Abstract
Phosphatidylcholine (PC), or lecithin, is the major phospholipid in eukaryotic membranes, whereas only 10% of all bacteria are predicted to synthesize PC. In Rhizobiaceae, including the phytopathogenic bacterium Agrobacterium tumefaciens, PC is essential for the establishment of a successful host-microbe interaction. A. tumefaciens produces PC via two alternative pathways, the methylation pathway and the Pcs pathway. The responsible genes, pmtA (coding for a phospholipid N-methyltransferase) and pcs (coding for a PC synthase), are located on the circular chromosome of A. tumefaciens C58. Recombinant expression of pmtA and pcs in Escherichia coli revealed that the individual proteins carry out the annotated enzyme functions. Both genes and a putative ABC transporter operon downstream of PC are constitutively expressed in A. tumefaciens. The amount of PC in A. tumefaciens membranes reaches around 23% of total membrane lipids. We show that PC is distributed in both the inner and outer membranes. Loss of PC results in reduced motility and increased biofilm formation, two processes known to be involved in virulence. Our work documents the critical importance of membrane lipid homeostasis for diverse cellular processes in A. tumefaciens.
Phosphatidylcholine (PC), or lecithin, is the most-abundant phospholipid in eukaryotic membranes. Apart from its structural function in membrane bilayers and lipoproteins, PC is involved in many signal transduction pathways (2). Meanwhile, PC has also been found in an increasing number of bacteria, in particular in species that interact with eukaryotic hosts (27). In bacteria, PC is synthesized either by the methylation pathway or by the Pcs pathway (Fig. 1). In the first pathway, the precursor phosphatidylethanolamine (PE) is methylated in three reactions by one or several phospholipid N-methyltransferases (Pmt enzymes) via the intermediates monomethylphosphatidylethanolamine (MMPE) and dimethylphosphatidylethanolamine (DMPE) to form PC. Each reaction step requires S-adenosylmethionine as the methyl donor. In the Pcs pathway, PC is produced via a direct condensation of choline and CDP-diacylglycerol. This reaction is catalyzed by the PC synthase (Pcs), an enzyme unique to prokaryotes (42).
FIG. 1.
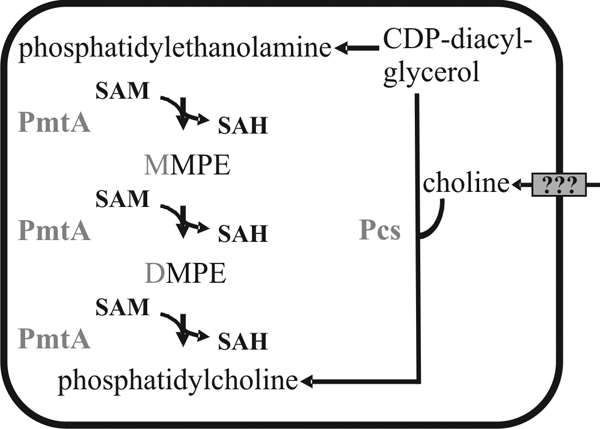
PC biosynthesis pathways in A. tumefaciens C58. As indicated by the question marks, the choline uptake system is unknown. SAM, S-adenosylmethionine; SAH, S-adenosylhomocysteine; PmtA, phospholipid N-methyltransferase; Pcs, phosphatidylcholine synthase.
The gram-negative alphaproteobacterium Agrobacterium tumefaciens is most commonly known for causing crown gall disease in plants. It synthesizes PC by using both the methylation pathway and the Pcs pathway (Fig. 1) (21, 23). The latter pathway requires choline in the medium. During growth in minimal medium, only marginal amounts of PC were formed by the Pcs enzyme (45). In Brucella abortus, PC production via the Pcs pathway has been reported to depend on the presence of choline provided by the host (5). The plant symbiont Sinorhizobium meliloti uses plant-exuded choline for PC biosynthesis (13). The finding that A. tumefaciens cannot produce choline de novo (41) implies that there must be a choline uptake system to supply the Pcs pathway with choline. The only evidence for a choline uptake system so far is that A. tumefaciens was shown to take up radiolabeled choline from the cultivation medium (41). The responsible transporter, however, remains elusive.
Several symbiotic and pathogenic bacteria depend on PC to establish a successful host-microbe interaction. We demonstrated previously that an A. tumefaciens mutant lacking both predicted PC biosynthesis pathways did not produce any PC and was incapable of tumor formation on plant leaves (45). This virulence defect was caused by the absence of the membrane-spanning type IV secretion system, which delivers the oncogenic T-DNA from the tumor-inducing (Ti) plasmid and effector proteins to plant cells (4, 47). In Bradyrhizobium japonicum, the nitrogen-fixing symbiont of the soybean Glycine max, PC is required to establish functional root nodules (31). PC also is necessary for full virulence of the human pathogens B. abortus (6) and Legionella pneumophila (7). Although this strongly suggests that PC is a critical determinant in host-microbe interactions, there still is little understanding regarding the exact role that PC might play in these processes. Moreover, it is largely unclear whether the expression of PC biosynthesis genes is regulated. The first hints that the expression of PC biosynthesis genes might be subject to environmental control were obtained in B. japonicum. This organism encodes a pmt multigene family comprised of pmtA and four additional pmt genes (18). Under normal conditions, only pmtA and pmtX1 are expressed. However, in a pmtA mutant, pmtX3 and, in particular, pmtX4 are expressed (17, 18).
In this study we examined the expression and physiological role of A. tumefaciens PC biosynthesis genes pmtA and pcs. The most important findings are that PC produced by these enzymes is distributed in the inner and outer membrane and that the absence of PC is associated with severe phenotypes in motility and biofilm formation.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All strains and plasmids used in this work are listed in Table 1. Oligonucleotides are listed in Table 2. Escherichia coli cells were grown at 37°C in Luria-Bertani (LB) medium (38), supplemented with kanamycin, streptomycin, and/or spectinomycin at a final concentration of 50 μg/ml if appropriate. E. coli DH5α was used as host for all cloning procedures. E. coli BL21(DE3), which contains the phage T7 polymerase gene under the control of the lacUV5 promoter (43), served as the host for overproduction of PmtA and Pcs from the corresponding pET24b-based expression plasmids. A. tumefaciens strain C58 (wild type) and its derivatives (ΔpmtA, Δpcs, ΔpmtA Δpcs, and ΔpmtA Δabc mutants) were routinely grown at 30°C in YEB complex or AB minimal medium (pH 5.5, 1% [wt/vol] glucose) (40), supplemented with 100 μg/ml ampicillin, 2 μg/ml tetracycline, 50 μg/ml kanamycin, 100 μg/ml streptomycin, and/or 300 μg/ml spectinomycin if necessary. For β-galactosidase assays, choline (Sigma-Aldrich, München, Germany) was added to the AB medium in final concentrations of 0.1 mM, 0.5 mM, or 1 mM. In support of virulence gene induction, Agrobacterium cells were precultivated to an optical density at 600 nm (OD600) of 0.2 at 30°C in liquid AB minimal medium (pH 5.5) with 1% (wt/vol) glucose prior to the addition of acetosyringone. Cells were further incubated for 16 to 20 h at 23°C. Acetosyringone was always used at a final concentration of 0.1 mM.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Reference or source |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| DH5α | Cloning host | 19 |
| BL21(DE3) | Expression host | 43 |
| Agrobacterium tumefaciens | ||
| C58 | Wild type | C. Baron, Montreal, Canada |
| C58 ΔpmtA | Derivative of the wild type with deletion of the pmtA gene | 45 |
| C58 Δpcs | Derivative of the wild type with deletion of the pcs gene | 45 |
| C58 ΔpmtA Δpcs | Derivative of the wild type with deletion of the pmtA and pcs genes | 45 |
| C58 ΔpmtA Δabc | Derivative of the wild type with deletion of the pmtA and abc1-4 genes | This study |
| Plasmids | ||
| pAC01 | Tcr Apr; transcriptional lacZ fusion vector containing promoterless lacZ gene | 26 |
| pET24b | Kmr; high-copy His-tag expression vector | Novagen, Darmstadt, Germany |
| pVS-BADNco | Spr Str; A. tumefaciens expression vector | 49 |
| pK19mobsacB | Kmr; suicide vector | 39 |
| pBO380 | Tcr Apr; pmtA-lacZ fusion in pAC01 | This study |
| pBO377 | Tcr Apr; pcs-lacZ fusion in pAC01 | This study |
| pBO1264 | Tcr Apr; abc-lacZ fusion in pAC01 | This study |
| pBO801 | Kmr; derivative of pET24b for overproduction of PmtA with a C-terminal His tag | This study |
| pBO803 | Kmr; derivative of pET24b for overproduction of Pcs with a C-terminal His tag | This study |
| pBO813 | Spr Str; derivative of pVS-BADNco carrying pmtA | This study |
| pBO814 | Spr Str; derivative of pVS-BADNco carrying pcs | This study |
| pBO308 | Kmr; derivative of pK19mobsacB carrying the upstream region of abc1-4 | This study |
| pBO309 | Kmr; derivative of pK19mobsacB carrying the up- and downstream regions of abc1-4 | This study |
| pLacTac-Gfp | lacIq Ptac::gfp-mut2 TcrrepA oriVpVSIoriVp15AoriT | K. Thormann, Marburg Germany |
Ap, ampicillin; Km, kanamycin; Sp, spectinomycin; St, streptomycin; Tc, tetracycline.
TABLE 2.
Oligonucleotides used in this study
| Purpose | Oligonucleotide | Sequence (5'→3')a |
|---|---|---|
| Transcriptional lacZ fusions | pmtA_Fw_KpnI | AAAAGGTACCGATTTTTCTGCTGGCG |
| pmtA_Rv_XhoI | AAAACTCGAGGATTCCCCTCGGATG | |
| pcs_Fw_KpnI | AAAAGGTACCCGGTTGTGTCGAGG | |
| pcs_Rv_XhoI | AAAACTCGAGGGTCAGCGCCTGC | |
| abc_Fw_KpnI | AAAAGGTACCCGCTTAATCTCCTCG | |
| abc_Rv_XhoI | AAAACTCGAGGAGTTCCCTCTCTC | |
| RT-PCR | pmtA_K1 (P1) | GCAACGGCTTGAACAAAAAT |
| pmtA_K2 (P2) | GAATATTGCGCACGATGAAA | |
| pmtA_Kontr_fw (N1) | GCTGATCGACCGGACGCAG | |
| pmtA_RT_PE3 (N2) | ATTTTTGTTCAAGCCGTTGC | |
| pmtA_do_fw_BamHI (A) | CGCGGATCCGGCGCAATTGTGGACCTAT | |
| pmtA_Kontr_rv (B) | GCCCAGGGAAGGCGGCTCG | |
| pcs_K1 (P3) | GCGCCTTTTCCGTTCATATC | |
| pcs_K2 (P4) | GTTTCGAAATGCATCAGCAG | |
| Sig219 (N3) | TTCAGGCTCAAGGTTTCCAG | |
| pcs_RT_PE (N4) | GTTTTTCCATCCGCCCTGTtca | |
| pcs_1 (C) | GCGAGCGGCATCTATCTTTA | |
| pcs_2 (D) | GAAATTGCCGAACAGCTTG | |
| Overproduction of PmtA and Pcs in E. coli | pmtA_Nde_fw | GGAATTCCATATGGCACTCAACCTGAAGCAAC |
| pmtA_Sal_Xho_rv | CCGGTCGACTCACTCGAGGGCCCGCGTATAGGTCCACAA | |
| pcs_Nde_fw | GGAATTCCATATGGCCGCATACCGGAATGAA | |
| pcs_Hind_Xho_rv | CCCAAGCTTTCACTCGAGTTTCGCGCCGAGCGAGGGGAA | |
| Overproduction of PmtA and Pcs in A. tumefaciens | pmtA_EcoR_fw | CCGGAATTCTGACAATGACCAATACATCCGAG |
| pmtA_Sal_rv | GCAGGTCGACTCAGGCCCGCGTATAGGTCCAC | |
| pcs_EcoR_fw2 | CCGGAATTCTGAGCGGATGGAAAAACGGGGCG | |
| pcs_Hind_rv | CCCAAGCTTTTATTTCGCGCCGAGCGAGGGG | |
| Construction of a markerless abc1-4 deletion mutant in A. tumefaciens °pmtA | abc_up_fw_EcoRI | CGGAATTCCGTCACCTATGTGCTCTTGCCCGCC |
| abc_up_rv_PstI | AACTGCAGGAACAGCTTGGTCAGCTTGCG | |
| abc_do_fw_PstI | AACTGCAGGCTCGGCCTCAACTTCTACGTT | |
| abc_do_rv_HindIII | CCCAAGCTTGCCTTGATCAACGAACCAACGG |
Restriction sites in the oligonucleotides are underlined.
Plasmid and mutant construction.
Recombinant DNA work was carried out according to standard protocols (38). PCR-generated fragments of the promoter regions of pmtA, pcs, and abc (16, 48) were digested with KpnI and XhoI and ligated into pAC01 (26) treated with the same enzymes to construct transcriptional fusions to the lacZ gene. For overproduction of the agrobacterial PmtA and Pcs in E. coli, the expression plasmids pET_PmtA and pET_Pcs were constructed. The pmtA and pcs genes were amplified by PCR. The fragments were cleaved with NdeI/SalI and NdeI/HindIII, respectively, and ligated into pET24b treated with the same enzymes. For overproduction of PmtA and Pcs in A. tumefaciens, the coding regions, including their own ribosome binding site, were amplified via PCR. The PCR products were digested with EcoRI/SalI and EcoRI/HindIII, respectively, and cloned into the corresponding sites of the vector pVS-BADNco (49), resulting in the plasmids pVS-PmtA and pVS-Pcs. The markerless abc1-4 deletion was introduced into the A. tumefaciens ΔpmtA strain as described previously (45) by using the suicide vector pK19mobsacB. The abc1-4 up- and downstream regions were amplified by PCR. The PCR product of the abc1-4 upstream region was digested with EcoRI/PstI and cloned into the vector pK19mobsacB, resulting in the plasmid pK19mobsacB_abc_up. The PCR product of the downstream region was digested with PstI/HindIII and cloned into pK19mobsacB_abc_up, resulting in the plasmid pK19mobsacB_abc_updo. The correct nucleotide sequences of all plasmids constructed were confirmed by automated sequencing. Plasmids were transferred into E. coli via transformation and into A. tumefaciens via electroporation.
Overproduction of PmtA and Pcs in E. coli.
E. coli BL21(DE3) carrying pET_PmtA or pET_Pcs was cultivated in LB medium at 37°C until the OD600 reached a value between 0.5 and 0.8. Then, the synthesis of PmtA or Pcs was induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 0.4 mM and the cultures were incubated for another 2 h at 30°C. Subsequently, 1 ml culture was harvested by centrifugation. Cell pellets were resuspended in 1× sodium dodecyl sulfate (SDS) loading buffer according to the OD600 (OD600 of 1 ≈ 100 μl 1× SDS loading buffer) and boiled for 10 min. Ten microliters of each sample was separated on 12.5% SDS-polyacrylamide gels, and the proteins were stained with Coomassie blue.
Lipid analysis by TLC.
The lipid composition of A. tumefaciens and E. coli strains was determined via thin-layer chromatography (TLC). Cells were cultivated as mentioned above, harvested by centrifugation, washed with 500 μl water, and resuspended in 100 μl water. The lipids were extracted according to the method of Bligh and Dyer (1), separated by one-dimensional thin-layer chromatography using HPTLC silica gel 60 plates (Merck, Darmstadt, Germany), and stained with molybdenum blue spray reagent (Sigma-Aldrich) or Cu2SO4 solution [300 mM copper(II)-sulfate-pentahydrate, 8.5% (vol/vol) phosphoric acid]. In the case of the separated inner and outer membranes, the lipid extraction was started directly from the collected fractions. PE, MMPE, DMPE, and PC were used as phospholipid standards (Sigma-Aldrich), and n-propanol-propionate-chloroform-water (3:2:2:1) as the running solvent.
RT-PCR.
A. tumefaciens strains were cultivated in YEB complex medium until exponential phase. Total RNA was isolated by using a Micro-to-Midi total RNA purification system (Invitrogen, Karlsruhe, Germany). The RNA was further treated with DNase I (amplification grade; Invitrogen) as specified by the manufacturer to remove contaminating chromosomal DNA. Reverse transcriptase (RT)-PCR and subsequent PCRs were performed according to the manufacturer's manual for the ThermoScript RT-PCR system (Invitrogen). The primers used for these experiments are listed in Table 2 (see also Fig. 3A and C). The reaction products were separated by electrophoresis in 2% (wt/vol) agarose gels and visualized by staining with ethidium bromide.
FIG. 3.
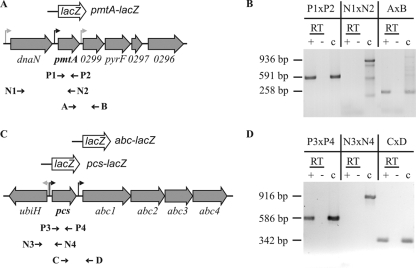
RT-PCR of the pmtA and pcs gene regions. The PCR strategy is outlined in panels A and C. The positions of primers used for the reverse transcription and PCRs are given below the corresponding gene regions. Putative promoters and constructed lacZ fusions are indicated. dnaN encodes β-chain of DNA polymerase III; pmtA encodes phospholipid N-methyltransferase; pyrF encodes orotidine 5′-monophosphate decarboxylase; atu0299, atu0297, and atu0296 are hypothetical open reading frames; ubiH encodes 2-octaprenyl-6-methoxyphenol hydroxylase; pcs encodes PC synthase; abc1 encodes a nucleotide binding ABC transporter; abc2 and abc3 encode membrane-spanning proteins forming an ABC transporter; and abc4 encodes a lipoprotein ABC transporter. In panels B and D, the results of RT-PCRs with RNA from wild-type A. tumefaciens C58 cells are presented. The primer pairs used and lengths of PCR products are indicated. c, PCR products using chromosomal DNA as template; +, standard RT-PCR; −, negative control in which no RT had been added to the reaction mixture.
β-Galactosidase assays.
The β-galactosidase activity of A. tumefaciens cells grown in liquid YEB complex medium or in AB minimal medium was measured according to standard protocols (30). The plasmid pAC01 (26) containing the promoterless lacZ gene was used as the negative control.
Separation of inner and outer membrane.
Membrane separation was performed as described previously (12), with minor modifications. Four hundred milliliters YEB cell culture was grown to an OD600 of 0.5 to 0.6 at 30°C and harvested by centrifugation at 10,000 × g, 4°C, for 10 min. The cells were resuspended in 24 ml lysis buffer (50 mM Tris-HCl, pH 7.5, 20% [wt/vol] sucrose, 0.2 M KCl, 0.2 mM dithiothreitol, 0.2 mg/ml DNase I, 0.2 mg/ml RNase A, 1 mM phenylmethylsulfonyl fluoride) and disrupted by two passes through a chilled French pressure cell at 16,000 lb/in2. The lysate was treated with 0.5 mg/ml lysozyme for 1 h on ice and centrifuged at 10,000 × g, 4°C, for 20 min to remove the unbroken cells. The supernatant was centrifuged at 150,000 × g (SW40Ti), 4°C, for 1 h in an ultracentrifuge to collect the membranes. The resulting membrane pellet was carefully resuspended in 2 ml of 20% (wt/vol) sucrose containing 5 mM EDTA, pH 7.5, and 0.2 mM dithiothreitol. The resuspended membranes were centrifuged for 5 min at 16,000 × g to remove the insoluble membranes. The gradient was prepared by layering 7.5 ml 53% (wt/vol) sucrose over a cushion of 2.5 ml 70% (wt/vol) sucrose. Both sucrose solutions contained 5 mM EDTA, pH 7.5. The membrane suspension was layered on the top of the gradient, and sucrose density gradient ultracentrifugation was carried out at 100,000 × g (SW40Ti), 4°C, for 16 h. After ultracentrifugation, the separated membranes were fractionated in 500-μl aliquots to analyze the protein concentration, the NADH activity, and the phospholipid pattern. The protein concentration was determined by using a Bradford assay (Bio-Rad Laboratories GmbH, München, Germany). The NADH oxidase activity was detected by the method of Osborn et al. (32).
Motility assay.
Motility assays were carried out on AB minimal medium solidified with 0.3% (wt/vol) agar (Difco, Lawrence, KS). A single colony from a YEB agar plate was inoculated onto the surface of the motility plates. The plates were examined after 48 h of incubation at 23°C. To check whether calcium and/or magnesium influence motility, MgSO4 was added in final concentrations of 0.24 mM, 1.2 mM, or 12 mM and CaCl2 was added in final concentrations of 0.014 mM, 0.07 mM, or 0.7 mM to AB medium.
SDS-PAGE and Western blotting.
Wild-type and mutant A. tumefaciens cells were cultivated as mentioned above, and 1 ml of each culture was harvested and resuspended in 1× SDS loading buffer in relation to the OD600 (OD600 of 1 ≈ 100 μl 1× SDS loading buffer). The samples were incubated for 10 min at 95°C, separated by 12.5% SDS-polyacrylamide gel electrophoresis (PAGE), and blotted onto polyvinylidene difluoride membranes (Bio-Rad). Detection was performed with a chemiluminescence-based system (Pierce Biotechnology, Rockford, IL) using A. tumefaciens flagellum protein-specific antiserum (1:30,000).
Flow cell biofilms.
Biofilms of A. tumefaciens strains carrying plasmid-encoded green fluorescent protein (GFP) were cultivated at 30°C in three-channel flow cells with individual channel dimensions of 1 by 4 by 40 mm. Each flow chamber was prepared by gluing a boron-silicate glass microscope coverslip, which served as a substratum for microbial attachment, onto the flow chamber with silicone and leaving it to dry for 24 h at room temperature prior to use. The assembly of the flow system was carried out essentially as described earlier (44).
The appropriate A. tumefaciens strains were cultivated until reaching the exponential or the stationary growth phase. The OD600 was then adjusted to 0.01 in AB minimal medium with 0.5% (vol/vol) glycerol. One milliliter of the diluted cell suspension was injected into each channel after the flow of medium was arrested, and the chambers were turned upside down to facilitate initial attachment. After 30 min of incubation at 30°C, the flow cells were inverted, and the flow of medium was started at a constant rate of 75 μl/min per channel, using a Watson-Marlow Bredel 205S peristaltic pump (Cornwall, United Kingdom). All biofilm characterizations were conducted in triplicate in at least two independent experiments.
Sixty minutes prior to microscopy, gfp expression from plasmid pLacTac-Gfp was induced by the addition of IPTG (1 mM). Microscopic visualization of biofilms was carried out using an inverted Zeiss LSM510 confocal laser scanning microscope (Carl Zeiss, Jena, Germany) equipped with the following objectives: 10×/0.3 W Plan-Neofluar, 20×/0.5 W Achroplan, and 40×/1.2 W C-Apochromat. For quantitative analysis, the image data were further processed using the IMARIS software package (Bitplane AG, Zürich, Switzerland) and Adobe Photoshop.
RESULTS AND DISCUSSION
Functional analysis of pmtA and pcs gene products.
We have previously shown that an A. tumefaciens ΔpmtA Δpcs mutant is no longer able to synthesize PC (45). To provide conclusive evidence that PmtA and Pcs act as phospholipid N-methyltransferase and PC synthase, respectively, we cloned pmtA and pcs into the expression vector pET24b, resulting in plasmids pET_PmtA and pET_Pcs. The analysis of crude protein extracts from E. coli cells containing the first plasmid by SDS-PAGE revealed an overproduced protein of 22 kDa (Fig. 2A), which is in close agreement with the calculated mass of the agrobacterial pmtA gene product. In contrast, overexpression of Pcs (calculated mass of 29 kDa), which is thought to be a membrane protein, was not observed (Fig. 2A). Either Pcs was not synthesized at all or it was synthesized in amounts below the detection limit.
FIG. 2.
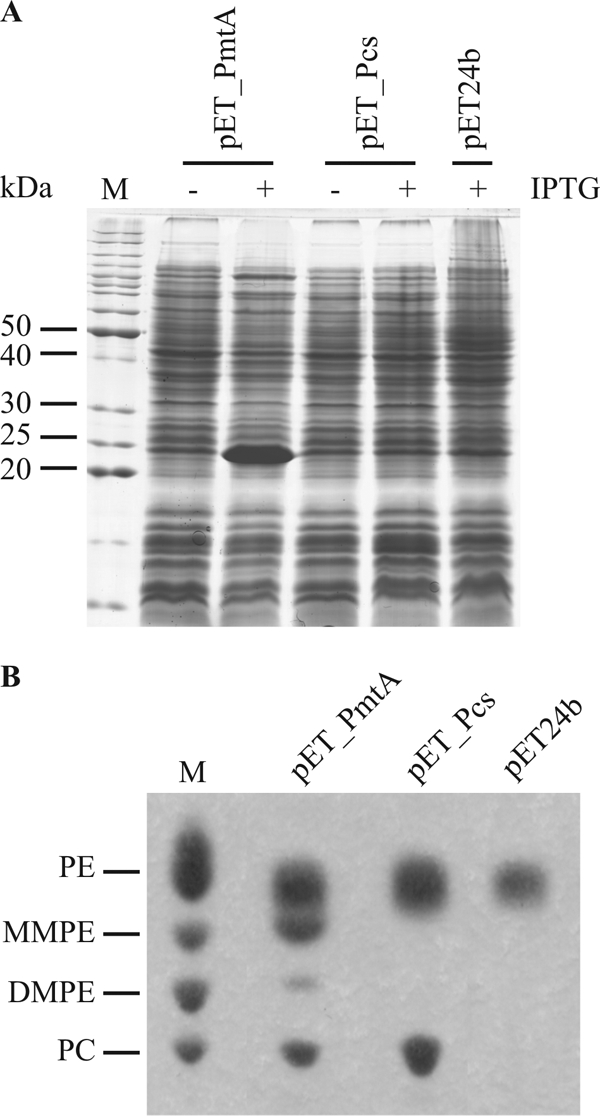
Activity of agrobacterial PmtA and Pcs after expression in E. coli. (A) Detection of agrobacterial phospholipid N-methyltransferase (PmtA) and PC synthase (Pcs) in crude extracts of E. coli cells via SDS-PAGE. E. coli BL21(DE3) cells with pET_PmtA, pET_Pcs, or pET24b were cultivated in LB complex medium, and protein expression was induced with IPTG. Protein bands were visualized by Coomassie blue staining. M, BenchMark protein standard (Invitrogen). +, present; −, absent. (B) Lipid formation after expression of agrobacterial PmtA and Pcs in E. coli BL21(DE3). Lipids of BL21(DE3) derivatives were extracted and separated by one-dimensional TLC. Phospholipids were specifically stained with molybdenum blue spray and compared to phospholipid standards in lane M (PE, MMPE, DMPE, and PC).
To obtain proof for the biochemical activity of both recombinant proteins, we compared the membrane lipid contents of E. coli BL21(DE3) cells carrying pET_PmtA, pET_Pcs, or the empty vector pET24b. E. coli membranes are known to contain the lipids PE, phosphatidylglycerol, and cardiolipin (8), which migrate as a single spot in one-dimensional TLC (Fig. 2B). As expected, E. coli BL21(DE3)/pET24b did not produce PC or methylated intermediates. The membranes of E. coli cells expressing pmtA consist of PE, MMPE, DMPE, and PC (Fig. 2B). This is consistent with the results described for a Sinorhizobium meliloti pmtA gene expressed in E. coli BL21(DE3) (31). Only PC and no methylated intermediates were produced when the Rhodobacter sphaeroides pmtA gene was expressed in E. coli (31). The expression of the B. japonicum pmtA gene in E. coli BL21(DE3) led predominantly to the formation of MMPE, with some DMPE but only marginal amounts of PC (18). Apparently, different Pmt enzymes possess different substrate and product specificities which cannot be predicted solely on the basis of their primary sequence.
As the Pcs enzyme uses choline directly to form PC, the E. coli strain containing pET_Pcs produces PE and PC but no mono- or dimethylated intermediates (Fig. 2B). It is notable that significant amounts of PC accumulated in this strain although the Pcs protein was only poorly expressed (Fig. 2A). Our results demonstrate that both recombinant proteins are active when expressed in E. coli and are probably responsible for the biosynthesis of PC in A. tumefaciens without the requirement of additional proteins.
Genetic organization of the Agrobacterium pmtA and pcs gene regions.
The pmtA gene is flanked by two open reading frames, namely dnaN and atu0299, which are oriented in the same direction as pmtA (Fig. 3A). The pmtA gene is separated from dnaN and atu0299 by 195 bp and 95 bp, respectively. To analyze whether pmtA is cotranscribed with dnaN and/or atu0299, RT-PCR experiments were carried out. Total RNA was isolated from a wild-type A. tumefaciens culture grown in YEB complex medium. Primers P2, N2, and B were used for RT reactions to produce cDNA. Primer pairs P1 and P2, N1 and N2, and A and B were applied for PCR amplification (Fig. 3A). As the positive control, PCRs were carried out with chromosomal DNA as template. Reaction mixtures without RT served as the control for the absence of chromosomal DNA in our RNA preparations and did not produce amplification products (Fig. 3B). The expected product of 591 bp featuring an internal pmtA fragment appeared only in the presence of RT, showing that the gene was expressed. The presence of a 258-bp PCR product with primer combination A and B suggests cotranscription of pmtA and atu0299. Since atu0299, pyrF, and atu0297 are separated by only 4 and 19 bp, respectively, it seems reasonable that these genes are cotranscribed. Thus, pmtA-atu0299-pyrF-atu0297 might form a tetracistronic operon. No PCR product was detectable with primer pair N1 by N2, indicating that dnaN and pmtA belong to different transcription units (Fig. 3B).
The second PC biosynthesis gene, pcs, is located 212 bp downstream of ubiH, encoding a predicted 2-octaprenyl-6-methoxyphenol hydroxylase which is transcribed in the opposite direction (Fig. 3C). Two hundred one base pairs further downstream of pcs are four genes oriented in the same direction. The gene products are annotated as an ABC transporter with unknown substrate. We tested whether the genes abc1 to abc4 might form an operon with pcs. The primer pair P3 and P4, covering an internal DNA fragment of pcs, yielded the expected product of 586 bp (Fig. 3D), demonstrating that the pcs gene is expressed under the tested conditions. In contrast, the primer set N3 and N4, designed to amplify a DNA fragment overlapping the borders of ubiH and pcs, did not produce a PCR product, as expected since the two genes are oriented in opposite directions. The primer pair C and D, designed to amplify a DNA fragment overlapping the gene borders of pcs and abc1, resulted in a PCR product of 342 bp (Fig. 3D). Hence, it appears that pcs and the abc1-4 genes form an operon.
Constitutive expression of A. tumefaciens PC biosynthesis genes.
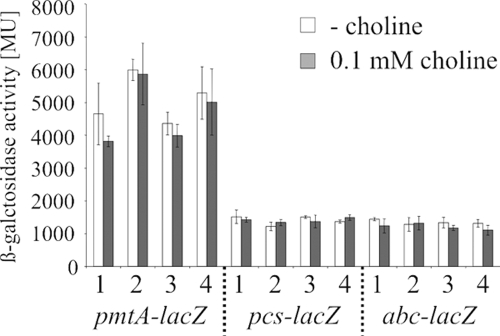
To further analyze the expression of pmtA and pcs, transcriptional lacZ fusions were constructed (Fig. 3A and C). In addition, an abc1-lacZ fusion was created to test whether abc expression depends exclusively on the pcs promoter (Fig. 3C). The resulting reporter gene plasmids, pBO380 (pmtA-lacZ), pBO377 (pcs-lacZ), and pBO1264 (abc1-lacZ), were electroporated into wild-type A. tumefaciens. To analyze if the presence or absence of PC influences the expression of PC biosynthesis genes and/or abc1, the lacZ fusions were also introduced into existing PC biosynthesis mutants. It is known that the two single deletion ΔpmtA and Δpcs mutants are able to synthesize PC through the remaining pathway, whereas the double deletion ΔpmtA Δpcs mutant is entirely PC deficient (45). All reporter strains were grown in YEB complex medium and in AB minimal medium at 30°C. The PC biosynthesis genes pmtA and pcs, as well as abc1, were clearly expressed under all conditions tested. The expression levels were three- to fourfold higher in minimal medium (data not shown). Expression was not altered in the PC biosynthesis mutants (Fig. 4), indicating that pmtA, pcs, and abc expression is independent of the presence of PC, MMPE, and DMPE. The significant β-galactosidase activity of the abc1-lacZ fusion demonstrates that the abc1-4 genes possess their own promoter in addition to the one upstream of pcs (Fig. 3D). In all cases, the level of expression of pmtA was higher than that of pcs (Fig. 4). This might be taken as an additional line of support for our previous assumption that the methylation pathway is more relevant than the Pcs pathway in A. tumefaciens. Stronger evidence for this conclusion derives from the following findings. Only traces of PC were detectable after growth in AB minimal medium in the presence of choline (45), indicating that the still-existing Pcs pathway cannot fully compensate for the methylation pathway. In addition, Kalanchoë leaves infected with the ΔpmtA mutant showed decreased tumor formation (45). The significance of the alternative PC biosynthesis pathways in prokaryotes differs. In contrast to A. tumefaciens, the Pcs pathway is of predominant importance in L. pneumophila (7) and Pseudomonas aeruginosa (46).
FIG. 4.
β-Galactosidase activities of plasmid-encoded transcriptional lacZ fusions in wild-type A. tumefaciens C58 and PC biosynthesis mutants. Cells were grown in AB minimal medium in the absence or presence of 0.1 mM choline at 30°C. The error bars indicate standard deviations of the results from three independent assays. The plasmid pAC01 containing the promoterless lacZ gene was used as negative control, and the background activity was below 3 Miller units (MU). 1, wild-type; 2, ΔpmtA; 3, Δpcs; 4, ΔpmtA Δpcs.
In S. meliloti, choline is taken up by an ABC transport system (14). Given the genetic organization of the abc1-4 genes immediately downstream of the pcs gene (Fig. 3C), we speculated that choline might be a substrate of this putative ABC transporter. In bacteria, the expression and activity of ABC transporters is often regulated via their substrates. The expression of the fructose uptake ABC transporter in S. meliloti is induced in the presence of this sugar (25). Likewise, the rhamnose uptake system in Rhizobium leguminosarum is controlled by rhamnose (36). To check if the expression of the abc operon is choline dependent, choline was added to the AB minimal medium at a final concentration of 0.1 mM. The pmtA-lacZ fusion was considered a negative control since the expression of the methylation pathway should not be dependent on choline. The expression of all three genes (abc1, pcs, and pmtA) was not altered in the presence of 0.1 mM choline (Fig. 4) or 0.5 mM or 1 mM choline (data not shown), indicating that the expression of the Pcs pathway and the putative ABC transporter is choline independent.
We have previously shown that the deletion of pmtA and pcs causes a drastic virulence defect (45). Genes involved in choline uptake and metabolism in S. meliloti are highly expressed under symbiotic conditions (14, 28). To further inspect the link between virulence and PC biosynthesis in A. tumefaciens, we tested if virulence conditions change the expression of the PC biosynthesis genes. The addition of 0.1 mM acetosyringone, an artificial virulence gene inducer, to the AB minimal medium did not affect the expression of pmtA, pcs, and abc1 (data not shown). We conclude that all three genes are constitutively expressed and not regulated by (i) the PC content, (ii) the availability of choline, and (iii) the presence of acetosyringone.
The abc1-4 genes do not encode a critical choline uptake system.
PC formation in an A. tumefaciens pmtA mutant depends on the Pcs pathway and the presence of choline in the growth medium (45). To address whether the predicted ABC transporter encoded downstream of the pcs gene is responsible for choline uptake, we monitored PC biosynthesis in a pmtA abc1-4 double mutant grown in YEB complex medium by TLC analysis. As expected, wild-type A. tumefaciens membranes consist of PE, MMPE, DMPE, and PC, whereas a ΔpmtA mutant produces only PC but not the intermediates MMPE and DMPE (Fig. 5). The ΔpmtA Δpcs mutant is no longer able to synthesize PC (45). Wild-type-like amounts of PC in the pmtA abc1-4 deletion strain (Fig. 5) indicate that the Pcs pathway is efficiently supplied with its substrate choline. We conclude that the abc1-4 genes are not responsible for choline uptake or that additional transporters compensate for the lack of the ABC transporter. Residual choline uptake activity has also been reported in the absence of the high-affinity ChoX system in S. meliloti (14). Bacillus subtilis contains two closely related ABC transport systems for the uptake of choline as precursor for the osmoprotectant glycine betaine (22). All these findings suggest that bacteria might be equipped with alternative routes for choline uptake. Based on its genome sequence, A. tumefaciens is predicted to contain 667 components of ABC transporters (16, 48). There clearly is potential for alternative choline transport systems.
FIG. 5.
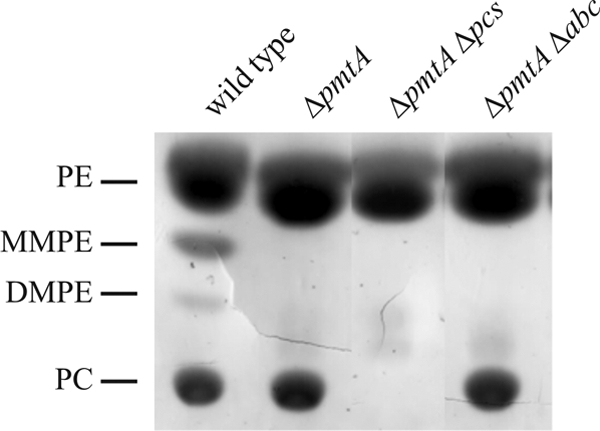
PC formation in the A. tumefaciens abc1-4 mutant. A. tumefaciens wild-type, ΔpmtA, ΔpmtA Δpcs, and ΔpmtA Δabc cells were grown in YEB complex medium at 30°C. Lipids were extracted, separated by one-dimensional TLC, and visualized by Cu2SO4 staining.
Membrane localization of PC.
The results shown in Fig. 5 and previous TLC analyses (23, 43) suggested that the relative PC content in A. tumefaciens membranes amounts to approximately 25%. The quantification of radiolabeled phospholipids revealed that PC levels indeed reach 23% of total phospholipids (data not shown). Since it was unknown whether PC is localized in the inner, the outer, or in both membranes of A. tumefaciens, we isolated membranes of wild-type A. tumefaciens and subsequently separated the inner and outer membranes by sucrose density gradient ultracentrifugation (Fig. 6A). As reported for R. leguminosarum (12), two major lipid fractions were visible as a diffuse cream-colored upper band and a distinct white lower band. Measuring the protein concentration of the individual fractions revealed two major peaks with maxima in fractions 3 to 7 and 19 to 21 (Fig. 6B). Both peaks contain a clearly distinguishable collection of proteins spanning the entire molecular-mass range (Fig. 6C). Fractions from the inner and outer membranes were clearly distinguishable. To assign the peaks to a membrane compartment, NADH oxidase activity, which is a marker enzyme for the inner membrane, was determined. As expected, the NADH oxidase activity was predominantly found in fractions 3 to 7 (data not shown).
FIG. 6.
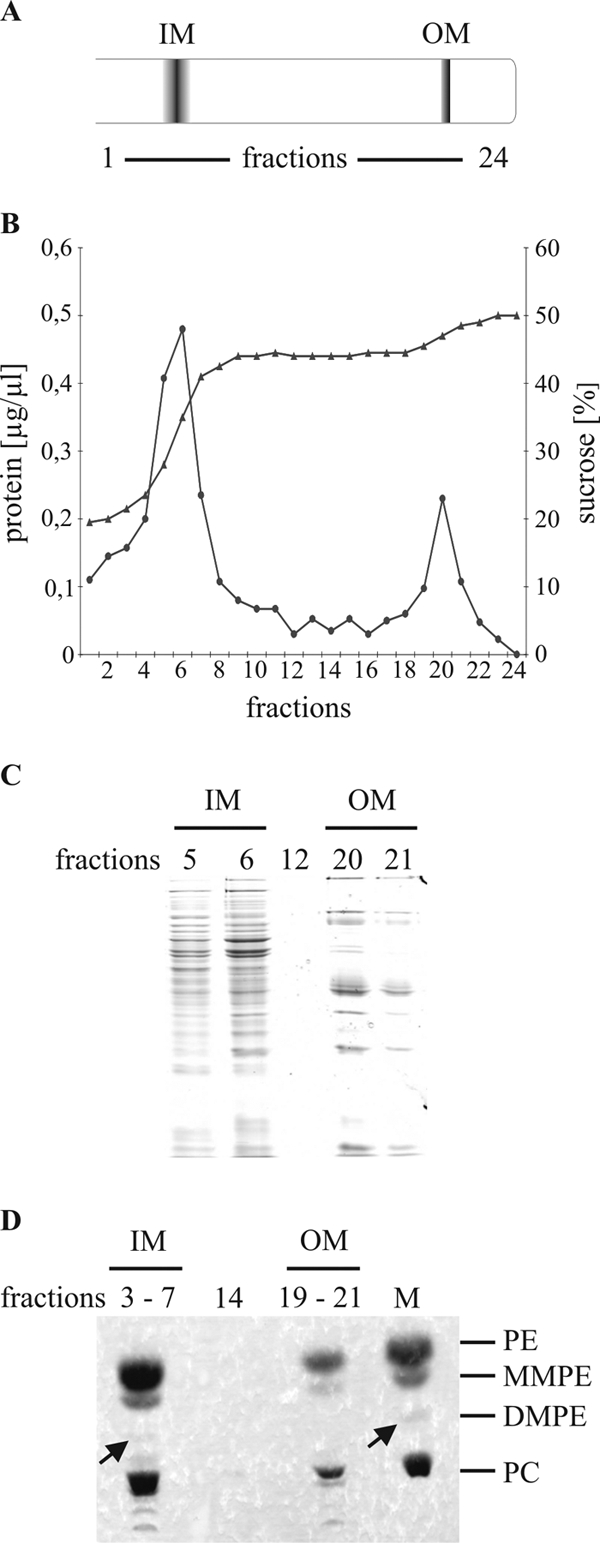
Localization of PC in membranes of wild-type A. tumefaciens C58. (A) Schematic representation of bands observed after centrifugation. (B) Separation of agrobacterial inner and outer membrane by discontinuous sucrose density gradient centrifugation. Fractions were collected as 500-μl aliquots from the top of the sucrose gradient. Protein concentrations (•) and sugar densities (▴) of the gradient fractions are shown. (C) Fractions 5 and 6 (IM), fraction 12, and fractions 20 and 21 (OM) were analyzed by SDS-PAGE and visualized with Coomassie blue staining. (D) Lipids were extracted, and fractions 3 to 7 and 19 to 21 were pooled, followed by one-dimensional TLC analysis and molybdenum blue staining. Barely detectable lipids are marked with arrows. PE, MMPE, DMPE, and PC were used as standards. M, phospholipid standard; IM, inner membrane; OM, outer membrane.
To determine the distribution of PC, membrane lipids from pooled fractions 3 to 7 and 19 to 21 were analyzed by TLC. The pooled inner membrane fraction mainly consists of PE and PC, with little MMPE and only traces of DMPE (Fig. 6D). The outer membrane is also composed of PE, MMPE, and PC. DMPE was not visible, probably because the amount was below the detection limit. We conclude that A. tumefaciens contains PC in both the inner and outer membrane.
PC and PE are also the most-abundant phospholipids in the pathogens L. pneumophila (20) and P. aeruginosa (46), and it has been shown that they are localized in both membrane compartments. The presence of PC is critical for adaptation to different environmental conditions in these organisms. The loss of PC in L. pneumophila led to reduced bacterial binding to macrophages during the infection process (7), and PC seems to be important for P. aeruginosa in specific responses to stress (46). An A. tumefaciens PC-deficient mutant showed sensitivity toward elevated temperature and SDS (45). The presence of PC in the outer membrane, which is in direct contact with the environment, might be important for coping with stressful conditions.
PC-deficient mutants are impaired in motility.
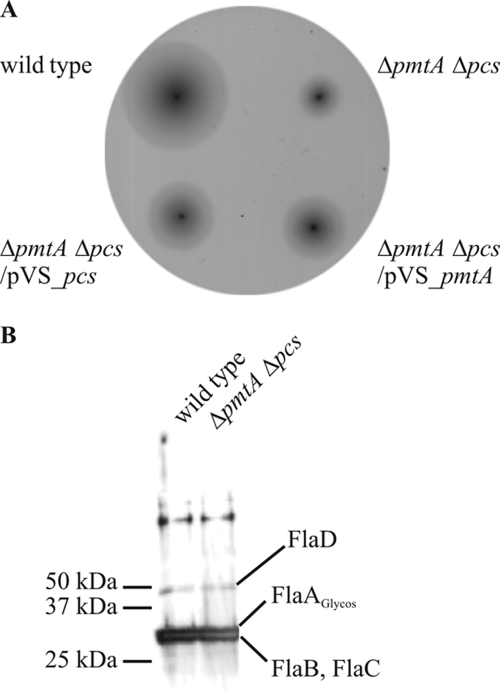
Since the lipid composition impacts bacterial fitness, we hypothesized that PC might be of particular importance for biological processes mediated through the membranes. The presence of PC is required for the induction of the type IV secretion system in A. tumefaciens (45). Another process that relies on a complex membrane-spanning system is bacterial motility. Swimming motility in A. tumefaciens is mediated by flagella which are typically localized as a small tuft at or around a single cell pole (3). To investigate the motility phenotype, the wild-type strain and the single deletion ΔpmtA and Δpcs mutants, as well as the PC-deficient mutant (ΔpmtA Δpcs), were plated on soft agar medium. Consistent with previous reports (29), A. tumefaciens is highly motile, as indicated by the formation of large concentric swim rings (Fig. 7A). Both single mutants showed swimming behavior comparable to that of the wild-type (data not shown). In contrast, the PC-deficient mutant was severely impaired in motility. The motility defect was partially restored when the ΔpmtA Δpcs mutant was complemented with either pmtA or pcs expressed from low-copy-number vector pVS_PmtA or pVS_Pcs. It is known that many members of the Rhizobiaceae are nonmotile in medium lacking divalent cations but retain good motility in medium containing calcium, magnesium, barium, or strontium (37). Therefore, we checked whether calcium and/or magnesium could rescue the motility defect of the double mutant and analyzed its motility in the absence or presence of different concentrations of both ions. Neither the addition of MgSo4 nor of CaCl2 restored the motility defect (data not shown). Motility was also inspected microscopically. The wild type showed normal motility, whereas the ΔpmtA Δpcs mutant showed uncoordinated circular movement (data not shown).
FIG. 7.
Motility assay of an agrobacterial PC-deficient mutant and complemented strains. (A) Motility of the wild type, the PC-deficient mutant (ΔpmtA Δpcs), and the mutant complemented with either pVS_pmtA or pVS_pcs was assayed on AB minimal medium plate containing 0.3% (wt/vol) agar. (B) Cell lysates of wild type and PC-deficient mutant were analyzed by SDS-PAGE and Western blotting using a flagellin antibody (1:30,000).
A L. pneumophila PC-deficient mutant has also been shown to be impaired in motility (7). This phenotype was due to the lack of flagellum proteins. Hence, we examined whether these proteins are present in the agrobacterial PC-deficient mutant. Total protein extracts of the wild type and the ΔpmtA Δpcs mutant were analyzed by Western blotting using a flagellum protein-specific antibody raised against flagellar proteins of A. tumefaciens (B. Scharf, personal communication). In contrast to the L. pneumophila PC mutant, the flagellar proteins FlaA (modified by glycosylation) (11), FlaB, FlaC, and FlaD were detected in the A. tumefaciens PC-deficient mutant (Fig. 7B), indicating that reduced motility is not caused by the absence of the flagellar proteins. The presence of flagellum filaments was confirmed by silver nitrate staining of intact cells (data not shown).
PC influences A. tumefaciens biofilm formation.
A. tumefaciens forms complex biofilms on abiotic surfaces and plant tissues, with an equilibrium between the sessile and motile lifestyle (10). When microbes directly encounter a surface, the outer membrane represents the interface between cell and substratum. It has been proposed that systems presumably involved in the cell envelope stress response, such as the Cpx or Rcs systems of E. coli, are participating in the transition from the planktonic to the attached life style (15, 34). In addition, it has been shown that motility often correlates with the colonization of surfaces. An aflagellate (Fla−) E. coli mutant is deficient in biofilm formation (35), and several pseudomonads show biofilm formation and architecture dependent on flagella, as well as type IV pili (24, 33). Therefore, we hypothesized that changes in the composition of the outer membrane might also influence the ability of A. tumefaciens to form microbial communities. To investigate biofilm formation in the absence of PC, we compared the wild-type and PC-deficient mutant (ΔpmtA Δpcs) strains in a hydrodynamic flow chamber system in which the attached cells were constantly provided with fresh AB minimal medium. After 24 h, the PC-deficient mutant formed thicker and denser communities than the wild type, with numerous large towering structures (Fig. 8A). The difference became even more obvious after 48 h. In addition to the enhanced structure formation, the mutant strains displayed almost-complete surface coverage, while the wild-type communities exhibited more-unsettled surface areas (Fig. 8B). Except for the increase in height and biomass, no alterations occurred in the basic architecture of the structures formed by the two strains. Cells harvested during the exponential and stationary growth phases showed similar behavior in biofilm formation. In summary, it appears that in the PC-deficient mutant, the equilibrium between the sessile and motile cells is shifted more toward the side of attachment, favoring the accumulation of larger amounts of biomass. Consistent with our results, a similar phenotype was described by Merritt et al. (29) for a nonmotile agrobacterial mutant (ΔflgE). The biofilms formed by the ΔflgE mutant showed increases in biomass, surface coverage, and average structure height compared to those of the wild type. Interestingly, the ΔflgE mutant biofilms grown in static culture were highly reduced relative to those of the wild type (29). In preliminary experiments, we detected a similar effect for the PC-deficient mutant when grown in static culture (data not shown). Several reasons might account for the biofilm phenotype of the ΔpmtA Δpcs mutant. First, it might be attributed to a motility defect similar to that of the ΔflgE mutant. Second, changes in the envelope stress response elicited by alterations in the membrane composition might play a role. Third, it is possible that the altered membrane composition affects biofilm matrix composition. Exopolysaccharides and cellulose are known to influence the attachment and biofilm formation of A. tumefaciens (9).
FIG. 8.
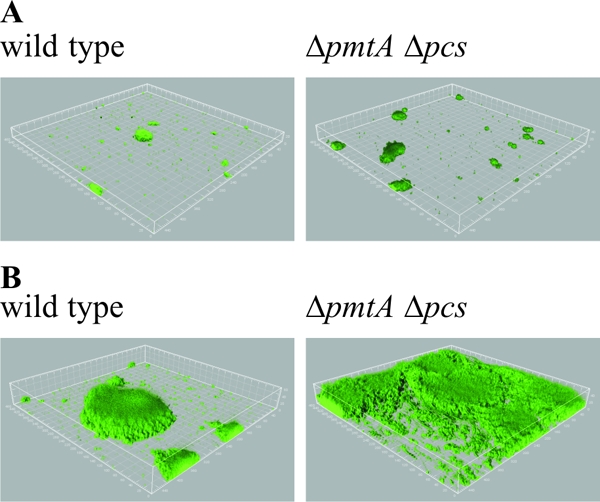
Biofilm formation of a PC-deficient mutant of A. tumefaciens C58. Flow cell biofilms of wild-type and mutant (ΔpmtA Δpcs) expressing GFP were grown in AB minimal medium at 30°C. Microscopic visualization of biofilms was carried out using an inverted Zeiss LSM510 confocal laser-scanning microscope equipped with 20×/0.5 W Achroplan objective. Image data were obtained after 24 h (A) and 48 h (B) and further processed using the IMARIS software package (Bitplane AG, Zürich, Switzerland) and Adobe Photoshop.
Although we are now beginning to appreciate the biological significance of bacterial PC biosynthesis, we are far from understanding the molecular details of how PC influences bacterial physiology and host-microbe interactions. We revealed that PC in the membrane of A. tumefaciens plays a critical role in seemingly diverse processes, such as stress response, motility, social behavior, and virulence. Further studies will be directed toward revealing the underlying mechanisms of PC action in bacterial membranes.
Acknowledgments
We are grateful to Ehr-Min Lai for plasmid pAC01, Yun-long Tsai for advice on membrane separation, and Birgit Scharf for antiflagella sera. We thank Christiane Fritz for excellent technical assistance, Stephanie Hacker for quantitative phospholipid analysis, Robbin Stantscheff for mutant construction, and Bernd Masepohl for helpful comments on the manuscript.
The work was in part supported by a grant from the German Research Foundation (DFG; SFB 480) to F.N. and a fellowship from the Promotionskolleg of the Ruhr-University Bochum to M.A.
Footnotes
Published ahead of print on 31 October 2008.
REFERENCES
- 1.Bligh, E. G., and W. J. Dyer. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37911-917. [DOI] [PubMed] [Google Scholar]
- 2.Canty, D. J., and S. H. Zeisel. 1994. Lecithin and choline in human health and disease. Nutr. Rev. 52327-339. [DOI] [PubMed] [Google Scholar]
- 3.Chesnokova, O., J. B. Coutinho, I. H. Khan, M. S. Mikhail, and C. I. Kado. 1997. Characterization of flagella genes of Agrobacterium tumefaciens, and the effect of a bald strain on virulence. Mol. Microbiol. 23579-590. [DOI] [PubMed] [Google Scholar]
- 4.Christie, P. J., and E. Cascales. 2005. Structural and dynamic properties of bacterial type IV secretion systems. Mol. Membr. Biol. 2251-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Comerci, D. J., S. Altabe, D. de Mendoza, and R. A. Ugalde. 2006. Brucella abortus synthesizes phosphatidylcholine from choline provided by the host. J. Bacteriol. 1881929-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conde-Alvarez, R., M. J. Grilló, S. P. Salcedo, M. J. de Miguel, E. Fugier, J. P. Gorvel, I. Moriyón, and M. Iriarte. 2006. Synthesis of phosphatidylcholine, a typical eukaryotic phospholipid, is necessary for full virulence of the intracellular bacterial parasite Brucella abortus. Cell Microbiol. 81322-1335. [DOI] [PubMed] [Google Scholar]
- 7.Conover, G. M., F. Martínez-Morales, M. I. Heidtman, Z. Q. Luo, M. Tang, C. Chen, O. Geiger, and R. R. Isberg. 2008. Phosphatidylcholine synthesis is required for optimal function of Legionella pneumophila virulence determinants. Cell. Microbiol. 10514-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cronan, J. E. 2003. Bacterial membrane lipids: where do we stand? Annu. Rev. Microbiol. 57203-224. [DOI] [PubMed] [Google Scholar]
- 9.Danhorn, T., and C. Fuqua. 2007. Biofilm formation by plant-associated bacteria. Annu. Rev. Microbiol. 61401-422. [DOI] [PubMed] [Google Scholar]
- 10.Danhorn, T., M. Hentzer, M. Givskov, M. R. Parsek, and C. Fuqua. 2004. Phosphorus limitation enhances biofilm formation of the plant pathogen Agrobacterium tumefaciens through the PhoR-PhoB regulatory system. J. Bacteriol. 1864492-4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deakin, W. J., V. E. Parker, E. L. Wright, K. J. Ashcroft, G. J. Loake, and C. H. Shaw. 1999. Agrobacterium tumefaciens possesses a fourth flagelin gene located in a large gene cluster concerned with flagellar structure, assembly and motility. Microbiology 1451397-1407. [DOI] [PubMed] [Google Scholar]
- 12.de Maagd, R. A., and B. Lugtenberg. 1986. Fractionation of Rhizobium leguminosarum cells into outer membrane, cytoplasmic membrane, periplasmic, and cytoplasmic components. J. Bacteriol. 1671083-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Rudder, K. E., C. Sohlenkamp, and O. Geiger. 1999. Plant-exuded choline is used for rhizobial membrane lipid biosynthesis by phosphatidylcholine synthase. J. Biol. Chem. 27420011-20016. [DOI] [PubMed] [Google Scholar]
- 14.Dupont, L., I. Garcia, M. C. Poggi, G. Alloing, K. Mandon, and D. Le Rudulier. 2004. The Sinorhizobium meliloti ABC transporter Cho is highly specific for choline and expressed in bacteroids from Medicago sativa nodules. J. Bacteriol. 1865988-5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferrieres, L., and D. J. Clarke. 2003. The RcsC sensor kinase is required for normal biofilm formation in Escherichia coli K-12 and controls the expression of a regulon in response to growth on a solid surface. Mol. Microbiol. 501665-1682. [DOI] [PubMed] [Google Scholar]
- 16.Goodner, B., G. Hinkle, S. Gattung, N. Miller, M. Blanchard, B. Qurollo, B. S. Goldman, Y. Cao, M. Askenazi, C. Halling, L. Mullin, K. Houmiel, J. Gordon, M. Vaudin, O. Iartchouk, A. Epp, F. Liu, C. Wollam, M. Allinger, D. Doughty, C. Scott, C. Lappas, B. Markelz, C. Flanagan, C. Crowell, J. Gurson, C. Lomo, C. Sear, G. Strub, C. Cielo, and S. Slater. 2001. Genome sequence of the plant pathogen and biotechnology agent Agrobacterium tumefaciens C58. Science 2942323-2328. [DOI] [PubMed] [Google Scholar]
- 17.Hacker, S., J. Gödeke, A. Lindemann, S. Mesa, G. Pessi, and F. Narberhaus. 2008. Global consequences of phosphatidylcholine reduction in Bradyrhizobium japonicum. Mol. Genet. Genomics 28059-72. [DOI] [PubMed] [Google Scholar]
- 18.Hacker, S., C. Sohlenkamp, M. Aktas, O. Geiger, and F. Narberhaus. 2008. Multiple phospholipid N-methyltransferases with distinct substrate specificities are encoded in Bradyrhizobium japonicum. J. Bacteriol. 190571-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166557-580. [DOI] [PubMed] [Google Scholar]
- 20.Hindahl, M. S., and B. H. Iglewski. 1984. Isolation and characterization of the Legionella pneumophila outer membrane. J. Bacteriol. 159107-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaneshiro, T., and J. H. Law. 1964. Phosphatidylcholine synthesis in Agrobacterium tumefaciens. I. Purification and properties of a phosphatidylethanolamine N-methyltransferase. J. Biol. Chem. 2391705-1713. [PubMed] [Google Scholar]
- 22.Kappes, R. M., B. Kempf, S. Kneip, J. Boch, J. Gade, J. Meier-Wagner, and E. Bremer. 1999. Two evolutionarily closely related ABC transporters mediate the uptake of choline for synthesis of the osmoprotectant glycine betaine in Bacillus subtilis. Mol. Microbiol. 32203-216. [DOI] [PubMed] [Google Scholar]
- 23.Karnezis, T., H. C. Fisher, G. M. Neumann, B. A. Stone, and V. A. Stanisich. 2002. Cloning and characterization of the phosphatidylserine synthase gene of Agrobacterium sp. strain ATCC 31749 and effect of its inactivation on production of high-molecular-mass (1→3)-β-D-glucan (curdlan). J. Bacteriol. 1844114-4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klausen, M., A. Heydorn, P. Ragas, L. Lambertsen, A. Aaes-Jørgensen, S. Molin, and T. Tolker-Nielsen. 2003. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol. 481511-1524. [DOI] [PubMed] [Google Scholar]
- 25.Lambert, A., M. Østerås, K. Mandon, M. C. Poggi, and D. Le Rudulier. 2001. Fructose uptake in Sinorhizobium meliloti is mediated by a high-affinity ATP-binding cassette transport system. J. Bacteriol. 1834709-4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, A. C., H. W. Shih, T. Hsu, and E. M. Lai. 2008. A citrate-inducible gene, encoding a putative tricarboxylate transporter, is downregulated by the organic solvent DMSO in Agrobacterium tumefaciens. J. Appl. Microbiol. 1051372-1383. [DOI] [PubMed] [Google Scholar]
- 27.López-Lara, I. M., C. Sohlenkamp, and O. Geiger. 2003. Membrane lipids in plant-associated bacteria: their biosyntheses and possible functions. Mol. Plant-Microbe Interact. 16567-579. [DOI] [PubMed] [Google Scholar]
- 28.Mandon, K., M. Østerås, E. Boncompagni, J. C. Trinchant, G. Spennato, M. C. Poggi, and D. Le Rudulier. 2003. The Sinorhizobium meliloti glycine betaine biosynthetic genes (betlCBA) are induced by choline and highly expressed in bacteroids. Mol. Plant-Microbe Interact. 16709-719. [DOI] [PubMed] [Google Scholar]
- 29.Merritt, P. M., T. Danhorn, and C. Fuqua. 2007. Motility and chemotaxis in Agrobacterium tumefaciens surface attachment and biofilm formation. J. Bacteriol. 1898005-8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 31.Minder, A. C., K. E. de Rudder, F. Narberhaus, H. M. Fischer, H. Hennecke, and O. Geiger. 2001. Phosphatidylcholine levels in Bradyrhizobium japonicum membranes are critical for an efficient symbiosis with the soybean host plant. Mol. Microbiol. 391186-1198. [PubMed] [Google Scholar]
- 32.Osborn, M. J., J. E. Gander, E. Parisi, and J. Carson. 1972. Mechanism of assembly of the outer membrane of Salmonella typhimurium. Isolation and characterization of cytoplasmic and outer membrane. J. Biol. Chem. 2473962-3972. [PubMed] [Google Scholar]
- 33.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30295-304. [DOI] [PubMed] [Google Scholar]
- 34.Otto, K., and T. J. Silhavy. 2002. Surface sensing and adhesion of Escherichia coli controlled by the Cpx-signaling pathway. Proc. Natl. Acad. Sci. USA 992287-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pratt, L. A., and R. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30285-293. [DOI] [PubMed] [Google Scholar]
- 36.Richardson, J. S., M. F. Hynes, and I. J. Oresnik. 2004. A genetic locus necessary for rhamnose uptake and catabolism in Rhizobium leguminosarum bv. trifolii. J. Bacteriol. 1868433-8442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robinson, J. B., O. H. Tuovinen, and W. D. Bauer. 1992. Role of divalent cations in the subunit associations of complex flagella from Rhizobium meliloti. J. Bacteriol. 1743896-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 39.Schäfer, A., A. Tauch, W. Jäger, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 14569-73. [DOI] [PubMed] [Google Scholar]
- 40.Schmidt-Eisenlohr, H., N. Domke, C. Angerer, G. Wanner, P. C. Zambryski, and C. Baron. 1999. Vir proteins stabilize VirB5 and mediate its association with the T pilus of Agrobacterium tumefaciens. J. Bacteriol. 1817485-7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sherr, S. I., and J. H. Law. 1965. Phosphatidylcholine synthesis in Agrobacterium tumefaciens. II. Uptake and utilization of choline. J. Biol. Chem. 2403760-3765. [PubMed] [Google Scholar]
- 42.Sohlenkamp, C., I. M. López-Lara, and O. Geiger. 2003. Biosynthesis of phosphatidylcholine in bacteria. Prog. Lipid Res. 42115-162. [DOI] [PubMed] [Google Scholar]
- 43.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189113-130. [DOI] [PubMed] [Google Scholar]
- 44.Thormann, K. M., R. M. Saville, S. Shukla, D. A. Pelletier, and A. M. Spormann. 2004. Initial phases of biofilm formation in Shewanella oneidensis MR-1. J. Bacteriol. 1868096-8104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wessel, M., S. Klüsener, J. Gödeke, C. Fritz, S. Hacker, and F. Narberhaus. 2006. Virulence of Agrobacterium tumefaciens requires phosphatidylcholine in the bacterial membrane. Mol. Microbiol. 62906-915. [DOI] [PubMed] [Google Scholar]
- 46.Wilderman, P. J., A. I. Vasil, W. E. Martin, R. C. Murphy, and M. L. Vasil. 2002. Pseudomonas aeruginosa synthesizes phosphatidylcholine by use of the phosphatidylcholine synthase pathway. J. Bacteriol. 1844792-4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Winans, S. C. 1992. Two-way chemical signaling in Agrobacterium-plant interactions. Microbiol. Rev. 5612-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wood, D. W., J. C. Setubal, R. Kaul, D. E. Monks, J. P. Kitajima, V. K. Okura, Y. Zhou, L. Chen, G. E. Wood, N. F. Almeida, Jr., L. Woo, Y. Chen, I. T. Paulsen, J. A. Eisen, P. D. Karp, D. Bovee, Sr., P. Chapman, J. Clendenning, G. Deatherage, W. Gillet, C. Grant, T. Kutyavin, R. Levy, M. J. Li, E. McClelland, A. Palmieri, C. Raymond, G. Rouse, C. Saenphimmachak, Z. Wu, P. Romero, D. Gordon, S. Zhang, H. Yoo, Y. Tao, P. Biddle, M. Jung, W. Krespan, M. Perry, B. Gordon-Kamm, L. Liao, S. Kim, C. Hendrick, Z. Y. Zhao, M. Dolan, F. Chumley, S. V. Tingey, J. F. Tomb, M. P. Gordon, M. V. Olson, and E. W. Nester. 2001. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 2942317-2323. [DOI] [PubMed] [Google Scholar]
- 49.Yeo, H. J., Q. Yuan, M. R. Beck, C. Baron, and G. Waksman. 2003. Structural and functional characterization of the VirB5 protein from the type IV secretion system encoded by the conjugative plasmid pKM101. Proc. Natl. Acad. Sci. USA 10015947-15952. [DOI] [PMC free article] [PubMed] [Google Scholar]