Abstract
Corneal epithelial stem cells are believed to be localized in the limbus, an annular zone between the cornea and the conjunctiva, but it has not been possible to identify individual stem cells in situ because of the lack of specific molecular markers. Description of stem cell distribution has also been ambiguous because limbal boundaries are ill defined. In this study, we investigated whether distribution of slow cycling, label-retaining cells (LRCs) could be determined precisely against a definable anatomical structure of an eye. We found that a boundary between the cornea and the limbus could be determined reliably by distinct epithelial nuclear staining patterns. Using this boundary line as a fiduciary marker, we determined that LRCs were located exclusively in the basal epithelium at the limbal side of the cornea–limbus boundary line along the entire circumference, within an annular zone of 100–200 μm wide. LRC density was highest in the superior temporal quadrant and lowest in the inferior nasal quadrant. These results show that LRCs are present asymmetrically in a narrow zone within the limbus that can be defined precisely in reference to a newly defined anatomical boundary line between the cornea and the limbus. (J Histochem Cytochem 57:177–185, 2009)
Keywords: limbus, corneal epithelium, epithelial homeostasis, label-retaining cells, nuclear staining
The corneal epithelium is a self-renewing tissue, and its stem cells are thought to reside primarily in the basal layer of the limbus, a transitional zone between the cornea and the conjunctiva (Dua and Azuara-Blanco 2000; Lavker et al. 2004; Wolosin et al. 2004; Pajoohesh-Ganji and Stepp 2005; Schlotzer-Schrehardt and Kruse 2005). The prevailing idea is that differentiated daughter cells are generated through asymmetrical divisions of stem cells in the limbus, enter the cornea, and move centripetally (Buck 1985; Nagasaki and Zhao 2003) and vertically (Beebe and Masters 1996) as part of epithelial homeostasis (Thoft and Friend 1983). As such, the importance of stem cells as the ultimate source of the tissue has been recognized, but there have been no reports of identifying individual limbal stem cells in situ. In the absence of definitive biochemical markers (Pajoohesh-Ganji and Stepp 2005; Schlotzer-Schrehardt and Kruse 2005), epithelial stem cells are frequently described as part of a population of slow cycling, label-retaining cells (LRCs), on the premise that stem cells divide less frequently than other differentiated cells (Bickenbach 1981; Cotsarelis et al. 1989). Although it is clear that all LRCs are not stem cells (Braun and Watt 2004), the nearly exclusive presence of LRCs in the limbus has been regarded as one of the evidence that stem cells are enriched in the limbus (Cotsarelis et al. 1989; Lehrer et al. 1998; Lavker et al. 2004). Previous studies suggest that stem cells are not evenly distributed along the limbus in human (Wiley et al. 1991; Pellegrini et al. 1999; Shanmuganathan et al. 2007) and mouse eyes (Pajoohesh-Ganji et al. 2006), but specific location of stem cells within the limbus against a known marker has not been reported.
One of the difficulties of determining stem cell location within the limbus is that boundaries of the limbus itself are not well defined (Van Buskirk 1989). In the mouse eye, an apparent lack of Bowman's membrane makes it even harder to draw the anatomical limbus. It seems necessary to first identify a universal structural marker in or in the immediate vicinity of the limbus that is present in all eyes, so that stem cell location could be referenced against it. Two of the major structural components in the limbus are capillary vessel arcades and stromal nerve trunks, but neither seems to be suited as a fiduciary marker because of heterogeneity of their patterns in different eyes.
Given this background, we set out to determine the precise location of LRCs in the limbus of a mouse eye, in the hope that this information will assist studies on limbal stem cells, including a search for their molecular markers. Our results show that an anatomical boundary between the cornea and the limbus could be determined reliably by a histological method and that this boundary line could be used as a fiduciary marker to describe the spatial distribution of individual LRCs.
Materials and Methods
Animals
All animal procedures were approved by the Columbia University Institutional Animal Care and Use Committee. CAG-EGFP mice [ubiquitous green fluorescent protein (GFP), stock number 003115] were obtained from the Jackson Laboratory (Bar Harbor, ME), and a colony was maintained in-house. Only male mice were used in this study. We chose to study this line because we have accumulated a body of quantitative data on epithelial cell movement and cell division in the ocular surface of these mice (Nagasaki and Zhao 2003,2005; unpublished data), and we intend to further analyze epithelial homeostasis with them. Histone H2B-EGFP mice (stock number 005418; Jackson Laboratory) were used in some experiments.
Metabolic Labeling of DNA for LRCs
Bromodeoxyuridine (BrdU; Sigma-Aldrich, St. Louis, MO) was injected into nine 1-day-old newborn mice subcutaneously at 20 mg/kg twice a day for 4 days (9:00 am and 9:00 pm, total 8 injections), and DNA synthesis was determined by BrdU incorporation into cell nuclei. Eyes from three 5-day-old animals were harvested at 12 hr after the last injection to assess the percentile of basal epithelial cells that were labeled with BrdU. The remaining six mice were sacrificed 9–11 weeks later by an intraperitoneal injection of pentobarbital (100 mg/kg). This chase period was chosen for a technical reason after we had examined corneas with a 6-week chase, which showed a wide range of BrdU immunostaining intensities in a wide area including central cornea. After lengthening the chase period, limbal BrdU staining became weak and fragmented, but a signal-to-noise ratio improved, making manual scoring of limbal LRCs much easier (see below).
Histology
A globe was fixed with freshly prepared 1% paraformaldehyde in PBS and dissected to prepare a flat-mount of an entire ocular surface containing the cornea and the conjunctiva up to the mucocutaneous junction, which was divided into a superior and an inferior half using inner and outer canthus as reference (Nagasaki and Zhao 2005). Whole mounts were stained with a nuclear fluorescence dye, 4′,6-diamidino-2-phenylindole (DAPI), or Hoechst 33258 (both from Sigma-Aldrich), both at 1 μM in PBS. A morphometric analysis of DAPI-stained nuclei was carried out with an open-source program, CellProfiler (Carpenter et al. 2006) (freely available at www.cellprofiler.org), using a segmentation routine based on adaptive Otsu thresholding.
Nuclear BrdU was detected by immunofluorescence. A fixed whole mount tissue was first treated with 2N HCl for 15 min at 37C with gentle shaking. After a rinse with PBS (four times, 5 min each), the tissue was incubated with PBS containing 10% donkey serum (Jackson ImmunoResearch Laboratories; West Grove, PA) and 1% saponin (Sigma-Aldrich) for 3 hr. It was incubated with rat anti-BrdU antibody (Serotec; Raleigh, NC) in PBS containing 4% donkey serum and 0.5% saponin overnight. After a rinse with PBS (four times, 10 min each), it was incubated with donkey anti-rat IgG-Cy3 conjugate (Jackson ImmunoResearch) in PBS containing 4% donkey serum and 0.5% saponin for 6 hr. The tissue was rinsed with PBS (four times, 30 min each), stained with 1 μM DAPI for 10 min, rinsed with PBS, and mounted for imaging with a fluorescence microscope. All antibody incubations were carried out at room temperature in a well of a 96-well plate with a volume of ∼100 μl on a rotary microplate shaker operated at 200–300 rpm.
After images of BrdU-positive cells were digitally recorded, some tissues were restained to identify limbal blood vessels by using rat MECA32 (pan-endothelial cell marker) antibody (developed by Dr. Eugene Butcher and obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biology, Iowa City, IA) and donkey anti-rat IgG-Cy2 conjugate (Jackson ImmunoResearch Laboratories) under the same incubation conditions for BrdU detection except that saponin was used at 0.1%. A very weak reaction of anti-BrdU antibody (that was bound to BrdU in the tissue) with anti-rat IgG-Cy2 did not interfere with imaging of blood vessels with MECA32 because the staining patterns were completely different between BrdU (nuclear) and MECA32 antigen (endothelial, non-nuclear).
For double immunofluorescence staining with keratins 12 and 15, GFP fluorescence of a whole mount tissue was first quenched completely by treatment with methanol before antibody incubation. A tissue was incubated with PBS containing 10% donkey serum and 0.2% saponin for 6 hr at room temperature. Incubation with primary antibodies, goat anti-keratin 12 antibody (Santa Cruz Biotechnology; Santa Cruz, CA), and rabbit anti-keratin 15 antibody (ProteinTech; Chicago, IL) was carried out in PBS containing 4% donkey serum and 0.1% saponin overnight at room temperature. After washing with PBS (four times, 20 min each), the tissue was incubated with secondary antibodies, donkey anti-goat IgG-conjugated with Cy2, and donkey anti-rabbit IgG-conjugated with Cy3 (both from Jackson Immunoresearch) for 6 hr at room temperature. The tissue was rinsed with PBS (four times, 20 min each), stained with 1 μM DAPI for 10 min, rinsed with PBS, and mounted for imaging. Control staining without primary antibody was all negligible.
Imaging and Mapping of LRCs
Ocular surface whole mounts were imaged under a fluorescence microscope (Axioskop2; Carl Zeiss, Oberkochen, Germany). Overlapping microscopic images of the basal epithelial cells were acquired digitally (Orca 100, Hamamatsu, Hamamatsu City, Japan; and Metamorph, Molecular Devices, Downingtown, PA) and assembled with Photoshop (Adobe Systems; San Jose, CA). To determine the number of basal epithelial cells that were labeled with BrdU before a chase period, we manually counted the number of BrdU-positive cells and that of DAPI-stained nuclei in a square area of 250 × 250 μm in each of four quadrants of the cornea.
Because BrdU staining of LRCs was dim and fragmented (see Results), digital images were not efficient in distinguishing BrdU signals from background noise. Confocal imaging was not helpful for surveying the entire cornea because the whole mount specimen was not perfectly flat. We therefore used a wide field fluorescence microscope (Zeiss Axioskop2) with a 40× objective (Achroplan, numerical aperture 0.8), which presented a convenient depth of field (roughly 1 μm), allowing discrimination of the basal epithelium (5–10 μm tall) from upper layers. Accordingly, BrdU-positive LRCs were identified individually under the microscope, inspecting the entire limbal circumference, and plotted manually on a printout of DAPI-stained corneal image outline. While LRCs were identified, the microscope was manually operated by focusing up and down and switching between BrdU and DAPI channels, to properly discriminate signals in the basal epithelium from background. Only those cells with at least one prominent fragment [similar to a “grain” seen in an autoradiogram of [3H]-thymidine-labeled nuclei, for example, in Cotsarelis et al. (1989)] of BrdU staining within the nucleus were counted as LRCs, which is similar to a BrdU score of 0.5 as defined by Pajoohesh-Ganji et al. (2006). The procedure was highly labor intensive but ensured that all, and only, epithelial LRCs in the basal layer were recorded. To avoid bias, two of the authors (JZ and VM) scored all specimens independently and blindly, and the results were averaged.
For spatial quantification, limbal demarcation lines were drawn based on the nuclear appearance of basal epithelial cells indicated by DAPI stain (see Results). A cornea–limbus demarcation line on the superior and inferior half of each eye was measured and further divided into temporal and nasal quarter by a line passing its midpoint and the center of the cornea. LRCs in four quadrants were counted from the manual plots.
Results
Defining the Boundaries of the Limbus
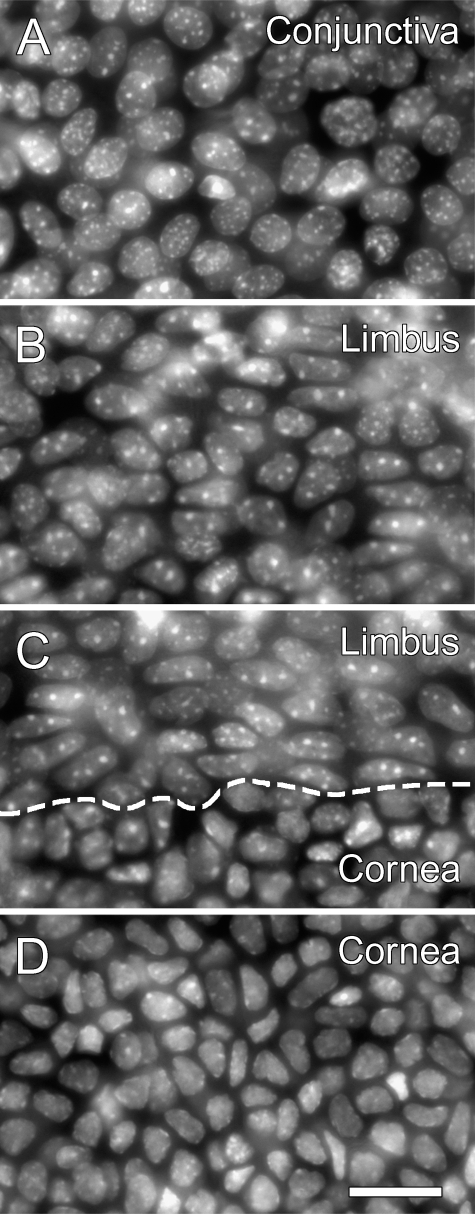
To define the location of limbal stem cells, the limbus itself must be defined precisely and reproducibly as a fixed anatomical structure. After examining histological specimens, we noted that the DAPI nuclear staining pattern appeared distinct among epithelial cells of the cornea, the limbus, and the conjunctiva (Figure 1). A morphometric analysis indicated that the nuclear shape could be distinguished quantitatively between them (Table 1). A close look at a DAPI-stained cornea showed that this feature of the basal epithelium could be used to identify at least one of the two boundaries of the limbus—the one bordering the cornea—in a reproducible manner (Figure 1C). Nuclei of the peripheral corneal cells (Figure 1D) were smaller, somewhat irregularly shaped, and exhibited barely detectable nucleoli. Limbal cell nuclei (Figure 1B) were clearly larger, an oval of a generally uniform shape, and exhibited prominent nucleoli. Because of these differences of the basal epithelial cells, a boundary line between the cornea and the limbus could be drawn. The transition zone occasionally contained two to three cells with ambiguous features that could be grouped into either the cornea or the limbus. Allowing this error margin, the transition zone could be identified unmistakably in all corneas we examined (>20).
Figure 1.
Anatomical distinction of the cornea, limbus, and conjunctiva by the epithelial nuclear appearance in a CAG-EGFP mouse eye. An ocular surface flat mount was stained with 4′,6-diamidino-2-phenylindole (DAPI) to show nuclear appearance, and the basal epithelium was imaged. (A) Conjunctiva. (B) Limbus. (C) Limbus and cornea. (D) Cornea. Dashed line in C represents approximate position of a cornea–limbus boundary line, based on the nuclear appearance. Bar = 20 μm.
Table 1.
Morphometric analysis of nuclei of basal epithelial cells
| Nuclear area (μm2) | Major axis length (μm) | Minor axis length (μm) | Eccentricity | |
|---|---|---|---|---|
| Conjunctiva | 82 ± 27 | 12.5 ± 2.9 | 8.7 ± 1.6 | 0.67 ± 0.15 |
| Limbus | 65 ± 24 | 11.8 ± 2.9 | 7.3 ± 1.4 | 0.74 ± 0.14 |
| Cornea | 45 ± 16 | 9.4 ± 2.1 | 6.2 ± 1.3 | 0.71 ± 0.15 |
4′,6-diamidino-2-phenylindole–stained nuclei in a flat whole mount were imaged and analyzed with CellProfiler. Average and SD are shown. The number of nuclei scored for the analysis was 133, 173, and 786 for conjunctiva, limbus, and cornea, respectively. Eccentricity is a ratio of the distance between the foci of the ellipse and its major axis length. A t-test showed that the data between conjunctiva, limbus, and cornea were significantly different from each other in all four data sets with criteria of p<0.05.
To verify that the line in this transition zone truly lies at the border of the limbus, we carried out two tests. First, we confirmed that the transition zone was in the exact area of clinical limbus, as judged by gross observation of an eye. Second, we compared the boundary line in the transition zone with immunofluorescence patterns of keratin 12 (cornea marker; Liu et al. 1993) and keratin 15 (conjunctiva and limbus marker; Yoshida et al. 2006) (Figure 2). Demarcation between keratin 12 and keratin 15 was clear with a low-power image (Figure 2C), but it was rather ambiguous at a single cell resolution because of the presence of several rows of cells that were apparently positive with keratin 12 and/or keratin 15 to different degrees (Figure 2G). Nevertheless, it was clear that the line in the transition zone, as determined by DAPI nuclear staining (Figure 2F), was right in the middle of the keratin12–keratin15 transition zone (Figure 2G). These results suggest that a transition zone of DAPI nuclear patterns can in fact define the boundary of the cornea and the limbus. An examination of the limbal area showed that, although the limbus is highly vascularized, tips of limbal capillary loops never reached the cornea–limbus boundary line in all corneas we examined. As a consequence, there was an avascular zone of 100–200 μm wide within the limbus adjacent to the cornea.
Figure 2.
Ocular surface staining patterns with keratins 12 and 15 in reference to a cornea–limbus boundary line. A flat whole mount tissue was processed for double immunofluorescence with keratin 12 (A,D) and keratin 15 (B,E) and also DAPI nuclear staining (F) to show a cornea–limbus boundary line, which is drawn with dashed white lines (D–G). C and G show merged images of keratins 12 and 15. A–C show an entire superior half of the ocular surface (central cornea at the bottom, a lid margin at the top, with slits to mount the tissue flat) from a 30-week-old mouse eye. D–G show a high-power image of the same tissue at an area of the cornea–limbus border. Original images were all in grayscale but converted to pseudo-color for presentation. For D–G, several high-power microscopic fields were patched together to generate each panel. Bar = 50 μm.
The cornea–limbus boundary could also be identified by nuclear GFP or Hoechst 33258 staining in an eye of a histone-H2B-EGFP mouse (Figure 3). This shows that this feature is not limited to DAPI staining or CAG-EGFP mice and suggests that a histone-H2B-EGFP mouse could be used in live imaging experiments without external nuclear labeling.
Figure 3.
Anatomical distinction of the cornea, limbus, and conjunctiva by the epithelial nuclear appearance in a histone H2B-EGFP mouse eye. The basal epithelium of an ocular surface flat-mount was imaged for intrinsic nuclear green fluorescent protein (GFP) (A,C,E,G) or Hoechst 33258 nuclear staining (B,D,F,H). Representative areas of conjunctiva (A,B), limbus (C,D), cornea–limbus border (E,F), and cornea (G,H) are shown. Dashed lines in E and F represent approximate position of a cornea–limbus boundary line. Bar = 20 μm.
A comparison of limbal cells and conjunctival cells in the respective basal epithelium showed that these two groups of cells also have some differences in DAPI staining patterns (Figures 1A and 1B). Compared with limbal nuclei, conjunctival nuclei were more circular and slightly larger. Nevertheless, the transition from the limbus to the bulbar conjunctiva was gradual and no distinct demarcation line, such as that between the cornea and the limbus, could be found. The results were essentially the same with histone-H2B-EGFP eyes, whose epithelial nuclei were shown by GFP or Hoechst staining (Figure 3). In combination with other features such as major trunks of blood vessels and nerves that run along the proximity of a limbus–conjunctiva boundary, our tentative assessment is that a limbus–conjunctiva boundary is at ∼250–350 μm from the cornea–limbus boundary. However, in the absence of clear anatomical features, we did not make any quantitative use of a limbus–conjunctiva boundary line in this study.
Spatial Distribution of LRCs
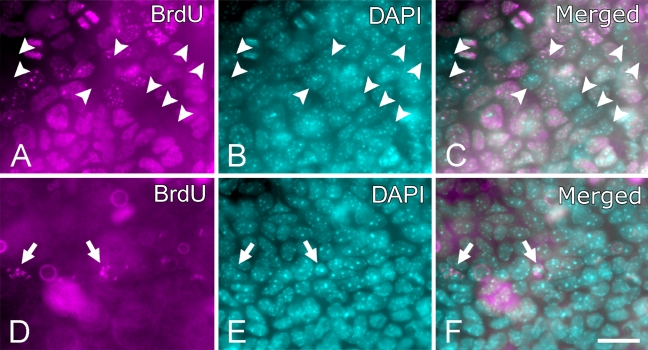
Immediately after BrdU labeling of pups, about three quarters (74.8 ± 4.9%, n=12,464) of basal epithelial cells were labeled with BrdU (Figure 4A). After a chase of 9–11 weeks, BrdU-positive cells (Figure 4B), or LRCs, in the basal epithelium were found exclusively within the limbus. The majority of them were located near the cornea–limbus boundary along the entire limbal circumference, inside a narrow annular zone of 100–200 μm wide (Figure 5). When compared against limbal blood vessels, LRCs were mostly found near the cornea side of the blood vessel arcades, which is an avascular part of the limbus, although there were occasional LRCs directly above the capillary loops (Figure 5). This general description was true in all 12 eyes we examined. A thorough study of entire corneas (n=12) showed that, outside the limbus, a handful of LRCs were present in the suprabasal layer of the epithelium at random locations but none in the basal layer (data not shown). Those LRCs may represent bone marrow–derived cells, but this was not determined.
Figure 4.
Bromodeoxyuridine (BrdU) staining in basal epithelium of the limbus. A and B show basal epithelium of a P4 mouse cornea labeled with BrdU for 4 days and fixed without a chase, double stained for BrdU (A) and DAPI (B). C shows a merged image of DAPI and BrdU. Arrowheads point to all cells that did not label with BrdU in the field. D–F show basal epithelium of the limbus after 4-day BrdU labeling and 9-week chase, double stained for BrdU (D) and DAPI (E). F is a merged image of DAPI and BrdU. Arrows indicate BrdU-positive cells [label-retaining cells (LRCs)], which could be clearly identified and located to the basal epithelium with a manual inspection under the microscope by changing the focal plane slightly up and down and by switching between the BrdU and DAPI channels. Original images were all in grayscale but converted to pseudo-color for presentation. Bar = 20 μm.
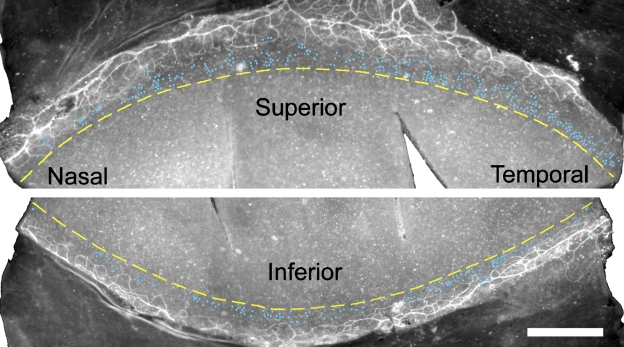
Figure 5.
Distribution of LRCs along the entire circumference of the limbus, presented as a superior and an inferior half. Each blue dot in the limbus represents an LRC that was manually identified under the microscope. Background is MECA32 immunofluorescence images that depict limbal capillary vessel arcades. Dashed yellow lines along the limbus indicate a cornea–limbus boundary as determined by the DAPI nuclear appearance of high-power images. This is a left eye and a representative of 12 eyes that showed similar patterns. Bar = 500 μm.
To describe the spatial distribution quantitatively, we counted the number of LRCs in the limbus; we did not count non-LRCs (BrdU-negative cells), but as a reference, an annular zone of 8.6 mm long and 100 μm wide would contain ∼8600 basal epithelial cells, assuming a density of 10,000 cells/mm2. The results (Table 2) indicated that there were more LRCs in the superior half than in the inferior half and more in the temporal half than the nasal half in both right eyes and left eyes. Among the four quadrants, the superior temporal region contained the highest number, and the inferior nasal had the least, in both eyes (Table 2).
Table 2.
Distribution of LRCs by quadrants
| Superior temporal | Superior nasal | Inferior temporal | Inferior nasal | Total | |
|---|---|---|---|---|---|
| Limbal arc length (μm) | 2110 ± 150 | 2110 ± 150 | 2180 ± 140 | 2180 ± 140 | 8590 ± 210 |
| Number of individual LRCs | 123 ± 35 | 89 ± 31 | 101 ± 30 | 48 ± 13 | 361 ± 78 |
| Percentage (%) | 34 ± 10 | 25 ± 9 | 28 ± 8 | 13 ± 4 | 100 |
The limbal arc length was measured along the cornea–limbus boundary. Average and SD of LRC numbers were determined from measurements with 12 eyes (six mice). Percentage distribution in four quadrants is shown. A t-test showed that LRC numbers in each quadrant were significantly different from each other with criteria of p<0.0005. LRC, label-retaining cell.
Discussion
We determined the distribution of individual LRCs in the entire limbal circumference in mouse eyes and confirmed previous reports that LRCs were mostly located in the limbus (Cotsarelis et al. 1989; Lehrer et al. 1998; Pajoohesh-Ganji et al. 2006). Major findings of this study are as follows: (a) we were able to define the cornea–limbus boundary by distinct appearances of DAPI-stained nuclei, (b) this cornea–limbus boundary line could be used to document the precise location of limbal LRCs, (c) LRCs were distributed asymmetrically along the limbal circumference—the highest number in the superior temporal and the lowest in the inferior nasal quadrant in both right and left eyes.
The general location of the limbus in the mouse eye can be pointed out at a gross level, but a cell level resolution of its definition has not been possible because the epithelium of the cornea–limbus–conjunctiva is continuous without any anatomical structures that would distinguish their boundaries against a neighboring unit. Our finding in this study indicates that there is an anatomical marker—a cornea–limbus boundary line—that can be reliably determined with a potential error margin of two to three cell lengths. This feature is useful not only with a fixed histological specimen but also with a cornea of a live mouse, raising various experimental possibilities. Our preliminary observation suggested that the cornea–limbus boundary can be identified in a cornea of a live and anesthetized mouse with a histone H2B-EGFP mouse (unpublished data).
The presence of a clear demarcation between the cornea and the limbus suggests that homeostatic movement of limbal cells into the cornea across this border is accompanied by an immediate and drastic change of a cellular phenotype. A question is whether this anatomical structure (a boundary line) is a cause of a limbal-to-corneal transition or whether it exists as a result of it. A study into this issue may reveal unique mechanisms of stem cell differentiation and epithelial homeostasis in the cornea.
LRCs are considered to be a population of cells enriched with stem cells (Bickenbach 1981; Cotsarelis et al. 1989; Potten and Booth 2002; Braun and Watt 2004), but some LRCs are decidedly not stem cells, and a correlation between LRCs and stem cells is not well established in the cornea. Nevertheless, a crude picture of stem cell distribution may be gleaned from LRC numbers. Assuming a density of 10,000 cells/mm2, we calculate from the data in Table 2 that roughly 1 in 24 basal epithelial cells (4.2%) is an LRC within the annular LRC-populated zone of the limbus (100 μm wide), which is comparable to the 0.94–3.6% that was reported by Pajoohesh-Ganji et al. (2006). The precise number of stem cells probably will only be determined after specific molecular markers become available.
On the other hand, the anatomical finding of LRC distribution may be useful in predicting stem cell distribution because it is likely that many of the LRCs are in fact stem cells (Bickenbach 1981; Cotsarelis et al. 1989; Potten and Booth 2002; Braun and Watt 2004). Our identification of the location of LRCs may facilitate a search for molecular markers for stem cells, because this area can be pointed out precisely with an epithelial nuclear transition line between the limbus and the cornea, as well as capillary loops as a secondary reference marker. The annular LRC-rich zone contains roughly 10,000–15,000 basal epithelial cells per eye, and these are the cells that future studies will target.
We found an asymmetric distribution of LRCs (Table 2), but regional heterogeneity in the limbal epithelium has been reported previously (Wiley et al. 1991; Pellegrini et al. 1999; Pajoohesh-Ganji et al. 2006; Shanmuganathan et al. 2007). Pellegrini et al. (1999) showed that the colony-forming efficiency varied among four quadrants of the human limbus, with the superior and temporal regions considerably higher than inferior and nasal regions. Our results on LRC distribution were similar (Table 2), although a species difference (human vs mouse) precludes a direct comparison. On the other hand, Pajoohesh-Ganji et al. (2006) reported that LRCs were distributed asymmetrically in Balb/c mouse eyes with superior and inferior quadrants containing more LRCs than nasal and temporal quadrants, which is slightly different from our results. The reason for the difference is not clear, but it may be because of the difference in the limbal anatomy in different mouse strains (Chan et al. 2004) or the difference in the length of the chase period for identifying LRCs (6 vs >9 weeks). Regardless of the cause, we think an approach such as this will provide a foundation for quantitative analysis of epithelial homeostasis for a better understanding of ocular surface biology.
In summary, our data showed that LRCs are located in a narrow zone of the limbus that can be defined precisely by anatomical markers. It is hoped that these data will be useful in a quantitative study of epithelial homeostasis in the mouse cornea, as well as in a search of molecular markers for stem cells and associated cells.
Acknowledgments
This work was supported by grants from the National Institute of Health (R01-EY-015835) and Research to Prevent Blindness.
References
- Beebe DC, Masters BR (1996) Cell lineage and the differentiation of corneal epithelial cells. Invest Ophthalmol Vis Sci 37:1815–1825 [PubMed] [Google Scholar]
- Bickenbach JR (1981) Identification and behavior of label-retaining cells in oral mucosa and skin. J Dent Res 60:1611–1620 [DOI] [PubMed] [Google Scholar]
- Braun KM, Watt FM (2004) Epidermal label-retaining cells: background and recent applications. J Investig Dermatol Symp Proc 9:196–201 [DOI] [PubMed] [Google Scholar]
- Buck RC (1985) Measurement of centripetal migration of normal corneal epithelial cells in the mouse. Invest Ophthalmol Vis Sci 26:1296–1299 [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, et al. (2006) CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol 7:R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CK, Pham LN, Chinn C, Spee C, Ryan SJ, Akhurst RJ, Hinton DR (2004) Mouse strain-dependent heterogeneity of resting limbal vasculature. Invest Ophthalmol Vis Sci 45:441–447 [DOI] [PubMed] [Google Scholar]
- Cotsarelis G, Cheng SZ, Dong G, Sun TT, Lavker RM (1989) Existence of slow-cycling limbal epithelial basal cells that can be preferentially stimulated to proliferate: implications on epithelial stem cells. Cell 57:201–209 [DOI] [PubMed] [Google Scholar]
- Dua HS, Azuara-Blanco A (2000) Limbal stem cells of the corneal epithelium. Surv Ophthalmol 44:415–425 [DOI] [PubMed] [Google Scholar]
- Lavker RM, Tseng SC, Sun TT (2004) Corneal epithelial stem cells at the limbus: looking at some old problems from a new angle. Exp Eye Res 78:433–446 [DOI] [PubMed] [Google Scholar]
- Lehrer MS, Sun TT, Lavker RM (1998) Strategies of epithelial repair: modulation of stem cell and transit amplifying cell proliferation. J Cell Sci 111:2867–2875 [DOI] [PubMed] [Google Scholar]
- Liu CY, Zhu G, Westerhausen-Larson A, Converse R, Kao CW, Sun TT, Kao WW (1993) Cornea-specific expression of K12 keratin during mouse development. Curr Eye Res 12:963–974 [DOI] [PubMed] [Google Scholar]
- Nagasaki T, Zhao J (2003) Centripetal movement of corneal epithelial cells in the normal adult mouse. Invest Ophthalmol Vis Sci 44:558–566 [DOI] [PubMed] [Google Scholar]
- Nagasaki T, Zhao J (2005) Uniform distribution of epithelial stem cells in the bulbar conjunctiva. Invest Ophthalmol Vis Sci 46:126–132 [DOI] [PubMed] [Google Scholar]
- Pajoohesh-Ganji A, Pal-Ghosh S, Simmens SJ, Stepp MA (2006) Integrins in slow-cycling corneal epithelial cells at the limbus in the mouse. Stem Cells 24:1075–1086 [DOI] [PubMed] [Google Scholar]
- Pajoohesh-Ganji A, Stepp MA (2005) In search of markers for the stem cells of the corneal epithelium. Biol Cell 97:265–276 [DOI] [PubMed] [Google Scholar]
- Pellegrini G, Golisano O, Paterna P, Lambiase A, Bonini S, Rama P, De Luca M (1999) Location and clonal analysis of stem cells and their differentiated progeny in the human ocular surface. J Cell Biol 145:769–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potten CS, Booth C (2002) Keratinocyte stem cells: a commentary. J Invest Dermatol 119:888–899 [DOI] [PubMed] [Google Scholar]
- Schlotzer-Schrehardt U, Kruse FE (2005) Identification and characterization of limbal stem cells. Exp Eye Res 81:247–264 [DOI] [PubMed] [Google Scholar]
- Shanmuganathan VA, Foster T, Kulkarni BB, Hopkinson A, Gray T, Powe DG, Lowe J, et al. (2007) Morphological characteristics of the limbal epithelial crypt. Br J Ophthalmol 91:514–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoft RA, Friend J (1983) The X, Y, Z hypothesis of corneal epithelial maintenance. Invest Ophthalmol Vis Sci 24:1442–1443 [PubMed] [Google Scholar]
- Van Buskirk EM (1989) The anatomy of the limbus. Eye 3:101–108 [DOI] [PubMed] [Google Scholar]
- Wiley L, SundarRaj N, Sun TT, Thoft RA (1991) Regional heterogeneity in human corneal and limbal epithelia: an immunohistochemical evaluation. Invest Ophthalmol Vis Sci 32:594–602 [PubMed] [Google Scholar]
- Wolosin JM, Budak MT, Akinci MA (2004) Ocular surface epithelial and stem cell development. Int J Dev Biol 48:981–991 [DOI] [PubMed] [Google Scholar]
- Yoshida S, Shimmura S, Kawakita T, Miyashita H, Den S, Shimazaki J, Tsubota K (2006) Cytokeratin 15 can be used to identify the limbal phenotype in normal and diseased ocular surfaces. Invest Ophthalmol Vis Sci 47:4780–4786 [DOI] [PubMed] [Google Scholar]