Abstract
Many Ser/Thr protein kinases are activated by autophosphorylation, but the mechanism of this process has not been defined. We determined the crystal structure of a mutant of the Ser/Thr kinase domain (KD) of the mycobacterial sensor kinase PknB in complex with an ATP competitive inhibitor and discovered features consistent with an activation complex. The complex formed an asymmetric dimer, with the G helix and the ordered activation loop of one KD in contact with the G helix of the other. The activation loop of this putative ‘substrate' KD was disordered, with the ends positioned at the entrance to the partner KD active site. Single amino-acid substitutions in the G-helix interface reduced activation-loop phosphorylation, and multiple replacements abolished KD phosphorylation and kinase activation. Phosphorylation of an inactive mutant KD was reduced by G-helix substitutions in both active and inactive KDs, as predicted by the idea that the asymmetric dimer mimics a trans-autophosphorylation complex. These results support a model in which a structurally and functionally asymmetric, ‘front-to-front' association mediates autophosphorylation of PknB and homologous kinases.
Keywords: autophosphorylation, bacterial protein kinase, G helix
Introduction
Tight regulation of Ser/Thr and Tyr protein kinases is required to control phospho-signaling pathways. The exquisite control of kinase activity is mediated by the convergence of multiple allosteric mechanisms that combine to establish kinase domain (KD) conformations (Huse and Kuriyan, 2002). These diverse activation signals include not only the binding of regulatory subunits but also the phosphorylation of the KD. In the Src family kinases, for example, the inactive form is maintained by a coupled network of interactions set up by phosphorylation of Tyr527, intramolecular binding of this pTyr residue by the Src SH2 domain, release of partner proteins by the Src SH3 domain, association of the SH3 domain with an extended linker that engages the KD and dephosphorylation of a characteristic motif called the activation loop. In contrast, activation-loop phosphorylation in Src and many homologous kinases promotes the assembly of a platform on the edge of the kinase catalytic site that engages the regulatory C helix in an active conformation and mediates binding of protein substrates. In some kinases, this phosphorylation also relieves a steric blockade of the ATP-binding site mediated by the unphosphorylated activation loop (Huse and Kuriyan, 2002; Biondi and Nebreda, 2003). Despite the central importance of activation-loop phosphorylation in kinase activation, the recognition mechanisms that mediate this reaction are not well defined.
Here, we investigate the autophosphorylation mechanism of Mycobacterium tuberculosis (Mtb) PknB, one of 11 Mtb Ser/Thr protein kinases (STPKs) that are structurally and mechanistically similar to eukaryotic homologues (Av-Gay and Everett, 2000; Greenstein et al, 2005). The primary sequences of the mycobacterial STPKs show up to 30% identity with those of human family members. The three-dimensional structures of the KDs of Mtb PknB, PknE and PknG resemble those of human kinases, with conserved sequence motifs having functionally analogous roles (Ortiz-Lombardia et al, 2003; Young et al, 2003; Gay et al, 2006; Scherr et al, 2007). By a mechanism with striking parallels to the activation of human double-stranded RNA-dependent protein kinase (PKR; Dar et al, 2005; Dey et al, 2005), the dephosphorylated Mtb PknD KD is activated by dimerization through an allosteric, ‘back-to-back' interface in the N-terminal lobe (Greenstein et al, 2007a). This apparently ancient allosteric interface shows high conservation among orthologous bacterial STPKs and promotes KD autophosphorylation, which activates the KDs for transphosphorylation of heterologous substrate proteins (Young et al, 2003; Greenstein et al, 2007a, 2007b).
Similar to many eukaryotic homologues, the mycobacterial KDs are activated by autophosphorylation of specific residues in the ‘activation loop', a conserved segment bordered in the amino-acid sequence by the DFG and APE tripeptides. The KDs from PknB, PknD, PknE and PknF, for example, show a conserved pattern of autophosphorylation on the activation loop and several other sites (Boitel et al, 2003; Young et al, 2003; Duran et al, 2005). Dephosphorylation of the KDs invariably causes a large reduction in kinase activity in vitro. Single Ala replacements for Thr171 and Thr173 in the PknB activation loop reduced kinase activity on the non-cognate substrate myelin basic protein (MyBP) by 15- and 20-fold, respectively, and the double mutant was 300-fold less active (Boitel et al, 2003). Phosphorylation of Thr171 and Thr173 was confirmed in PknB overexpressed in Mtb, suggesting that these sites are autophosphorylated in vivo (Kang et al, 2005). Overexpression of the full-length PknD kinase in Mtb led simultaneously to the accumulation of phosphorylated PknD and to increased phosphorylation of cellular proteins (Greenstein et al, 2007a). Addition of the PknD inhibitor SP600125 reduced both autophosphorylation and transphosphorylation, supporting the idea that autophosphorylation activates the kinase in vivo. These studies established that the activation loops of several mycobacterial KDs are autophosphorylated similar to those of numerous eukaryotic kinases, and these phosphorylation reactions activate the bacterial STPKs in vitro and in vivo.
Here, we investigate the mechanisms of inhibitor recognition and autophosphorylation of Mtb STPKs. Because these kinases afford validated targets for new anti-infectives (Wehenkel et al, 2006), we sought to define the basis for Mtb PknD recognition by the eukaryotic protein kinase inhibitor, KT5720. To overcome our failure to cocrystallize the KD–inhibitor complex, we mutated the ATP-binding site of the Leu33Asp variant of PknB, which crystallizes readily, to closely resemble the active site of PknD. Here, we show that this ‘PknD surrogate' acquired enhanced affinity for KT5720 and staurosporine and yielded cocrystals with KT5720 suitable for structural analysis. In addition to revealing the mechanism of KT5720 selectivity, this structure showed unanticipated features that suggested clues about the mechanism of KD autophosphorylation. The two PknD surrogate KDs formed an unusual offset dimer in which the activation loop of only one monomer was ordered, whereas that of the other monomer was disordered and positioned to extend into the active site of the other KD. Asymmetric contacts between G-helix residues in both KDs formed most of the dimer interface. Supporting the idea that this structure represents an autophosphorylation ES complex, activation-loop autophosphorylation occurred in an intermolecular reaction and mutations in helix G altered the efficiency of autophosphorylation. We used a new strategy to map the phosphorylation states of the activation loops of the PknB G-helix mutants, and the results suggested that autophosphorylation may occur in a preferred order. These results support a model in which two distinct interfaces mediate PknB activation and suggest that asymmetric G-helix recognition has a key function in the activation of STPKs in prokaryotes and eukaryotes.
Results
Structure of PknD surrogate bound to KT5720 revealed an asymmetric interface
To identify selective inhibitors of Mtb STPKs, we screened a variety of commercially available inhibitors of eukaryotic kinases. Staurosporine and the staurosporine analogue, KT5720, inhibited the purified KD of Mtb PknD with IC50 values in the mid-nanomolar range (Figure 1). Because efforts to crystallize the complex of KT5720 with the PknD KD failed, we made mutations in the PknB KD to mimic the ATP-binding site of PknD and to promote crystallization. With two mutations, Met145Leu and Met155Val, we converted the sequence of the PknB ATP-binding site to that of PknD (Figure 1A). Importantly, this PknD surrogate KD phosphorylated the non-cognate substrate MyBP with activity comparable to the wild-type (WT) PknB KD (data not shown) and gained the ability to bind KT5720 and staurosporine with IC50 values below 1 μM (Figure 1A). To aid in crystallizing the KD–inhibitor complex, we also introduced the Leu33Asp mutation in the ‘back-to-back' dimer interface that mediates activation of the unphosphorylated KD. This single mutation reduces the strength of the back-to-back interface, preserves significant autophosphorylation activity and promotes crystallization of the PknB KD (T Noelle Lombana et al, unpublished results).
Figure 1.
Complex of PknD surrogate KD with KT5720. (A) Autoradiograms showing MyBP phosphorylation catalysed by the KDs (100 nM) of PknD, PknB and the Met145Leu/Met155Val double mutant of PknB. In this double mutant ‘PknD surrogate', all the ATP contact residues except Tyr94 (bottom) matched those in PknD. Each set of three lanes shows the reactions without inhibitor and in the presence of 10 μM staurosporine and 10 μM KT5720, respectively. The double substitution to PknB allows KT5720 to bind at 10 μM. (B) Ribbon diagram showing the KT5720–PknD surrogate KD structure. Monomers A (yellow) and B (blue) form an asymmetric dimer stabilized by contacts of the G helices (highlighted in transparent surface) and the activation loop (red) of monomer B. (C) Superposition of the active site of the KT5720–PknD surrogate KD (blue) with wild-type PknB (grey). The Met145Leu and Met155Val substitutions derived from PknD create room in the active site for the inhibitor. (D) Superposition of the two independent PknD surrogate monomers. The two monomers adopt similar overall structures (0.43 Å r.m.s.d.). The activation loop (residues 161–179) of monomer A was disordered, but this segment (red surface) except for residues 174–177 could be placed in the electron density in monomer B. (E) View of G-helix interface. The dimer is not two-fold symmetric, putting identical residues in the monomers in distinct environments. Van der Waals contacts include Val222 (B, blue)-Val229 (A, yellow), Ala225 (A)-Val222 (B), Leu183 (B)-Val222 (A) (not shown). (F) Interactions in the G-helix interface. Several backbone interactions occur between the EF loop (monomer A) and the activation loop (monomer B). Arg161 and Asp219 form an ion pair, and Leu183 also contacts the opposing G helix. The only phosphorylated residue with clear density, pThr171 (B), is exposed and contacts Thr217 (A) in the EF loop. (G) Sequence alignment of 90 PknB orthologues mapped onto surface of PknB. Conserved regions (red >95% identical, blue least conserved) include the ATP site, the putative protein–substrate-binding site, the N-lobe interface (not shown) and the G helix.
As expected, the back-to-back dimer found in three crystal forms of the WT PknB KD (Ortiz-Lombardia et al, 2003; Young et al, 2003; Wehenkel et al, 2006) was not formed in the crystals of the KT5720 complex of the Leu33Asp PknD surrogate (Table I). Instead, the surrogate KD crystallized as an unusual asymmetric dimer (Figure 1B). The monomers adopted similar conformations (backbone root mean square deviation (r.m.s.d.)=0.43 Å), and each monomer bound KT5720 in the ATP-binding site. KT5720 bound in a position similar to that seen earlier in the complexes of staurosporine or its analogues with numerous eukaryotic protein kinases (Supplementary Figure 1) (Lawrie et al, 1997). The structure revealed that the Met145Leu and Met155Val mutations in the PknB ATP-binding site created space to accommodate the inhibitor (Figure 1C).
Table 1.
Data collection and refinement statistics
| PknB L33D/M145L/M155V-KT5720 | PknB L33D/V222D-ADP | |
|---|---|---|
| Data collection | ||
| Space group | F4132 | P 212121 |
| Unit cell dimensions | ||
| a, b, c (Å) | 297.56, 297.56, 297.56 | 38.98, 51.54, 152.54 |
| Resolution (Å)a | 50–2.8 (2.9–2.8) | 50–1.8 (1.86–1.8) |
| Rmerge | 9.5 (84.9) | 5.6 (36.0) |
| I/σI | 29.7/0.6 (2.7/0.6) | 136.1/2.8 (6.4/1.7) |
| Completeness (%) | 99.2 (99.9) | 99.3 (93.4) |
| Redundancy | 32.3 (32.7) | 8.0 (6.4) |
| Refinement | ||
| Resolution (Å) | 39.8–2.80 | 37.8–1.80 |
| Unique reflections | 26 637 | 27 635 |
| Rwork/Rfree | 21.8/26.8 | 20.3/22.8 |
| Atoms | 4267 | 2326 |
| Protein | 4147 | 2111 |
| Ligand/ion | 90 | 31 |
| Water | 30 | 184 |
| B-factors (overall) | 25.5 | 27.4 |
| r.m.s.d. | ||
| Bond lengths (Å) | 0.010 | 0.012 |
| Bond angles (deg) | 1.41 | 1.46 |
| Backbone dihedral angles preferred/allowed (%) | 91.0/9.0 | 93.5/6.5 |
| aValues in parentheses describe the highest resolution shell. | ||
Unlike previous structures of the PknB KD (Ortiz-Lombardia et al, 2003; Young et al, 2003; Gay et al, 2006; Wehenkel et al, 2006), the activation loop of monomer B was largely visible in the electron density map (Figure 1D). The phosphorylated Thr171 made intramolecular contacts to the CD loop and intermolecular contacts to the opposing EF loop. In contrast, the activation loop of monomer A was disordered. Unexpectedly, monomers A and B were not related to each other by a simple two-fold rotation axis. Instead, a rotation of 132° and a translation of 4.4 Å were required to optimally superimpose the two KDs. This offset resulted from the zipper-like interdigitation of residues in helix G of each monomer. The offset placed the ends of the disordered activation loop of monomer A into the entrance to the kinase active site of monomer B.
This asymmetric dimer interface buried a total of 1492 Å2, sequestering a number of hydrophobic residues from solvent (Figure 1E; Supplementary Figure 2). Val222, Ala225, Tyr226 and Val229 in the G helix and other residues, including Leu183 and Arg189, at the margins of the activation loop made the principal intermolecular contacts. The relative translation of the monomers placed identical residues in distinct environments. For instance, Val222 in monomer B makes contacts with Ala225 and Val229 in monomer A, whereas Val222 in monomer A makes contacts with Ala225, Ala180 and Leu183 in monomer B. Additional interactions included a van der Waals contact between Arg189 (B) and Tyr226 (A), a salt bridge between Asp219 (A) and Arg161 (B), and a possible cation–pi interaction between Arg230 (A) and Tyr226 (B) (Figure 1F). Asymmetric backbone interactions also occur throughout the interface. Most contact residues are well conserved in PknB orthologues from 90 species (Figure 1G), supporting the hypothesis that the observed interactions are not simply the adventitious consequence of crystallization, but instead may mediate important functions.
Substitutions in PknB G-helix dimer alter autophosphorylation and kinase activation
To explore the function of the helix G interface, mutations were introduced into the WT PknB KD, and the effects on the rates of auto-activation and transphosphorylation were analysed. In these experiments, the purified KDs were first dephosphorylated using the Mtb Ser/Thr phosphatase, PstP, and autophosphorylation was measured using γ-32P-ATP. Single amino-acid replacements in the hydrophobic G-helix interface reduced the rate of autophosphorylation in vitro (Figure 2A). Double substitutions such as Val222Asp/Val229Asp and Ala225Glu/Val229Asp further reduced the autophosphorylation rate. In contrast, replacement of Ala225 in helix G with Leu increased the autophosphorylation of the KD, supporting the idea that this site is buried during the autophosphorylation reaction.
Figure 2.
Helix G substitutions reduced autophosphorylation of the PknB KD. (A) Autoradiogram and plot showing autophosphorylation activities of dephosphorylated PknB KD variants. The Val222Asp, Ala225Glu, Ala225Leu and Val229Asp substitutions (cyan) were made in the G helix. The acidic substitutions of residues 222, 225 and 229 individually reduced autophosphorylation 3.7-, 4.3- and 4.5-fold after 15 min, respectively (values are tabulated in Supplementary Figure 3). The Ala225Leu PknB KD showed 2.4-fold increased autophosphorylation activity at 15 min compared with the wild-type KD. The Leu33Asp substitution (orange) in the conserved regulatory N-lobe interface on the opposite side of the KD and the Val222Asp G-helix substitution showed synergistic effects. (B) Relative activities of the PknB 1–308 KD variant populations purified from E. coli. Activities were determined by measuring in vitro phosphorylation of MyBP. The relative transphosphorylation activities of each variant correlated well with the degree of intrinsic autophosphorylation (Table II) and the autophosphorylation activity observed in vitro.
To assess the autophosphorylation activities of the KDs in a cellular context, electrospray ionization mass spectrometry (ESI-MS) was used to define the populations of phosphates on the recombinant KDs purified from Escherichia coli. E. coli was used as a host organism because of the ease of analysis and the absence of any other STPKs capable of phosphorylating the KD variants. Single mutations such as Val222Asp and Val229Asp in the G-helix interface reduced the number of phosphates on the PknB KD (Table II). Moreover, these reductions in autophosphorylation were associated with decreased in vitro activity against MyBP (Figure 2B). Double or triple substitutions in helix G successively reduced phosphorylation of the KD during expression of the protein. These hypophosphorylated KD variants showed minimal autophosphorylation activity and, as anticipated from the deficit of activating phosphoryl groups, also had negligible activity on MyBP in vitro. The Ala225Glu/Val229Asp PknB KD, for example, was predominantly unphosphorylated and the Leu183Asp/Val222Asp/Val229Asp KD was 90–95% unphosphorylated. Ala225Glu/Val229Asp phosphorylated MyBP 30-fold more slowly than the WT KD. The interface triple mutant, Leu183Asp/Val222Asp/Val229Asp, showed negligible autophosphorylation in E. coli and in vitro kinase activity on MyBP. Paralleling the results of the in vitro activation of the dephosphorylated KDs, however, the Ala225Leu variant was hyperphosphorylated in E. coli (4–7 phosphates versus 3–6 phosphates for WT PknB; Table II). The hyperphosphorylated Ala225Leu variant was more active than the WT KD in MyBP transphosphorylation (Figure 2). These results suggested that PknB autophosphorylation is mediated by the G-helix interface.
Table 2.
G-helix substitutions alter the extent of PknB KD phosphorylation
| PknB KD | Phosphates (major species) |
|---|---|
| Wild type | 3–6 (5) |
| Val222Asp | 2–5 (4) |
| Ala225Leu | 4–7 (5, 6) |
| Val229Asp | 2–5 (3, 4) |
| Val222Asp/Val229Asp | 0–2 (1) |
| Ala225Glu | 2–4 (3) |
| Ala225Glu/Val229Asp | 0–2 (0) |
| Leu183Asp/Val222Asp/Val229Aspa | 0–1 (0) |
| Leu33Asp | 2–3 (3) |
| Leu33Asp/Val222Asp | 0 |
| aPredominant species (90–95%) was unphosphorylated. | |
| Phosphorylation states were determined from the mass spectra of intact PknB variants expressed in E. coli. The predominant species in the LC-MS spectrum are shown in parentheses. Of the mutants tested, rates of autophosphorylation and activity on MyBP correlate with the number of phosphates present on each PknB variant. The mutations were made in the G-helix except for Leu33Asp, which disrupts the conserved N-lobe dimer interface. Up to four phosphates were found in the wild-type PknB activation loop, and two phosphates have been mapped to the C-terminal region of the kinase domain (Boitel et al, 2003; Young et al, 2003; Duran et al, 2005). | |
Interestingly, mutations in the G-helix interface synergized with the Leu33Asp mutation in the previously identified N-lobe interface that mediates activation of the unphosphorylated kinase (Young et al, 2003; Greenstein et al, 2007a). PknB orthologues in over 90 bacterial species contain Leu33 in this ‘back-to-back' interface. When we combined the Leu33Asp mutation with the moderately attenuating Val222Asp mutation in the G-helix interface, the double mutant KD showed negligible autophosphorylation and MyBP transphosphorylation in vitro (Figure 2). Moreover, this double mutant expressed in E. coli lacked phosphoryl groups (Table II). This synergy revealed that both helix G in the C-lobe and the back-to-back, allosteric interface containing Leu33 in the N-lobe contribute to the activation process.
Site-specific activation-loop phosphorylation
To define the products of PknB autophosphorylation, tandem mass spectrometry (MS/MS) was used to map the phosphorylation sites on the activation loops of the variant KDs. Because traditional MS/MS results in neutral losses of phosphates, mapping the phosphorylated residues is problematic, especially for the activation loop of PknB, where up to four phosphates are present on a tryptic peptide of 28 residues (Boitel et al, 2003; Young et al, 2003; Duran et al, 2005). An alternative strategy has been reported in which the phosphorylated residues are chemically modified to a lysine mimetic to enable tryptic or LysC digestion at the modified site (Knight et al, 2003). We used a variation of this bottom–up method in which the PknB KD was digested with trypsin and the peptides were chemically modified on pSer and pThr residues. As the lysine mimetic is stable under typical ESI/MS/MS conditions, the positions of the modifications were deduced unambiguously. This top–down approach has advantages over digesting the modified sites, as each phosphorylated residue is mapped within a single fragment that may contain additional phospho-residues. This chemical context provides additional information about the potential coupling of phosphoryl modifications.
As reported earlier, 2–4 modifications were observed in the activation loop of WT PknB 1–308 (Figure 3) (Boitel et al, 2003; Young et al, 2003; Duran et al, 2005). The earlier reported major phosphorylation sites, Thr171 and Thr173, were identified, and two minor sites were found at Ser169 and Thr179. The partial phosphorylation of Thr179 has not been reported earlier. Consistent with earlier data (Boitel et al, 2003), we found that the Ser169Ala mutant produced minimal effects on autophosphorylation and MyBP phosphorylation (data not shown).
Figure 3.
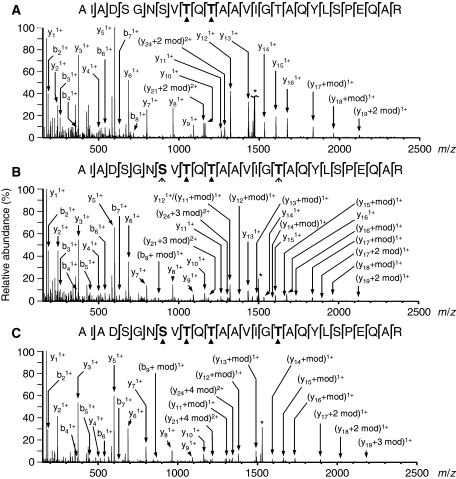
Tandem mass spectrum (MS/MS) showing modifications of the 28-residue activation-loop peptide. (A) Mass spectrum showing ions formed by electrospray ionization. The inset shows detail for the (M+2H)2+ ions of the activation-loop fragment of PknB (residues 162–189) with two, three and four modifications. The asterisks denote ions that are 18 Da lower in mass, presumably due to losses of water molecules. (B) Quadrupole isolation mass spectrum of the (M+2H)2+ ion with three modifications. (C) MS/MS spectrum of the (M+2H)2+ ion with three modifications showing fragment ions formed by collisionally activated dissociation.
To explore the site-specific effects of G-helix mutations, we mapped the phosphorylation sites on the PknB KD variants purified from E. coli (Figure 4) and measured the activity of the KD variants in vitro. Unexpectedly, each G-helix mutation not only reduced the autophosphorylation activity (Figure 2A) but also preferentially reduced phosphorylation of specific sites. The activation loop of the Val229Asp variant, for example, was phosphorylated nearly exclusively at Thr171 and Thr173, with fewer modifications at other sites in the activation loop. In comparison to WT PknB 1–308, less triply modified activation loop was observed by LC/MS, and no tetra-phosphopeptide was detected (Supplementary Figure 4). Helix-G double mutants, such as Ala225Glu/Val229Asp, which showed reduced autophosphorylation and MyBP phosphorylation activities (Figure 2), were partially phosphorylated at only one site, Thr171. Interestingly, for variants where only one modification was detected, Thr171 was exclusively modified. When two modifications were observed in the activation loop, Thr171 and Thr173 were modified exclusively. In the case of three or more activation-loop phosphorylations, Thr179 and/or Ser169 were the additional sites of modification. For the WT KD, Ser169 was the preferred third phosphorylation site detected. For Val229Asp PknB, Thr179 was the predominant third modification site.
Figure 4.
MS/MS spectra and cleavage maps of the (M+2H)2+ ions of the activation-loop fragment of PknB (residues 162–189) with (A) two, (B) three and (C) four modifications. Chemical modification increases the mass of each phospho-residue by 60 Da. The asterisks in the spectra denote residual precursor ion and the arrowheads in the cleavage maps denote modification sites (bold). The MS/MS spectrum of the ion with three modifications (B) indicates a mixture of isoforms with ∼80% having the third modification on Ser169 and ∼20% on Thr179.
Structure of the PknB Leu33Asp/Val222Asp variant bound to ADP
To test whether substitutions in the G-helix interface inhibit PknB activation directly by weakening the asymmetric KD association or indirectly through long-range conformational changes, we determined the 1.8-Å resolution crystal structure of the unphosphorylated Val222Asp/Leu33Asp PknB KD bound to ADP. This inactive, double-mutant protein crystallized in a distinct habit in which neither the G-helix nor the N-lobe surface containing Asp33 made extensive intermolecular contacts (Figure 5). The Val222Asp/Leu33Asp PknB KD was completely unphosphorylated when purified from E. coli (Table II), and typical of an unphosphorylated KD, it had negligible phosphotransfer activity on MyBP (Figure 2B). Nonetheless, the unphosphorylated KD bound ADP at the concentration (3 mM) used in the crystallization. The structure of the double interface mutant PknB KD resembled that of the PknB Leu33Asp variant bound to ATPγS (backbone r.m.s.d.=0.31 Å; T Noelle Lombana, Nathaniel Echols, unpublished results). No electron density was observed for the activation loop in either structure. The lack of a conformational perturbation associated with the Val222Asp mutation suggested that changes in the G helix exert an effect not by altering the kinase conformation, but rather by directly inhibiting helix-G interactions.
Figure 5.
The Val222Asp substitution causes minimal changes in the PknB KD structure. Leu33Asp PknB KD bound to ATPγS (light pink) superimposed on the Leu33Asp/Val222Asp PknB KD bound to ADP (dark blue). Spheres show the locations of Leu33Asp in the N-lobe and Val222 in the G-helix.
Helix G mediates intermolecular phosphorylation of PknB
The hypothesis that the KT5720–KD dimer represents an autophosphorylation complex predicts that PknB activates through an intermolecular interaction involving the G helix of both the substrate and enzymatic KDs. PknB has been observed earlier to autophosphorylate in trans (Kang et al, 2005). We confirmed this result by measuring phosphorylation of the inactive Asp138Asn PknB KD by the WT protein. Because Asp138 functions as the catalytic base in the phosphotransfer reaction, the Asp138Asn mutation inactivates the enzyme. WT (1–308) and Asp138Asn (1–279) PknB KD variants of different lengths were used to distinguish the phosphorylated products using SDS–PAGE. In a phosphoryl transfer assay, the inactive Asp138Asn variant was only phosphorylated in the presence of the WT KD, indicating that the modification(s) occurred in an intermolecular reaction (Figure 6). To test whether this reaction was mediated by helix G, the Val229Asp replacement was engineered into the inactive PknB substrate. The mutation inhibited phosphorylation by the WT KD 6.3 (±1.1)-fold at 1 μM substrate KD. The same trend also was observed when the Val229Asp replacement was made in the active PknB KD (10-fold reduction in activity). Combining the Val229Asp mutation in both the active PknB KD and the inactive substrate KD produced a larger reduction (∼50-fold) in substrate phosphorylation (Figure 6). Similarly, the Ala225Glu and Leu183Asp substitutions in the G-helix interface of the substrate KD reduced phosphorylation by the WT PknB KD by 43 (±3.5)- and 8.4 (±0.9)-fold, respectively (Supplementary Figure 5). These results demonstrated directly that mutations in the helix-G interfaces in both the active and inactive KDs inhibited autophosphorylation in trans.
Figure 6.
Intermolecular PknB autophosphorylation involves G-helix contacts. Autoradiograms showing intermolecular phosphorylation of inactive Asp138Asn PknB (1–279) KD substrates by active PknB (1–308) KD variants. Corresponding SDS–PAGE gels are shown as loading controls. (A) The Val229Asp mutation in helix G of both the active kinase (lanes 6–8) and inactive substrate KD (lanes 5 and 8) reduced trans-autophosphorylation of the Asp138Asn KD substrate. (B) The Leu33Asp mutation reduced intermolecular autophosphorylation (by 60%) when engineered into the active kinase domain. However, PknB (1–308) efficiently phosphorylated both the PknB (1–279) Asp138Asn KD substrate with and without the Leu33Asp mutation.
Analogous experiments were performed to investigate the contribution by the N-lobe interface to intermolecular autophosphorylation. The Leu33Asp mutation reduces autophosphorylation (Figure 2), reflecting the role of the back-to-back, N-lobe interface in activating the KD (Greenstein et al, 2007a). When the Leu33Asp mutation was engineered into the inactive Asp138Asn substrate, however, efficient phosphorylation by active WT PknB was observed (Figure 6B). This phosphorylation was comparable to the level observed for the substrate containing the Asp138Asn mutation alone. These results suggest that the active, dimeric KD can efficiently phosphorylate a monomeric KD. Thus, N-lobe dimerization is important for activating the unphosphorylated KD, but is not required in the substrate KD.
Discussion
Earlier studies using synthetic peptides identified the motif Thr-Gln-X-hydrophobic–hydrophobic as favouring phosphorylation by PknB and supported the idea that the Thr-Gln-Thr-Ala-Ala sequence in the PknB activation loop functions as a local determinant of autophosphorylation (Kang et al, 2005). In contrast, here we identified distinct structural recognition interfaces that mediate autophosphorylation of the WT PknB KD. The structure of the KT5720 complex of the PknD surrogate KD suggested not only that the larger volume of the PknD ATP-binding site (compared with that of PknB) mediates inhibitor selectivity but also revealed an unprecedented asymmetric dimer involving offset G-helix contacts. Although the inhibitor occupied the nucleotide-binding site in both monomers, this asymmetric dimer stabilized in the crystals displayed several features expected in an authentic kinase ES or EP complex.
Characteristic of the active conformation of the KD (Huse and Kuriyan, 2002), the activation loop of one monomer was largely ordered in the dimer interface. The conformation of this loop resembled that seen in activated PKA (Madhusudan et al, 2002) and other active eukaryotic protein kinases (Huse and Kuriyan, 2002). In contrast, the other KD monomer resembled a protein substrate. In particular, a segment of 16 residues in the activation loop was disordered with the ends positioned at the mouth of the active site of the ‘enzymatic' monomer (Figure 1B). A disordered phosphorylation target site was also identified earlier in the protein substrate in the complex of the eIF2α bound to the cognate kinase, PKR (Dar et al, 2005). Thus, the specific asymmetry of the KT5720–KD complex reconciles the apparently contradictory requirements for the activation loop to fold in an active kinase and to unfold to position the multiple target Thr (and Ser) residues in the partner kinase active site.
The asymmetry of the PknB KD dimer is consistent with distinct roles for the two kinase molecules. In this sense, the PknB KD structure resolves a paradox apparent in studies of p21-activated kinase-2 (PAK2) autophosphorylation inspired by the crystal structure of the activated homologue, PAK1 (Pirruccello et al, 2006). Similar to the PknD surrogate KD complex, the PAK1 KD formed a front-to-front dimer mediated primarily by hydrophobic contacts of the G (and E) helices. Unlike the PknD surrogate KD, however, the PAK1 KDs were related by a crystallographic two-fold rotation axis that places the G helices in register, and the activation loops of both molecules were ordered and oriented away from the active site of the partner KD. Even though this presumptive autophosphorylation complex required structural remodelling to place the phosphorylation site of one PAK1 monomer in the partner active site, amino-acid substitutions in the G-helix interface reduced intermolecular autophosphorylation in the homologous KD of PAK2 (Pirruccello et al, 2006). Transient unfolding of one activation loop was proposed to resolve this paradox. Alternatively, if PAK2 formed an asymmetric complex in solution akin to the offset PknD surrogate dimer, the target activation loop would be positioned to access the partner kinase active site and G-helix substitutions would reduce autophosphorylation. Whether the functional PAK2 interaction is equivalent to or distinct from that of the PknB KD, analogous G-helix substitutions in the two KDs cause qualitatively similar reductions in autophosphorylation. Such functional conservation in distant homologues predicts that the G helix may mediate activation-loop autophosphorylation in prokaryotic and eukaryotic STPK family members.
Recently, symmetric dimer interfaces distinct from the E- and G-helix contacts of the PAK KDs were observed in the structures of checkpoint kinase 2 (CHK2), STE20-like kinase (SLK), lymphocyte-originated kinase (LOK) and death-associated protein kinase 3 (DAPK3) (Oliver et al, 2006; Pike et al, 2008). Each of these KDs forms homodimers detectable in solution, and they crystallize as dimers with swapped activation loops projecting into the active site of the neighbouring monomer (Oliver et al, 2006; Pike et al, 2008). N-lobe contacts through the C helix occur in some of these dimers, making them fundamentally different from the associations seen for either PAK1 or PknB. Similar to the PAK1 dimer, however, the activation-loop exchange complexes are generally two-fold symmetric, and structural fluctuations would be required to reposition the target residue(s) for phosphoryl transfer by the catalytic machinery of the partner KD. In contrast, one activation loop (monomer A) in the PknB KD pair is poised to access the opposing active site and the disorder of this activation-loop segment recapitulates the mechanism by which the Ser51 phosphorylation site of eIF2α accesses the cognate kinase, PKR (Dar et al, 2005).
Mutational analysis of the WT PknB KD supported the idea that the G helix mediates KD autophosphorylation. Substitutions of conserved, external residues in every turn of the G helix reduced autophosphorylation, and multiple G-helix changes eliminated autophosphorylation (Figure 2 and Table II). Several lines of evidence suggest that these reductions were due to direct disruption of the kinase interactions rather than indirect effects of conformational changes in the active KD. First, mutations in the G helix of an inactive substrate KD reduced intermolecular autophosphorylation by the WT PknB KD (Figure 6, Supplementary Figure 5). Second, the Ala225Leu substitution in the G helix increased autophosphorylation, whereas the Ala225Glu substitution slowed activation (Figure 2). This pattern is consistent with burial of residue 225 on the edge of the G-helix interface. Third, the Val222Asp mutation in the PknB G helix caused only small structural changes in the Leu33Asp variant and directly eliminated G-helix contacts in the crystals (Figure 5). Fourth, the Val229Asp substitution in the G helix of the active KD further reduced transphosphorylation of the inactive Val229Asp/Asp138Asn substrate (Figure 6). These results support the idea that the G helix mediates PknB interactions on both sides of the autophosphorylation interface, as seen in the asymmetric KT5720–PknD-surrogate structure.
To analyse autophosphorylation in a cellular context that lacks other STPKs, we mapped the activation-loop phosphates in the mutant KDs purified from E. coli. A chemical modification strategy (Knight et al, 2003) was adapted to define multiply phosphorylated species. The G-helix variants revealed cumulative effects that followed a particularly simple pattern. Single mutations in the G helix reduced total autophosphorylation, and combining these substitutions caused larger reductions (Table II). One double and one triple G-helix mutant were >90% unphosphorylated when purified from E. coli. The Thr-Gln-Thr motif was retained in the activation loop of all of these G-helix variants, indicating that this local sequence motif in the activation loop is insufficient by itself to mediate KD autophosphorylation. Instead, the G-helix variants showed that this structural interface can influence directly or indirectly all autophosphorylation of the PknB KD.
The four activation-loop phosphorylation sites showed different sensitivities to G-helix mutations (Table III). Mutations first eliminated Ser169 and Thr179 phosphorylation, followed by Thr173. Thr171 was the last activation-loop phosphorylation site to be eliminated by G-helix substitutions (Table III). These effects were recapitulated in the pattern of phosphorylation of the WT KD, in which Thr171 and Thr173 were fully phosphorylated, whereas Ser169 and Thr179 were partially phosphorylated. Our results suggest that sites in the activation loop may be phosphorylated in a specific order. In the simplest interpretation of the results, the autophosphorylation sequence is Thr171 followed by Thr173 followed by the other two activation-loop sites.
Table 3.
MS/MS measurements of the chemically modified PknB activation loop
| Variant | Number of modifications | Location of modification |
|---|---|---|
| WT | 2 | Thr171, Thr173 |
| WT | 3 | Thr171, Thr173, Thr179 or Ser169a |
| WT | 4 | Thr171, Thr173, Thr179, Ser169 |
| Val229Asp | 2 | Thr171, Thr173 |
| Val229Asp | 3 | Thr171, Thr173, Thr179a or Ser169 |
| Ala225Glu/Val229Asp | 1 | Thr171 |
| Val222Asp/Val229Asp | 1 | Thr171 |
| aPredominant site of third modification. | ||
| Substitutions in the G helix decreased phosphorylation of the activation loop at preferential sites. | ||
Only the Thr171 and Thr173 phosphorylations have been reported to occur in vivo (Kang et al, 2005) and to mediate activation in vitro (Boitel et al, 2003). Thus, the fraction of the population doubly phosphorylated on Thr171 and Thr173 may control specific activity. However, PknB variants with single G-helix replacements were doubly phosphorylated on Thr171 and Thr173 regardless of whether they were more (Ala225Leu) or less (Val222Asp, Ala225Glu and Val229Asp) active in phosphorylating MyBP than the WT KD. Moreover, the hypophosphorylated G-helix mutants were less active on MyBP than the WT PknB KD, and the Ala225Leu mutant is more phosphorylated and more active. Thus, it is possible that additional phosphorylation sites may contribute to full kinase activity. Skepticism is needed, however, in considering functional roles for the minor phosphorylation sites in the activation loop. The Ser169Ala mutation does not alter activity (Boitel et al, 2003), and Thr179 is analogous to Thr201 in PKA, which has a key function in positioning the catalytic loop in the active conformation of the kinase (Pirruccello et al, 2006). Alternatively, the parallel effects of G-helix mutations on KD and MyBP phosphorylation could be explained if this structural interface mediated direct substrate interactions or controlled KD specific activity in auto- and trans-phosphorylation reactions. Additional studies are needed to distinguish these models.
The synergistic effects of mutations in the G helix and the back of the N-lobe also indicate that these distinct interfaces mediate PknB autophosphorylation. The Leu33Asp mutation in the N-lobe dimer interface and the Val229Asp mutation in the G helix individually reduced PknB KD autophosphorylation from 3–6 phosphates on the WT KD to an average of 3–4 phosphates, respectively (Table II). In contrast, the combination of these two substitutions, which are located >48 Å apart in the PknB KD structure, eliminated autophosphorylation (Table II). Interestingly, when the same changes were made only in the inactive KD substrates, the Val229Asp substitution was inhibitory, whereas the Leu33Asp replacement was not. These distinct effects suggest that, although the N-lobe interactions are essential to activate the unphosphorylated KD (Greenstein et al, 2007a), the G-helix contacts mediate recognition of monomeric and dimeric KD substrates. Consistent with this model, the superposition of monomers from the WT and PknD surrogate KD structures produces no steric clashes (Figure 7). Because the phosphorylated kinase can be fully active as a monomer (Greenstein et al, 2007a), phosphorylation of the monomer activation loop provides a mechanism to amplify the activating signal.
Figure 7.
Model of the PknB intermolecular autophosphorylation complex. Active, wild-type PknB KD dimer (grey) superimposed on the KT5720–PknD surrogate complex (monomers A and B represented by yellow and blue, respectively). This ‘two-on-one' model is consistent with mutational effects and the structural organization of the N-lobe and G-helix (C-lobe) interfaces. The wild-type N-lobe dimer superimposes on each monomer in the asymmetric C-lobe dimer with an average r.m.s.d. of 1.59 Å with no overlapping contacts. For the putative substrate monomer (yellow), the disordered activation loop is modelled (grey surface). The ordered activation loop of the putative active monomer (blue) is highlighted in red. A KD monomer is phosphorylated efficiently, but the substrate KD may also be phosphorylated as a dimer. Because the monomeric KD is not directly activated by an extracellular cue and the phosphorylated monomer is active, phosphorylation can amplify the signal.
The PknD surrogate structure, however, does not readily explain how reported phosphorylation sites (Thr294, Ser295 and Thr309) outside the activation loop can access the catalytic site (Boitel et al, 2003; Young et al, 2003; Duran et al, 2005). Global relative reorientation of the KDs would seem to be necessary for these sites in the juxtamembrane segment C-terminal to the KD to access the kinase active site. Considering the many combinations of phosphorylation states (Boitel et al, 2003; Young et al, 2003; Duran et al, 2005), the sequential autophosphorylation we observed in PknB may afford a mechanism to regulate the activities of prokaryotic STPKs. Our studies implicate two interfaces with different roles in this process—the conserved N-lobe interface orients the catalytic sites in the dimer away from each other and activates the unphosphorylated KDs by an allosteric mechanism (Greenstein et al, 2007a), whereas the G-helix interface mediates intermolecular activation-loop autophosphorylation.
Materials and methods
Protein expression and purification
PknB (1–308) with a thrombin-cleavable, N-terminal His6 tag was expressed and purified as described (Young et al, 2003). KD variants were made by site-directed mutagenesis using the QuikChange method (Stratagene). WT and mutant KDs were purified using Ni-NTA chromatography (GE Healthcare) using 25 mM Tris pH 8.0, 300 mM NaCl, 1 mM TCEP, 10% glycerol, 10 mM imidazole as wash buffer and proteins were eluted in wash buffer containing 300 mM imidazole. To remove the His tag, 10 U thrombin (Sigma) was added, and the reaction was dialysed against 25 mM Tris pH 8.0, 200 mM NaCl, 1 mM TCEP, 5% glycerol, 10 mM imidazole and 1 mM CaCl2. This solution was passed over a Ni-NTA affinity column, and the flow-through was dialysed against 25 mM Tris pH 7.5, 125 mM NaCl, 1 mM TCEP and 5% glycerol. Glycerol was added to 40%, and the samples were flash frozen in liquid N2 and stored at −80°C for 5–15 days before use in kinase activity assays. For crystallization, proteins were further purified on a MonoQ column (GE Healthcare) eluted using a NaCl gradient. Proteins were concentrated to >10 mg/ml and dialysed against 20 mM Tris pH 7.5, 75 mM NaCl and 1 mM TCEP.
Crystallization and structure determination
KT5720:PknD-surrogate The inhibitor complex of PknB Leu33Asp/Met145Leu/Met155Val KD was crystallized using the vapour diffusion method. A PknD surrogate solution (10 mg/ml) containing 400 μM KT5720 (Calbiochem) was mixed with an equal volume of 100 mM Tris pH 8.5, 25% PEG 3350 and 200 mM ammonium sulphate and equilibrated against precipitant at 4°C. Crystals that formed in 1–3 weeks were harvested by transferring to mother liquor containing 37% PEG 3350 and flash frozen in liquid N2. X-ray data were collected at the Lawrence Berkeley National Laboratory Advanced Light Source Beamline 8.3.1 at 100°K and 11 111 eV and indexed using ELVES (Holton and Alber, 2004). Data were processed and scaled using HKL2000 (Otwinowski and Minor, 1997). Molecular replacement was carried out using Phaser with the structure of the Leu33Asp PknB KD bound to ATPγS as the search model (Potterton et al, 2003; Storoni et al, 2004). Two molecules were located in the asymmetric unit. Models were modified using Coot and refined with REFMAC (Murshudov et al, 1997; Emsley and Cowtan, 2004). Structures were superimposed using SSM-based methods (Krissinel and Henrick, 2004). Coordinates for KT5720 were generated using the PRODRG server (Schuttelkopf and van Aalten, 2004). The quality of the models was evaluated using PROCHECK (Laskowski et al, 1993). Structures were displayed using PYMOL (DeLano, 2002). Solvent-accessible surface area was calculated using Arealmol (Potterton et al, 2003). Surface conservation mapping was performed using sequence alignments generated by ClustalW (Thompson et al, 1994).
PknB Leu33Asp/Val222Asp:ADP The Leu33Asp/Val222Asp PknB KD was expressed and purified as described earlier for WT PknB 1–308 (Young et al, 2003). Crystals were grown from 20% PEG 3000, 3 mM ADP, 0.2 M NaF and 100 mM Tris pH 8.5. Crystals were flash frozen in 30% PEG 3000, 0.2 M NaF, 100 mM Tris pH 8.5 and 5% glycerol. Data collection, phasing and refinement were carried out as described above for the PknD surrogate structure.
Kinase activity assays
Autophosphorylation assays were carried out using completely dephosphorylated, purified WT and mutant PknB KDs. The KDs were dephosphorylated using a 1:10 ratio of PstP phosphatase (with a non-cleavable N-term His6 tag) to PknB KD overnight at 25°C in 20 mM Tris pH 7.5, 25 mM NaCl, 0.5 mM TCEP and 1 mM MnCl2. PstP and PknB were separated using Ni-NTA chromatography and dephosphorylation of PknB was verified using SDS–PAGE (Invitrogen) and intact MS. Kinase assays were carried out at 25°C in 20 mM Tris pH 7.5, 25 mM NaCl, 0.1 mM MgCl2, 2 mM MnCl2, 0.5 mM TCEP using 2 μM dephosphorylated KD. Reactions were initiated by adding 50 μM ATP containing γ-32P-ATP (0.2 μCi/μl). Reactions were quenched using SDS–PAGE loading buffer containing 20 mM EDTA.
Transphosphorylation assays were performed in the same buffer by incubating active PknB KD (500 nM) with 1 mg/ml MyBP or active PknB (1 μM) with the Asp138Asn PknB 1–279 KD (1 μM) in 100 μM ATP with γ-32P-ATP added (0.2 μCi/μl). KD concentrations were determined by measuring the absorbance at 280 nm (Perkin Elmer) and verified by SDS–PAGE analysis. Reaction products were quenched at 5 min. For both auto- and transphosphorylation assays, products were separated using SDS–PAGE and blotted onto a PVDF membrane (Perkin Elmer). Dried membranes were exposed to a Storage Phospho Screen (Molecular Dynamics), imaged using a Molecular Dynamics Typhoon scanner and analysed quantitatively using Imagequant software. Values reported are from triplicate sets of data.
Chemical modification of phosphopeptides and mass spectrometry
PknB phosphopeptides were prepared by tryptic digestion of purified PknB (25 mM Tris, pH 8.5) followed by enrichment of phosphopeptides through Fe-IMAC chromatography using Chelating Sepharose Fastflow (GE Healthcare) charged with FeCl3 (Knight et al, 2003). Phosphopeptides were eluted in 100 mM Na2HPO4 and desalted using Spec C18 desalting tips (Varian). Chemical modification of pThr and pSer residues using 2-aminoethanethiol was performed as described (Knight et al, 2003).
Mass spectra of chemically modified PknB peptides were acquired using a quadrupole time-of-flight (Q-TOF) mass spectrometer equipped with a Z-spray source (Q-TOF Premier™; Waters). Ions were formed by nano-ESI from emitters made from borosilicate capillary tubes (1.0 mm o.d./0.78 mm i.d; Sutter Instruments) pulled to a tip with an inner diameter of ∼5 μm. The flow rates were ∼50–200 nl/min. For MS/MS experiments, the quadrupole resolution was adjusted to encompass the ion of interest and the accelerating voltage into the argon-filled collision cell was 60 V. External mass calibration was performed using sodium formate immediately prior to measuring samples. Mass spectra were processed using MassLynx software (v4.1; Waters). Scaling of mass spectral ions and assignment of fragment ions were performed as described earlier (Roepstorff and Fohlman, 1984; Marshall et al, 2002).
The Stanford University Mass Spectrometry Laboratory performed intact mass analysis (LC/MS) of purified PknB KDs. The LC-MS system (Waters) consisted of an Alliance 2795 HPLC with inline 2487 UV detector and Micromass ZQ single quadrupole mass spectrometer. Protein samples were separated on a 2.1x 40 mm C8 Targa Sprite column (Higgins Analytical) at 300 μl/min, using a linear gradient of 2–95% B; solvent A was 0.1% formic acid in H2O, solvent B was 0.1% formic acid in acetonitrile. UV response was monitored at 254 and 280 nm. MS data were collected up to 2000 m/z and raw spectra were deconvoluted using the MaxEnt1 algorithm.
Accession codes
Coordinates and structure factors were deposited in the Protein Data Bank under accession codes 3F69 (Leu33Asp/Met145Leu/Met155Val PknB-KT5720 complex) and 3F61 (Leu33Asp/Val222Asp PknB-ADP complex).
Supplementary Material
Supplementary Figures
Acknowledgments
We thank James Fraser, Allis Chien and Lindsay Comeaux for technical assistance; T Noelle Lombana, Christine Gee, Nathaniel Echols and Christina Baer for many discussions and the reviewers for helpful insights. We are grateful to Evan Williams for discussions about nano-ESI methods. A mass spectrometer used in these studies was acquired with support from the National Institutes of Health (1S10RR022393). This study was supported by NIH grants R01GM48598 and GM70962 to TA.
Competing interests statement The authors declare that they have no competing financial interests.
References
- Av-Gay Y, Everett M (2000) The eukaryotic-like Ser/Thr protein kinases of Mycobacterium tuberculosis. Trends Microbiol 8: 238–244 [DOI] [PubMed] [Google Scholar]
- Biondi RM, Nebreda AR (2003) Signalling specificity of Ser/Thr protein kinases through docking-site-mediated interactions. Biochem J 372: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boitel B, Ortiz-Lombardia M, Duran R, Pompeo F, Cole ST, Cervenansky C, Alzari PM (2003) PknB kinase activity is regulated by phosphorylation in two Thr residues and dephosphorylation by PstP, the cognate phospho-Ser/Thr phosphatase, in Mycobacterium tuberculosis. Mol Microbiol 49: 1493–1508 [DOI] [PubMed] [Google Scholar]
- Dar AC, Dever TE, Sicheri F (2005) Higher-order substrate recognition of eIF2alpha by the RNA-dependent protein kinase PKR. Cell 122: 887–900 [DOI] [PubMed] [Google Scholar]
- DeLano WL (2002) The PyMOL Molecular Graphics System. San Carlos, CA: DeLano Scientific [Google Scholar]
- Dey M, Cao C, Dar AC, Tamura T, Ozato K, Sicheri F, Dever TE (2005) Mechanistic link between PKR dimerization, autophosphorylation, and eIF2alpha substrate recognition. Cell 122: 901–913 [DOI] [PubMed] [Google Scholar]
- Duran R, Villarino A, Bellinzoni M, Wehenkel A, Fernandez P, Boitel B, Cole ST, Alzari PM, Cervenansky C (2005) Conserved autophosphorylation pattern in activation loops and juxtamembrane regions of Mycobacterium tuberculosis Ser/Thr protein kinases. Biochem Biophys Res Commun 333: 858–867 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Gay LM, Ng HL, Alber T (2006) A conserved dimer and global conformational changes in the structure of apo-PknE Ser/Thr protein kinase from Mycobacterium tuberculosis. J Mol Biol 360: 409–420 [DOI] [PubMed] [Google Scholar]
- Greenstein AE, Echols N, Lombana TN, King DS, Alber T (2007a) Allosteric activation by dimerization of the PknD receptor Ser/Thr protein kinase from Mycobacterium tuberculosis. J Biol Chem 282: 11427–11435 [DOI] [PubMed] [Google Scholar]
- Greenstein AE, Grundner C, Echols N, Gay LM, Lombana TN, Miecskowski CA, Pullen KE, Sung PY, Alber T (2005) Structure/function studies of Ser/Thr and Tyr protein phosphorylation in Mycobacterium tuberculosis. J Mol Microbiol Biotechnol 9: 167–181 [DOI] [PubMed] [Google Scholar]
- Greenstein AE, MacGurn JA, Baer CE, Falick AM, Cox JS, Alber T (2007b) M. tuberculosis Ser/Thr protein kinase D phosphorylates an anti-anti-sigma factor homolog. PLoS Pathog 3: e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holton J, Alber T (2004) Automated protein crystal structure determination using ELVES. Proc Natl Acad Sci USA 101: 1537–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse M, Kuriyan J (2002) The conformational plasticity of protein kinases. Cell 109: 275–282 [DOI] [PubMed] [Google Scholar]
- Kang CM, Abbott DW, Park ST, Dascher CC, Cantley LC, Husson RN (2005) The Mycobacterium tuberculosis serine/threonine kinases PknA and PknB: substrate identification and regulation of cell shape. Genes Dev 19: 1692–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight ZA, Schilling B, Row RH, Kenski DM, Gibson BW, Shokat KM (2003) Phosphospecific proteolysis for mapping sites of protein phosphorylation. Nat Biotechnol 21: 1047–1054 [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr 60: 2256–2268 [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallog 26: 283–291 [Google Scholar]
- Lawrie AM, Noble ME, Tunnah P, Brown NR, Johnson LN, Endicott JA (1997) Protein kinase inhibition by staurosporine revealed in details of the molecular interaction with CDK2. Nat Struct Biol 4: 796–801 [DOI] [PubMed] [Google Scholar]
- Madhusudan, Akamine P, Xuong NH, Taylor SS (2002) Crystal structure of a transition state mimic of the catalytic subunit of cAMP-dependent protein kinase. Nat Struct Biol 9: 273–277 [DOI] [PubMed] [Google Scholar]
- Marshall AG, Hendrickson CL, Shi SD (2002) Scaling MS plateaus with high-resolution FT-ICRMS. Anal Chem 74: 252A–259A [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Oliver AW, Paul A, Boxall KJ, Barrie SE, Aherne GW, Garrett MD, Mittnacht S, Pearl LH (2006) Trans-activation of the DNA-damage signalling protein kinase Chk2 by T-loop exchange. EMBO J 25: 3179–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Lombardia M, Pompeo F, Boitel B, Alzari PM (2003) Crystal structure of the catalytic domain of the PknB serine/threonine kinase from Mycobacterium tuberculosis. J Biol Chem 278: 13094–13100 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. In Macromolecular Crystallography Carter Jr CW, Sweet RM (eds), Vol. 276, pp 307–326. San Diego: Academic Press [DOI] [PubMed] [Google Scholar]
- Pike AC, Rellos P, Niesen FH, Turnbull A, Oliver AW, Parker SA, Turk BE, Pearl LH, Knapp S (2008) Activation segment dimerization: a mechanism for kinase autophosphorylation of non-consensus sites. EMBO J 27: 704–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirruccello M, Sondermann H, Pelton JG, Pellicena P, Hoelz A, Chernoff J, Wemmer DE, Kuriyan J (2006) A dimeric kinase assembly underlying autophosphorylation in the p21 activated kinases. J Mol Biol 361: 312–326 [DOI] [PubMed] [Google Scholar]
- Potterton E, Briggs P, Turkenburg M, Dodson E (2003) A graphical user interface to the CCP4 program suite. Acta Crystallogr D Biol Crystallogr 59: 1131–1137 [DOI] [PubMed] [Google Scholar]
- Roepstorff P, Fohlman J (1984) Proposal for a common nomenclature for sequence ions in mass spectra of peptides. Biomed Mass Spectrom 11: 601. [DOI] [PubMed] [Google Scholar]
- Scherr N, Honnappa S, Kunz G, Mueller P, Jayachandran R, Winkler F, Pieters J, Steinmetz MO (2007) Structural basis for the specific inhibition of protein kinase G, a virulence factor of Mycobacterium tuberculosis. Proc Natl Acad Sci USA 104: 12151–12156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuttelkopf AW, van Aalten DM (2004) PRODRG: a tool for high-throughput crystallography of protein–ligand complexes. Acta Crystallogr D Biol Crystallogr 60: 1355–1363 [DOI] [PubMed] [Google Scholar]
- Storoni LC, McCoy AJ, Read RJ (2004) Likelihood-enhanced fast rotation functions. Acta Crystallogr D Biol Crystallogr 60: 432–438 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehenkel A, Fernandez P, Bellinzoni M, Catherinot V, Barilone N, Labesse G, Jackson M, Alzari PM (2006) The structure of PknB in complex with mitoxantrone, an ATP-competitive inhibitor, suggests a mode of protein kinase regulation in mycobacteria. FEBS Lett 580: 3018–3022 [DOI] [PubMed] [Google Scholar]
- Young TA, Delagoutte B, Endrizzi JA, Falick AM, Alber T (2003) Structure of Mycobacterium tuberculosis PknB supports a universal activation mechanism for Ser/Thr protein kinases. Nat Struct Biol 10: 168–174 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures