Abstract
Free-living miracidia of Schistosoma mansoni, upon penetration of the their snail intermediate host, undergo dramatic morphological and physiological changes as they transform to the parasitic sporocyst stage. During this transformation process, developing larvae release a diverse array of proteins, herein referred to as larval transformation proteins (LTPs), some of which are postulated to serve a parasite protective function. In the present study, nanoLC-tandem MS analysis was performed on all proteins represented in entire 1-dimensional SDS-PAGE-separated samples in order to gain a more comprehensive picture of the protein constituents associated with miracidium-to-sporocyst transformation and thus, their potential role in influencing establishment of intramolluscan infections. Of 127 proteins with sufficient peptide/sequence information, specific identifications were made for 99, while 28 represented unknown or hypothetical proteins. Nineteen percent of identified proteins possessed signal peptides constituting a cohort of classical secretory proteins, while 22% were identified as putative nonclassically-secreted leaderless proteins based on SecretomeP analysis. Proteins comprising these groups consisted mainly of proteases/protease inhibitors, small HSPs, redox/antioxidant enzymes, ion-binding proteins including those with anti-oxidant Fe-binding activities (ferritins, heme-binding protein), and venom allergen-like (VAL) proteins. A polyclonal antibody generated against whole LTPs recognized proteins primarily associated with the cilia, ciliated epidermal plates and intercellular ridges of miracidia and the tegument of fully-transformed sporocysts, identifying these structures as sources of a subset of LTPs. Thus lysis of plates and/or leakage during formation of the sporocyst syncytium likely represent significant contributors to the overall LTP makeup, especially identified nonsecretory proteins. However, as plate release/degradation and tegument formation are part of the normal developmental process, all LTPs regardless of tissue origin, would be expected at the parasite-host interface upon infection. This study significantly expands the repertoire of LTPs associated with larval transformation and identifies several, e.g., those involved in stress responses, proteolysis/inhibition, antioxidant and detoxication, and immune modulation, that may play a parasite protective role during this crucial period of transition.
Keywords: Schistosoma mansoni, miracidium/sporocyst, proteomics, in vitro culture, larval transformation proteins (LTP), excretory-secretory proteins (ESP)
1. Introduction
Intramolluscan development of larval digenetic trematodes is complex process involving initial infection of the snail host by the free-swimming miracidium, its subsequent transformation to a parasitic primary sporocyst stage, followed by asexual reproduction and release of secondary sporocysts or rediae, and finally the eventual formation and release of cercariae, the next free-swimming stage in the life cycle. In Schistosoma mansoni, one of the causative agents of human intestinal schistosomiasis, miracidial penetration of the snail intermediate host, Biomphalaria spp., results in the rapid shedding of ciliary epidermal plates from the larval surface during formation of the new tegumental syncytium of the developing sporocyst [1,2]. It has been postulated that proteins released during miracidium-to-sporocyst transformation, previously referred to as excretory-secretory proteins (ESP; [3]) or secretory-excretory products (SEP; [4]) may function in either stimulating the innate immune system resulting in the activation of circulating host hemocytes, or modulating the immune response through direct interference with host hemocyte effector functions or serving as a molecular deterrent through release of host-like proteins [5-7].
Given the potential importance of larval products introduced to the host during the initial establishment of infections, few studies have focused on identifying molecules released from S. mansoni miracidia as this critical period of development to the primary or mother sporocyst stage following larval penetration. SDS-PAGE analyses of proteins released from in vitro cultured schistosome miracidia, referred to here as larval transformation proteins or LTPs, have revealed a complex mixture [3] consisting of cysteine proteinase [8], hemocyte mitogenic [9] and hydrogen peroxide scavenging [10] activities. Schistosome LTPs capable of disrupting various hemocyte functions including motility [11], superoxide formation [12] and cell signaling [13] under in vitro conditions also have been documented. S. mansoni-specific peroxiredoxins found in LTP samples are believed to be responsible, at least in part, for its hydrogen peroxide-scavenging activity [10]. Recently, a proteomic analysis of proteins released from in vitro cultured S. mansoni miracidia was conducted in which 7 proteins were identified [14] including 2 glycolytic enzymes (triosephosphate isomerase, GAPDH), 2 antioxidants (Cu/Zn SOD, GST), calreticulin, histone H4 and a trematode hemoglobin. However, because previous 1- and 2-dimensional (1- and 2-D) SDS-PAGE analyses of concentrated S. mansoni LTPs revealed a highly complex protein mixture [3,15], we undertook a more comprehensive analysis of proteins released during the first 24-hr following in vitro miracidium-to-sporocyst transformation by analyzing the entire proteome separated by 1-D SDS-PAGE. In addition, a polyclonal anti-S. mansoni LTP antiserum was used to localize the source of immunoreactive LTPs released during early larval development. Our results indicate that >120 different proteins are released during the first 24-hr of in vitro larval cultivation, and that a substantial subset of those proteins appear to originate from ciliated epidermal plates being shed from the miracidial surface and/or the developing sporocyst tegument. The relevance of these findings in evaluating the role of LTP in regulating in vivo larval schistosome-snail interactions is discussed.
2. Materials and Methods
2.1. Parasite cultivation and preparation of larval transformation proteins (LTPs)
Miracidia of S. mansoni were hatched from eggs recovered from infected mouse livers and axenically placed into wells of a sterile 24-well cell culture cluster (Corning Glass, Corning, NY) containing sterile Chernin's balanced salt solution (CBSS; [16]) to which 1 g/L each of glucose and trehalose and penicillin/streptomycin had been added (CBSS+; [17]). Miracidia were cultured at 26C under normoxic conditions for 24 hr, after which time nearly all miracidia (>88%) had completely “transformed” (i.e., shed their ciliated epidermal plates). Viability, based on larval motility and morphology, was consistently >98%.
The culture medium (containing LTPs) was carefully pipetted from each well, pooled in a 15-ml sterile tube, filtered with a 0.22 μm syringe filter (Thermo Fisher Sci, Rochester, NY) to remove detached ciliated epidermal plates and other cellular debris and transferred to an Icon Concentrator (9 kDa nominal MW cut-off; Pierce, IL) where the culture LTP was concentrated ∼10-fold by centrifugation at 3000 × g and at 4 C. The protein concentration was estimated by direct measurement at OD280 (BioMate 3 Spectrophotometer; Thermo Spectronic, Rochester, NY). Following introduction of a proteinase inhibitor cocktail (Novagen, Madison, WI), proteins were precipitated by addition of three volumes of cold acetone (Fisher Scientific, IL), incubation at -20°C for 1 hr, and centrifugation at 14k rpm for 15 min. The pellet was then washed with 100% acetone, centrifuged at 14k rpm for 5 min, air dried at 22 C and store at -80 C for further use.
2.2. One-dimensional (1-D) SDS-PAGE
In order to “simplify” the protein mixture being subjected to mass spectrometric analyses, LTPs were first separated by 1-D SDS-PAGE, followed by dividing the entire gel lane into 15 slices (fractions) representing essentially all proteins present in the sample. To accomplish this separation, approximately 200 μg acetone-precipitated LTP from 3 pooled larval cultivations were dissolved in 40 μL snail Tris-buffered saline (sTBS), followed by addition of 10 μL of 4× SDS sample buffer (Novagen, Madison, WI) and boiling for 5 min. Samples were then centrifuged at 12,000 × g for 5 min, loaded onto a 12.5% polyacryamide gel assembled into a GT series electrophoresis unit (SE 600; Hoefer Scentific Instruments, San Francisco, CA) and subjected to standard SDS-PAGE as described by Castillo et al. [18]. Gels were stained with SimplyBlue SafeStainR (Invitrogen, Carlsbad, CA) to visualize protein bands prior to gel slicing.
2.3. Mass spectroscopy and peptide identification
In-gel digestion and mass spectrometric (MS) analyses were performed at the Mass Spectrometry Facility (Biotechnology Center, University of Wisconsin-Madison) as outlined on the Center website; http://www.biotech.wisc.edu/ServicesResearch/MassSpec/ingel.htm, and the data generated represent the pooled results of two separate LTP analyses. Using SimplyBlue-stained protein bands as guides, the lane containing separated transformation proteins was sliced into 15 portions, each destained in 50% MeOH/100mM NH4HCO3, dehydrated in 50% acetonitrile (ACN)/25 mM NH4HCO3 and then 100% ACN, and dried in a Speed-Vac (Savant Instruments, Holbrook, NY) for 5 min. Protein-containing gel fragments were then reduced in 25 mM dithiothreitol (DTT) in 25 mM NH4HCO3 for 30 min at 56C, alklyated with 55 mM iodoacetamide (IAA) in 25 mM NH4HCO3 in darkness for 30 min at 22C, washed twice with dH2O and again dehydrated as above in ACN and vacuum centrifugation. Protein-containing gel samples were then rehydrated in a trypsin solution (20 ng/μl trypsin; Sequence Grade; Promega, Madison, WI), digested at 37C for 18 hr, followed by acidification with 2.5% trifluoroacetic acid (TFA), followed by extractions first in 0.1% TFA and then ACN/H2O/TFA (70%:25%:5%). Finally collected peptide solutions were dried in a Speed-Vac resuspended in 50 μl of 0.1% TFA and solid phase-extracted using either ZipTip® μC18 pipette tips (Millipore, Billerica, MA) for nanoLC-MS/MS Ion Trap analysis.
2.3.1. NanoLC-MS/MS Ion Trap Mass Spectrometric Analysis
Due to the potentially large number of proteins contained in single bands or gel slices of 1-D SDS-PAGE-separated protein mixtures, the LC-tandem MS approach was used. Following tryptic digestion of a protein sample, an initial peptide separation step involving reverse-phase hydrophobic interaction chromatography results in a further reducing of peptide complexity prior to MS-MS spectrometric analysis. Chromatography of peptides was accomplished using a C18 reverse-phase HPLC trap column (Zorbax 300SB-C18, 5μM, 5×0.3mm; Agilent Technologies, PaloAlto, CA) and separation column (Zorbax 300SB-C18, 3.5μm, 0.075×150mm; Agilent Technologies) onto which 40 μl of each extracted peptide fraction was automatically loaded. Peptides from extracted fractions were individually analyzed by nanoLC-MS/MS using a 1100 series LC/MSD Trap SL spectrometer (Agilent Technologies). An Agilent 1100 series HPLC delivered solvents A: 0.1% (v/v) formic acid in water and B: 95% (v/v) CAN/0.1% (v/v) formic acid, at either 10 μL/min to load sample or 0.28 μL/min to elute peptides directly into a nano-electrospray apparatus over a 60 min 20% (v/v) B to 80% (v/v) B gradient. As peptides eluted from the HPLC-column/electrospray source, MS/MS spectra were collected over 4 channels from 300 to 2200 m/z with redundancy being limited by dynamic exclusion.
2.3.2. Sequence analysis and protein database searches
Raw data generated from LC-MS/MS analyses were deconvoluted using Data Analysis software ((Agilent Technologies) and submitted for peptide mapping and MS/MS ion search analysis against the non-redundant NCBI database and the Schistosoma mansoni sequence database (http://www.genedb.org/genedb/smansoni/) using an in-house licensed MASCOT search engine (Matrix Science, London, UK) with methionines oxidized, cysteines carbamidomethylated, and glutamate and aspartate deamidated as variable modifications with a mass tolerance of ± 0.1 Da. Protein identifications, based on sequence data from at least 2 unique peptides per protein with Mascot scores of ≥ 50 or a single peptide with Mascot scores of ≥ 100, were considered significant matches to query databases. In addition, sequences of identified proteins were further evaluated for their secretory potential using SecretomeP 2.0 (http://www.cbs.dtu.dk/services/SecretomeP/), which predicts those proteins/peptides capable of classical (signal sequences; SignalP 2.0 HMM) or nonclassical protein secretion [19]. For these analyses, all identified proteins obtained from the database were estimated to be full-length and possessed putative methionine start sites. Also, because SecretomeP was entrained on mammalian secretory protein sequences, a more rigorous neural network (NN) score of ≥ 0.600, rather than 0.500, was used as a cut-off value to identify candidate proteins capable of secretion via nonclassical secretory processes [20]. Gene ontology assignments for identified proteins were made using Blast2GO (www.blast2go.de) and AmiGO (http://amigo.geneontology.org/cgi-bin/amigo/go.cgi).
2.4. Anti-LTP antibody production
In order to determine the origin of the LTP components released during larval transformation and to investigate the time-course of larval protein release during in vitro cultivation, a polyclonal antiserum was generated in rabbits (Panigen, Blanchardville,WI) that was used to immunolocalize reactive LTP epitopes in intact miracidia and in vitro transformed primary sporocysts. In addition, the antiserum was used in Western blot analyses of larval extracts and LTP. Culture supernatants, collected 24-48 hr after S. mansoni miracidia were first placed into culture wells, were collected and concentrated by acetone precipitation as described above, and injected SC (250 μg total protein; multiple sites) into each of 2 rabbits using Freund's complete adjuvant for the primary injection series followed by Freund's incomplete adjuvant for the next two booster injections. The IgG fraction was purified from approximately 2 ml of anti-LTP antiserum by incubating the antiserum with 600 μL of packed Protein G Plus/Protein A agarose beads (Calbiochem, La Jolla, CA) for 1 hr at 22C under constant rotation. IgG-bound beads were packaged into Poly-Prep chromatography columns (Bio-Rad Laboratories; Richmond, CA), washed 3 times, each with 2.5mL sPBS to remove unbound proteins, followed by elution of bound IgG using 250μL of sPBS, pH2.5. Twenty-five μL of 1M Tris-HCl, pH8.0, was immediately added to readjust the pH to 7.5. This purified IgG preparation was stored in 1% sodium azide and used at working antibody dilutions of 1:7500 for Western blot, and 1:5000 for immunocytochemical localization.
2.5. Western blot analysis
In order to visualize anti-LTP antibody-reactive proteins, 1-D SDS-PAGE was used to separate LTP followed by Western blot analysis. This was accomplished by subjecting LTP to SDS-PAGE using 12.5% slab gels, followed by transfer to nitrocellulose (NC) membranes for 60 min at 30 mA at 4C using a Semi-Dry transfer unit (Hoefer TE 70; Amersham Biosciences, Piscataway, NJ). After transfer, NC membranes were blocked for 2 hr at 22C in TBS/0.03% Tween-20 (TBST), pH 7.4, containing 5% BSA, followed by overnight incubation in IgG-fractionated anti-LTP antibody (1:7500 dilution in blocking buffer) at 4C. Membranes were then washed in TBST followed by incubation in a 1:10,000 dilution of alkaline phosphatase (AP)-conjugated goat anti-rabbit IgG (GAR-AP; Promega, Madison, WI) for 1 hr at 22C, 5× washes in PBST, once in AP buffer (pH 9.5) and finally incubation in AP substrate (NBT/BCIP). In addition, to determine if antibodies contained in the anti-LTP antisera were reacting with carbohydrates (CHOs) associated with glycoproteins, LTP-blotted membranes were subjected to 3 successive treatments with 10 mM meta-periodate (20 min each) at 22C, followed by rinsing 3× with PBS and anti-LTP staining as outlined above.
2.6 Immunofluorescence localization of LTP proteins
Miracidia were obtained from eggs isolated from the livers of S. mansoni-infected mice as described previously [17]. Newly hatched miracidia were either immediately fixed in a mixture of 2% buffered paraformaldehyde and 1% Triton X-100 (PFA-TX100, pH 7.2) (Sigma-Aldrich, St.Louis, MO) or introduced into culture for 24-48 hr to obtain in vitro developing primary sporocysts prior to PFA-TX100 fixation. After overnight fixation at 4C, larvae were washed 2× with sPBS. 4× with 2% BSA in sPBS, followed by overnite incubation in 5% BSA blocking buffer containing 0.02% sodium azide. Fixed, blocked and Triton-permeablized larvae were treated with IgG-purified anti-LTP antibodies diluted 1:10,000 in blocking buffer overnight under constant rotation at 4C. This was followed by washing 5× in 1% BSA in sPBS, once in blocking buffer and then incubation in a mixture of goat anti-rabbit IgG- Alexafluor488 (GAR-Alexa488) (Invitrogen, 1:500 dilution), 7.5 U/ml phalloidin-Alexafluor546 (Invitrogen) and 50 μg/ml Hoechst 33258 dye overnite at 4C. Control larvae were incubated with the pre-immune rabbit serum diluted 1:10,000 and GAR-Alexa488. Specimens were transferred into glass coverslips in mounting medium (Vectashield; Vector Labs; Burlingame, CA) and examined with a Bio-Rad Radiance 2100 MP Rainbow confocal system. Images were processed using Adobe Photoshop CS software.
3. Results
In vitro culture of free-swimming miracidia results in a dramatic morphological transformation characterized by the cessation of swimming activity, followed by loosening and detachment of ciliated epidermal plates that is coincident with the expansion of intercellular ridges and the formation of the sporocyst tegumental syncytium [2, 21]. One-dimensional SDS-PAGE separation of concentrated larval LTP released during the initial 24-hr of in vitro miracidial-sporocyst cultivation revealed a complex population of proteins ranging in molecular mass from <10 to >200 kDa (Fig. 1). From LC-MS/MS analyses of excreted larval proteins contained in gel slices comprising entire SDS-PAGE-separated samples, a diverse assemblage of 99 proteins were identified in the S. mansoni protein database (http://www.genedb.org/genedb/smansoni) (Tables 1 and 2), as well as an additional 28 hypothetical or expressed unknown proteins that met our scoring criteria (data not shown). Analysis by SecretomeP 2.0 (www.cbs.dtu.kd/services/SecretomeP) further demonstrated that 19 (19.2%) identified proteins possessed predicted signal sequences, while an additional 22 leaderless proteins (22.2%) were assigned neural network scores (NN#) ≥ 0.600 (Table 1) indicating their potential for secretion by a nonclassical secretory pathway(s) [20]. Therefore, in total, ∼41% of identified proteins in the LTP possessed molecular characteristics consistent with possible release through a secretory process(es), while the remaining majority of proteins (59%) lacked any molecular signatures for secretion.
Figure 1.
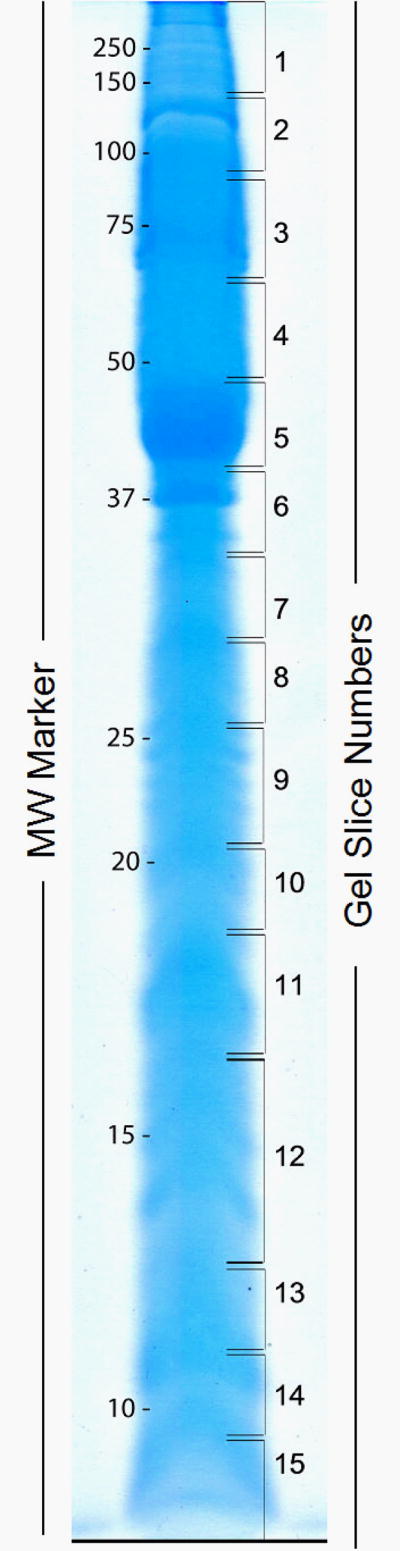
TABLE 1.
Summary of putative secretory proteins contained in Schistosoma mansoni larval transformation culture supernatants based on neural network scores (NN No.) ≥ 0.600 and/or the presence of a signal peptide (+) as determined by SecretomeP and SignalP analyses. For Gel Band No. refer to Fig. 1. TM, putative transmembrane domain
| PROTEIN ID | Accession No. | Score | #Peptides | Gel band No. | NN No. | Signal peptide | TM domain |
|---|---|---|---|---|---|---|---|
| Venom allergen-like 3/23 (SmVAL3/23) | Sm00763 | 1440 | 7 | 10,11 | 0.784 | + | |
| Major egg antigen (p40) (HSP16/20) | Smp_049230/50 | 1076 | 8 | 5-9,11 | 0.786 | 1,aa72/78-94/100 | |
| Venom allergen-like 9 (SmVAL9) | Smp_176180 | 1015 | 4 | 7-9,10 | 0.281 | + | 1, aa5-24 |
| Venom allergen-like 2 (SmVAL2) | Smp_002630 | 969 | 5 | 8-10 | 0.666 | ||
| DJ-1/PfpI family protein | Smp_082030 | 905 | 8 | 11 | 0.498 | + | |
| 20.7 kDa tegumental protein | Smp_077310 | 881 | 8 | 9,10,12,13 | 0.612 | ||
| Annexin | Smp_164100 | 838 | 4 | 6,7,10,11 | 0.618 | ||
| Calpain 4, 6, 7 | Smp_157500 | 765 | 9 | 4,11 | 0.933 | ||
| Exonuclease SbcC (secretory glycoprotein k5) | Sm03050 | 692 | 8 | 4-8 | 0.432 | + | |
| Venom allergen-like 15 (SmVAL15) | Smp_070250 | 649 | 4 | 6,7 | 0.564 | + | 1, aa7-26 |
| Adenylate kinase | Smp_071390 | 639 | 8 | 9 | 0.729 | ||
| 21.9 kDa expressed protein (SPO-1) | Smp_096790 | 610 | 2 | 11-13 | 0.916 | + | |
| α-2-macroglobulin-related | Smp_089670 | 574 | 10 | 1 | 0.485 | + | |
| 14 kDa fatty acid binding protein (sm14) | Smp_095360.1 | 539 | 6 | 12 | 0.798 | ||
| Voltage-gated potassium channel | Smp_136440 | 405 | 1 | 3,4 | 0.735 | 4, aa84-103, 188-210, 252-274,281-303 | |
| Glutathione-S-transferase omega | Smp_152710.1 | 402 | 4 | 8,9 | 0.835 | + | |
| α-galactosidase/α-N-acetylgalactominidase | Smp_179250 | 316 | 2 | 4,5 | 0.671 | + | 1, aa 7-29 |
| Copper/zinc superoxide dismutase, putative | Smp_176200.1/.2 | 314 | 4 | 12 | 0.682 | ||
| Tetraspannin/CD63 receptor | Smp_140000 | 283 | 3 | 7,8 | 0.819 | + | 4, aa12-34, 77-99, 106-128, 253-275 |
| Glutenin, high MW subunit 12 precursor-related | Smp_098420 | 257 | 2 | 11,13 | 0.848 | + | 1, aa7-24 |
| Endoglycoceramidase | Smp_120050 | 232 | 1 | 2,3 | 0.529 | + | |
| Protein inhibitor of NOS (PIN); dynein lg chain | Smp_040680 | 218 | 3 | 13-15 | 0.712 | ||
| Venom allergen-like 26/28 (SmVAL26/28) | Smp_176160 | 214 | 3 | 7-10 | 0.758 | + | |
| Egg protein C122-like | Smp_032670.2 | 705 | 4 | 8 | 0.539 | + | |
| Thioredoxin peroxidase/peroxiredoxin | Smp_158110.1/2 | 201 | 2 | 14 | 0.660 | ||
| Venom allergen-like 27 (SmVAL27) | Smp_154260/90 | 197 | 3 | 11 | 0.716 | + | |
| 200-kDa GPI-anchored surface glycoprotein | Smp_017730 | 189 | 3 | 1 | 0.667 | + | |
| Leucine-rich transmembrane protein | Smp_147470 | 184 | 3 | 6 | 0.064 | + | 1, aa342-364 |
| Eukaryotic phosphomannomutase | Smp_087860 | 181 | 3 | 8,9 | 0.683 | ||
| Heme binding protein 2-related | Smp_016730 | 166 | 3 | 10 | 0.632 | ||
| HSP70 | Smp_049550 | 150 | 3 | 3 | 0.546 | + | |
| Program cell death protein (sorcin-related) | Smp_136640 | 141 | 3 | 11,12 | 0.668 | ||
| Calmodulin (EF-hand domain) | Smp_026560.1 | 129 | 2 | 12 | 0.883 | ||
| Ubiquitin (ribosomal L40e family domain) | Smp_089430 | 118 | 3 | 15 | 0.673 | ||
| WD-repeat protein | Smp_073940 | 200 | 2 | 1 | 0.608 | ||
| Peptidyl-gly α-amidating monooxygenase | Smp_010820 | 103 | 3 | 3,4 | 0.557 | + | |
| Glutathione-S-transferase 26 kDa (SmGST26) | Smp_163610 | 100 | 2 | 9 | 0.638 | ||
| Immunophilin FK506 binding protein | Smp_079230 | 99 | 2 | 12 | 0.622 | ||
| Ferritin light chain | Smp_047650 | 97 | 2 | 10,11 | 0.676 | ||
| Major egg antigen (HSPβ1) | Smp_049240 | 73 | 2 | 1,10 | 0.798 | ||
| Placental protein 11-related (endoribonuclease) | Smp_051640 | 69 | 2 | 7 | 0.816 |
TABLE 2.
Summary of putative nonsecreted proteins contained in Schistosoma mansoni larval transformation culture supernatants based on neural network scores (NN No.) < 0.600 and the absence of signal peptides as determined by SecretomeP and SignalP analyses. For Gel Band No. refer to Fig. 1.
| PROTEIN ID | Accession No. | Score | #Peptides | Gel band No. | NN No. |
|---|---|---|---|---|---|
| HSP 70 homologue (major surface antigen) | Smp_106930 | 2467 | 20 | 1,3,6,8, | 0.254 |
| Phosphoenolpyruvate carboxykinase (PepCK) | Smp_005880 | 2011 | 22 | 3 | 0.460 |
| Fructose-bisphosphate aldolase class I | Smp_042160.2 | 1407 | 16 | 5,6 | 0.317 |
| Leishmanolysin-like protease | Smp_167070 | 1180 | 10 | 3,4 | 0.308 |
| Triosephosphate isomerase | Smp_003990 | 1175 | 9 | 8,9 | 0.508 |
| α-N-acetylgalactosaminidase | Sm06626 | 1171 | 10 | 4,5 | 0.540 |
| 22.6 kDa tegumental antigen | Smp_086470 | 1171 | 8 | 10-15 | 0.452 |
| Phosphopyruvate hydratase/enolase | Smp_024110 | 863 | 9 | 4,5 | 0.471 |
| ATP:guanidino kinase smc74 | Smp_194770 | 774 | 12 | 2,3 | 0.402 |
| Thioredoxin | Smp_008070 | 768 | 6 | 13,14 | 0.585 |
| Nucleotide diphosphate kinase | Smp_092750 | 763 | 4 | 11,12 | 0.384 |
| Cytosolic malate dehydrogenase | Smp_035270.2 | 750 | 6 | 7 | 0.343 |
| Phosphoglyerate mutase | Smp_096760 | 731 | 5 | 8,9 | 0.293 |
| Malate dehydrogenase, NAD-dependent | Smp_047370 | 678 | 8 | 6,7 | 0.500 |
| Peptidylglycine α-hydroxylating monooxygenase (PHM) | Smp_145300 | 670 | 2 | 1-3 | 0.563 |
| 14-3-3 episilon protein | Smp_034840.1/.2 | 627 | 11 | 8,11 | 0.303/0.225 |
| 14-3-3 protein homologue 1 | Smp_009760/80 | 476 | 6 | 8,9,11 | 0.246 |
| Glutathione-S-transferase 26 kDa (SmGST26) | Smp_102070 | 611 | 8 | 9 | 0.445 |
| Cytochrome c | Smp_033400 | 610 | 4 | 12,13 | 0.423 |
| Glutathione-S-transferase 28 kDa (SmGST28) | Smp_054160 | 597 | 5 | 8,9 | 0.300 |
| L-lactate dehydrogenase | Smp_038950 | 538 | 7 | 7,10 | 0.492 |
| Ferritin-1 light chain | Smp_087760 | 475 | 7 | 11 | 0.522 |
| Noncanonical purine NTP pyrophosphatase | Smp_063120.1 | 422 | 6 | 10 | 0.571 |
| β-tubulin | Smp_030730 | 407 | 6 | 4 | 0.535 |
| Mannosidase, endo-α-isoform 1 | Smp_016880 | 403 | 5 | 9,10 | 0.427 |
| Glyceraldehyde-3-phosphate dehydrogenase | Smp_056970.1 | 368 | 4 | 6 | 0.412 |
| Hydroxyacylglutathione hydrolase | Smp_091010 | 364 | 6 | 8 | 0.386 |
| NUDIX family hydrolase, putative | Smp_138760 | 361 | 5 | 7,8 | 0.532 |
| 90-kDa heat shock protein (HSP83) | Smp_072330 | 352 | 7 | 2,3 | 0.192 |
| Histone H4 | Smp_053290 | 350 | 4 | 12 | 0.408 |
| Protein kinase (cAMP-depend. catalytic subunit) | Smp_152330 | 349 | 1 | 1, 10-14 | 0.338 |
| Venom allergen-like 5/15 (SmVAL 5/15); CRISP-like | Smp_120670 | 330 | 2 | 8-10 | 0.456 |
| Phosphoglycerate kinase | Smp_018890 | 322 | 4 | 5,6 | 0.422 |
| Myotonic dystrophy s/t kinase-related | Smp_151140 | 293 | 4 | 12 | 0.280 |
| 16 kDa calcium-binding protein (SME16) | Smp_096390.2 | 281 | 4 | 12 | 0.448 |
| Actin-1 | Smp_046590.2 | 279 | 3 | 5 | 0.495 |
| Uridine phosphorylase | Smp_082420 | 234 | 3 | 8 | 0.417 |
| Methylthioadenosine phosphorylase-related | Smp_171620 | 227 | 3 | 7 | 0.457 |
| Transketolase | Smp_059790.2 | 224 | 4 | 3 | 0.514 |
| Carbonyl reductase (short-chain dehydrogenase) | Smp_033540 | 218 | 7,8 | 4 | 0.387 |
| P53 inducible protein | Smp_150980.2 | 214 | 1 | 1, 7,8 | 0.479 |
| Cyclophilin-type (peptidylpolyprolyl cis-trans Isomerase) | Smp_040130 | 208 | 1 | 11,12 | 0.543 |
| Myoglobin 1-related | Smp_162360 | 197 | 2 | 11 | 0.378 |
| DnaK family protein (HSP70-related) | Smp_069130.1 | 195 | 3 | 2,12 | 0.319 |
| Phosphoglycerate kinase | Smp_187370 | 178 | 2 | 3,5,6 | 0.352 |
| Glycerol 3-phosphate dehydrogenase | Smp_030500.1 | 174 | 2 | 6,7 | 0.511 |
| Dipeptidylpeptidase III | Smp_019010 | 156 | 2 | 3 | 0.597 |
| Purine nucleoside phosphorylase | Smp_090520 | 144 | 3 | 7 | 0.546 |
| Proteosome subunit (α-type) | Smp_070930 | 141 | 2 | 8 | 0.289 |
| Proteosome subunit (β-type) | Smp_025800 | 136 | 2 | 9 | 0.439 |
| α-tubulin | Smp_016780 | 133 | 3 | 4 | 0.457 |
| Cystatin B, putative | Smp_006390 | 129 | 1 | 12,13 | 0.597 |
| Glucose-6-phosphate isomerase | Smp_022400 | 126 | 2 | 4 | 0.596 |
| Transaldolase | Smp_070600 | 124 | 2 | 6 | 0.493 |
| Proteosome subunit (β-type) | Smp_034490 | 124 | 2 | 9 | 0.435 |
| Proteosome subunit (α-type) | Smp_067890 | 118 | 2 | 9 | 0.443 |
| Cell division protein 49 (cdc48) | Smp_018240.1 | 105 | 2 | 2 | 0.431 |
| Glycogen phosphorylase | Smp_143840 | 105 | 2 | 6 | 0.467 |
In order to gain further insight into possible functions of larval released proteins, gene ontology (GO) annotations using Blast2GO were applied to putative secretory and nonsecretory proteins, resulting in groupings according to GO-assigned molecular function. As noted in Fig. 2, the prevalence of secretory vs. nonsecretory proteins differed in their representation within a number of the higher level molecular function categories. For example, proteins characterized as ion binding (22%) and protein binding (33%) were more highly represented in the “secretory” LTP subset than those of the nonsecretory group (3% and 12%, respectively). By contrast, the nonsecretory protein group was more highly represented by nucleotide binding proteins (19% vs. 7% for secretory proteins) and those possessing hydrolase (16% vs. 5%), transferase (14% vs. 5%) and oxidoreductase (14% vs. 2%) activities. The relatively large representation of nonsecretory redox enzymes is due mainly to the presence of cytosolic dehdrogenases and cytochrome c. A complete listing of all identified proteins and their GO anotations are listed in supplemental Table 1 (Suppl. Table 1). In addition, since entries in the Schistosoma mansoni GeneDB (http://www.genedb.org/genedb/smansoni/) are under continual revision, amino acid sequences of peptides identified by tandem MS analyses for all proteins to which identifications were made are given in supplementary Table 2 (Suppl. Table 2).
Figure 2.

Table 3 provides a more detailed view of specific genes highly represented within molecular or biological function categories according to gene term assignments from AmiGO with adjustments based on recent literature. These functional categories included proteins involved in protein-protein interactions (protein binding, folding, proteolysis), electron transport/redox/anti-oxidant activities, kinase/phosphorylase activities, carbohydrate metabolism; motility, ion-binding and the like. Although placement of proteins into “secretory” and “nonsecretory” categories was quite variable within groups, in general it was noted that many of the proteins involved in protein interactions (“Protease” and “Protein folding” categories), metal ion-binding (“Ion binding” category) and the venom allergen-like group were highly represented in “secretory” proteins, whereas those mainly involved in glycolysis, gluconeogenesis, or the TCA cycle (“Carbohydrate metabolism” group ) or associated with kinase/phosphorylating activities were generally characterized as “nonsecretory” (Table 3). Although many of the proteins/enzymes listed in the “Redox/anti-oxidant/electron transport” category are nonsecretory, most of these are associated with CHO or energy metabolism (namely the dehydrogenases and cytochrome c), while those with putative secretory features are associated with anti-oxidant/detoxification functions (GST, peroxiredoxin, SOD). Among the ion-binding proteins identified as “secretory” in nature, those with Ca-binding activities (possessing EF hand domains) were most prevalent including 2 dynein light chain members, a calpain-related enzyme (calpain 4,6,7), a calmodulin and an annexin-related protein. Two proteins associated with Fe binding/sequestration (ferritin 1 light chain, heme-binding protein 2), a Cu/Zn-binding superoxide dismutase (SOD) and a putative potassium channel protein also were included in the secretory protein category (Table 3). Likewise, all but one of S. mansoni venom allergen-like proteins (SmVAL2, 3/23, 9, 15, 26/28, and 27) identified in our proteomic screen possessed signal peptides and/or significant NN values (≥ 0.600) that are characteristic of secretory proteins. Of the putative “secretory protein” group, VAL proteins represented a prominent constituent, having 3 of the 4 highest Mascot scores among the secretory proteins identified (Table 1). A final highly represented group of proteins found in the larval transformation proteome were those associated with motility, especially those involved in microtubule assembly/motor activity. These included several calcium-binding dynein light chains (8 kDa cytoplasmic DLC; 20.7 kDa tegumental protein; 22.6 kDa antigen), an actin and actin-binding protein, and α and β tubulins (Table 3).
Table 3.
Hypothetical biological/molecular functions of identified larval proteins released during in vitro transformation of Schistosoma mansoni miracidia to primary sporocysts at 24 hr of cultivation in snail saline (CBSS). S, putative secretory protein; N, putative nonsecretory protein.
| Protease/protein degradation/inhibition | |||
| Calpain 4, 6, 7 | Smp_157500 | S | endopeptidase, cys-type; Ca-dependent |
| Alpha-2-macroglobulin related | Smp_089670 | S | endopeptidase inhibitor |
| DJ-1/PfpI family protein | Smp_082030 | S | serine-type peptidase (S51 family) |
| Ubiquitin (ribosomal L40e family domain) | Smp_089430 | S | ubiquitin-proteosome protease complex |
| Dipeptidylpeptidase III | Smp_019010 | N | exopeptidase; cytoplasmic |
| Cystatin B, putative | Smp_006390 | N | cysteine protease inhibitor |
| Proteosome subunit (β-type); | Smp_025800 | N | ubiquitin-proteosome protease complex |
| Proteosome subunit (β-type) | Smp_034490 | N | ubiquitin-proteosome protease complex |
| Proteosome subunit (alpha-type) | Smp_067890 | N | ubiquitin-proteosome protease complex |
| Proteosome subunit (alpha-type) | Smp_070930 | N | ubiquitin-proteosome protease complex |
| Leishmanolysin-like protease | Smp_167070 | N | metalloendopeptidase; M8 family |
| Protein folding/stress/chaperone | |||
| Major egg antigen (p40) (HSP20) | Smp_049230/50 | S | heat-shock protein; alpha-crystallin-like |
| Major egg antigen (HSPβ1; HSP27) | Smp_049240 | S | heat-shock protein; stress response; chaparone |
| HSP70 | Smp_049550 | S | heat-shock protein; stress response; chaparone |
| Immunophilin (FKBP12 binding) | Smp_079230 | S | peptidyl-prolyl cis-trans isomerase; protein folding |
| Leucine-rich transmembrane protein | Smp_147470 | S | protein-protein interaction; adhesion |
| Egg protein C122-like | Smp_032670.2 | S | R-spondin-like; stress response |
| Cyclophilin (CS-A binding) | Smp_040130 | N | peptidyl-prolyl cis-trans isomerase; protein folding |
| Major surface antigen (HSP70 homologue) | Smp_106930 | N | ATP-binding; stress response; chaparone |
| HSP90 (83 kDa HSP) | Smp_072330 | N | ATP-binding; stress response; chaparone |
| HSP70-related | Smp_069130.1 | N | ATP-binding; stress; DnaK family protein |
| Redox/anti-oxidant/electron transport | |||
| Glutathione-S-transferase omega | Smp_152710.1 | S | redox; anti-oxidant |
| Glutathione-S-transferase 26 kDa | Smp_163610 | S | redox; anti-oxidant; xenobiotic metabolism |
| Copper/zinc superoxide dismutase, putative | Smp_176200.1/2 | S | redox; anti-oxidant |
| Thioredoxin peroxidase/peroxiredoxin | Smp_158110.1/2 | S | redox; anti-oxidant |
| Protein inhibitor of NOS (PIN); dynein lg chain | Smp_040680 | S | microtubule; motor activity; anti-oxidant |
| Heme binding protein 2-related | Smp_016730 | S | heme binding; anti-oxidant |
| Peptidyl-gly α-amidating monooxygenase | Smp_010820 | S | redox; protein metabolism |
| Thioredoxin | Smp_008070 | N | redox; anti-oxidant |
| Peptidyl-gly α-hydroxylating monooxygenase | Smp_145300 | N | redox; protein metabolism |
| Malate dehydrogenase, NAD-dependent | Smp_047370 | N | redox; glycolysis |
| Glycerol 3-phosphate dehydrogenase | Smp_030500.1 | N | redox; CHO metabolism |
| Carbonyl reductase (short-chain dehydrogenase) | Smp_033540 | N | redox; antioxidant |
| Glutathione-S-transferase 26 kDa | Smp_102070 | N | redox; anti-oxidant; xenobiotic metabolism |
| Glutathione-S-transferase 28 kDa | Smp_054160 | N | redox; anti-oxidant; xenobiotic metabolism |
| Hydroxyacylglutathione hydrolase | Smp_091010 | N | redox; detoxication; xenobiotic metabolism |
| Cytochrome c | Smp_033400 | N | redox; electron transport |
| Cytosolic malate dehydrogenase | Smp_035270.2 | N | redox; glycolysis |
| Glyceraldehyde-3-phosph. dehydrogenase | Smp_056970.1 | N | redox; glycolysis |
| Microtubule/motility | |||
| 20.7 kDa tegumental protein; dynein light chain | Smp_077310 | S | microtubule; motor activity; EF-hand domain |
| Protein inhibitor of NOS (PIN); dynein lg chain | Smp_040680 | S | microtubule; motor activity; anti-oxidant |
| WD-repeat protein | Smp_073940 | S | actin-binding, putative |
| β-tubulin | Smp_030730 | N | microtubule; motor activity; GTP-binding |
| 22.6 kDa antigen; dynein light chain | Smp_086470 | N | microtubule; motor activity; EF-hand domain |
| α-tubulin | Smp_016780 | N | microtubule; motor activity; GTP-binding |
| Actin-1 | Smp_046590.2 | N | cytoskeletal protein; motility; ATP-binding |
| Carbohydrate (CHO) metabolism | |||
| α-galactosidase/α-N-acetylgalactominidase | Smp_179250 | S | carbohydrase; CHO binding |
| Endoglycoceramidase | Smp_120050 | S | glycosidase; CHO binding |
| Egg protein C122-like | Smp_032670.2 | S | heparin-binding, putative |
| Glucose-6-phosphate isomerase | Smp_022400 | N | CHO metabolism; glycolysis; gluconeogenesis |
| Eukaryotic phosphomannomutase | Smp_087860 | N | hexose metabolism; mannose biosynthesis |
| Triosephosphate isomerase | Smp_003990 | N | CHO metabolism; glycolytic activity |
| Transketolase | Smp_059790.2 | N | CHO metabolism; glycolysis; pentose-P shunt |
| α-N-acetylgalactosaminidase | Sm06626 | N | carbohydrase activity; CHO binding |
| Glycogen phosphorylase | Smp_143840 | N | glucose phosphorylation; Glc-1-P synthesis |
| Phosphoenolpyruvate carboxykinase (PepCK) | Smp_005880 | N | CHO metabolism; gluconeogenesis; GTP binding |
| Fructose-bisphosphate aldolase class I | Smp_042160.2 | N | CHO metabolism; glycolysis |
| Transaldolase | Smp_070600 | N | CHO metabolism; glycolysis-pentose-P shunt |
| Lactate dehydrogenase | Smp_038950 | N | CHO metabolism; glycolysis |
| Phosphopyruvate hydratase (enolase) | Smp_024110 | N | CHO metabolism; glycolysis |
| Phosphoglycerate mutase | Smp_096760 | N | CHO metabolism; glycolysis |
| Malate dehydrogenase (cytosolic) | Smp_035270.2 | N | CHO metabolism; glycolysis; TCA cycle |
| Glyceraldehyde-3-phosphate dehydrogenase | Smp_056790.1 | N | CHO metabolism; glycolysis |
| Kinases/phosphorylases | |||
| Adenylate kinase | Smp_071390 | S | phosphotransferase; ATP binding |
| ATP:guanidine kinase smc74 | Smp_194770 | N | creatine kinase-like phosphotransferase |
| Nucleotide diphosphate kinase | Smp_092750 | N | NTP biosynthesis (CTP, GTP, UTP) |
| Guanylate kinase (mannosidase, endo-a iso.1) | Smp_016880 | N | Nucleotide binding; GMP cycling; cGMP synth. |
| Uridine phosphorylase | Smp_082420 | N | glycosyltransferase; uridine binding |
| Methylthioadenosine phosphorylase-related | Smp_171620 | N | transferase; purine metabolism |
| Purine nucleoside phosphorylase | Smp_090520 | N | phosphorylase family 2 |
| Protein kinase | Smp_152330 | N | cAMP-dependent; catalytic subunit |
| Myotonic dystrophy s/t kinase-related | Smp_151140 | N | Serine-threonine kinase |
| Phosphoenolpyruvate carboxykinase (PepCK) | Smp_005880 | N | CHO metabolism; gluconeogenesis; GTP binding |
| Glycogen phosphorylase | Smp_143840 | N | CHO metabolism; Glu-1-phosphate synthesis |
| Phosphoglycerate kinase | Smp_018890 | N | CHO metabolism; glycolysis |
| Ion-binding proteins | |||
| 20.7 kDa tegumental protein (dynein light chain) | Smp_077310 | S | Ca-binding; EF-hand domain; motor activity |
| Annexin | Smp_164100 | S | Ca-binding; EF-hand dom.; phospholipid-binding |
| Calmodulin | Smp_026560.1 | S | Ca-binding; EF-hand dom.; regulation |
| Calpain 4, 6, 7 | Smp_157500 | S | Ca-dependent protease |
| Program cell death protein (sorcin-related) | Smp_136640 | S | Ca-binding; EF-hand dom.; regulation |
| Superoxide dismutase, putative | Smp_176200.1/2 | S | Cu/Zn-binding; dismutase activity; anti-oxidant |
| Heme binding protein 2-related | Smp_016730 | S | Fe-heme binding; transport; storage |
| Ferritin 1 light chain | Smp_047650 | S | Fe-binding; cellular iron transport; storage |
| Voltage-gated potassium channel | Smp_136440 | S | K-binding; ion channel; K transport |
| Ferritin light chain | Smp_087760 | N | Fe-binding; cellular iron transport; storage |
| 22.6 kDa antigen (dynein light chain) | Smp_086470 | N | Ca-binding; EF-hand dom.; motor activity |
| 16 kDa egg antigen (SME16) | Smp_096390.2 | N | Ca-binding protein; EF-hand dom. |
| Myoglobin 1-related | Smp_162360 | N | Fe-heme binding protein; oxygen transport |
| Venom allergen-like proteins | |||
| Venom allergen-like 2 (SmVAL2) | Smp_002630 | S | Sperm-coating protein (SCP)/TAPS domain family |
| Venom allergen-like 3/23 (SmVAL 3/23) | Sm00763 | S | SCP-TAPS domain; cyst-rich proteinase inhibitor |
| Venom allergen-like 9 (SmVAL 9) | Smp_176180 | S | SCP-TAPS domain family |
| Venom allergen-like 15 (SmVAL15) | Smp_070250 | S | SCP-TAPS domain family |
| Venom allergen-like 26/28 (SmVAL26/28) | Smp_154260 | S | SCP-TAPS domain family |
| Venom allergen-like 27 (SmVAL27) | Smp_154290 | S | SCP-TAPS domain family |
| Venom allergen-like 5/15 (SmVAL 5/15) | Smp_120670 | N | SCP-TAPS domain; CRISP-like subfam. glioma |
In order to gain further insight into the cellular origins of LTP released during early larval development, a polyclonal antibody generated to concentrated LTP was used to immunolocalize that subset of immunoreactive proteins within miracidia and 24-hr cultured sporocysts. Initial Western blot analyses of isolated LTP revealed a complex banding pattern of anti-LTP antibody-reactive proteins ranging from >100 to ∼20 kDa (Fig. 3). Following periodate treatment, which destroys exposed carbohydrate (CHO) epitopes, separated immunoreactive LTPs exhibited a similar banding profile as the untreated blot, with the exception of the loss of reactivity to several lower molecular mass proteins (< ∼25 kDa) (Fig. 3). Loss of immunoreactivity in these bands were likely the result of periodate-destruction of CHO epitopes and not protein desorption from the blot since both periodate-treated and untreated Simply-Blue-stained blots retained identical protein banding profiles (Fig. 3). When the anti-LTP antibody was applied to intact fixed and detergent-permeablized miracidia and in vitro cultured primary sporocysts, confocal epifluorescence microscopy revealed highly localized reactivity mainly at the surface of both stages. For miracidia, immunoreactivity was concentrated on or in the ciliated epidermal plates and cilia themselves, while less reactivity was observed at the intercellular ridges (Fig. 4). Primary sporocysts exhibited a similar distribution of anti-LTP immunoreactivity, being localized exclusively on and/or within the fully-formed sporocyst tegument (Fig. 5). In the case of both stages, the majority of reactivity appeared distal to a phalloidin-reactive layer (red), mainly comprised of actin-rich subtegumental muscles or other actin-containing structures. Although CHO may be the source of some reactivity, the restricted immunolocalization associated with the surface structures of permeablized miracidia and sporocysts suggests that protein epitopes, rather than common, widely-distributed CHO epitopes, are primarily recognized by this antiserum.
Figure 3.
Figure 4.
Figure 5.

4. Discussion
Following penetration of the snail host, transformation of S. mansoni miracidia to parasitic primary or mother sporocysts represents a critical period in the establishment of successful larval infections. It is during this time (∼24-hr post-infection), when miracidia are shedding their ciliated epidermal plates and simultaneously are in the process of forming the new sporocyst tegumental syncytium [21], that larvae are most vulnerable to potentially hostile elements of the host environment such as circulating immune cells (hemocytes), cytotoxic humoral factors, free O2/N2 radicals, and the like [5,23]. One mechanism by which these early developing stages may be able to counteract or suppress deleterious host factors is the production and release of parasite products capable of “neutralizing” or eliminating such factors. In the present study we focused on the proteins released during in vitro transformation of miracidia to sporocysts (termed larval transformation proteins or LTPs) since we, and others [5,7, 14,23], have hypothesized that such larval products released during this crucial period of development may exert a protective effect allowing for establishment of infections within naturally susceptible B. glabrata snails. Previous studies have shown that culture-derived proteins can bind both plasma [24] and hemocytes [25] and have documented their disruptive effects on various hemocyte functions including chemokinesis, superoxide production, reactive oxygen species and ERK signaling [10-13].
Given the cellular and physiological modulating effects of LTP, an important initial step in analyzing molecular structure-function relationships in this host-parasite system is a comprehensive evaluation of the LTP proteome to provide insights into the types of proteins being released during early miracidial-to-sporocyst development. As reported, tandem MS analysis of LTP yielded a total of 99 proteins, identified from exact peptide matches in the Sanger S. mansoni GeneDB that met our selection criteria. By contrast, a similar proteomic analysis recently performed on 24-hr S. mansoni larval excretory-secretory proteins (ESP) recovered from cultures essentially identical to that described in our study yielded only 7 identified proteins [14], 5 of which (TPI, GAPDH. Cu/Zn SOD, GST26, histone H4) were identified in the present study. Employing different methods in initial sample preparation and the use of 2-D, rather than 1-D, SDS-PAGE separations prior to subsequent protein isolation and MS analysis [14] may have contributed to our differing results. In addition the relatively low protein content, visible as silver-stained spots in their separated S. mansoni protein sample, contrasts with the complex banding pattern exhibited in the 1-D SDS-PAGE gels used in the present study. Starting with higher protein loads (concentration or volume) and subjecting entire gels (versus only a subset of protein spots) to proteomic analysis likely accounts for our greater yield and wider diversity of identified proteins. A disadvantage of 1-D “shotgun” approach is the reduced resolution of distinct protein groups, although the application of nanoLC-MS/MS is highly capable of efficiently separating and identifying tryptic peptides from complex protein mixtures [26].
A surprising finding of the present investigation was the high representation (∼60%) of identified proteins that lacked molecular signatures of secretory proteins. Most likely these proteins were released as a result of events transpiring during normal larval development. As mentioned earlier, transformation of the miracidium to sporocyst stage involves a dynamic expansion of intercellular syncytial ridges resulting in the “rounding up”, pinching off and detachment of ciliated epidermal plates from the subtending basement membrane [21,22]. It is highly probable that leakage of cytoplasmic proteins may occur as plates are released and ridge membranes fuse to form the contiguous tegumental syncytium [21]. Moreover, detached ciliated epidermal plates themselves also may serve as a significant source of nonsecreted proteins. Under in vitro and presumably in vivo [2,27] conditions, released ciliated epidermal plates degenerate, losing their cilia, and undergo spontaneous autolysis. Thus it appears that both somatic “leakage” during tegument formation and degeneration of released epidermal plates and cilia may be responsible for the presence in culture supernatants of nonsecretory proteins such as proteosome complexes, glycolytic enzymes, phosphorylating enzymes (kinases) and microtubule/cytoskeleton-associated proteins (e.g., dynein light chain members, actin, tubulin) (Table 3). Immunocytochemical localization of anti-LTP reactivity almost exclusively in epidermal plates, cilia and intercellular ridges of miracidia, and the tegument of transformed sporocysts strongly support the notion that the dramatic structural changes associated with miracidial transformation are significant contributors to the protein composition of LTP, particularly those of the nonsecretory group. This being the case, it is suggested that the terminology “larval transformation proteins” (LTP) be adopted in place of “excretory-secretory proteins/products”, previously used to describe proteins released during miracidium-to-sporocyst transformation in vitro (see Lodes and Yoshino [3]), in order to more accurately reflect the true nature of its protein composition.
Another unexpected finding of this study was the relatively high number of leaderless proteins (22%) identified by SecretomeP 2.0 as potentially capable of secretion via nonclassical secretory pathways. Because the structural features used by SecretomeP to identify nonclassically secreted proteins were “trained” on model mammalian proteins, we adopted a more stringent NN score of 0.600 (rather than 0.500) as our cut-off value as described by Bendtsen et al. [20]. In their analysis, they predicted nonclassical secretion for an expanded array of proteins, some of which were identified in our study; namely thioredoxin, SCP-like extracellular protein and Cu-Zn SOD. Although we assigned S. mansoni thioredoxin as nonsecretory proteins, its NN score (0.585) was well above the traditional 0.500 cut-off. Other leadless proteins in our LTP samples also have been identified in other systems as being nonclassically secreted including HSP70 in tumor cells [28], annexin in swine enterocytes [29], and calmodulin and ferritin in Hydra [30]. Based on these findings and our NN scoring criterion, it is highly probable that these, and other, leaderless proteins identified in LTP are representative of the “nonclassical” secretory protein group.
Results of the present study closely parallel a recent proteomic analysis of secretions from in vitro cultured S. mansoni eggs in which 188 proteins were identified using MudPIT (Multidimensional Protein Identification Technology) [31]. Consistent with their results, 65% of the identified egg-secreted proteins showing highest abundance also were represented in our LTP samples, including 7 of the 10 most abundant egg proteins (secretory proteins: Sm-p40 egg antigen, SmGST26, Prx and nonsecretory proteins: HSP70 surface antigen, fructose-bisphosphate aldolase, PHM, PepCK). Also, as in our analysis, ∼48% of high abundance proteins in the egg secretome consisted proteins predicted to be nonsecretory. Although sequence truncation or mammalian-based SecretomeP entrainment procedures may have contributed to some mis-assignment, based on our current findings, release of cytoplasmic proteins from damaged or degrading cellular components in ovo cannot be excluded as a source of these proteins. Interestingly, >50% of uniquely identified S. mansoni proteins reported in cercarial secretions [32,33] also were found in our LTP samples, and of these, ∼ 20% were classed as secretory proteins including Sm16 (Spo-1), GST26, VAL proteins (SCP-related), Sm14 (fatty acid-binding protein), immunophilin, calmodulin, calpain, SOD and thioredoxin peroxidase. These finding indicate that intramolluscan developing larval stages share a common subset of secretory proteins, suggesting similar functional roles in establishing infections within their respective hosts.
The anatomical or tissue origin of the secretory protein subset of LTP remains unclear, although as has been suggested for cercaria-schistosomula transformation, proteins may be secreted by or shed from the tegument [32]. Anti-LTP reactivity with the intact sporocyst tegument also supports this structure as one potential source of secreted proteins. Another anatomical source of secretory proteins are the penetration glands that function in cercariae to facilitate host entry by elaboration of hydrolytic enzymes, especially an elastase-type serine protease [34], This enzyme was well represented in the cercarial secretory proteome [32,33], as was a calpain [32], a metalloproteinase (SmPepM8), and a dipeptidyl peptidase IV (DPP IV) [33]. While elastases were conspicuously absent from our secretory LTP group, a calpain and serine peptidase (DJ-1/Pfpl) were identified suggesting a degree of commonality between cercarial vs. miracidial/sporocyst protease repertoires. However, whether or not these latter proteases participate in snail host penetration remains unknown as no detectable anti-LTP immunoreactivity was observed in miracidial penetration glands. Lack of glandular immunoreactivity may reflect the possibility that these, or other identified proteases (e.g., DPP III and leishmanolysin), are not associated with glands, or that other tissues (e.g., the tegument or epidermis) serve as alternative source(s) for these proteins. It should also be noted that in addition to proteases, protease inhibitors also were detected in LTP (α2-macroglobulin, cystatin B), and this finding is consistent with the cercarial secretory proteome [32,33]. Functionally, these inhibitors may be involved in regulating the activity of their own proteases [35] or counteracting specific host proteases that may cause harm to developing larvae. Protease activity [36,37] and expression of protease genes [38-40] have been demonstrated in hemocytes and other tissues of the host snail B. glabrata.
Regardless of the origins of LTP or how they are released, it is important to keep in mind that elaboration of proteins, whether a result of secretory processes, breakdown and degrading of cast-off ciliated epidermal plates or leakage during sporocyst tegument formation, is an integral part of the normal larval developmental process. However, there still remains a question of the functional role, if any, being played by LTP [5]. Free-swimming miracidia, upon penetration into its snail host may be confronted with a potentially hostile environment requiring a myriad of physiological adjustments in transitioning to its parasitic lifestyle as a primary sporocyst. In addition to the potential involvement of proteases in larval penetration, other proteins released during larval transformation may serve to enhance larval survival by enhancing motility within the host, creating a more suitable microenvironment for growth/development, or counteracting noxious host reactions. For example another prominent group of proteins include those involved in oxidation-reduction reactions and/or electron-transport. Many, such as the dehydrogenases, cytochrome c, and the peptidyl monooxygenases most likely function in intracellular CHO/energy or protein metabolic processes. However, others of this group possess redox activity that serve, either directly or indirectly, in antioxidant or detoxification functions [41], and as a result, may provide a parasite-protective role during early infection of the snail host [10,14,23]. These include Trx, a Cu/Zn SOD, the H2O2-scavenger Prx, several GSTs and a hydroxyacylglutathione hydrolase [42]. Previous studies have demonstrated that S. mansoni sporocysts are highly sensitive to the damaging effects of ROS [43,44], and it is envisaged that antioxidant-containing LTP elaborated from newly-infecting larvae developing within an oxygen-rich host environment (hemolymph) or confronted by oxidative stress during hemocyte immune responses may contribute to enhanced survivorship [10,14,23],. In addition, because free iron ions (Fe2+ or 3+) or iron-containing compounds may have toxic effects, especially in conjunction with ROS [23,45], the elaboration of Fe-binding proteins (e.g., ferritins, myoglobin, heme-binding proteins) also may have larval survival value.
One of the more striking features of the S. mansoni larval LTP proteome was the large representation of sperm-coating protein (SCP)-related, venom allergen-like (VAL) proteins, recently discovered in S. mansoni cercarial secretions [33] and characterized in detail by Chalmers et al. [46]. SmVAL proteins are related to the large gene family that contain the conserved SCP domain [47] commonly associated with venom proteins of insects [48] and mollusks [49], and secretions of parasitic nematodes [50-52]. Consistent with the findings of Cass et al. [31] and Chalmers et al. [46], early larval LTP contained SmVAL2, 3, 5 and 9, providing support for fully-formed miracidia as the likely source of these proteins in egg secretions. Our finding of several additional SmVAL proteins (SmVAL26, 27 and 28) further highlights the diversity represented by this protein group at a time of critical larval development. The functions of SmVALs are not known, although nematode VALs have been implicated in mechanisms of larval infection [50] and host immunomodulation [53]. Whether or not SmVAL proteins released during early S. mansoni development in the snail serve similar roles has yet to be determined.
Additional proteins with potential immunomodulatory potential have been identified in larval LTP. For example, a calcium-binding dynein light chain (DLC; Smp_040680), which is highly homologous to the Chlamydomonas (EDO96760.1) and Caenorhabditis (AAC77510.1) 8 kDa DLC, also shares significant homology with the fish, Apinephelus (AAP40019) and human (P63167.1) PIN or protein inhibitor of nitric oxide synthase, an essential enzyme involved in NO formation and NO-mediated larval killing by B. glabrata hemocytes [54]. Two other LTP proteins with potential for interacting with host cells and modulating their functions are the phospholipid-binding 21.9 kDa Sm16 (SPO-1; Smp_096790) and an annexin (Smp_164100). Both are capable of membrane binding and cell entry, and effecting various cell signaling activities [55,56]. In the case of Sm16 this included the regulation of caspase-dependent cellular apoptosis [55] and anti-inflammatory responses [57].
In summary, the present proteomic analysis of LTP released during the first 24-hr of S. mansoni larval development in vitro has revealed a diverse array of proteins representing many of the major functional GO categories including those involved in binding/folding activities (nucleotide, protein, lipid and ion binding proteins) and catalytic reactions (oxidoreductases, transferases, hydrolases, lyases and isomerases). This finding contrasts a previous proteomic analysis of miracidial transformation proteins [14] and suggests that LTP content of in vitro developing S. mansoni may be comparable both quantitatively and qualitatively to that of other S. mansoni stages [32,33] or other trematode species, e.g., Echinostomes [14]. Based on results of our “shotgun” proteomic analysis, it is concluded that upon miracidial entry into B. glabrata snails and subsequent transformation to the primary sporocyst stage, the host is confronted with what can be described as a “cloud” of larval proteins, many of which may possess functions or activities with potential for promoting parasite survival in susceptible snail strains [5,23]. These include anti-oxidant/detoxifying enzymes, potential immunomodulating proteins, proteases and protease inhibitors, heme- or iron-binding protein, carbohydrases, and others. On the other hand, the likelihood of larval survival would decrease for parasites infecting resistant snails possessing higher endogenous levels of chemical stressors or greater immune capacity (e.g., oxidant-generating capacity or proteolytic activities [37,44]) than can be overcome by counteracting LTP molecules. Thus it is postulated that the “susceptible” or “resistant” phenotype exhibited in a given snail-parasite encounter is based on the relative balance between host environmental “stressors” (including immune reactivity) and the efficacy of larval counter-measures to those stressors. We are in general agreement with Guillou et al. [14] that S. mansoni most likely uses both passive mimetic, possibly through shared complex CHO structures [58], and active molecular counter-measures in the initial establishment of infections within it snail host. However, our finding that a subset of immunoreactive LTPs continues to persist in the sporocyst tegument suggests that within susceptible hosts, larvae S. mansoni potentially are capable of actively modulating the host environment, including immune stressors, throughout their development.
Acknowledgments
This work was supported by NIH grant AI015503 to TPY and NIH schistosomiasis supply contract AI30026 (F. Lewis, Biomedical Research Institute, Rockville, MD). Thanks to John Kunert for providing technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Basch PF, DiConza JJ. The miracidium-sporocyst transition in Schistosoma mansoni: surface changes in vitro with ultrastructural correlation. J Parasitol. 1974;60:935–41. [PubMed] [Google Scholar]
- 2.Pan CT. Schistosoma mansoni: the ultrastructure of larval morphogenesis in Biomphalaria glabrata and of associated hot-parasite interactions. Jpn J Med Sci Biol. 1996;49:129–49. doi: 10.7883/yoken1952.49.129. [DOI] [PubMed] [Google Scholar]
- 3.Lodes MJ, Yoshino TP. Characterization of excretory-secretory proteins synthesized in vitro by Schistosoma mansoni primary sporocysts. J Parasitol. 1989;75:853–62. [PubMed] [Google Scholar]
- 4.DeGaffe G, Loker ES. Susceptibility of Biomphalaria glabrata to infection with Echinostoma parensei: correlation with the effect of parasite secretory-excretory products on host hemocyte spreading. J Invertebr Pathol. 1998;71:64–72. doi: 10.1006/jipa.1997.4710. [DOI] [PubMed] [Google Scholar]
- 5.Bayne CJ, Yoshino TP. Determinants of compatibility in mollusk-trematode parasitism. Amer Zool. 1989;29:399–407. [Google Scholar]
- 6.Yoshino TP, Vasta GR. Parasite-invertebrate host immune interactions. Adv Comp Environ Physiol. 1996;24:125–67. [Google Scholar]
- 7.Adema CM, Loker ES. Specificity and immunobiology of larval digenean-snail associations. In: Fried B, Graczyk TK, editors. Advances in Trematode Biology. Boca Raton: CRC Press; 1997. pp. 229–64. [Google Scholar]
- 8.Yoshino TP, Lodes MJ, Rege AA, Chappell CL. Proteinase activity in miracidia, transformation excretory-secretory products, and primary sporocysts of Schistosoma mansoni. J Parasitol. 1993;79:23–31. [PubMed] [Google Scholar]
- 9.Sullivan JT. Mitotic responses to injected extracts of larval and adult Schistosoma mansoni in Biomphalaria glabrata: effects of dose and colchicine treatment. J Parasitol. 2007;93:213–5. doi: 10.1645/GE-956R.1. [DOI] [PubMed] [Google Scholar]
- 10.Vermeire JJ, Yoshino TP. Antioxidant gene expression and function in in vitro-developing Schistosoma mansoni mother sporocysts: possible role in self-protection. Parasitology. 2007;134:1369–78. doi: 10.1017/S0031182007002697. [DOI] [PubMed] [Google Scholar]
- 11.Lodes MJ, Yoshino TP. The effect of schistosome excretory-secretory products on Biomphalaria glabrata hemocyte motility. J Invertebr Pathol. 1990;56:75–85. doi: 10.1016/0022-2011(90)90147-x. [DOI] [PubMed] [Google Scholar]
- 12.Connors VA, Yoshino TP. In vitro effect of larval Schistosoma mansoni excretory-secretory products on phagocytosis-stimulated superoxide production in hemocytes from Biomphalaria glabrata. J Parasitol. 1990;76:895–902. [PubMed] [Google Scholar]
- 13.Zahoor Z, Davies AJ, Kirk RS, Rollinson D, Walker AJ. Disruption of ERK signaling in Biomphalaria glabrata defence cells by Schistosoma mansoni: Implications for parasite survival in the snail host. Dev Comp Immunol. 2008;32:1561–71. doi: 10.1016/j.dci.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 14.Guillou F, Roger E, Mone Y, Rognon A, Grunau C, Theron A, Mitta G, Coustau C, Gourbal BEF. Excretory-secretory proteome of larval Schistosoma mansoni and Echinostoma caproni, two parasites of Biomphalaria glabrata. Mol Biochem Parasitol. 2007;155:45–56. doi: 10.1016/j.molbiopara.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 15.Lodes MJ, Connors VA, Yoshino TP. Isolation and functional characterization of snail hemocyte-modulating polypeptides from primary sporocysts of Schistosoma mansoni. Mol Biochem Parasitol. 1991;49:1–10. doi: 10.1016/0166-6851(91)90124-o. [DOI] [PubMed] [Google Scholar]
- 16.Chernin E. Observations on hearts explanted in vitro from the snail Australorbis glabratus. J Parasitol. 1963;39:353–64. [PubMed] [Google Scholar]
- 17.Yoshino TP, Laursen JR. Production of Schistosoma mansoni daughter sporocysts from mother sporocysts maintained in synxenic culture with Biomphalaria glabrata embryonic (Bge) cells. J Parasitol. 1995;81:714–22. [PubMed] [Google Scholar]
- 18.Castillo MG, Wu XJ, Dinguirard N, Nyame AK, Cummings RD, Yoshino TP. Surface membrane proteins of Biomphalaria glabrata embryonic cells bind fucosly determinants on the tegumental surface of Schistosoma mansoni primary sporocysts. J Parasitol. 2007;93:832–40. doi: 10.1645/GE-954R.1. [DOI] [PubMed] [Google Scholar]
- 19.Nickel W. Unconventional secretory routes: direct protein export across the plasma membrane of mammalian cells. Traffic. 2005;6:607–14. doi: 10.1111/j.1600-0854.2005.00302.x. [DOI] [PubMed] [Google Scholar]
- 20.Bendtsen JD, Jensen LJ, Blom N, von Heijne G, Brunak S. Feature-based prediction of non-classical and leaderless protein secretion. Prot Eng Design Sel. 2004;17:349–56. doi: 10.1093/protein/gzh037. [DOI] [PubMed] [Google Scholar]
- 21.Samuelson JC, Caulfield JP. Role of pleated septate junctions in the epithelium of miracidia of Schistosoma mansoni during transformation to sporocysts in vitro. Tiss Cell. 1985;17:667–82. doi: 10.1016/0040-8166(85)90003-5. [DOI] [PubMed] [Google Scholar]
- 22.Dunn TS, Yoshino TP. Schistosoma mansoni: origin and expression of a tegumental surface antigen on the miracidium and primary sporocyst. Exp Parasitol. 1988;67:67–81. doi: 10.1016/0014-4894(88)90064-1. [DOI] [PubMed] [Google Scholar]
- 23.Bayne CJ, Hahn UK, Bender RC. Mechanisms of molluscan host resistance and of parasite strategies for survival. Parasitology. 2001;123:S159–S67. doi: 10.1017/s0031182001008137. [DOI] [PubMed] [Google Scholar]
- 24.Davids BJ, Yoshino TP. Schistosoma mansoni: Excretory-secretory polypeptides exhibit selective binding to plasma components of the snail Biomphalaria glabrata. Exp Parasitol. 1995;81:292–301. doi: 10.1006/expr.1995.1120. [DOI] [PubMed] [Google Scholar]
- 25.Johnston LA, Yoshino TP. Larval Schistosoma mansoni excretory-secretory glycoproteins (ESPs) Bind to hemocytes of Biomphalaria glabrata (Gastropoda) via surface carbohydrate binding receptors. J Parasitol. 2001;87:786–93. doi: 10.1645/0022-3395(2001)087[0786:LSMESG]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 26.Wymelenberg AV, Sabat G, Mozuch M, Kersten PJ, Cullen D, Blanchette RA. Structure, organization, and transcriptional regulation of a family of copper radical oxidase genes in the lignin-degrading basidiomycete Phanerochaete chrysosporium. Appl Environ Microbiol. 2006;72:4871–7. doi: 10.1128/AEM.00375-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loker ES, Bayne CJ, Buckley PM, Kruse KT. Ultrastructure of encapsulation of Schistosoma mansoni mother sporocysts by hemocytes of juveniles of the 10-R2 strain of Biomphalaria glabrata. J Parasitol. 1982;68:84–94. [PubMed] [Google Scholar]
- 28.Mambula SS, Calderwood SK. Heat shock protein 70 is secreted from tumor cells by a nonclassical pathway involving lysosomal endosomes. J Immunol. 2006;177:7849–57. doi: 10.4049/jimmunol.177.11.7849. [DOI] [PubMed] [Google Scholar]
- 29.Danielsen EM, van Deurs B, Hansen GH. “Nonclassical” secretion of annexin A2 to the luminal side of the enterocyte brush border membrane. Biochemistry. 2003;42:14670–76. doi: 10.1021/bi0355239. [DOI] [PubMed] [Google Scholar]
- 30.Bottger A, Strasser D, Alexandrova O, Levin A, Fischer S, Lasi M, Rudd S, David CN. Genetic screen for signal peptides in Hydra reveal novel secreted proteins and evidence for non-classical protein secretion. Eur J Cell Biol. 2006;85:1107–17. doi: 10.1016/j.ejcb.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 31.Cass CL, Johnson JR, Califf LL, Xu T, Hernandez HJ, Stadecker MJ, Yates JR, III, Williams DL. Proteomic analysis of Schistosoma mansoni egg secretions. Mol Biochem Parasitol. 2007;155:84–93. doi: 10.1016/j.molbiopara.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knudsen GM, Medzihradszky KF, Lim KC, Hansell E, McKerrow JH. Proteomic analysis of Schistosoma mansoni ceercarial secretions. Mol Cell Proteomics. 2005;4:1862–75. doi: 10.1074/mcp.M500097-MCP200. [DOI] [PubMed] [Google Scholar]
- 33.Curwen RS, Ashton PD, Sundaralingam S, Wilson RA. Identification of novel proteases and immunomodulators in the secretions of schistosome cercariae that facilitate host entry. Mol Cell Proteomics. 2006;5:5. 835–44. doi: 10.1074/mcp.M500313-MCP200. [DOI] [PubMed] [Google Scholar]
- 34.Salter JP, Lim KC, Hansell E, Hsieh I, McKerrow JH. Schistosome invasion of human skin and degradation of dermal elastin are mediated by a single serine protease. J Biol Chem. 2000;275:38667–73. doi: 10.1074/jbc.M006997200. [DOI] [PubMed] [Google Scholar]
- 35.Armstrong PB. Proteases and protease inhibitors: balance of activities in host-pathogen interaction. Immunobiology. 2006;211:263–81. doi: 10.1016/j.imbio.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 36.Bahgat M, Doenhoff M, Kirschfink M, Ruppel A. Serine protease and phenoloxidase activities in hemocytes of Biomphalaria glabrata snails with varying susceptibility to infection with the parasite Schistosoma mansoni. Parasitol Res. 2002;88:489–94. doi: 10.1007/s00436-002-0595-6. [DOI] [PubMed] [Google Scholar]
- 37.Myers J, Ittiprasert W, Raghavan N, Miller A, Knight M. Differences in cysteine protease activity in Schistosoma mansoni-resistant and –susceptible Biomphalaria glabrata and characterization of the hepatopancreas cathepsin B full-length cDNA. J Parasitol. 2008;94:659–668. doi: 10.1645/GE-1410R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitta G, Galinier R, Tissevre P, Allienne JF, Girerd-Chambaz Y, Guillou F, Bouchut A, Coustau C. Gene discovery and expression analysis of immune-relevant genes from Biomphalaria glabrata hemocytes. Dev Comp Immunol. 2005;29:393–407. doi: 10.1016/j.dci.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 39.Lockyer AE, Spinks JN, Walker AJ, Kane RA, Noble LR, Rollinson D, Dias-Neto E, Jones CS. Biomphalaria glabrata transcriptome: Identification of cell-signalling, transcription control and immune-related genes from open reading frame expressed sequence tags (ORESTES) Dev Comp Immunol. 2007;31:763–82. doi: 10.1016/j.dci.2006.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hanelt B, Lun CM, Adema CM. Comparative ORESTES-sampling of transcriptomes of immune-challenged Biomphalaria glabrata snails. J Invertebr Pathol. 2008;99:192–203. doi: 10.1016/j.jip.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alger HM, Williams DL. The disulfide redox system of Schistosoma mansoni and the importance of a multifunctional enzyme, thioredoxin glutathione reductase. Mol Biochem Parasitol. 2002;121:129–39. doi: 10.1016/s0166-6851(02)00031-2. [DOI] [PubMed] [Google Scholar]
- 42.Cordell PA, Futers S, Grant PJ, Pease RJ. The human hydroxyacylglutathione hydrolase (HAGH) gene encodes both cytosolic and mitochondrial forms of glyoxalase II. J Biol Chem. 2004;279:28653–61. doi: 10.1074/jbc.M403470200. [DOI] [PubMed] [Google Scholar]
- 43.Hahn UK, Bender RC, Bayne CJ. Killing of Schistosoma mansoni sporocysts by hemocytes from resistant Biomphalaria glabrata: Role of reactive oxygen species. J Parasitol. 2001;87:292–9. doi: 10.1645/0022-3395(2001)087[0292:KOSMSB]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 44.Bender RC, Broderick EJ, Goodall CP, Bayne CJ. Respiratory burst of Biomphalaria glabrata hemocytes: Schistosoma mansoni-resistant snails produce more extracellular H2O2 than susceptible snails. J Parasitol. 2005;91:275–9. doi: 10.1645/GE-415R. [DOI] [PubMed] [Google Scholar]
- 45.Hintze KJ, Theil EC. Cellular regulation and molecular interactions of the ferritins. Cell Mol Life Sci. 2006;63:591–600. doi: 10.1007/s00018-005-5285-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chalmers IW, McArdle AJ, Coulson RM, Wagner MA, Schmid R, Hirai H, Hoffmann KF. Developmentally regulated expression, alternative splicing and distinct sub-groupings in members of the Schistosoma mansoni venom allergen-like (SmVAL) gene family. BMC Genomics. 2008;9:89. doi: 10.1186/1471-2164-9-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yeats C, Bentley S, Bateman A. New knowledge from old: In silico discovery of novel protein domains in Streptomyces coelicolor. BMC Microbiol. 2003;3:3. doi: 10.1186/1471-2180-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoffman DR. Structural biology of allergens from stinging and biting insects. Curr Opin Allergy Clin Immunol. 2008;8:338–42. doi: 10.1097/ACI.0b013e3283036a7d. [DOI] [PubMed] [Google Scholar]
- 49.Milne TJ, Abbenante G, Tyndall JD, Halliday J, Lewis RJ. Isolation and characterization of a cone snail protease with homology to CRISP proteins of the pathogenesis-related protein superfamily. J Biol Chem. 2003;278:31105–10. doi: 10.1074/jbc.M304843200. [DOI] [PubMed] [Google Scholar]
- 50.Hawdon JM, Narasimhan S, Hotez PJ. Ancylostoma secreted protein 2: cloning and characterization of a second member of a family of nematode secreted proteins from Ancylostoma caninum. Mol Biochem Parasitol. 1999;99:149–65. doi: 10.1016/s0166-6851(99)00011-0. [DOI] [PubMed] [Google Scholar]
- 51.Murray J, Gregory WF, Gomez-Escobar N, Atmadia AK, Maizels RM. Expression and immune recognition of Brugia malayi VAL-1, a homologue of vespid venom and Ancylostoma secreted proteins. Mol Biochem Parasitol. 2001;118:89–96. doi: 10.1016/s0166-6851(01)00374-7. [DOI] [PubMed] [Google Scholar]
- 52.Yatsuda AP, Krijgsveld J, Cornelissen AW, Heck AJ, de Vries E. Comprehensive analysis of the secreted proteins of the parasite Haemochus contortus reveals extensive variation and differential immune recognition. J Biol Chem. 2003;278:16941–51. doi: 10.1074/jbc.M212453200. [DOI] [PubMed] [Google Scholar]
- 53.Ali F, Brown A, Stanssens P, Timothy LM, Soule HR, Pritchard DI. Vaccination with neutrophil inhibitory factor reduces the fecundity of the hookworm Ancylostoma ceylanicum. Parasite Immunol. 2001;23:237–49. doi: 10.1046/j.1365-3024.2001.00383.x. [DOI] [PubMed] [Google Scholar]
- 54.Hahn UK, Bender RC, Bayne CJ. Involvement of nitric oxide in killing of Schistosoma mansoni sporocysts by hemocytes from resistant Biomphalaria glabrata. J Parasitol. 2001;87:778–85. doi: 10.1645/0022-3395(2001)087[0778:IONOIK]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 55.Holmfeldt P, Brannstrom K, Sellin ME, Segerman B, Carlsson SR, Gullberg M. The Schistosoma mansoni protein Sm16/SmSLP/SmSPO-1 is a membrane-binding protein that lacks the proposed microtubule-regulatory activity. Mol Biochem Parasitol. 2007;156:225–34. doi: 10.1016/j.molbiopara.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 56.Moss SE, Morgan RO. The annexins. Genome Biol. 2004;5:219. doi: 10.1186/gb-2004-5-4-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rao KV, Ramaswamy K. Cloning and expression of a gene encoding Sm16, an anti-inflammatory protein from Schistosoma mansoni. Mol Biochem Parasitol. 2000;108:101–8. doi: 10.1016/s0166-6851(00)00209-7. [DOI] [PubMed] [Google Scholar]
- 58.Lehr T, Geyer H, Maaβ K, Doenhoff MJ, Geyer R. Structural characterization of N-glycans from the freshwater snail Biomphalaria glabrata cross-reacting with Schistosoma mansoni glycoconjugates. Glycobiology. 2007;17:82–103. doi: 10.1093/glycob/cwl048. [DOI] [PubMed] [Google Scholar]




