Abstract
The regulation of gene transcription is critical for the proper development and growth of an organism. The transcription of protein-coding genes initiates at the RNA polymerase II core promoter, which is a diverse module that can be controlled by many different elements such as the TATA box and downstream core promoter element (DPE). To understand the basis for core promoter diversity, we explored potential biological functions of the DPE. We found that nearly all of the Drosophila homeotic (Hox) gene promoters, which lack TATA-box elements, contain functionally important DPE motifs that are conserved from Drosophila melanogaster to Drosophila virilis. We then discovered that Caudal, a sequence-specific transcription factor and key regulator of the Hox gene network, activates transcription with a distinct preference for the DPE relative to the TATA box. The specificity of Caudal activation for the DPE is particularly striking when a BREu core promoter motif is associated with the TATA box. These findings show that Caudal is a DPE-specific activator and exemplify how core promoter diversity can be used to establish complex regulatory networks.
Keywords: Core promoter, RNA polymerase II transcription, DPE, Hox, Caudal, BREu
Sequence-specific DNA-binding transcription factors play a key role in the control of many biological phenomena, such as in the development of the body plan of Drosophila melanogaster (Nüsslein-Volhard and Wieschaus 1980; Akam 1987; Gehring 1987; Pearson et al. 2005; Lappin et al. 2006; Lemons and McGinnis 2006; Ochoa-Espinosa and Small 2006). A simple view of this process is as follows: Maternal genes such as bicoid and caudal establish the anteroposterior axis of the embryo; gap genes such as hunchback, giant, Krüppel, and knirps subdivide the embryo into broad regions; pair-rule genes such as fushi tarazu specify the number of segments; segment polarity genes such as engrailed determine the anteroposterior polarity of each segment; and homeotic (Hox) genes such as Antennapedia specify the identity of the segments. Remarkably, the vast majority of these genes encode sequence-specific DNA-binding transcription factors that bind to promoter and enhancer elements and regulate transcription by RNA polymerase II. It remains important to investigate, however, whether these developmental regulators act with specificity not only at promoters and enhancers, but also at the site of transcription initiation, the core promoter.
The RNA polymerase II core promoter is a diverse transcriptional module that directs the initiation of transcription (for review, see Smale 2001; Smale and Kadonaga 2003; Juven-Gershon et al. 2008). Core promoters may contain one or more core promoter motifs, which include DNA sequence elements such as the TATA box, TFIIB recognition elements (BREu and BREd), initiator (Inr), motif 10 element (MTE), and the downstream core promoter element (DPE). There are no universal core promoter motifs.
The existence of many different types of core promoters suggests that their functions extend beyond the specification of transcription initiation. Consistent with this notion, it has been found that core promoter motifs can contribute to enhancer-promoter specificity (for review, see Smale 2001; Butler and Kadonaga 2002). For instance, the Drosophila AE1 and IAB5 enhancers exhibit a preference for activation of the TATA-containing even-skipped core promoter relative to the TATA-less and DPE-containing white core promoter (Ohtsuki et al. 1998). In addition, enhancer trapping studies in Drosophila led to the discovery of DPE-specific transcriptional enhancers (Butler and Kadonaga 2001). Thus, core promoter diversity provides an additional dimension to the constellation of mechanisms by which genes are regulated. At the level of basal transcription, it has been found that factors such as NC2, Mot1, and TBP affect DPE-dependent versus TATA-dependent transcription (for example, see Hsu et al. 2008). However, for regulated transcription, the enhancer-binding factors that mediate core promoter specificity have not yet been identified.
To identify biological functions of core promoter motifs, we explored the potential role of the DPE in transcriptional regulatory networks. The DPE was originally discovered as a TFIID recognition site that is located downstream from the Inr element (Burke and Kadonaga 1996). The DPE is conserved from Drosophila to humans (Burke and Kadonaga 1997). TFIID binds cooperatively to the Inr and DPE, which is located precisely at +28 to +32 relative to A + 1 in the Inr consensus sequence. The correct spacing between the Inr and DPE is critical for optimal transcriptional activity (Kutach and Kadonaga 2000). Current evidence suggests that the DPE is present in ∼2.1%–22% of Drosophila core promoters (Ohler et al. 2002; FitzGerald et al. 2006; Gershenzon et al. 2006).
In this study, we identify a critical role of the DPE motif in the transcription of genes that are involved in the early embryonic development of Drosophila. We further discovered that Caudal, a master regulator of the Hox genes, is a DPE-specific enhancer-binding factor. These findings reveal the use of the DPE as a component of the regulatory circuitry of an important biological process and uncover a new mechanism with which sophisticated patterns of gene expression are established to achieve organismal complexity (for example, see Levine and Tjian 2003).
Results
The core promoters of nearly all Drosophila Hox genes contain conserved DPE motifs
To uncover a specific biological role of the DPE, we looked for patterns or related themes in Drosophila genes whose core promoters have been shown to possess or are believed to contain DPE motifs. This analysis led us to the genes that are involved in the early development of the embryo. In the course of our previous studies, Antennapedia (Antp; downstream P2 promoter), engrailed (en), and caudal (cad; zygotic promoter) were among the genes whose promoters had been found to contain functionally important DPE motifs (Burke and Kadonaga 1996; Kutach and Kadonaga 2000), but a correlation between the DPE and developmental genes was not yet apparent. However, with the current Drosophila genome data (http://www.fruitfly.org; http://flybase.net; http://genome.ucsc.edu/cgi-bin/hgGateway), we found that the core promoters of many developmentally important genes, particularly the Hox genes, appear to contain DPE motifs. The Hox genes encode sequence-specific transcription factors that bind to DNA via their conserved homeodomain and are key regulators of the development of the embryonic body plan. The promoters of the Hox genes are TATA-less; hence, the DPE motifs could support Hox gene transcription in the absence of the TATA box.
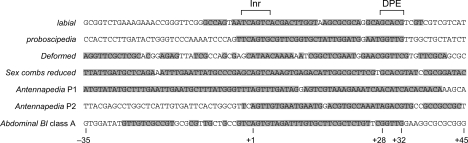
To assess the potential biological significance of the DPE motifs in the Hox genes, we examined the conservation of the DPE sequences in eight different Drosophila species. The Inr and DPE consensus sequences as well as the Inr-to-DPE spacing are conserved across species in the labial (lab), proboscipedia (pb), Deformed (Dfd), Sex combs reduced (Scr), Antennapedia P1 (Antp P1), Antennapedia P2 (Antp P2), and Abdominal-B (Abd-B) core promoters (Supplemental Fig. 1; data not shown). The conservation of these core promoter sequences between D. melanogaster and Drosophila virilis, which are separated by an evolutionary period of ∼40–60 million years, is shown in Figure 1. Only two Drosophila Hox genes, Abdominal-A (Abd-A) and Ultrabithorax (Ubx), do not contain DPE consensus motifs. However, Abd-A and Ubx have arisen more recently than the other Hox genes (Grenier et al. 1997; Brooke et al. 1998); hence, all of the more ancient Hox genes contain DPE consensus sequences.
Figure 1.
The core promoters of Drosophila Hox genes contain a conserved DPE motif. The core promoter sequences of the indicated D. melanogaster Hox genes are shown. We initially analyzed the sequence conservation of the promoters in eight different Drosophila species by using VISTA tools and CLUSTALW (Supplemental Fig. 1; data not shown). Based on this sequence alignment, positions that are identical between D. melanogaster and D. virilis (in terms of sequence and spacing relative to the A + 1 in the Inr) are shaded. D. melanogaster and D. virilis are estimated to be separated by an evolutionary period of ∼40–60 million years.
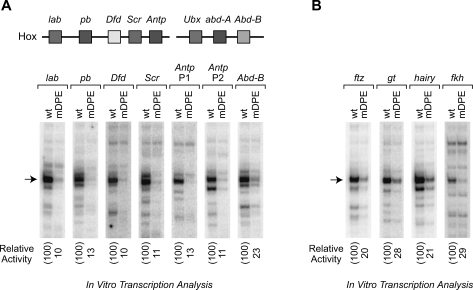
We then tested whether the DPE consensus sequences in the Hox genes are functionally important for transcription. To this end, we constructed and analyzed wild-type and mutant DPE (mDPE) versions of the Hox gene core promoters with putative DPE sequences. These experiments showed that all of the D. melanogaster Hox genes, except for Abd-A and Ubx, contain functionally important DPE motifs (Fig. 2A; Supplemental Fig. 2).
Figure 2.
The DPE is present in many genes that regulate the development of the Drosophila body plan. (A) Most of the Hox genes contain DPE-dependent core promoters. A schematic diagram of the Hox gene cluster is shown at the top. (lab) labial; (pb) proboscipedia; (Dfd) Deformed; (Scr) Sex combs reduced; (Antp P1) Antennapedia upstream promoter; (Antp P2) Antennapedia downstream promoter; (Ubx) Ultrabithorax; (abd-A) abdominal-A; (Abd-B) Abdominal-B. The wild-type (wt) and mutant DPE (mDPE) versions of the indicated core promoters (from −10 to +40 relative to the A + 1 start site) were subjected to in vitro transcription analysis with a Drosophila embryo nuclear extract. The resulting transcripts were detected by primer extension-reverse transcription analysis. (B) The core promoters of Caudal target genes contain functional DPE motifs. (ftz) fushi tarazu; (gt) giant; (h) hairy; (fkh) forkhead. In vitro transcription analysis was performed as in A.
Caudal, a key regulator of the Hox gene network, activates DPE-dependent promoters and some but not all TATA-dependent promoters
The prevalence of the DPE in the core promoters of the Hox genes suggests that some of the transcriptional regulatory factors that activate Hox gene expression are DPE-specific activators. To investigate this hypothesis, we examined the protein encoded by the caudal (cad) gene. cad is a paralog of the Hox genes and is thus termed a ParaHox gene (Brooke et al. 1998). cad was first identified in D. melanogaster and is expressed both maternally and zygotically (Mlodzik et al. 1985; Mlodzik and Gehring 1987). It is required for the specification of the posterior embryo and patterning of the anteroposterior axis (Levine et al. 1985; Mlodzik et al. 1985; Hoey et al. 1986; Macdonald and Struhl 1986; Mlodzik and Gehring 1987). Caudal protein contains a homeodomain and is a sequence-specific DNA-binding activator. In vertebrates, the Caudal proteins (Cdx1, Cdx2, and Cdx4) are master regulators of Hox gene expression (for example, see Charité et al. 1998; Davidson et al. 2003; Lohnes 2003; Davidson and Zon 2006).
In D. melanogaster, Caudal has been observed to regulate the developmentally important genes fushi tarazu (ftz) (Dearolf et al. 1989), giant (gt) (Rivera-Pomar et al. 1995; Schulz and Tautz 1995; Rivera-Pomar and Jäckle 1996), hairy (h) (Rivera-Pomar et al. 1995; Rivera-Pomar and Jäckle 1996), and forkhead (fkh) (Wu and Lengyel 1998). Notably, the core promoters of these Caudal target genes appear to contain DPE motifs. We constructed and analyzed the wild-type and mutant DPE versions of the ftz, gt, h, and fkh core promoters and found that they all contain functional DPE motifs (Fig. 2B). These findings suggest that Caudal might be a DPE-specific activator.
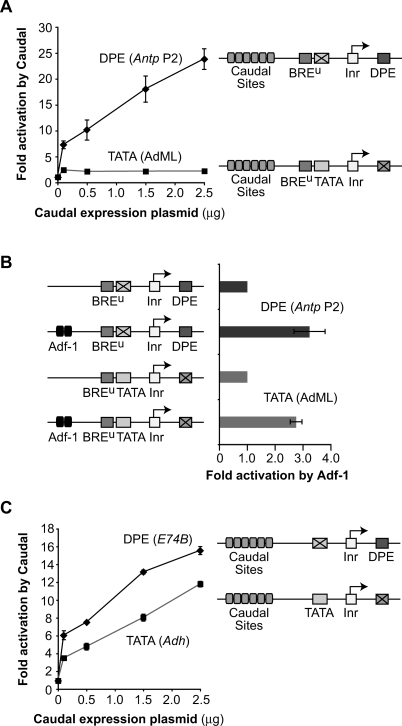
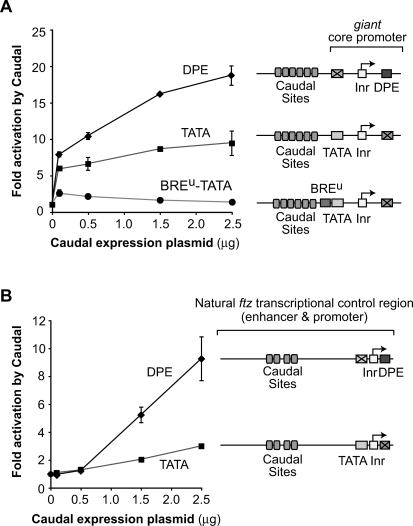
To test whether Caudal selectively activates transcription from DPE-dependent core promoters, we developed an assay system (Fig. 3A) that involves cotransfection of a Caudal expression plasmid along with a DPE- or TATA-dependent reporter gene in D. melanogaster Schneider (S2) cells, which contain negligible levels of cad transcripts (data not shown). The DPE and TATA reporter constructs contain well-characterized TATA and DPE motifs along with six Caudal-binding sites from the ftz enhancer (Dearolf et al. 1989) and are identical except for the sequences at the DPE and TATA regions. These experiments revealed that Caudal strongly activates transcription with the Antp P2 DPE, but only weakly activates transcription with the adenovirus major late (AdML) TATA box (Fig. 3A).
Figure 3.
Caudal, a conserved master regulator of the Hox gene network, is a core-promoter-specific activator. (A) Caudal activates transcription with the Antp P2 DPE motif but not with the AdML TATA element. The two reporter constructs are identical except for the TATA and DPE sequences. Drosophila S2 cells were transfected with firefly luciferase reporter constructs as well as a Caudal expression plasmid, where indicated. To normalize for transfection efficiency, cells were cotransfected with a Pol III-Renilla luciferase control plasmid and assayed for dual luciferase activity. Error bars represent the SEM. (B) The Adh distal enhancer activates transcription with both the AdML TATA box and the Antp P2 DPE motif. The four reporter constructs are identical except for the presence or absence of the TATA, DPE, and Adh distal enhancer, as indicated. The Adh distal enhancer contains two binding sites for the Adf-1 transcription factor, which is present in S2 cells. Transfection assays were performed with the indicated constructs as in A. Error bars represent the SEM. (C) Caudal activates transcription with the E74B DPE motif and, to a lesser extent, the Adh proximal TATA element. Transfection assays were performed with the indicated constructs as in A. Error bars represent the SEM.
The differential effect of Caudal on the two reporter genes is not due to differences in core promoter strength (Supplemental Fig. 3). We also observed that strong activation by Caudal is dependent on the presence of Caudal-binding sites in the reporter gene (Supplemental Fig. 4). In addition, the inability of Caudal to activate the TATA-dependent construct is not due to a defect in the ability of the promoter to respond to activators, because reporter constructs with the Antp P2 DPE or the AdML TATA box are both activated by the Alcohol dehydrogenase (Adh) distal enhancer (Fig. 3B).
To investigate the generality of Caudal activation of DPE-dependent core promoters, we constructed and analyzed a different set of DPE- and TATA-dependent reporter genes with the E74B DPE and the Adh proximal promoter TATA box. We found that Caudal activates transcription not only with the E74B DPE motif, but also with the Adh TATA box (Fig. 3C). Given the results with the AdML TATA box (Fig. 3A), the activation of the Adh TATA-dependent reporter by Caudal was surprising. We therefore tested different combinations of the Adh and AdML TATA boxes and the Antp P2 and E74B DPE motifs and found that Caudal activates transcription with either DPE motif as well as with the Adh TATA box, but not with the AdML TATA box (Fig. 3; Supplemental Fig. 5). Therefore, Caudal activates transcription from DPE-dependent promoters and some but not all TATA-dependent promoters.
The BREu core promoter motif suppresses the ability of Caudal to activate TATA-dependent promoters
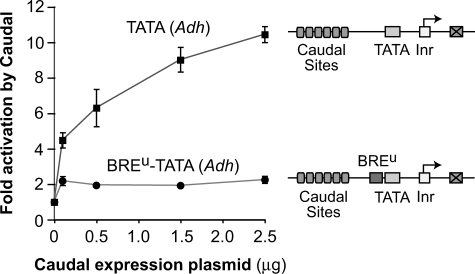
To determine the basis for Caudal activation with the Adh TATA but not the AdML TATA, we examined the promoter sequences and observed that the AdML TATA is flanked by BREu and BREd motifs, whereas the Adh TATA region lacks both BRE motifs. BRE sequences flank a subset of TATA-box elements and are sites of interaction with TFIIB (Lagrange et al. 1998; Deng and Roberts 2005). Depending on the context, the BRE motifs can have a positive or negative influence upon transcription (Deng and Roberts 2007).
We tested whether the addition of BREu or BREd sequences to the Adh TATA box affects the ability of Caudal to activate transcription. The addition of a BREu motif to the Adh TATA box renders the TATA box unresponsive to activation by Caudal (Fig. 4). In contrast, the presence or absence of the BREd (with or without a BREu) had little effect on Caudal activation (data not shown). These experiments show that transcriptional activation by Caudal is suppressed by the presence of a BREu motif upstream of the TATA box. This novel activity of the BREu core promoter motif may be an additional means of preventing undesired gene activation by Caudal.
Figure 4.
Transcriptional activation by Caudal is suppressed by the presence of a BREu motif upstream of the TATA box. The two reporter constructs are identical except for the absence or presence of the AdML BREu motif immediately upstream of the TATA box, and were analyzed as in Figure 3. Error bars represent the SEM.
Caudal is a DPE-specific activator with the natural ftz enhancer-promoter region
Next, we sought to characterize the ability of Caudal to activate transcription with core promoters from natural Caudal target genes instead of hybrid core promoters. We first analyzed the gt core promoter. gt is a gap gene that is zygotically expressed in the early embryo and is involved in the initial steps in the segmentation of the embryo (Rivera-Pomar et al. 1995; Schulz and Tautz 1995; Rivera-Pomar and Jäckle 1996). The gt core promoter contains TATA and DPE motifs and is activated by Caudal in S2 cells (Supplemental Fig. 6). To determine the relative contribution of the DPE versus TATA motifs to the activation of transcription by Caudal, we compared a DPE-dependent version of the gt promoter (which has a mutant TATA box) with a TATA-dependent version of the promoter (which has a mutant DPE). These experiments show that Caudal preferentially activates transcription through the DPE in the gt core promoter (Fig. 5A). We also found, consistent with the results in Figure 4, that the selectivity of Caudal for activation of DPE- relative to TATA-dependent promoters is further enhanced by the presence of a BREu motif upstream of the TATA box (Fig. 5A). In contrast, the BREu motif does not affect the ability of Caudal to activate transcription with the DPE motif (Supplemental Fig. 6). As with the hybrid promoter constructs (Supplemental Fig. 3), the differential ability of Caudal to activate the reporter genes is not due to differences in core promoter strength (Supplemental Fig. 6).
Figure 5.
Caudal is a DPE-specific transcriptional activator. (A) Caudal preferentially activates transcription through the DPE in the gt core promoter. The reporter constructs contain six Caudal-binding sites upstream of the gt core promoter, which has both DPE and TATA motifs but lacks a BREu. The constructs are identical except for the mutation of the TATA and DPE motifs and the addition of the AdML BREu, where indicated. The transfection experiments were performed as in Figure 3. Error bars represent the SEM. (B) Caudal primarily activates through the DPE in the natural ftz transcriptional control region. The ftz core promoter contains both DPE and TATA motifs. The reporter constructs contain ftz enhancer and promoter sequences from −988 to +40 relative to the +1 start site, and are identical except for mutation of the DPE or TATA, as depicted. The transfection experiments were performed as in Figure 3. Error bars represent the SEM.
Lastly, we examined the ability of Caudal to activate transcription with the natural enhancer and promoter region of the ftz gene, which is a direct target of Caudal with known binding sites (Dearolf et al. 1989). ftz is a pair-rule gene that is involved in the specification of the number of segments in the embryo. We analyzed the ftz enhancer and promoter region to characterize Caudal activation in a natural context, as it is possible that Caudal activation with six tandem binding sites upstream of the TATA box (as in Figs. 3, 4, 5A) is more potent than that in a typical Caudal response gene. The ftz core promoter contains both TATA and DPE motifs that are functional in vivo (Supplemental Fig. 7). To determine the relative contributions of the DPE versus the TATA box to Caudal activation, we compared the properties of DPE-dependent (with mutant TATA) versus TATA-dependent (with mutant DPE) versions of the ftz transcriptional control region from −988 to +40 relative to the RNA start site. These studies reveal that Caudal-mediated activation of the ftz gene occurs primarily via the DPE motif in the core promoter (Fig. 5B). Therefore, in the context of a natural transcriptional control region, Caudal is a DPE-specific transcriptional activator.
Caudal activates the TATA-less, DPE-dependent Antp P2 and Scr promoters
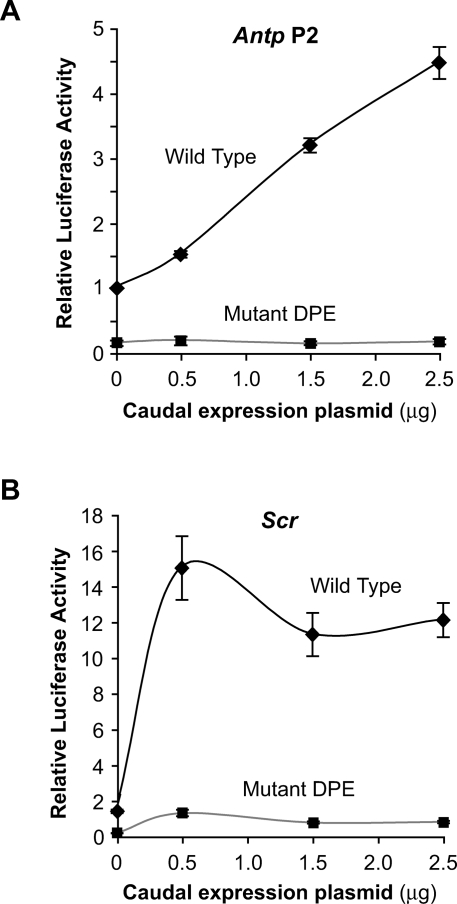
We observed that most of the Drosophila Hox genes contain DPE motifs and that Caudal is a DPE-specific activator. We therefore tested whether Caudal activates transcription from DPE-containing Hox genes. To this end, we isolated Antp P2 and Scr genomic DNA fragments that contain at least 3 kb of sequence upstream of the A + 1 site in the Inr. Both the Antp P2 and Scr core promoters contain DPE and Inr motifs and lack TATA boxes. To determine the effect of the DPE on transcription from these promoters, we made a parallel set of constructs in which the DPE sequences are mutated. Transfection analysis of the wild-type and mutant DPE versions of the Antp P2 and Scr promoters revealed that both promoters are activated by Caudal and are strongly dependent on the DPE motif in cultured cells (Fig. 6). Thus, the DPE is a critical component in Caudal-mediated activation of the natural Antp P2 and Scr promoters.
Figure 6.
The natural Antp P2 and Scr promoters are activated by Caudal and are dependent on the DPE core promoter motif in S2 cells. (A) Antp P2 promoter. Constructs containing the wild-type and mutant versions of the Antp P2 promoter region (from −3033 to +61 relative to A + 1 in the Inr) were analyzed as in Figure 3. The activities are reported relative to the wild-type Antp P2 promoter in the absence of cotransfected Caudal expression plasmid, which was defined to be 1. Error bars represent the SEM. (B) Scr promoter. Constructs containing the wild-type and mutant versions of the Scr promoter region (from −3103 to +110 relative to A + 1 in the Inr) were analyzed as in Figure 3. The activities are reported relative to the wild-type Antp P2 promoter in the absence of cotransfected Caudal expression plasmid, which was defined to be 1. Error bars represent the SEM.
Discussion
The DPE is a transcriptional element shared by most Hox genes
In this study, we found that the DPE is used extensively in the network of genes that are involved in the development of the early Drosophila embryo. Nearly all of the Drosophila Hox gene promoters, which have been known to be TATA-less, contain functionally essential DPE motifs that are conserved from D. melanogaster to D. virilis. The two Hox genes lacking DPE motifs are also the most recent Hox genes from an evolutionary standpoint. Thus, the DPE is a critical yet previously unrecognized component of the Hox genes.
Caudal is a DPE-specific enhancer-binding factor
The DPE is not only in the Hox genes, but is also present in ftz, gt, h, fkh, cad (zygotic promoter), and en, which are involved in early embryonic development. This finding suggests that the DPE is used broadly throughout the network of genes that mediate the development of the embryo. We tested this hypothesis by analyzing the transcriptional properties of Caudal, a ParaHox protein and sequence-specific DNA-binding factor that regulates ftz, gt, h, and fkh. These studies revealed that Caudal is a DPE-specific activator. The preference of Caudal for activating transcription from DPE- versus TATA-dependent core promoters is seen most distinctly either with the natural ftz enhancer-promoter region (Fig. 5B) or with a core promoter containing a BREu motif associated with the TATA box (Figs. 3A, 5A). We also examined the effect of Caudal on transcription of two Hox genes, Antp and Scr, and found that Caudal activates the TATA-less, DPE-dependent Antp P2 and Scr promoters (Fig. 6). These findings thus provide a direct link between Caudal, a DPE-specific activator, and the DPE-containing Hox genes.
The discovery that Caudal is a DPE-specific activator provides new insight into the basic mechanisms of transcriptional regulation. Previous enhancer-trapping experiments have shown that there are enhancers that activate DPE-dependent promoters but not TATA-dependent promoters (Butler and Kadonaga 2001); however, neither the cis-acting elements nor the trans-acting factors that are responsible for the DPE-specific activation had been identified. Therefore, these studies demonstrate the existence of a DPE-specific enhancer-binding factor. Moreover, it is likely that other core-promoter-specific enhancer-binding factors, such as TATA-specific activators, will be discovered.
A novel activity of the BREu core promoter motif
These experiments uncovered a novel activity of the BREu core promoter motif. The BREu is a 5′ extension of the TATA box that is bound by the TFIIB basal/general transcription factor (Lagrange et al. 1998). Depending on the context, the BREu has been found to have either a positive or a negative effect on transcription (Lagrange et al. 1998; Evans et al. 2001). In this study, we found that the BREu motif has little effect on basal/unactivated transcription, but potently suppresses the ability of Caudal to activate transcription via the TATA box. In contrast, the BREu in its normal upstream location has no effect on Caudal-mediated activation via the DPE (Supplemental Fig. 6). These findings indicate that the BREu can contribute to core-promoter-element-mediated transcriptional regulation. Hence, there is a positive linkage between Caudal and the DPE as well as a negative interaction between Caudal and the BREu-TATA element. The combination of both positive (DPE) and negative (BREu-TATA) interactions yields maximal specificity of Caudal function.
The use of core promoter motif specificity in transcriptional circuits
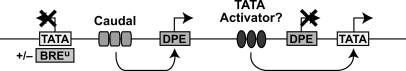
The new findings lead to the model that transcriptional regulation involves the combined action of sequence motifs in both the core promoter and the enhancer. The ability of Caudal to discriminate among DPE, TATA, and BREu-TATA motifs regulates the flow of information from the enhancer-bound activator to the core promoter—the site of transcription initiation. In this manner, core promoter elements can be viewed as a component of transcriptional circuits (Fig. 7). In these transcriptional circuits, connections between enhancers and core promoters are established and modulated according to the properties of the activators and the sequence motifs in the core promoter. Thus, the discovery that Caudal is a core-promoter-specific activator reveals a new strategy with which complex transcriptional networks can be established. Hence, in a broader sense, these findings show how diversity in core promoter structure can contribute to organismal diversity.
Figure 7.
A model for DPE-specific activation of transcription by Caudal. This model depicts a hypothetical segment of a regulatory network that comprises specific connections between activators and core promoter motifs. Caudal is a DPE-specific activator. TATA-specific activators might also exist. The ability of Caudal to function with a TATA box is further reduced by the presence of a BREu motif, which is located immediately upstream of a subset of TATA elements.
Materials and methods
Sequence conservation analysis
Sequence conservation analysis was performed using VISTA tools and CLUSTALW.
In vitro transcription assays
Double-stranded oligonucleotides comprising sequences from −10 to +40 of the core promoters (relative to A + 1 in the Inr) were inserted into the PstI and XbaI sites of pUC119. Mutation of the DPE in the core promoters was identical to that used previously (Lim et al. 2004), where the mutant DPE has CATA at +30 to +33 relative to A + 1. In vitro transcription reactions were carried out as described previously (Wampler et al. 1990) by using 250 ng of supercoiled DNA templates with Drosophila high salt nuclear extracts (Soeller et al. 1988). The resulting transcripts were subjected to primer extension analysis with M13 reverse sequencing primer (AGCGGATAACAATTTCA CACAGGA). Quantitation of reverse transcription products was carried out with a PhosphorImager (Molecular Dynamics). All experiments were carried out a minimum of three independent times to ensure reproducibility of the data.
Transient transfection and reporter gene assays
Drosophila Schneider S2 adherent cells were cultured in Shields & Sang M3 media (Sigma) prepared with yeast extract (Sigma) and bactopeptone (Difco) that was supplemented with 10% heat-inactivated FBS. Cells were transfected in 24-well plates by using the Transfectol reagent (Gene Choice). For dual luciferase assays, cells were plated at 0.6 × 106 cells per each well of a 24-well plate 1 d prior to transfection. Cells were transfected with the indicated amounts of a Caudal expression vector that was supplemented, where necessary, with pAC control expression vector to give a total of 2.5 μg of DNA of expression vector. In addition, the firefly luciferase reporter constructs (162 ng) were cotransfected with the Pol III-Renilla luciferase reporter (50 ng) (obtained from N. Perrimon, Harvard Medical School). Cells were harvested 20–26 h post-transfection and assayed for dual luciferase activities, as specified by the manufacturer (Promega). To correct for transfection efficiency, the firefly luciferase activity of each sample was normalized to the corresponding Renilla luciferase activity. Each transfection was performed in duplicate. The graphs represent an average of two to three independent experiments.
Acknowledgments
We thank W. McGinnis, T. Yusufzai, J. Theisen, B. Rattner, H. Ishii, D. Urwin, S. Torigoe, and S. Pitak for critical reading of the manuscript. We thank F. Furnari and W. Cavenee (Ludwig Institute for Cancer Research; University of California at San Diego) for the use of their luminometer; T. Yusufzai, D. Urwin, and W. McGinnis for helpful advice; and Norbert Perrimon (Harvard Medical School) for the generous gift of the Renilla luciferase control plasmid. This work was supported by a grant from the NIH to J.T.K. (GM041249).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1698108.
References
- Akam M. The molecular basis for metameric pattern in the Drosophila embryo. Development. 1987;101:1–22. [PubMed] [Google Scholar]
- Brooke N.M., Garcia-Fernàndez J., Holland P.W. The ParaHox gene cluster is an evolutionary sister of the Hox gene cluster. Nature. 1998;392:920–922. doi: 10.1038/31933. [DOI] [PubMed] [Google Scholar]
- Burke T.W., Kadonaga J.T. Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes & Dev. 1996;10:711–724. doi: 10.1101/gad.10.6.711. [DOI] [PubMed] [Google Scholar]
- Burke T.W., Kadonaga J.T. The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes & Dev. 1997;11:3020–3031. doi: 10.1101/gad.11.22.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler J.E.F., Kadonaga J.T. Enhancer-promoter specificity mediated by DPE or TATA core promoter motifs. Genes & Dev. 2001;15:2515–2519. doi: 10.1101/gad.924301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler J.E.F., Kadonaga J.T. The RNA polymerase II core promoter: A key component in the regulation of gene expression. Genes & Dev. 2002;16:2583–2592. doi: 10.1101/gad.1026202. [DOI] [PubMed] [Google Scholar]
- Charité J., de Graaff W., Consten D., Reijnen M.J., Korving J., Deschamps J. Transducing positional information to the Hox genes: Critical interaction of cdx gene products with position-sensitive regulatory elements. Development. 1998;125:4349–4358. doi: 10.1242/dev.125.22.4349. [DOI] [PubMed] [Google Scholar]
- Davidson A.J., Zon L.I. The caudal-related homeobox genes cdx1a and cdx4 act redundantly to regulate hox gene expression and the formation of putative hematopoietic stem cells during zebrafish embryogenesis. Dev. Biol. 2006;292:506–518. doi: 10.1016/j.ydbio.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Davidson A.J., Ernst P., Wang Y., Dekens M.P., Kingsley P.D., Palis J., Korsmeyer S.J., Daley G.Q., Zon L.I. cdx4 mutants fail to specify blood progenitors and can be rescued by multiple hox genes. Nature. 2003;425:300–306. doi: 10.1038/nature01973. [DOI] [PubMed] [Google Scholar]
- Dearolf C.R., Topol J., Parker C.S. The caudal gene product is a direct activator of fushi tarazu transcription during Drosophila embryogenesis. Nature. 1989;341:340–343. doi: 10.1038/341340a0. [DOI] [PubMed] [Google Scholar]
- Deng W., Roberts S.G. A core promoter element downstream of the TATA box that is recognized by TFIIB. Genes & Dev. 2005;19:2418–2423. doi: 10.1101/gad.342405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W., Roberts S.G. TFIIB and the regulation of transcription by RNA polymerase II. Chromosoma. 2007;116:417–429. doi: 10.1007/s00412-007-0113-9. [DOI] [PubMed] [Google Scholar]
- Evans R., Fairley J.A., Roberts S.G. Activator-mediated disruption of sequence-specific DNA contacts by the general transcription factor TFIIB. Genes & Dev. 2001;15:2945–2949. doi: 10.1101/gad.206901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FitzGerald P.C., Sturgill D., Shyakhtenko A., Oliver B., Vinson C. Comparative genomics of Drosophila and human core promoters. Genome Biol. 2006;7:R53. doi: 10.1186/gb-2006-7-7-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring W.J. Homeo boxes in the study of development. Science. 1987;236:1245–1252. doi: 10.1126/science.2884726. [DOI] [PubMed] [Google Scholar]
- Gershenzon N.I., Trifonov E.N., Ioshikhes I.P. The features of Drosophila core promoters revealed by statistical analysis. BMC Genomics. 2006;7:161. doi: 10.1186/1471-2164-7-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenier J.K., Garber T.L., Warren R., Whitington P.M., Carroll S. Evolution of the entire arthropod Hox gene set predated the origin and radiation of the onychophoran/arthropod clade. Curr. Biol. 1997;7:547–553. doi: 10.1016/s0960-9822(06)00253-3. [DOI] [PubMed] [Google Scholar]
- Hoey T., Doyle H.J., Harding K., Wedeen C., Levine M. Homeo box gene expression in anterior and posterior regions of the Drosophila embryo. Proc. Natl. Acad. Sci. 1986;83:4809–4813. doi: 10.1073/pnas.83.13.4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu J.Y., Juven-Gershon T., Marr M.T., Wright K.J., Tjian R., Kadonaga J.T. TBP, Mot1, and NC2 establish a regulatory circuit that controls DPE-dependent versus TATA-dependent transcription. Genes & Dev. 2008;22:2353–2358. doi: 10.1101/gad.1681808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juven-Gershon T., Hsu J.Y., Theisen J.W.M., Kadonaga J.T. The RNA polymerase II core promoter – the gateway to transcription. Curr. Opin. Cell Biol. 2008;20:1–7. doi: 10.1016/j.ceb.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutach A.K., Kadonaga J.T. The downstream promoter element DPE appears to be as widely used as the TATA box in Drosophila core promoters. Mol. Cell. Biol. 2000;20:4754–4764. doi: 10.1128/mcb.20.13.4754-4764.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagrange T., Kapanidis A.N., Tang H., Reinberg D., Ebright R.H. New core promoter element in RNA polymerase II-dependent transcription: Sequence-specific DNA binding by transcription factor IIB. Genes & Dev. 1998;12:34–44. doi: 10.1101/gad.12.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappin T.R., Grier D.G., Thompson A., Halliday H.L. HOX genes: Seductive science, mysterious mechanisms. Ulster Med. J. 2006;75:23–31. [PMC free article] [PubMed] [Google Scholar]
- Lemons D., McGinnis W. Genomic evolution of Hox gene clusters. Science. 2006;313:1918–1922. doi: 10.1126/science.1132040. [DOI] [PubMed] [Google Scholar]
- Levine M., Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–151. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- Levine M., Harding K., Wedeen C., Doyle H., Hoey T., Radomska H. Expression of the homeo box gene family in Drosophila. Cold Spring Harb. Symp. Quant. Biol. 1985;50:209–222. doi: 10.1101/sqb.1985.050.01.027. [DOI] [PubMed] [Google Scholar]
- Lim C.Y., Santoso B., Boulay T., Dong E., Ohler U., Kadonaga J.T. The MTE, a new core promoter element for transcription by RNA polymerase II. Genes & Dev. 2004;18:1606–1617. doi: 10.1101/gad.1193404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohnes D. The Cdx1 homeodomain protein: An integrator of posterior signaling in the mouse. Bioessays. 2003;25:971–980. doi: 10.1002/bies.10340. [DOI] [PubMed] [Google Scholar]
- Macdonald P.M., Struhl G. A molecular gradient in early Drosophila embryos and its role in specifying the body pattern. Nature. 1986;324:537–545. doi: 10.1038/324537a0. [DOI] [PubMed] [Google Scholar]
- Mlodzik M., Gehring W.J. Expression of the caudal gene in the germ line of Drosophila: Formation of an RNA and protein gradient during early embryogenesis. Cell. 1987;48:465–478. doi: 10.1016/0092-8674(87)90197-8. [DOI] [PubMed] [Google Scholar]
- Mlodzik M., Fjose A., Gehring W.J. Isolation of caudal, a Drosophila homeo box-containing gene with maternal expression, whose transcripts form a concentration gradient at the pre-blastoderm stage. EMBO J. 1985;4:2961–2969. doi: 10.1002/j.1460-2075.1985.tb04030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nüsslein-Volhard C., Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- Ochoa-Espinosa A., Small S. Developmental mechanisms and cis-regulatory codes. Curr. Opin. Genet. Dev. 2006;16:165–170. doi: 10.1016/j.gde.2006.02.014. [DOI] [PubMed] [Google Scholar]
- Ohler U., Liao G.C., Niemann H., Rubin G.M. Computational analysis of core promoters in the Drosophila genome. Genome Biol. 2002;3:RESEARCH0087. doi: 10.1186/gb-2002-3-12-research0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuki S., Levine M., Cai H.N. Different core promoters possess distinct regulatory activities in the Drosophila embryo. Genes & Dev. 1998;12:547–556. doi: 10.1101/gad.12.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson J.C., Lemons D., McGinnis W. Modulating Hox gene functions during animal body patterning. Nat. Rev. Genet. 2005;6:893–904. doi: 10.1038/nrg1726. [DOI] [PubMed] [Google Scholar]
- Rivera-Pomar R., Jäckle H. From gradients to stripes in Drosophila embryogenesis: Filling in the gaps. Trends Genet. 1996;12:478–483. doi: 10.1016/0168-9525(96)10044-5. [DOI] [PubMed] [Google Scholar]
- Rivera-Pomar R., Lu X., Perrimon N., Taubert H., Jäckle H. Activation of posterior gap gene expression in the Drosophila blastoderm. Nature. 1995;376:253–256. doi: 10.1038/376253a0. [DOI] [PubMed] [Google Scholar]
- Schulz C., Tautz D. Zygotic caudal regulation by hunchback and its role in abdominal segment formation of the Drosophila embryo. Development. 1995;121:1023–1028. doi: 10.1242/dev.121.4.1023. [DOI] [PubMed] [Google Scholar]
- Smale S.T. Core promoters: Active contributors to combinatorial gene regulation. Genes & Dev. 2001;15:2503–2508. doi: 10.1101/gad.937701. [DOI] [PubMed] [Google Scholar]
- Smale S.T., Kadonaga J.T. The RNA polymerase II core promoter. Annu. Rev. Biochem. 2003;72:449–479. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- Soeller W.C., Poole S.J., Kornberg T. In vitro transcription of the Drosophila engrailed gene. Genes & Dev. 1988;2:68–81. doi: 10.1101/gad.2.1.68. [DOI] [PubMed] [Google Scholar]
- Wampler S.L., Tyree C.M., Kadonaga J.T. Fractionation of the general RNA polymerase II transcription factors from Drosophila embryos. J. Biol. Chem. 1990;265:21223–21231. [PubMed] [Google Scholar]
- Wu L.H., Lengyel J.A. Role of caudal in hindgut specification and gastrulation suggests homology between Drosophila amnioproctodeal invagination and vertebrate blastopore. Development. 1998;125:2433–2442. doi: 10.1242/dev.125.13.2433. [DOI] [PubMed] [Google Scholar]