Abstract
Cellular signaling mediated by the epidermal growth factor receptor (EGFR or ErbB) family of receptor tyrosine kinases plays an important role in regulating normal and oncogenic cellular physiology. While structures of isolated EGFR extracellular domains and intracellular protein tyrosine kinase domains have suggested mechanisms for growth factor-mediated receptor dimerization and allosteric kinase domain activation, understanding how the transmembrane and juxtamembrane domains contribute to transmembrane signaling requires structural studies on intact receptor molecules. In this report, recombinant EGFR constructs containing the extracellular, transmembrane, juxtamembrane, and kinase domains are overexpressed and purified from human embryonic kidney 293 cell cultures. The oligomerization state, overall structure, and functional stability of the purified EGF-bound receptor are characterized in detergent micelles and phospholipid bilayers. In the presence of EGF, catalytically active EGFR dimers can be isolated by gel filtration in dodecyl maltoside. Visualization of the dimeric species by negative stain electron microscopy and single particle averaging reveals an overall structure of the extracellular domain that is similar to previously published crystal structures and is consistent with the C-termini of domain IV being juxtaposed against one another as they enter the transmembrane domain. Although detergent-soluble preparations of EGFR are stable as dimers in the presence of EGF, they exhibit differential functional stability in Triton X-100 versus dodecyl maltoside. Furthermore, the kinase activity can be significantly stabilized by reconstituting purified EGF-bound EGFR dimers in phospholipid nanodiscs or vesicles, suggesting that the environment around the hydrophobic transmembrane and amphipathic juxtamembrane domains is important for stabilizing the tyrosine kinase activity in vitro.
The epidermal growth factor receptor (EGFR)1is a type I transmembrane receptor tyrosine kinase (RTK) that binds extracellular polypeptide growth factor ligands, such as epidermal growth factor, to stimulate an array of intracellular signaling cascades that regulate normal cellular growth and proliferation. In one model for growth factor-mediated EGFR activation, ligand binding induces receptor dimerization with a concomitant activation of intracellular protein tyrosine kinase activity (1-3), while other models suggest that receptors are predimerized on the cell surface and ligand binding alters the dimer configuration (4-6). Once activated, subsequent autophosphorylation of tyrosine residues in the intracellular C-terminal domain provides docking sites for SH2 and PTB domain-containing adaptor proteins that couple extracellular signals and receptor activation to downstream signaling pathways (7, 8). Finally, once activated, EGFR signal transduction is attenuated by a variety of mechanisms that result in receptor internalization and downregulation (9).
The mechanisms regulating growth factor-dependent activation of EGFR and related members of the ErbB family, ErbB2, ErbB3, and ErbB4, are currently of significant clinical importance, as deregulated ErbB receptor signaling and constitutive activation have been identified in several types of cancers (10). As such, EGFR and ErbB2 are important targets of anticancer therapeutics, with a number of antagonistic monoclonal antibodies and small molecule tyrosine kinase inhibitors currently available to inhibit EGFR and ErbB2 signaling (11). However, the restricted responsiveness by only select patient populations and the emergence of drug-resistant mutations necessitate the continued development of ErbB-targeted therapeutic strategies and a better understanding of the structural mechanisms of EGFR activation and regulation (11-13).
Recent structural studies of the isolated EGFR extracellular domain (14-17) and kinase domain (18, 19) have provided mechanistic insight into growth factor-mediated receptor dimerization and allosteric kinase domain activation, respectively. However, it is still poorly understood how the transmembrane domains associate in the plasma membrane and how their structure is propagated through the intracellular juxtamembrane domain to properly position the tyrosine kinase domains for activation (20, 21). Biochemical and cell-based studies utilizing fragments of EGFR that include the transmembrane domain or isolated transmembrane domains have provided conflicting results on the role of the membrane spanning region in mediating receptor dimerization (22-24). In addition, the juxtamembrane domain is suggested to play an important role in regulating receptor activation. Models have been proposed in which the membrane-proximal portion of the juxtamembrane domain interacts with the inner leaflet of the plasma membrane in a phosphorylation-dependent manner (25, 26), while the membrane-distal region comprises a portion of the dimerization interface observed in the kinase domain crystal structure (19) and is required for allosteric activation (27). However, significant structural heterogeneity observed in the NMR structure of the isolated juxtamembrane domain prevented definition of the relative orientations of the membrane-proximal and -distal regions (28). Thus, determining how the structures and the dynamics of the transmembrane and juxtamembrane domains contribute to EGFR activation is best suited for characterization in the context of intact receptor molecules.
Structural characterization of intact EGFR requires systems for reconstituting EGFR that provide structural and functional stability in vitro. In the pioneering studies by Cohen, Schlessinger, Gill, and others, biochemical mechanisms of EGFR signaling were initially established working with Triton X-100 solubilized preparations of EGFR extracted from A431 cell membranes (29). Furthermore, partial purification of endogenously expressed EGFR from A-431 cells was achieved by immunoaffinity chromatography and other means (29-33). While these approaches provided sufficient quantities of wild-type protein preparations for biochemical characterization, they do not provide sufficient quantities or a means to isolate mutant constructs for biophysical characterization of intact receptor molecules. In addition, the stability of highly purified detergent-soluble EGFR preparations has not been characterized in detail.
In this report we present a system for the high yield expression and purification of recombinant EGFR constructs from mammalian cell cultures and screen detergents for their ability to solubilize functional and monodisperse preparations of EGFR. We characterize the oligomeric state, overall structure and domain organization, and functional stability of purified EGF-bound EGFR in detergent micelles and phospholipid bilayers. In dodecyl maltoside detergent micelles, the extracellular domain of the intact receptor dimer has a conformation in electron microscopy projection averages that is similar to that seen in crystal structures. Although the EGF-bound EGFR dimer is a stable structural unit in dodecyl maltoside and is functionally active, the kinase activity is markedly less stable than in Triton X-100. Furthermore, the kinase activity can be significantly stabilized by reconstituting purified EGF-bound EGFR dimers in phospholipid bilayer nanodiscs or vesicles, suggesting that the environment around the hydrophobic transmembrane and amphipathic juxtamembrane domains is important for stabilizing the tyrosine kinase activity in vitro.
EXPERIMENTAL PROCEDURES
Materials and Reagents
All detergents were from Anatrace as 10% solutions stored under argon gas at 4 °C or in powder form stored at −20 °C. Egg phosphatidylcholine and brain total lipid extract were purchased from Avanti Polar Lipids in powder form and stored under argon gas at −20 °C. [γ-32P]ATP was from GE Healthcare. Recombinant EGF was from PeproTech, reconstituted in water at a final concentration of 100 μM and stored at −20 °C.
EGFR Expression Constructs
DNA sequences coding for the full-length, mature human EGFR (amino acids 1−1186) or deletion construct Δ998-EGFR (amino acids 1−998) were PCR amplified from an EGFR expression vector and sub-cloned into the XhoI and MluI restriction sites of the ExpressTag-1 (ET-1) expression vector. The ET-1 expression vector is a modified version of the pIRES2-GFP vector (Clontech, Inc.) engineered for the expression of secreted or cell surface recombinant proteins in mammalian cells from a CMV promoter. EGFR constructs expressed from the ET-1 vector contain an N-terminal murine immunoglobulin κ chain signal peptide, a C-terminal protein C epitope (34), and C-terminal hexahistadine and streptavidin binding protein (SBP) affinity tags. The SBP tag was designed from sequences selected in vitro by Szostack and colleagues (35) and is of higher affinity than the commonly used streptactin-binding peptide. All experiments herein were carried out on cysteine light variants of wild-type EGFR in which the following surface-exposed cysteines in the kinase domain (full length and Δ998) and C-terminal domain (full length) have been mutated to serine or alanine: C751S, C757S, C773S, C1025A, C1034A, and C1121A. Mutations were introduced by site-directed mutagenesis using the QuikChange strategy (Stratagene). An EGFR-GFP fusion construct was generated by digesting EGFR-ET1 plasmid DNA with MluI and BstXI restriction enzymes, which cut immediately after the EGFR coding sequence (before the C-terminal tags) and immediately before the GFP coding sequence, respectively. The digested vector was gel purified and religated in the presence of a synthetic fragment coding for a Gly-Gly-Gly-Ala amino acid linker with compatible MluI/BstXI restriction sites.
Cell Culture and Protein Expression
Human embryonic kidney (HEK) 293T (36) or 293S GnTI−/− (37) monolayers were cultured in Dulbecco's modified Eagle's medium (DMEM), HEPES modification (Sigma) supplemented with 10% fetal bovine serum (Omega Scientific, Inc.), 4 mM l-glutamine (Sigma), and MEM nonessential amino acids.
EGFR constructs were expressed in 293T or 293S GntI−/− cell cultures by transient transfection. Confluent monolayers were split 1:4 in 15 cm dishes approximately 24 h prior to transfection. Cultures were transfected at approximately 65−75% confluency with plasmid DNA (50 μg per 175 cm2 monolayer surface area) complexed to linear polyethylenimine (PEI) at a DNA:PEI ratio of 1:11 (w/w). Three hours posttransfection, the culture medium was changed to ExCell 293 serum free medium (SAFC Biosciences) supplemented with 6 mM l-glutamine. Cells were harvested 48 h posttransfection, washed once with phosphate-buffered saline (pH 7.4), and either used immediately or stored at −80 °C.
Detergent Solubility Screen
Cells transfected with EGFR-GFP were thawed, resuspended in buffer (25 mM Tris, pH 8.0, 400 mM NaCl, 10% glycerol, 1× Complete protease inhibitor cocktail (Roche), 1 mM Na3VO4), and lysed in the presence of 1% (w/v) detergent on ice for 1 h. Cell lysates were cleared by centrifugation at 50000g for 15 min, and the cleared supernatants were fractionated by gel filtration on Superose 6HR equilibrated in 20 mM Tris, pH 8.0, 400 mM NaCl, 1 mM EDTA, and 0.4 mM dodecyl maltoside. Fractions containing EGFR-GFP were detected by monitoring GFP fluorescence at excitation and emission wavelengths of 488 and 510 nm, respectively, in a Perkin-Elmer LS-50 luminescence spectrometer. Unfractionated cleared lysates were also assayed for kinase activity as described below.
Purification of EGFR by Streptactin–Sepharose Affinity Chromatography
Cells transfected with EGFR or Δ998-EGFR constructs were solubilized in lysis buffer (5 mL/15 cm plate; 20 mM Tris, pH 8.0, 400 mM NaCl, 10% glycerol, 0.2% Triton X-100, 1 mM EDTA, 1× Complete protease inhibitor cocktail) and lysed at 4 °C for 1 h. Lysates were cleared by centrifugation at 50000g for 30 min at 4 °C, and the cleared lysate was applied to a streptactin–Sepharose column (1 mL bed volume per four plates of transfected cells) equilibrated in wash buffer (20 mM Tris, pH 8.0, 400 mM NaCl, 10% glycerol, 0.1% Triton X-100, 1 mM EDTA) at a flow rate of 0.1−0.2 mL/min. The column was washed with 20 column volumes of wash buffer, and bound sample was eluted with wash buffer containing 2.5 mM desthiobiotin. Eluate fractions were assayed for EGFR by SDS–PAGE and staining with Coomassie blue. Fractions containing EGFR were aliquoted and stored at −80 °C.
In Vitro Kinase Assay
Samples were assayed for tyrosine kinase activity using the peptide-based in vitro phosphorylation assay described elsewhere (33, 38). The synthetic peptide substrate (RR1173; H3N+-RRKGSTAENAEY1173LRVCOO−) corresponds to the Y1173 major autophosphorylation site of EGFR. Briefly, reactions were carried out in 50 μL reaction volumes at room temperature with the final reaction conditions of 100 mM HEPES, pH 7.4, 1% glycerol, 0.1 mg/mL BSA, 5 mM MgCl2, 1.25 mM MnCl2, 0.1 mM Na3VO4, 1 mM RR1173 substrate, 20 μM ATP, and 2 μCi of [γ-32P]ATP. Samples (either cell lysates or purified material) were treated with EGF or buffer as necessary in a volume of 30 μL and incubated at room temperature for 5 min. To each sample was added 10 μL of 5× kinase buffer (500 mM HEPES, pH 7.4, 5% glycerol, 0.5 mg/mL BSA, 25 mM MgCl2, 12.5 mM MnCl2, 500 μM Na3VO4), and the resultant solution was incubated for an additional 5 min. Kinase reactions were initiated by the addition of 10 μL of a 5× substrate/ATP mixture (5 mM RR1173, 100 μM ATP, 2 μCi of [γ-32P]ATP) and incubated at room temperature for 10 min. Reactions were terminated by addition of 20 μL of 25% trichloroacetic acid and 5 μL of 10 mg/mL BSA to facilitate protein precipitation and incubated on ice for 30 min. Precipitates were spun down, and 10 μL of the supernatant was spotted on 2 cm × 2 cm squares of phosphocellulose filter paper. The filter paper was washed 4× with 75 mM phosphoric acid and the remaining bound radioactivity counted. A control sample of buffer without EGFR was used for measuring the background radioactivity.
Negative Stain Electron Microscopy and Image Analysis
Purified EGFR constructs (2−5 μM) were treated with or without 20 μM EGF for 30 min on ice in the presence of 1 mM ATP and subjected to gel filtration on Superose 6HR equilibrated in running buffer containing 0.4 mM dodecyl maltoside. Following gel filtration, peak fractions corresponding to EGF-bound EGFR dimers were diluted to approximately 1−5 μg/mL (5−30 nM), treated with 10 μM gefitinib, immediately adsorbed to glow-discharged carbon-coated grids, and stained with uranyl formate (39). Specimens were inspected with a Tecnai T12 electron microscope (FEI, Hillsboro, OR) equipped with an LaB6 filament and operated at 120 kV. Images were recorded on imaging plates at a nominal magnification of 67000× using low-dose procedures. Imaging plates were scanned and digitized with a Ditabis micron imaging plate scanner (DITABIS Digital Biomedical Imaging System, AG, Pforzheim, Germany) using a step size of 15 μm, and 2 × 2 pixels were averaged to yield a final pixel size of 4.46 Å at the specimen level. Following digitization of electron micrographs, 2990 single particles were manually selected from 56 images, windowed into 75 × 75 pixel individual images, and grouped into 100 classes using multireference alignment and K-means classification procedures implemented in SPIDER (40). Alignment, classification, and averaging were iteratively repeated 10 times to obtain the final set of class averages. To assign regions of density to particular EGFR domains, two-dimensional projections were calculated from extracellular or kinase domain crystal structures and cross-correlated with masked regions of EM class averages. Briefly, crystal structures were low-pass filtered to 25 Å, and a series of two-dimensional projections were calculated at 4 deg intervals. Each of these projections was aligned with a class average to calculate a cross-correlation coefficient, and the projection with the highest coefficient was chosen. The cross-correlation coefficients were normalized by dividing by the coefficient for the class average correlated with itself.
Reconstitution of Purified EGFR in Phospholipid Nanodiscs
Egg phosphatidylcholine (egg PC) lipids were dissolved in chloroform and dried in a glass tube under a gentle stream of argon while rotating the tube to form a thin lipid film. Residual chloroform was removed under vacuum for 3 h. Lipids were rehydrated with buffer (10 mM Tris, pH 7.4, 100 mM NaCl, 20 mM Triton X-100) at a final lipid concentration of 10 mM. During the rehydration step, tubes were sealed with a rubber septum and purged with argon gas to minimize lipid oxidation. Rehydrated lipid was stored on ice until use.
All disc assembly reactions were performed with membrane scaffold protein construct MSP1T2 (generously provided by S. Sligar, University of Illinois at Urbana–Champaign) (41). In some experiments, the N-terminal heptahistidine tag of MSP1T2 was removed by cleavage with TEV protease; this construct will be referred to as MSP1T2(−). Assembly of discs with egg PC was optimized as described previously for other lipid compositions (41, 42). Briefly, in 200 μL assembly reactions, 10 μM MSP1T2 was incubated with varying amounts of lipid for 40 min on ice. Assembly was initiated by detergent removal with washed SM-2 Bio-Beads (Bio-Rad; 20 mg of beads/mg of detergent) for 16 h on a rotator at 4 °C under argon. At each step, reaction tubes were purged with argon gas to minimize lipid oxidation. Following detergent removal, beads were allowed to settle, and the assembly reaction was transferred to a new tube, purged with argon, and stored at 4 °C. Disc assembly was assayed by gel filtration on Superose 6HR equilibrated in 10 mM Tris, pH 7.4, and 100 mM NaCl. Optimal disc formation was observed for an MSP1T2:egg PC molar ratio of 1:75.
EGFR was reconstituted into egg PC nanodiscs by incorporating purified Triton X-100 solubilized EGFR into the disc assembly mixture. Samples of EGFR were pretreated with a 20-fold excess of EGF for 10 min on ice to induce dimer formation. Disc assembly mixtures containing (final concentrations) 1 μM EGFR, 20 μM EGF, 10 μM MSP1T2, 750 μM egg PC, and 2.0−2.5 mM Triton X-100 were incubated on ice for 40 min. Assembly was initiated by removing detergent with SM-2 Bio-Beads (20 mg of beads/mg of detergent) for 16 h on a rotator at 4 °C under argon. Following assembly, beads were spun down, and the assembly mixture was transferred to a new tube, purged with argon, and stored at −80 °C.
Reconstitution of Purified EGFR in Phospholipid Vesicles
Dried lipids were rehydrated at 10 mg/mL final lipid concentration in 20 mM Tris, pH 8.0, 10% glycerol, 1 mM EDTA, and 400 mM NaCl. The lipid suspension was passed through an extruder (Lipex) equipped with a 0.8 μM filter (2×), 0.4 μM filter (2×), and 0.1 μM filter (11×) to form small unilamellar vesicles. Purified EGFR in 0.1% Triton X-100 was treated with 20 μM EGF for 5 min on ice. Preformed liposomes (50 μL) were added to the EGF-treated sample and incubated for 1 h at 4 °C. The final detergent/lipid mass ratio was 1. Detergent was removed by incubation with Bio-Rad SM-2 Bio-Beads for 16 h at 4 °C. Following detergent removal, beads were spun down and reconstituted proteoliposomes transferred to new tubes and stored at −80 °C.
RESULTS
Solubility, Monodispersity, and Activity of EGFR in Detergent-Soluble Cell Lysates
Cohen and co-workers initially showed that 1% Triton X-100 and 10% glycerol could extract 75−90% of the EGF-dependent kinase activity from preparations of A-431 cell membranes (29). In addition, this could be achieved with a number of other industrial detergents. While Triton X-100 has been historically the detergent of choice for solubilizing EGFR, we sought to determine if other detergents were suitable for functional reconstitution and structural characterization of EGFR by monitoring their effect on receptor solubility, monodispersity, and activity.
The ability of a particular detergent to solubilize EGFR from transfected 293 cells was assayed by SDS–PAGE of detergent-soluble cell lysates and Western blotting with antiprotein C mAb (34, 43), which reacts with the C-terminal tags of the recombinant EGFR constructs expressed here. Most detergents tested were able to solubilize EGFR to a similar extent as Triton X-100 (Table 1).
Table 1.
Solubility, Monodispersity, and Activity of EGFR in Detergent-Soluble Cell Lysates
| detergenta | solubilityb | monodispersityc | activity (−EGF/+EGF)d |
|---|---|---|---|
| Triton X-100 | + | m | 1.0/4.2 |
| Triton X-114 | + | m | |
| Triton X-305 | − | ||
| Triton X-405 | − | ||
| Brij 35 | − | ||
| Brij 58 | + | m | |
| Tween 20 | + | m | |
| Tween 80 | − | ||
| C12E10 | + | 2.8/4.4 | |
| C12E9 | + | 2.1/4.1 | |
| C12E8 | + | m | 2.1/4.9 |
| C10E9 | + | m | 2.2/1.7 |
| C10E6 | + | p | 1.2/1.4 |
| dodecyl maltoside (DDM) | + | m | 3.6/7.5 |
| octyl β-glucoside | + | a | |
| diheptanoylphosphatidylcholine (DHPC) | + | p | 0.3/0.1 |
| dodecylphosphocholine (DPC) | + | p | 0.0/0.4 |
| lauryldimethylamine N-oxide (LDAO) | + | m | 1.4/0.8 |
| anzergent 3−8 | + | m | 3.5/5.6 |
| anzergent 3−10 | + | m | 1.0/1.1 |
| anzergent 3−12 | + | m | 1.0/0.9 |
| CYMAL-6 | + | a | |
| CHAPSO | + | a |
Detergents were used at a final concentration of 1% (w/v) to solubilize recombinant EGFR in lysates of transfected cells.
Solubility was judged by SDS–PAGE of detergent-soluble cell lysates and Western blot with antiprotein C mAb to detect recombinant EGFR. Detergents that solubilize EGFR to a comparable or greater extent than Triton X-100 are indicated by a (+) and to a lesser extent by a (−).
The monodispersity of detergent-soluble EGFR-GFP was judged by gel filtration to be (m) monodisperse, (p) polydisperse, or (a) aggregated if the majority of the GFP fluorescence eluted from gel filtration in a single peak, multiple peaks, or the void volume, respectively.
The kinase activities measured for detergent-soluble cell lysates are shown relative to the kinase activity of EGFR in Triton X-100 solubilized cell lysates in the absence of EGF.
Next, the monodispersity of the EGFR-GFP fusion construct was assayed using gel filtration chromatography of detergent-soluble cell lysates. In these experiments, elution of EGFR is followed in the presence of other detergent-soluble material by monitoring the fluorescence of the GFP fusion protein (44). Attachment of GFP to the C-terminus of EGFR does not affect the in vitro or in situ tyrosine kinase activity (data not shown). Of the 21 detergents screened, Triton X-100 is representative of a “good” detergent, with the majority of the GFP fluorescence eluting as a relatively sharp peak, compared to protein standards, at an elution volume of approximately 14 mL (Figure 1, solid line); this position corresponds to a receptor monomer (see below). In contrast, octyl β-d-glucopyranoside (β-OG) is representative of a “poor” detergent causing most of the GFP fluorescence to elute in the void volume of the column (Figure 1, dashed line). The peaks at 17−18 mL in both chromatograms correspond to free GFP resulting from proteolytic degradation of the fusion construct. The results for 15 other detergents are listed in Table 1.
Figure 1.
Solubility and monodispersity of recombinant EGFR in detergent-solubilized cell lysates. EGFR-GFP fusion protein was expressed by transient transfection in 293S GntI−/− cells and solubilized in buffer containing (solid line) 1% Triton X-100 or (dashed line) 1% octyl β-d-glucopyranoside. Lysates were subjected to gel filtration on Superose 6HR (24 mL bed volume). GFP fluorescence of eluate fractions was monitored at excitation and emission wavelengths of 488 and 512 nm, respectively.
For detergents that yield a relatively monodisperse preparation of EGFR-GFP in detergent-soluble cell lysates, the kinase activity in the absence and presence of EGF was assayed by monitoring the incorporation of 32P into an exogenous peptide substrate. Relative to the activity in Triton X-100 lysates, comparable basal and stimulated activities are observed for the homogeneous polyoxyethylene dodecyl ethers C12E10, C12E9, and C12E8; dodecyl maltoside yields similar levels of fold stimulation although with higher basal activity; and LDAO and the anzergent series of zwitterionic detergents yield similar basal activities without any further stimulation by EGF. These results are summarized in Table 1.
Purification of EGFR by Affinity Chromatography
Recombinant EGFR constructs expressed from our ET1 vector by transient transfection in 293T or 293S GntI−/− cell cultures are purified by streptactin–Sepharose affinity chromatography in the presence of 0.1% Triton X-100. SDS–PAGE analysis of eluate fractions from a representative purification (from cleared lysate of 4 × 15 cm plates of transfected cell culture) is shown in Figure 2a. After this single purification step, EGFR is >90% pure as judged by SDS–PAGE and Coomassie staining. A minor contaminant that migrates at ∼70 kDa molecular mass on reducing SDS–PAGE is invariably observed in all preparations. This contaminant was identified by mass spectrometry to be Hsc70/Hsp70 molecular chaperones (data not shown). The typical yield of purified EGFR from 4 × 15 cm plates of transfected cell culture is approximately 200−400 μg in 0.5 mL of the most concentrated eluate fraction (lane 3 in Figure 2a).
Figure 2.
EGF-dependent dimerization and kinase activation of purified, detergent-soluble EGFR. (a) Δ998-EGFR was expressed by transient transfection in 293S GntI−/− cells and purified from detergent-solubilized cell lysates by streptactin–Sepharose affinity chromatography in the presence of 0.1% Triton X-100. Lanes 1−5 show 5 μL of the corresponding 0.5 mL fractions eluted with 2.5 mM desthiobiotin and subjected to reducing SDS–PAGE and Coomassie blue staining. (b, c) Gel filtration chromatograms of EGFR samples from lane 3 in (a) treated (b) without or (c) with 20 μM EGF and subjected to gel filtration on Superose 6HR equilibrated in 0.4 mM dodecyl maltoside. Protein elution was monitored by absorbance at 280 nm (solid line), and kinase activity of eluate fractions was determined by monitoring the incorporation of 32P into an exogenous peptide substrate (dashed line, circles).
EGF-Stimulated Dimerization and Activation of EGFR Tyrosine Kinase Activity
The oligomerization state of EGFR purified in 0.1% Triton X-100 was assayed by gel filtration in 0.4 mM dodecyl maltoside. Dodecyl maltoside was chosen as the detergent in the gel filtration running buffer for the superior quality of EM images that could be obtained in comparison to images for samples prepared in Triton X-100, for example (data not shown). In the absence of EGF, purified Δ998-EGFR predominantly elutes in a sharp peak with an apparent Stokes radius of 5.9 nm (Figure 2b). When assayed for kinase activity, this dominant peak in absorbance at 280 nm exhibits very low kinase activity; however, a relatively small peak of kinase activity elutes at a higher molecular mass with an apparent Stokes radius of 7.6 nm. In contrast, when purified Δ998-EGFR is pretreated with 20 μM EGF and fractionated by gel filtration, the dominant peak in the absorbance at 280 nm and in kinase activity coelutes with an apparent Stokes radius of 7.6 nm (Figure 2c). In the context of the ligand-induced dimerization model, these results suggest that the species eluting from gel filtration with apparent Stokes radii of 5.9 and 7.6 nm correspond to the inactive Δ998-EGFR monomer and EGF-bound Δ998-EGFR dimer, respectively.
Global Structure and Domain Organization of the EGF-Bound Δ 998-EGFR Dimer
The overall structure and domain organization of the detergent-soluble, EGF-bound Δ998-EGFR dimer isolated by gel filtration in 0.4 mM dodecyl maltoside were characterized by negative stain electron microscopy. From 56 images of negatively stained samples, 2990 particles were selected and grouped into 100 class averages (Supporting Information Figure S1); the averages of the three most populated classes are shown in Figure 3a. In these class averages, three major regions of stain exclusion, which represent protein domains, can be observed. The top portion, which exhibits an approximate 2-fold symmetry, is followed by a small region of density that connects the top portion to a larger globular density on the bottom.
Figure 3.

Overall structure and domain organization of EGFR determined by negative stain electron microscopy. (a) Δ998-EGFR samples purified by streptactin–Sepharose affinity chromatography were treated with 4 μM EGF for 30 min on ice and subjected to gel filtration on Superose 6HR equilibrated in 0.4 mM dodecyl maltoside. Samples of the peak fraction corresponding to dimeric EGF-bound EGFR were imaged by negative stain EM and used to calculate class averages. The averages from the most populated classes are shown. (b) Masked region of density corresponding to the extracellular domain in (a) that was used for cross-correlation with two-dimensional projections calculated from crystal structures of the dimeric, liganded EGFR extracellular domain (c and d). Two-dimensional projections and cross-correlation coefficients from (c) the extracellular domain EGF-bound crystal structure that contains domains I–III and (d) the model of the entire extracellular domain including domain IV. (e) Ribbon diagram of the dimeric EGF-bound EGFR extracellular domain crystal structure containing domains I–III. (f) Model of the entire EGFR extracellular domain generated by aligning domain IV from the unliganded structure of the extracellular domain (14) with the first two Cys-rich modules of domain IV (residues 480−512) that were resolved in the liganded structure (17). The ribbon diagrams (e and f) are shown in approximately the same orientations as the two-dimensional projections shown in (c) and (d).
In the class averages of the EGF-bound EGFR dimer (Figure 3a), the 2-fold symmetric density at the top of the images is similar in shape and size to the crystal structures of the dimeric ligand-bound EGFR extracellular domain (14, 15). To test the hypothesis that this density corresponds to the extracellular domain, two-dimensional projections were calculated from the crystal structure of the EGF-bound EGFR extracellular domain and cross-correlated with masked regions of the class averages (Figure 3b). Using only domains I–III from the previously published crystal structures (Figure 3e), projections are obtained that are in excellent visual agreement with the top portion seen in the averages (Figure 3c), suggesting that this portion corresponds to domains I–III.
The small, middle region of density in the class averages is not accounted for by two-dimensional projections of domains I–III and may correspond to extracellular domain IV, which is not observed in either of the two published crystal structures of the ligated extracellular domain dimer (14, 15). To test the hypothesis that this density corresponds to domain IV of the extracellular domain, two-dimensional projections were calculated for a model of the EGF-bound EGFR extracellular domain that contains domains I–IV. The model was built by superposing the structure of domain IV observed in the tethered (undimerized) conformation (17) onto the first two cysteine-rich modules of domain IV (residues 480−512) observed in the EGF-bound dimer structure (14). This superposition places the C-terminus of domain IV for each monomer in very close proximity in the context of the dimer (Figure 3f). Projections calculated from this model yield higher cross-correlation coefficients than projections that do not include domain IV and are of similar size and shape as the masked EM images. These results suggest that the C-termini of the two domains IV are very close to one another as they approach the transmembrane segment in the intact receptor.
By elimination, the large globular density at the bottom of the class averages of Δ998-EGFR in Figure 3a must consist of the transmembrane, intracellular juxtamembrane, and intracellular tyrosine kinase domains. This region is less well defined (as compared to density for the extracellular domain) in class averages and exhibits a range of shapes and orientations with respect to the extracellular domain density. When solubilized in detergent, flexibility of the transmembrane and juxtamembrane regions likely results in a range of orientations between extracellular and intracellular domains that are adsorbed onto the EM grids.
In addition to EGF-bound Δ998-EGFR dimers, images and class averages were also obtained for the monomeric Δ998-EGFR in the absence of EGF. In these class averages (data not shown), three distinct regions of spherical density were observed that are considerably smaller in size than the densities observed for the dimeric material. However, the lack of distinguishing features and considerable variability in spatial organization of the densities among class averages precluded the assignment of corresponding domains for Δ998-EGFR monomers.
Stability of EGFR Kinase Activity in Detergent Micelles
The stability of the EGF-bound Δ998-EGFR dimer in dodecyl maltoside detergent micelles was assayed by monitoring the kinase activity as a function of time incubated on ice. The fraction corresponding to the EGF-bound EGFR dimer that eluted from gel filtration (Figure 2c) was aliquoted and frozen at −80 °C. Samples were thawed and incubated for varying times at 0 °C before measuring kinase activity. As shown in Figure 4a (solid line), upon exchange from 0.1% Triton X-100 into 0.4 mM dodecyl maltoside by gel filtration, the kinase activity of purified Δ998-EGFR decreases rapidly with time incubated at 0 °C. The kinase activity is lost with a rapid initial decay followed by a slower decay at longer times. Kinase activity decays less rapidly when the dodecyl maltoside concentration in gel filtration is lowered to 0.2 mM (Figure 4a, dashed line), although less than 20% of the initial kinase activity remains after 6 h at 0 °C.
Figure 4.
Stability of EGFR kinase activity in dodecyl maltoside. (a) Δ998-EGFR samples purified by streptactin–Sepharose affinity chromatography in the presence of 0.1% Triton X-100 were treated with EGF and subjected to gel filtration on Superose 6HR equilibrated in (filled circles, solid line) 0.4 mM dodecyl maltoside or (open circles, dashed line) 0.2 mM dodecyl maltoside. The peak fraction corresponding to dimeric EGF-bound EGFR was dispensed into aliquots and frozen at −80 °C. Samples were thawed and stored on ice for the indicated amount of time and assayed for kinase activity by monitoring the incorporation of 32P into an exogenous peptide substrate. (b) Peak fractions corresponding to Δ998-EGFR dimer species eluted from gel filtration in 0.2 mM dodecyl maltoside were refractionated by gel filtration either (top) immediately or (bottom) following incubation on ice for 10 h. (c) Kinase activity of the purified EGFR kinase domain (residues 672−998) in the (open circles, dashed line) absence or (filled circles, solid line) presence of 0.2 mM dodecyl maltoside. (d) Kinase activity of Δ998-EGFR in the presence of 0.2 mM dodecyl maltoside and brain whole lipid extract at detergent:lipid weight ratios of (circles, solid line) 4:1, (squares, dashed line) 1:1, and (triangles, dashed-dotted line) 1:10.
To test if the decrease in kinase activity with time at 0 °C is due to dissociation of the EGF-bound EGFR dimer in dodecyl maltoside, the peak dimer fraction eluted from gel filtration in 0.2 mM dodecyl maltoside was divided into two aliquots and frozen at −80 °C. The aliquots were thawed on ice and refractionated on gel filtration either immediately after thawing or after incubation on ice for 10 h. In both cases, nearly identical chromatograms are observed with the main peak in absorbance at 280 nm eluting with an apparent Stokes radius of 7.6 nm (Figure 4b), suggesting that EGF-bound EGFR remains dimeric even after 10 h at 0 °C in 0.2 mM dodecyl maltoside.
To test if detergent has a direct inhibitory effect on the kinase domain itself, the kinase activity of the isolated kinase domain (residues 672−998) was assayed in the absence and presence of 0.2 mM DDM. As shown in Figure 4c, the activity of the isolated kinase domain is stable for at least 10 h when incubated on ice in the absence or presence of 0.2 mM dodecyl maltoside.
Finally, phospholipids are sometimes required to yield biologically active preparations of purified membrane proteins. To test if the addition of lipid can stabilize Δ998-EGFR kinase activity following elution from gel filtration in 0.2 mM dodecyl maltoside, brain whole lipid extract was added to samples at detergent:lipid mass ratios of 4:1 (Figure 4d, circles), 1:1 (squares), and 1:10 (triangles) and the kinase activity measured as a function of time incubated at 0 °C (Figure 4d). Although increasing the amount of lipid appears to stabilize the kinase activity to some extent, only 25% of the initial kinase activity remains after 10 h for the highest amount of lipid tested.
Reconstitution of EGFR in Phospholipid Bilayer Nanodiscs
The requirement of a phospholipid bilayer for maintaining the stability of EGFR kinase activity was tested by reconstituting purified EGFR into phospholipid bilayer nanodiscs. Nanodiscs are reconstituted high-density lipoprotein particles composed of a lipid bilayer surrounded by two amphipathic helical membrane scaffold protein (MSP) molecules derived from human apolipoprotein A1 (42). Sligar and co-workers have demonstrated that nanodiscs, in addition to providing a more physiological membrane bilayer compared to detergent, allow membrane proteins to be reconstituted in a more controlled and stoichiometric manner compared to phospholipid vesicles (45-47).
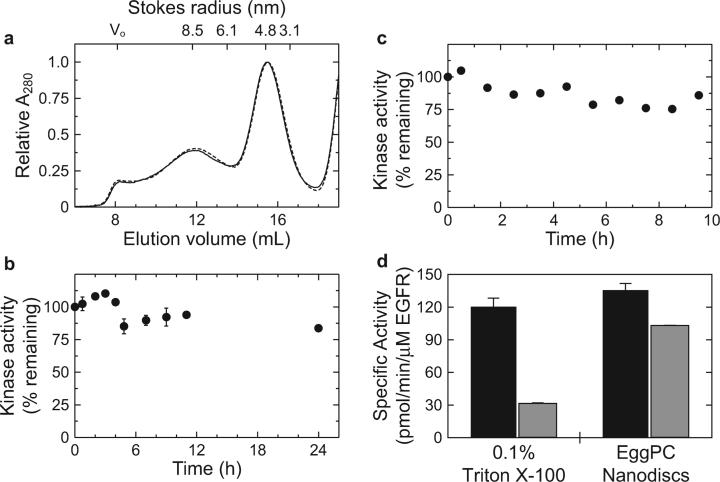
In the absence of EGFR, nanodiscs formed with membrane scaffold protein construct MSP1T2 and a mixture of phosphatidylcholine lipids extracted from chicken egg yolk (egg PC) elute from gel filtration with an apparent Stokes radius of approximately 4.8 nm (Figure 5a). These results are consistent with the 10 nm diameter discs formed with MSP1T2 and other lipid compositions reported previously (41, 42).
Figure 5.
Reconstitution of EGFR dimers in phospholipid nanodiscs. Phospholipid nanodiscs were assembled in the (a) absence or (b) presence of purified WT-EGFR pretreated with EGF. Assembly mixtures consisted of 10 μM membrane scaffold protein MSP1T2 and 750 μM egg phosphatidylcholine without or with 1 μM EGFR plus 20 μM EGF. Disc formation and receptor incorporation were analyzed by gel filtration on Superose 6HR equilibrated in 10 mM Tris, pH 7.4, and 100 mM NaCl. Protein elution was monitored by absorbance at 280 nm (solid line), and kinase activity of eluate fractions was determined by monitoring the incorporation of 32P into an exogenous peptide substrate (dashed line, circles). (c) Ni-NTA purification of EGFR-incorporated nanodiscs monitored by SDS–PAGE and Coomassie blue staining of input, flow-through, wash, and eluate fractions. The molar ratio of EGFR:MSP1T2(−) in eluate fractions was estimated by comparing the staining intensities of EGFR and MSP1T2(−) bands relative to the staining intensities of the 150 and 20 kDa molecular mass standards, respectively.
To incorporate EGFR into nanodiscs, purified EGFR in 0.1% Triton X-100 was pretreated with EGF and reconstituted in 10 nm discs under conditions where MSP1T2 is present at a 10-fold molar excess over EGFR. Under these conditions, the majority of discs formed will not contain any membrane protein (“empty” discs) and will elute from gel filtration at a similar position as discs formed in the absence of EGFR. However, approximately 10% of the discs formed will contain, on average, one EGF-bound EGFR dimer per disc and elute from gel filtration with a higher apparent molecular mass. The result of this experiment is shown in Figure 5b, where two major species elute from gel filtration. The dominant peak in the absorbance at 280 nm elutes with an apparent Stokes radius of 4.8 nm corresponding to the “empty” nanodiscs. The second, minor, peak in the absorbance at 280 nm elutes with a significantly higher Stokes radius of 8.5 nm. When the gel filtration fractions are assayed for tyrosine kinase activity, the peak in kinase activity coelutes with the minor peak of 8.5 nm Stokes radius (Figure 5b), suggesting that this species corresponds to EGFR-containing nanodiscs.
To determine the stoichiometry of EGFR molecules incorporated per disc under these assembly conditions, EGFR was reconstituted in nanodiscs formed with MSP1T2 in which the N-terminal His7 tag was removed from the membrane scaffold construct by TEV protease digestion, denoted MSP1T2(−); removal of the tags does not affect disc assembly (ref 41 and data not shown). Following disc assembly, EGFR-containing discs were separated from empty discs by Ni-NTA affinity chromatography using the C-terminal His6 tag in our recombinant EGFR constructs. While the majority of MSP1T2(−) does not bind Ni-NTA, SDS–PAGE and Coomassie staining indicate that EGFR and MSP1T2(−) coelute in eluate fractions with an EGFR: MSP1T2(−) mole ratio of approximately 0.9:1.1 (Figure 5c). Since there are 2 mol of MSP1T2(−) per disc, this result suggests that, on average, one EGF-bound EGFR dimer is incorporated per disc under these assembly conditions.
The structural and functional stability of EGF-bound Δ998-EGFR dimers reconstituted in phosphatidylcholine nanodiscs was assayed as a function of time incubated at 4 °C. As shown in Figure 6a, EGF/EGFR/MSP1T2 assembly mixtures exhibit nearly identical gel filtration chromatograms when fractionated immediately following disc assembly (t = 0) or following incubation at 4 °C for 48 h. In addition, over 80% of the initial kinase activity is retained following incubation at 4 °C for 24 h (Figure 6b); a similar stabilization of kinase activity compared to detergents is obtained when purified Δ998-EGFR is reconstituted in phosphatidylcholine liposomes in the presence of EGF (Figure 6c). As a further comparison, purified EGF-bound Δ998-EGFR dimers in 0.1% Triton X-100 have an initial specific activity of 120 ± 8 pmol of phosphate transferred min−1 (μM EGFR)−1 that decays with apparent single exponential kinetics to 27.5 ± 0.5% of the initial activity after 24 h at 4 °C (Figure 6d and data not shown). In contrast, after the 16 h disc assembly procedure, essentially all of the original specific activity that was present in 0.1% Triton X-100 is recovered when EGF-bound Δ998-EGFR dimers are reconstituted in phosphatidylcholine nanodiscs (135 ± 7 pmol/min/μM EGFR) and is maintained after an additional 24 h incubation at 4 °C (103.1 ± 0.3 pmol min−1 (μM EGFR)−1, Figure 6d). These results demonstrate that reconstitution of EGF-bound EGFR dimers in phosphatidylcholine nanodiscs or liposomes provides significantly increased functional stability compared to dodecyl maltoside or Triton X-100 detergent micelles.
Figure 6.
Stability of EGFR reconstituted in phospholipid bilayer nanodiscs and vesicles. (a) Purified WT-EGFR reconstituted in phospholipid bilayer nanodiscs in the presence of EGF was assayed for structural stability by gel filtration (solid line) immediately following disc assembly or (dashed line) following a 48 h incubation at 4 °C. The absorbance at 280 nm for each trace is shown as a relative value with respect to the peak in absorbance that elutes at 15.51 mL to account for different sample volumes injected on the gel filtration column. (b, c) The functional stability of Δ998-EGFR reconstituted in (b) phosphatidylcholine nanodiscs or (c) vesicles was assayed by monitoring tyrosine kinase activity as a function of time incubated at 4 °C following disc or vesicle assembly. (d) Specific activity of Δ998-EGFR reconstituted in 0.1% Triton X-100 or phosphatidylcholine nanodiscs measured after (black bars) 0 h or (gray bars) 24 h incubation at 4 °C. Note that 0 h for the samples reconstituted in nanodiscs is relative to once disc assembly has been completed.
DISCUSSION
In this report we present optimized protocols for the expression, purification, and reconstitution of intact EGFR. Through the expression of recombinant EGFR constructs in 293 cell culture and single step purification by affinity chromatography, we can obtain highly purified preparations of intact EGFR and EGFR mutants in sufficient quantities for biochemical and biophysical characterization. The material obtained by these methods is predominantly monomeric in dodecyl maltoside detergent micelles in the absence of EGF. Although primarily a monomer, there is a small equilibrium population of unligated dimeric material that retains catalytic activity toward an exogenous peptide substrate even in the absence of EGF. The addition of EGF shifts the equilibrium in favor of the ligated EGFR dimer with concomitant stimulation of kinase activity. Interestingly, unligated and ligated EGFR dimers have identical specific activities toward the RR1173 peptide substrate, suggesting that ligand does not stimulate the kinase activity but shifts the equilibrium toward the ligated dimer in our preparations.
Although our preparations of EGFR in Triton X-100 and other detergents clearly exhibit EGF-stimulated kinase activity, the level of stimulation does not likely represent the level of stimulation that would be achieved on the cell surface due to a potentially nonphysiological mixture of oligomeric states present in our cell lysates. Unligated dimers reported previously (1, 2) and herein do not necessarily correspond to physiological unligated dimers that have been debated to be populated on the surface of unstimulated cells (5). The unligated dimeric material reported here may be the result of overexpression of recombinant EGFR constructs in transfected cell lines, where the absence of endogenous regulatory mechanisms and the high concentrations of receptor may promote dimerization in the absence of ligand and increase the basal activity.
The EM class averages of our negatively stained detergent-soluble EGFR preparations represent the first molecular images of the overall EGFR structure and domain organization for intact receptor molecules. The correlation between regions in the class averages and extracellular domain crystal structures suggests that the extracellular domain of intact detergent-soluble EGFR adopts a similar structure to that observed in crystals. In addition, our EM class averages support a molecular model for the orientation of domains IV within the extracellular domain dimer, which has not been visualized in the crystal structures of the ligand-bound dimer (14, 15); a similar model has been proposed previously (17). In this model the C-termini of domains IV are juxtaposed against one another such that two polypeptide chains would be in close approach to one another as they enter the lipid bilayer. This model derived from the EM class averages is consistent with unpublished cross-linking results from our laboratory, in which cysteine residues introduced in the extracellular juxtamembrane region are capable of forming disulfide cross-links in the presence of EGF (C. Lu and T. A. Springer, unpublished results). Thus, the orientation of extracellular domain IV in the ligated dimer places spatial restraints on the transmembrane helices as they enter the plane of the membrane.
Although ligated EGFR appears to be stable as dimers and catalytically active when reconstituted in detergent micelles, the detergent environment does not appear to be sufficient for maintaining the kinase activity over prolonged incubations (>2 h in dodecyl maltoside, >8 h in Triton X-100) at 0 or 4 °C. We demonstrated that this is not due to structural instability of the ligated dimer or by a direct inhibitory effect of detergents on the kinase domain itself. Furthermore, addition of phospholipids to the purified detergent-soluble preparations does not appear to significantly stabilize the kinase activity.
As an alternative to detergent micelles, the kinase activity of purified EGF-bound EGFR dimers is significantly stabilized when reconstituted in a phospholipid bilayer environment. Ligated EGFR dimers reconstituted in phosphatidylcholine lipid nanodiscs are structurally and functionally stable over periods of 24−48 h when stored at 4 °C. Although the precise structural mechanism for the increased functional stability of EGFR kinase activity in a phospholipid bilayer is not known, we do not believe that the stabilizing effect is mediated through stabilization of the transmembrane domain itself. The introduction of a cysteine at position A623 near the extracellular face of the transmembrane domain results in EGF-induced disulfide cross-linked (and catalytically active) dimers on the surface of transfected cells (C. Lu and T. A. Springer, unpublished results). If proper transmembrane domain interactions are not maintained in the detergent micelle, one might propose that stabilizing these interactions through a disulfide cross-link may stabilize the kinase activity. However, the stability of purified A623C-EGFR disulfide-bonded dimers in dodecyl maltoside is very similar to that of the wild-type protein (L.-Z. Mi and T. A. Springer, unpublished results), suggesting that it is not destabilization of transmembrane domain interactions that contribute to loss of activity in detergent micelles.
Instead of providing structural stability for maintaining transmembrane domain interactions, the importance of a lipid bilayer for maintaining functional stability of EGFR in vitro is consistent with previously proposed models involving interactions between the intracellular juxtamembrane domain and the phospholipid bilayer (25, 26). In this case it is interactions of regions outside the transmembrane domain with the phospholipid headgroups that are proposed to be functionally important, and these interactions presumably cannot be recapitulated by the detergent micelles. Furthermore, since dodecylphosphocholine was found to be a very poor detergent for solubilizing and reconstituting functional EGFR in cell lysates, we can propose that it is not interactions with the phosphocholine headgroups per se but rather interactions with the headgroups in the context of the planar lipid bilayer that may be important for stabilizing EGFR kinase activity in vitro.
Supplementary Material
ACKNOWLEDGMENT
L.-Z.M., C.L., and T.A.S. express deep gratitude to John Kuriyan and Xuewu Zhang (University of California, Berkeley) for their support and intellectual contributions, which enabled this project to be initiated during a sabbatical in the Kuriyan laboratory in 2004−2005. We are grateful to Stephen G. Sligar, Timothy H. Bayburt, and Yelena V. Grinkova (University of Illinois at Urbana–Champaign) for providing purified MSP1T2 membrane scaffold protein and protocols for assembly of and reconstitution of membrane proteins in nanodiscs; Michael J. Eck and Cai-Hong Yun (Dana Farber Cancer Institute and Harvard Medical School) for providing purified EGFR kinase domain; Laura Harmacek (Immune Disease Institute) for assistance with cell culture; and Thomas Schürpf (Immune Disease Institute) for critical reading of the manuscript.
Footnotes
This work has been supported by grants from the National Institutes of Health and the Guggenheim Foundation awarded to T.A.S. and a postdoctoral fellowship from the American Cancer Society awarded to M.J.G. The molecular electron microscopy facility at Harvard Medical School was established with a generous donation from the Giovanni Armenise Harvard Center for Structural Biology and is maintained by funds from the National Institutes of Health awarded to Stephen C. Harrison.
Abbreviations: EGF, epidermal growth factor; EGFR, epidermal growth factor receptor; DDM, dodecyl maltoside; EM, electron microscopy; MSP, membrane scaffold protein.
SUPPORTING INFORMATION AVAILABLE
A figure of EGF-bound Δ998-EGFR particle class averages determined by negative stain electron microscopy. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Yarden Y, Schlessinger J. Self-phosphorylation of epidermal growth factor receptor: evidence for a model of intermolecular allosteric activation. Biochemistry. 1987;26:1434–1442. doi: 10.1021/bi00379a034. [DOI] [PubMed] [Google Scholar]
- 2.Yarden Y, Schlessinger J. Epidermal growth factor induces rapid, reversible aggregation of the purified epidermal growth factor receptor. Biochemistry. 1987;26:1443–1451. doi: 10.1021/bi00379a035. [DOI] [PubMed] [Google Scholar]
- 3.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 4.Yu X, Sharma KD, Takahashi T, Iwamoto R, Mekada E. Ligand-independent dimer formation of epidermal growth factor receptor (EGFR) is a step separable from ligand-induced EGFR signaling. Mol. Biol. Cell. 2002;13:2547–2557. doi: 10.1091/mbc.01-08-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clayton AH, Walker F, Orchard SG, Henderson C, Fuchs D, Rothacker J, Nice EC, Burgess AW. Ligand-induced dimer-tetramer transition during the activation of the cell surface epidermal growth factor receptor-A multidimensional microscopy analysis. J. Biol. Chem. 2005;280:30392–30399. doi: 10.1074/jbc.M504770200. [DOI] [PubMed] [Google Scholar]
- 6.Liu P, Sudhaharan T, Koh RM, Hwang LC, Ahmed S, Maruyama IN, Wohland T. Investigation of the dimerization of proteins from the epidermal growth factor receptor family by single wavelength fluorescence cross-correlation spectroscopy. Biophys. J. 2007;93:684–698. doi: 10.1529/biophysj.106.102087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat. Rev. Mol. Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 8.Singh AB, Harris RC. Autocrine, paracrine and juxtacrine signaling by EGFR ligands. Cell Signalling. 2005;17:1183–1193. doi: 10.1016/j.cellsig.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 9.Wiley HS. Trafficking of the ErbB receptors and its influence on signaling. Exp. Cell Res. 2003;284:78–88. doi: 10.1016/s0014-4827(03)00002-8. [DOI] [PubMed] [Google Scholar]
- 10.Holbro T, Hynes NE. ErbB receptors: directing key signaling networks throughout life. Annu. Rev. Pharmacol. Toxicol. 2004;44:195–217. doi: 10.1146/annurev.pharmtox.44.101802.121440. [DOI] [PubMed] [Google Scholar]
- 11.Hynes NE, Lane HA. ErbB receptors and cancer: the complexity of targeted inhibitors. Nat. Rev. Cancer. 2005;5:341–354. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 12.Arteaga CL. ErbB-targeted therapeutic approaches in human cancer. Exp. Cell Res. 2003;284:122–130. doi: 10.1016/s0014-4827(02)00104-0. [DOI] [PubMed] [Google Scholar]
- 13.Baselga J. Targeting tyrosine kinases in cancer: the second wave. Science. 2006;312:1175–1178. doi: 10.1126/science.1125951. [DOI] [PubMed] [Google Scholar]
- 14.Ogiso H, Ishitani R, Nureki O, Fukai S, Yamanaka M, Kim JH, Saito K, Sakamoto A, Inoue M, Shirouzu M, Yokoyama S. Crystal structure of the complex of human epidermal growth factor and receptor extracellular domains. Cell. 2002;110:775–787. doi: 10.1016/s0092-8674(02)00963-7. [DOI] [PubMed] [Google Scholar]
- 15.Garrett TP, McKern NM, Lou M, Elleman TC, Adams TE, Lovrecz GO, Zhu HJ, Walker F, Frenkel MJ, Hoyne PA, Jorissen RN, Nice EC, Burgess AW, Ward CW. Crystal structure of a truncated epidermal growth factor receptor extracellular domain bound to transforming growth factor R. Cell. 2002;110:763–773. doi: 10.1016/s0092-8674(02)00940-6. [DOI] [PubMed] [Google Scholar]
- 16.Cho HS, Leahy DJ. Structure of the extracellular region of HER3 reveals an interdomain tether. Science. 2002;297:1330–1333. doi: 10.1126/science.1074611. [DOI] [PubMed] [Google Scholar]
- 17.Ferguson KM, Berger MB, Mendrola JM, Cho HS, Leahy DJ, Lemmon MA. EGF activates its receptor by removing interactions that autoinhibit ectodomain dimerization. Mol. Cell. 2003;11:507–517. doi: 10.1016/s1097-2765(03)00047-9. [DOI] [PubMed] [Google Scholar]
- 18.Stamos J, Sliwkowski MX, Eigenbrot C. Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. J. Biol. Chem. 2002;277:46265–46272. doi: 10.1074/jbc.M207135200. [DOI] [PubMed] [Google Scholar]
- 19.Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 20.Hubbard SR. EGF receptor activation: push comes to shove. Cell. 2006;125:1029–1031. doi: 10.1016/j.cell.2006.05.028. [DOI] [PubMed] [Google Scholar]
- 21.Lemmon MA, Ferguson KM. A new twist in the transmembrane signaling tool-kit. Cell. 2007;130:213–215. doi: 10.1016/j.cell.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 22.Tanner KG, Kyte J. Dimerization of the extracellular domain of the receptor for epidermal growth factor containing the membrane-spanning segment in response to treatment with epidermal growth factor. J. Biol. Chem. 1999;274:35985–35990. doi: 10.1074/jbc.274.50.35985. [DOI] [PubMed] [Google Scholar]
- 23.Mendrola JM, Berger MB, King MC, Lemmon MA. The single transmembrane domains of ErbB receptors self-associate in cell membranes. J. Biol. Chem. 2002;277:4704–4712. doi: 10.1074/jbc.M108681200. [DOI] [PubMed] [Google Scholar]
- 24.Stanley AM, Fleming KG. The transmembrane domains of ErbB receptors do not dimerize strongly in micelles. J. Mol. Biol. 2005;347:759–772. doi: 10.1016/j.jmb.2005.01.059. [DOI] [PubMed] [Google Scholar]
- 25.Hunter T, Ling N, Cooper JA. Protein kinase C phosphorylation of the EGF receptor at a threonine residue close to the cytoplasmic face of the plasma membrane. Nature. 1984;311:480–483. doi: 10.1038/311480a0. [DOI] [PubMed] [Google Scholar]
- 26.McLaughlin S, Smith SO, Hayman MJ, Murray D. An electrostatic engine model for autoinhibition and activation of the epidermal growth factor receptor (EGFR/ErbB) family. J. Gen. Physiol. 2005;126:41–53. doi: 10.1085/jgp.200509274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thiel KW, Carpenter G. Epidermal growth factor receptor juxtamembrane region regulates allosteric tyrosine kinase activation. Proc. Natl. Acad. Sci. U.S.A. 2007;104:19238–19243. doi: 10.1073/pnas.0703854104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choowongkomon K, Carlin CR, Sonnichsen FD. A structural model for the membrane-bound form of the juxtamembrane domain of the epidermal growth factor receptor. J. Biol. Chem. 2005;280:24043–24052. doi: 10.1074/jbc.M502698200. [DOI] [PubMed] [Google Scholar]
- 29.Cohen S, Carpenter G, King L., Jr. Epidermal growth factor-receptor-protein kinase interactions. J. Biol. Chem. 1980;255:4834–4842. [PubMed] [Google Scholar]
- 30.Hock RA, Nexo E, Hollenberg MD. Solubilization and isolation of the human placenta receptor for epidermal growth factor-urogastrone. J. Biol. Chem. 1980;255:10737–10743. [PubMed] [Google Scholar]
- 31.Cohen S, Ushiro H, Stoscheck C, Chinkers M. A native 170,000 epidermal growth factor receptor-kinase complex from shed plasma membrane vesicles. J. Biol. Chem. 1982;257:1523–1531. [PubMed] [Google Scholar]
- 32.Yarden Y, Harari I, Schlessinger J. Purification of an active EGF receptor kinase with monoclonal antireceptor antibodies. J. Biol. Chem. 1985;260:315–319. [PubMed] [Google Scholar]
- 33.Weber W, Bertics PJ, Gill GN. Immunoaffinity purification of the epidermal growth factor receptor. Stoichiometry of binding and kinetics of self-phosphorylation. J. Biol. Chem. 1984;259:14631–14636. [PubMed] [Google Scholar]
- 34.Stearns DJ, Kurosawa S, Sims PJ, Esmon NL, Esmon CT. The interaction of a Ca2+-dependent monoclonal antibody with the protein C activation peptide region. Evidence for obligatory Ca2+ binding to both antigen and antibody. J. Biol. Chem. 1988;263:826–832. [PubMed] [Google Scholar]
- 35.Keefe AD, Wilson DS, Seelig B, Szostak JW. One-step purification of recombinant proteins using a nanomolaraffinity streptavidin-binding peptide, the SBP-Tag. Protein Expression Purif. 2001;23:440–446. doi: 10.1006/prep.2001.1515. [DOI] [PubMed] [Google Scholar]
- 36.Heinzel SS, Krysan PJ, Calos MP, DuBridge RB. Use of simian virus 40 replication to amplify Epstein-Barr virus shuttle vectors in human cells. J. Virol. 1988;62:3738–3746. doi: 10.1128/jvi.62.10.3738-3746.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reeves PJ, Callewaert N, Contreras R, Khorana HG. Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc. Natl. Acad. Sci. U.S.A. 2002;99:13419–13424. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Casnellie JE. Assay of protein kinases using peptides with basic residues for phosphocellulose binding. Methods Enzymol. 1991;200:115–120. doi: 10.1016/0076-6879(91)00133-h. [DOI] [PubMed] [Google Scholar]
- 39.Nishida N, Xie C, Shimaoka M, Cheng Y, Walz T, Springer TA. Activation of leukocyte b2 integrins by conversion from bent to extended conformations. Immunity. 2006;25:583–594. doi: 10.1016/j.immuni.2006.07.016. [DOI] [PubMed] [Google Scholar]
- 40.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 41.Denisov IG, Grinkova YV, Lazarides AA, Sligar SG. Directed self-assembly of monodisperse phospholipid bilayer Nanodiscs with controlled size. J. Am. Chem. Soc. 2004;126:3477–3487. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- 42.Bayburt TH, Grinkova YV, Sligar SG. Self assembly of discoidal phospholipid bilayer nanoparticles with membrane scaffold proteins. Nano Lett. 2002;2:853–856. [Google Scholar]
- 43.Rezaie AR, Esmon CT. The function of calcium in protein C activation by thrombin and the thrombin-thrombomodulin complex can be distinguished by mutational analysis of protein C derivatives. J. Biol. Chem. 1992;267:26104–26109. [PubMed] [Google Scholar]
- 44.Kawate T, Gouaux E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure. 2006;14:673–681. doi: 10.1016/j.str.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 45.Bayburt TH, Sligar SG. Self-assembly of single integral membrane proteins into soluble nanoscale phospholipid bilayers. Protein Sci. 2003;12:2476–2481. doi: 10.1110/ps.03267503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boldog T, Grimme S, Li M, Sligar SG, Hazelbauer GL. Nanodiscs separate chemoreceptor oligomeric states and reveal their signaling properties. Proc. Natl. Acad. Sci. U.S.A. 2006;103:11509–11514. doi: 10.1073/pnas.0604988103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bayburt TH, Grinkova YV, Sligar SG. Assembly of single bacteriorhodopsin trimers in bilayer nanodiscs. Arch. Biochem. Biophys. 2006;450:215–222. doi: 10.1016/j.abb.2006.03.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.