Abstract
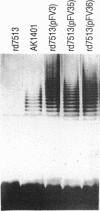
Most strains of Pseudomonas aeruginosa can express two chemically and immunologically distinct types of lipopolysaccharide (LPS), an antigenically conserved form called A band and the serotype-specific form called B band. To study the molecular controls regulating expression of the A-band LPS antigen, we have cloned the genes involved with A-band LPS expression. Strain AK1401, a phage-resistant mutant of PAO1 which was shown previously to produce only A-band LPS and not the O-antigen-containing B-band LPS, was mutagenized by using ethyl methanesulfonate to generate an A-band-deficient mutant called rd7513. A cosmid clone bank of P. aeruginosa PAO1 whole genomic DNA was constructed in Escherichia coli. The gene bank was mobilized en masse into strain rd7513, and detection of complementation of synthesis of A band was done by screening transconjugants in a colony immunoblot assay with the A-band-specific monoclonal antibody N1F10. One recombinant cosmid, pFV3, complemented synthesis of A-band polysaccharide in rd7513. Silver-stained polyacrylamide gel and Western immunoblot analyses of LPS extracted from the transconjugant rd7513(pFV3) showed that the A band produced had a higher molecular weight than the A band of AK1401. Analysis of the plasmid pFV3 showed that it contained a chromosomal insert of 27 kb. Two subclones of pFV3, namely, pFV35 and pFV36, containing chromosomal inserts of 5.3 and 4.2 kb, respectively, also complemented A-band expression in rd7513. The LPS banding profile of rd7513(pFV35) was similar to that of AK1401, while the LPS profile of rd7513(pFV36) more closely resembled that of rd7513(pFV3). pFV3 complemented A-band expression in five of the six P. aeruginosa O serotypes which lack A band as well as in rough strain AK44 but failed to complement A-band expression in core mutants AK1012 and AK1282, suggesting that pFV3 contains genes for A-band expression and that synthesis of a complete core region in isogenic mutant strains is required for A-band synthesis.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berry D., Kropinski A. M. Effect of lipopolysaccharide mutations and temperature on plasmid transformation efficiency in Pseudomonas aeruginosa. Can J Microbiol. 1986 May;32(5):436–438. doi: 10.1139/m86-082. [DOI] [PubMed] [Google Scholar]
- Berst M., Lüderitz O., Westphal O. Studies on the structure of T1 lipopolysaccharides. Eur J Biochem. 1971 Feb 1;18(3):361–368. doi: 10.1111/j.1432-1033.1971.tb01251.x. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnette W. N. "Western blotting": electrophoretic transfer of proteins from sodium dodecyl sulfate--polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem. 1981 Apr;112(2):195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- Cryz S. J., Jr, Pitt T. L., Fürer E., Germanier R. Role of lipopolysaccharide in virulence of Pseudomonas aeruginosa. Infect Immun. 1984 May;44(2):508–513. doi: 10.1128/iai.44.2.508-513.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzins A., Chakrabarty A. M. Cloning of genes controlling alginate biosynthesis from a mucoid cystic fibrosis isolate of Pseudomonas aeruginosa. J Bacteriol. 1984 Jul;159(1):9–18. doi: 10.1128/jb.159.1.9-18.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G., Schmidhauser T., Yakobson E., Lu P., Liang X. W., Finlay D. R., Guiney D., Helinski D. R. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and for monitoring gene expression. Plasmid. 1985 Mar;13(2):149–153. doi: 10.1016/0147-619x(85)90068-x. [DOI] [PubMed] [Google Scholar]
- Dubray G., Bezard G. A highly sensitive periodic acid-silver stain for 1,2-diol groups of glycoproteins and polysaccharides in polyacrylamide gels. Anal Biochem. 1982 Jan 15;119(2):325–329. doi: 10.1016/0003-2697(82)90593-0. [DOI] [PubMed] [Google Scholar]
- Engels W., Endert J., Kamps M. A., van Boven C. P. Role of lipopolysaccharide in opsonization and phagocytosis of Pseudomonas aeruginosa. Infect Immun. 1985 Jul;49(1):182–189. doi: 10.1128/iai.49.1.182-189.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figurski D. H., Helinski D. R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman A. M., Long S. R., Brown S. E., Buikema W. J., Ausubel F. M. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene. 1982 Jun;18(3):289–296. doi: 10.1016/0378-1119(82)90167-6. [DOI] [PubMed] [Google Scholar]
- Goldberg J. B., Ohman D. E. Cloning and expression in Pseudomonas aeruginosa of a gene involved in the production of alginate. J Bacteriol. 1984 Jun;158(3):1115–1121. doi: 10.1128/jb.158.3.1115-1121.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock R. E., Carey A. M. Outer membrane of Pseudomonas aeruginosa: heat- 2-mercaptoethanol-modifiable proteins. J Bacteriol. 1979 Dec;140(3):902–910. doi: 10.1128/jb.140.3.902-910.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock R. E., Mutharia L. M., Chan L., Darveau R. P., Speert D. P., Pier G. B. Pseudomonas aeruginosa isolates from patients with cystic fibrosis: a class of serum-sensitive, nontypable strains deficient in lipopolysaccharide O side chains. Infect Immun. 1983 Oct;42(1):170–177. doi: 10.1128/iai.42.1.170-177.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchcock P. J., Brown T. M. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J Bacteriol. 1983 Apr;154(1):269–277. doi: 10.1128/jb.154.1.269-277.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrell K. F., Kropinski A. M. Isolation and characterization of a bacteriophage specific for the lipopolysaccharide of rough derivatives of Pseudomonas aeruginosa strain PAO. J Virol. 1981 May;38(2):529–538. doi: 10.1128/jvi.38.2.529-538.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrell K. F., Kropinski A. M. Pseudomonas aeruginosa bacteriophage phi PLS27-lipopolysaccharide interactions. J Virol. 1981 Nov;40(2):411–420. doi: 10.1128/jvi.40.2.411-420.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrell K., Kropinski A. M. Identification of the cell wall receptor for bacteriophage E79 in Pseudomonas aeruginosa strain PAO. J Virol. 1977 Sep;23(3):461–466. doi: 10.1128/jvi.23.3.461-466.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kido N., Ohta M., Iida K., Hasegawa T., Ito H., Arakawa Y., Komatsu T., Kato N. Partial deletion of the cloned rfb gene of Escherichia coli O9 results in synthesis of a new O-antigenic lipopolysaccharide. J Bacteriol. 1989 Jul;171(7):3629–3633. doi: 10.1128/jb.171.7.3629-3633.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knirel YuA, Vinogradov E. V., Kocharova N. A., Paramonov N. A., Kochetkov N. K., Dmitriev B. A., Stanislavsky E. S., Lányi B. The structure of O-specific polysaccharides and serological classification of Pseudomonas aeruginosa (a review). Acta Microbiol Hung. 1988;35(1):3–24. [PubMed] [Google Scholar]
- Kropinski A. M., Chan L. C., Milazzo F. H. The extraction and analysis of lipopolysaccharides from Pseudomonas aeruginosa strain PAO, and three rough mutants. Can J Microbiol. 1979 Mar;25(3):390–398. doi: 10.1139/m79-060. [DOI] [PubMed] [Google Scholar]
- Kropinski A. M., Jewell B., Kuzio J., Milazzo F., Berry D. Structure and functions of Pseudomonas aeruginosa lipopolysaccharide. Antibiot Chemother (1971) 1985;36:58–73. doi: 10.1159/000410472. [DOI] [PubMed] [Google Scholar]
- Kuzio J., Kropinski A. M. O-antigen conversion in Pseudomonas aeruginosa PAO1 by bacteriophage D3. J Bacteriol. 1983 Jul;155(1):203–212. doi: 10.1128/jb.155.1.203-212.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam J. S., MacDonald L. A., Lam M. Y., Duchesne L. G., Southam G. G. Production and characterization of monoclonal antibodies against serotype strains of Pseudomonas aeruginosa. Infect Immun. 1987 May;55(5):1051–1057. doi: 10.1128/iai.55.5.1051-1057.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam M. Y., McGroarty E. J., Kropinski A. M., MacDonald L. A., Pedersen S. S., Høiby N., Lam J. S. Occurrence of a common lipopolysaccharide antigen in standard and clinical strains of Pseudomonas aeruginosa. J Clin Microbiol. 1989 May;27(5):962–967. doi: 10.1128/jcm.27.5.962-967.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P. V., Wang S. Three new major somatic antigens of Pseudomonas aeruginosa. J Clin Microbiol. 1990 May;28(5):922–925. doi: 10.1128/jcm.28.5.922-925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo R. Y., Cameron L. A. A simple immunological detection method for the direct screening of genes from clone banks. Biochem Cell Biol. 1986 Jan;64(1):73–76. doi: 10.1139/o86-012. [DOI] [PubMed] [Google Scholar]
- Marolda C. L., Welsh J., Dafoe L., Valvano M. A. Genetic analysis of the O7-polysaccharide biosynthesis region from the Escherichia coli O7:K1 strain VW187. J Bacteriol. 1990 Jul;172(7):3590–3599. doi: 10.1128/jb.172.7.3590-3599.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum K. L., Schoenhals G., Laakso D., Clarke B., Whitfield C. A high-molecular-weight fraction of smooth lipopolysaccharide in Klebsiella serotype O1:K20 contains a unique O-antigen epitope and determines resistance to nonspecific serum killing. Infect Immun. 1989 Dec;57(12):3816–3822. doi: 10.1128/iai.57.12.3816-3822.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palva E. T., Mäkelä P. H. Lipopolysaccharide heterogeneity in Salmonella typhimurium analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis. Eur J Biochem. 1980;107(1):137–143. doi: 10.1111/j.1432-1033.1980.tb04634.x. [DOI] [PubMed] [Google Scholar]
- Potter A. A., Loutit J. S. Exonuclease activity from Pseudomonas aeruginosa which is missing in phenotypically restrictionless mutants. J Bacteriol. 1982 Sep;151(3):1204–1209. doi: 10.1128/jb.151.3.1204-1209.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera M., Bryan L. E., Hancock R. E., McGroarty E. J. Heterogeneity of lipopolysaccharides from Pseudomonas aeruginosa: analysis of lipopolysaccharide chain length. J Bacteriol. 1988 Feb;170(2):512–521. doi: 10.1128/jb.170.2.512-521.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera M., McGroarty E. J. Analysis of a common-antigen lipopolysaccharide from Pseudomonas aeruginosa. J Bacteriol. 1989 Apr;171(4):2244–2248. doi: 10.1128/jb.171.4.2244-2248.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe P. S., Meadow P. M. Structure of the Core oligosaccharide from the lipopolysaccharide of Pseudomonas aeruginosa PAC1R and its defective mutants. Eur J Biochem. 1983 May 2;132(2):329–337. doi: 10.1111/j.1432-1033.1983.tb07366.x. [DOI] [PubMed] [Google Scholar]
- Ruvkun G. B., Ausubel F. M. A general method for site-directed mutagenesis in prokaryotes. Nature. 1981 Jan 1;289(5793):85–88. doi: 10.1038/289085a0. [DOI] [PubMed] [Google Scholar]
- Schiller N. L., Hatch R. A. The serum sensitivity, colonial morphology, serogroup specificity, and outer membrane protein of Pseudomonas aeruginosa strains isolated from several clinical sites. Diagn Microbiol Infect Dis. 1983 Jun;1(2):145–157. doi: 10.1016/0732-8893(83)90044-5. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valvano M. A., Crosa J. H. Molecular cloning and expression in Escherichia coli K-12 of chromosomal genes determining the O7 lipopolysaccharide antigen of a human invasive strain of E. coli O7:K1. Infect Immun. 1989 Mar;57(3):937–943. doi: 10.1128/iai.57.3.937-943.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollin R., Stocker B. A., Lindberg A. A. Lysogenic conversion of Salmonella typhimurium bacteriophages A3 and A4 consists of O-acetylation of rhamnose of the repeating unit of the O-antigenic polysaccharide chain. J Bacteriol. 1987 Mar;169(3):1003–1009. doi: 10.1128/jb.169.3.1003-1009.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota S., Kaya S., Sawada S., Kawamura T., Araki Y., Ito E. Characterization of a polysaccharide component of lipopolysaccharide from Pseudomonas aeruginosa IID 1008 (ATCC 27584) as D-rhamnan. Eur J Biochem. 1987 Sep 1;167(2):203–209. doi: 10.1111/j.1432-1033.1987.tb13324.x. [DOI] [PubMed] [Google Scholar]