Abstract
Olfactory receptor (OR) genes constitute the basis for the sense of smell. It has long been observed that a subset of mammalian OR genes are expressed in nonolfactory tissues, in addition to their expression in the olfactory epithelium. However, it is unknown whether OR genes have alternative functions in the nonolfactory tissues. Using a dedicated microarray, we surveyed OR gene expression in olfactory epithelium as well as a number of nonolfactory tissues, in human and chimpanzee. Our observations suggest that ectopically expressed OR orthologous genes are expressed in the same nonolfactory tissues in human and chimpanzee more often than expected by chance alone. Moreover, we found that the subset of orthologous OR genes with conserved ectopic expression evolve under stronger evolutionary constraint than OR genes expressed exclusively in the olfactory epithelium. Thus, although we cannot provide direct functional data, our observations are consistent with the notion that a subset of ectopically expressed OR genes have additional functions in nonolfactory tissues.
Keywords: olfaction, olfactory receptors, primate evolution
Nearly two decades ago, mammalian olfactory receptor (OR) genes were first identified, based partly on the observation that these genes were expressed solely in olfactory epithelium (Buck and Axel 1991). Subsequently, however, the expression of several predicted OR genes was also detected in nonolfactory tissues, notably in testis (Parmentier et al. 1992; Vanderhaeghen et al. 1997; Feldmesser et al. 2006). The observation of ectopic expression of putative OR genes raised the possibility that a subset of mammalian OR genes may have additional functions, which they carry out in nonolfactory tissues.
As the complete repertoire of OR genes became available for a number of mammalian species (Glusman et al. 2001; Zhang and Firestein 2002; Quignon et al. 2003; Gilad et al. 2005), evidence for rampant ectopic expression of OR genes began to emerge (Vanderhaeghen et al. 1997; Branscomb et al. 2000; Zhang et al. 2004, 2007; Feldmesser et al. 2006). However, the hypothesis that ectopically expressed OR genes carry additional functions in nonolfactory tissues remains largely unsupported. In sperm, functional studies of a small number of ectopically expressed OR genes revealed a possible role for OR genes in sperm chemotaxis (Spehr et al. 2003). Beyond these observations, the functional data on ectopically expressed OR genes are sparse, probably due to the difficulty of expressing OR proteins in order to functionally characterize them.
Although direct evidence for additional functions of ectopically expressed OR genes awaits the development of improved experimental approaches, we set out to examine this question by using evolutionary analysis. Our approach relies on the rationale that functionally important traits are expected to evolve under evolutionary constraint.
To evaluate the selective forces that shape the evolution of ectopically expressed OR genes, we collected expression data from olfactory epithelium as well as four nonolfactory tissues from chimpanzee (liver, testis, lung, and heart), using a dedicated OR gene microarray. We compared the chimpanzee expression data with a previously published analogous data set, which was collected from human tissues (Zhang et al. 2007). We analyzed the chimpanzee OR gene expression data using an approach that is similar to the one we previously applied for the human data (Zhang et al. 2007).
We detected the expression of 308 (89%) chimpanzee OR genes in olfactory epithelium (P < 0.05; at this statistical cutoff only 17 genes are expected to be detected as expressed by chance alone; see table 1 for results with alternative statistical cutoffs). Additionally, in agreement with previous observations in mouse (Zhang et al. 2004) and human (Feldmesser et al. 2006; Zhang et al. 2007), we detected expression of chimpanzee OR genes in all four nonolfactory tissues studied here (table 2 and supplementary table S1, Supplementary Material online). All chimpanzee OR genes that were found to be expressed in the nonolfactory tissues are also expressed in the olfactory epithelium—hence, we tentatively refer to the expression of these genes in nonolfactory tissues as “ectopic expression.”
Table 1.
Number of Expressed OR Genes in Chimpanzee Olfactory Epithelium
The statistical cutoff used to identify OR genes as expressed.
The number and percentage (in parentheses) of chimpanzee OR genes that were detected as expressed in olfactory epithelium. The array includes probe sets for 343 predicted intact chimpanzee OR genes.
Table 2.
Number of Expressed OR Genes in Nonolfactory Tissues (at Uncorrected P < 0.05)
| Liver | Heart | Testis | Lung | |
| In human | 58 | 108 | 83 | 93 |
| In chimpanzee | 74 | 24 | 11 | 6 |
| Overlap (P value)a | 19 (P < 0.03) | 10 (P < 0.11) | 6 (P < 0.03) | 4 (P < 0.09) |
The overlap in genes expressed in the same tissue in human and chimpanzee, as well as the P value testing the null that the overlap results by chance alone (by hypergeometric distribution). The combined P value for the overlap across all tissues is P < 0.01.
We note that only one chimpanzee OR gene was found to be expressed in more than one nonolfactory tissue (supplementary table S1, Supplementary Material online). This is likely, at least in part, due to the conservative approach we used to detect ectopic expression, which relies on differences in expression level between tissues. Indeed, an important caveat of this approach, as discussed in Zhang et al. (2007), is the expected high false negative rate, in particular when genes are expressed at similar levels across multiple tissues. Consistently, when we used reverse transcriptase PCR (RT-PCR) to validate the inference from the microarray data, the results (text S1, Supplementary Material online) indicated that although our false positive rate is low (we successfully confirmed gene expression in all 23 cases), the false negative rate is markedly higher (7/52 = 13.4% with 95% confidence intervals [CIs] of 5.8–26.7%). Importantly, our conclusions (below) are expected to be conservative with respect to the relatively high false negative rate.
After validating the results from the microarray, we proceeded to analyze patterns of expression across species. As orthologous genes in human and chimpanzee may be expected to perform similar functions, if OR genes have alternative functions, it may be expected that orthologous OR genes in human and chimpanzee will have a similar pattern of ectopic expression. To test this, we combined the chimpanzee data with the previously published human data (Zhang et al. 2007) and compared the patterns of ectopic expression of orthologous OR genes in the four nonolfactory tissues (table 2 and supplementary table S1, Supplementary Material online). We analyzed results from each nonolfactory tissue separately, counting the number of orthologous genes that are expressed in the each tissue in both species (i.e., we did not require a pattern of ectopic expression across tissues to be similar between orthologs). We found that orthologous genes are expressed in the same tissues in human and chimpanzee more often than expected by chance alone (combined P < 0.01; see table 2 for P values for testing the overlap across species in individual tissues). When only OR pseudogenes are considered, namely—when we do not expect selection to maintain the same expression pattern across orthologs, we see much less evidence for conservation of expression patterns: Although the combined P value is still significant (P = 0.036), it is mainly driven in this case by high overlap in the expression of OR pseudogene orthologs in the liver (P < 0.01), as no indication for overlap exist in the other tissues (P = 0.78, P = 0.72, and P = 0.18, for overlap in the expression of OR pseudogene orthologs in heart, lung, and testis, respectively).
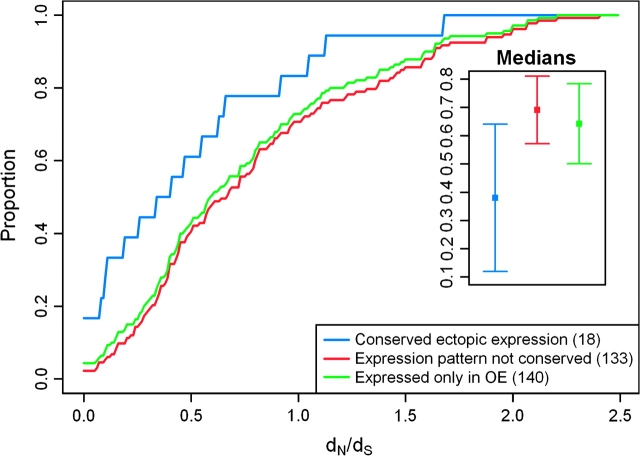
Further, if orthologous OR genes with conserved pattern of ectopic expression have additional functions, it may be expected that these genes evolve at a slower rate compared with other OR genes, due to the evolutionary constraint imposed by the requirement of additional functions (Duret and Mouchiroud 2000; Winter et al. 2004; Khaitovich et al. 2005). We tested this by using dN/dS ratios as a measure of protein evolution, that is, the ratio of the rates of nonsynonymous to synonymous substitutions. To do so, we estimated dN/dS ratios from human–chimpanzee–rhesus macaque three-way alignments of OR genes from Zhang et al. (2007). Using this approach, we found evidence that OR genes with conserved pattern of ectopic expression evolve slower (median dN/dS = 0.38) compared with OR genes that are expressed exclusively in the olfactory epithelium (median dN/dS = 0.69; P = 0.01 by permutation; fig. 1). This observation suggests that at least a subset of OR genes, which have conserved ectopic expression patterns, evolve under stronger evolutionary constraint.
FIG. 1.—
Cumulative distributions of dN/dS values (x-axis) of ectopically expressed OR genes with conserved expression pattern (blue), OR genes that are expressed in at least one nonolfactory tissues, but do not have a conserved expression pattern (red), and OR genes that are expressed exclusively in olfactory epitheliums (green). The smaller panel shows the dN/dS medians in the three groups. The error bars represent 95% CIs calculated using bootstrapping (1,000 repetitions).
It should be noted that previously we failed to identify evidence for additional constraint on OR genes that are ectopically expressed, using the same gene expression data from human (the pattern was consistent, but the result was not statistically significant; Zhang et al. 2007). The difference in the current analysis is the addition of the chimpanzee gene expression data, which allowed us to focus on ectopically expressed OR genes that also have a conserved expression pattern across species—a much better indication that these OR genes may perform additional functions in nonolfactory tissues.
In conclusion, although we cannot provide direct functional data, our evolutionary analysis supports the hypothesis that (at least) a subset of mammalian OR genes that are ectopically expressed carry out additional functions in nonolfactory tissues. Consequently, referring to expression of OR genes in nonolfactory tissues as ectopic expression may be misleading, as is probably the case for OR gene expression in the testis (Spehr et al. 2003). Future developments in our ability to directly express and assay OR genes will allow us to provide more direct evidence for this hypothesis.
Methods
Samples and Controls
We extracted total RNA from two samples of chimpanzee olfactory epithelium tissues, which were collected for us by the Yerkes Primate Center. We confirmed that RNA was extracted from olfactory epithelium tissues by amplification of the odorant binding protein 2B (OBP2B) gene, and by testing for the presence of olfactory sensory neurons in each sample by amplifying the olfactory marker protein (Buiakova et al. 1994). Once the source of the RNA was confirmed, we proceeded by labeling and hybridizing each RNA sample, in two technical replicates, to the custom human–chimpanzee OR gene microarray (see below). Similarly, we hybridized RNA from chimpanzee liver, lung, heart, and testis tissue samples to the microarray, in two technical replicates each.
The Human–Chimpanzee OR Gene Microarray
The custom Affymetrix microarray used has probes for nearly all human and chimpanzee putatively functional OR genes. The human OR gene probe sets (for 578 predicted human OR genes and pseudogenes) have been described previously (Zhang et al. 2007). The array also contains probe sets for 343 predicted chimpanzee intact OR genes. Because many OR genes are similar to each other at the coding region level, cross hybridization may be an issue. To avoid this problem, the expression of each OR gene is measured by probe sets designed in predicted 3′ untranslated regions of the gene (Zhang et al. 2007).
Hybridization and Analysis of the Data
Hybridizations and scanning of the arrays were performed at the University of Chicago core facility. The data were analyzed using a similar approach to the one we detailed for the analysis of the human OR gene expression data (Zhang et al. 2007). Briefly, we used the robust multiarray average (RMA) algorithm (Irizarry et al. 2003) to obtain one expression estimate for each probe sets and performed quantile normalization on the raw intensity values of either the olfactory epithelium or the nonolfactory epithelium tissues separately, followed by an experimentwide adjustment, based only on the 13 Affymetrix control probe sets for putatively “house keeping” genes. The purpose of the adjustment is to normalize together the data from the hybridizations of olfactory epithelium and nonolfactory tissues, without artificially reducing the intensity of OR gene expression in olfactory epithelium. To estimate the adjustment, we used an iterative reweighting procedure, as previously described (Zhang et al. 2007). Using this approach, we estimated that an adjustment of 0.164 (on a log scale) is required.
We note that the application of the adjustment was only needed for the analysis of the olfactory epithelium hybridizations. We did not use an adjustment for the analysis of ectopic expression, as we did not include the data from the olfactory epithelium in this analysis.
The normalized data were analyzed using the following mixed effects model:
where we have suppressed the probe-set labels. Here, Rijk is the normalized log transformed RMA value (of a particular probe set) for technical replicate k of a particular tissue sample j; the label i is used to indicate the tissue(s) used in the comparison (e.g., olfactory epithelia compared with nonolfactory tissues). The term β is a random effect for the tissue sample j, assumed to be uncorrelated with mean zero and variance σβ2. The term εijk is the residual error term (technical variance) and is assumed to be uncorrelated with mean zero and variance σε2. We used this model to estimate whether the difference in gene expression, α1−α2, between olfactory epithelium and nonolfactory tissues is significantly greater than zero (using a one-tailed t-test). We used the same procedure to compare gene expression only among the nonolfactory tissues.
Electronic Database Information
All expression data and original CEL files were submitted to the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) under the series accession number [GSE11156].
Supplementary Material
Supplementary table S1 and text S1 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
We thank the Yerkes Primate Center for collecting for us the chimpanzee tissue samples. We thank D. Lancet for helpful discussions and A. Oshlack, Z. Gauhar, O. Man, and J. Marioni for comments on the manuscript. Y.G. is supported by the Alfred P. Sloan foundation and S.F. by a grant from NIDCD.
References
- Branscomb A, Seger J, White RL. Evolution of odorant receptors expressed in mammalian testes. Genetics. 2000;156:785–797. doi: 10.1093/genetics/156.2.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- Buiakova OI, Krishna NS, Getchell TV, Margolis FL. Human and rodent OMP genes: conservation of structural and regulatory motifs and cellular localization. Genomics. 1994;20:452–462. doi: 10.1006/geno.1994.1200. [DOI] [PubMed] [Google Scholar]
- Duret L, Mouchiroud D. Determinants of substitution rates in mammalian genes: expression pattern affects selection intensity but not mutation rate. Mol Biol Evol. 2000;17:68–74. doi: 10.1093/oxfordjournals.molbev.a026239. [DOI] [PubMed] [Google Scholar]
- Feldmesser E, Olender T, Khen M, Yanai I, Ophir R, Lancet D. Widespread ectopic expression of olfactory receptor genes. BMC Genomics. 2006;7:121. doi: 10.1186/1471-2164-7-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad Y, Man O, Glusman G. A comparison of the human and chimpanzee olfactory receptor gene repertoires. Genome Res. 2005;15:224–230. doi: 10.1101/gr.2846405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glusman G, Yanai I, Rubin I, Lancet D. The complete human olfactory subgenome. Genome Res. 2001;11:685–702. doi: 10.1101/gr.171001. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Khaitovich P, Hellmann I, Enard W, Nowick K, Leinweber M, Franz H, Weiss G, Lachmann M, Paabo S. Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science. 2005;309:1850–1854. doi: 10.1126/science.1108296. [DOI] [PubMed] [Google Scholar]
- Parmentier M, Libert F, Schurmans S, Schiffmann S, Lefort A, Eggerickx D, Ledent C, Mollereau C, Gerard C, Perret J, et al. (11 co-authors) Expression of members of the putative olfactory receptor gene family in mammalian germ cells. Nature. 1992;355:453–455. doi: 10.1038/355453a0. [DOI] [PubMed] [Google Scholar]
- Quignon P, Kirkness E, Cadieu E, Touleimat N, Guyon R, Renier C, Hitte C, Andre C, Fraser C, Galibert F. Comparison of the canine and human olfactory receptor gene repertoires. Genome Biol. 2003;4:R80. doi: 10.1186/gb-2003-4-12-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spehr M, Gisselmann G, Poplawski A, Riffell JA, Wetzel CH, Zimmer RK, Hatt H. Identification of a testicular odorant receptor mediating human sperm chemotaxis. Science. 2003;299:2054–2058. doi: 10.1126/science.1080376. [DOI] [PubMed] [Google Scholar]
- Vanderhaeghen P, Schurmans S, Vassart G, Parmentier M. Molecular cloning and chromosomal mapping of olfactory receptor genes expressed in the male germ line: evidence for their wide distribution in the human genome. Biochem Biophys Res Commun. 1997;237:283–287. doi: 10.1006/bbrc.1997.7043. [DOI] [PubMed] [Google Scholar]
- Winter EE, Goodstadt L, Ponting CP. Elevated rates of protein secretion, evolution, and disease among tissue-specific genes. Genome Res. 2004;14:54–61. doi: 10.1101/gr.1924004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, De la Cruz O, Pinto JM, Nicolae D, Firestein S, Gilad Y. Characterizing the expression of the human olfactory receptor gene family using a novel DNA microarray. Genome Biol. 2007;8:R86. doi: 10.1186/gb-2007-8-5-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Firestein S. The olfactory receptor gene superfamily of the mouse. Nat Neurosci. 2002;5:124–133. doi: 10.1038/nn800. [DOI] [PubMed] [Google Scholar]
- Zhang X, Rogers M, Tian H, Zou DJ, Liu J, Ma M, Shepherd GM, Firestein SJ. High-throughput microarray detection of olfactory receptor gene expression in the mouse. Proc Natl Acad Sci USA. 2004;101:14168–14173. doi: 10.1073/pnas.0405350101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.