–12475. doi: 10.1074/jbc.273.20.12466. [DOI] [PubMed] [Google Scholar]
- 46.Westphal O, Jann K. Bacterial lipopolysaccharides: Extraction with phenol-water and further applications of the procedure. Methods Carbohydr Chem. 1965;5:83–91. [Google Scholar]
- 47.Fischer W, Koch HU, Hass R. Improved preparation of lipoteichoic acids. Eur J Biochem. 1983;133:523–530. doi: 10.1111/j.1432-1033.1983.tb07495.x. [DOI] [PubMed] [Google Scholar]
- 48.Caroff M, Tacken A, Szabo L. Detergent-accelerated hydrolysis of bacterial endotoxins and determinations of the anomeric configuration of the glycosyl phosphate present in the “isolated lipid A” fragment of the Bordetella pertussis endotoxin. Carbohydr Res. 1988;175:273–282. doi: 10.1016/0008-6215(88)84149-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.