Abstract
Antibiotic resistance determination of Ureaplasma spp. (Ureaplasma parvum and Ureaplasma urealyticum) usually requires predetermination of bacterial titer, followed by antibiotic interrogation using a set bacterial input. This 96-well method allows simultaneous quantification of bacteria in the presence and absence of antibiotics. A method for determining precise MICs and a method for screening against multiple antibiotics using breakpoint thresholds are detailed. Of the 61 Ureaplasma-positive clinical isolates screened, one (1.6%) was resistant to erythromycin (MIC, >64 mg/liter) and clarithromycin (MIC, 4 mg/liter), one to ciprofloxacin (1.6%), and one to tetracycline/doxycycline (1.6%). Five isolates were also consistently found to have an elevated MIC of 8 mg/liter for erythromycin, but this may not represent true antibiotic resistance, as no mutations were found in the 23S rRNA operons or ribosome-associated L4 and L22 proteins for these strains. However, two amino acids (R66Q67) were deleted from the L4 protein of the erythromycin-/clarithromycin-resistant strain. The tetM genetic element was detected in the tetracycline-resistant clinical isolate as well as in the positive control Vancouver strain serotype 9. The tetM gene was also found in a fully tetracycline-susceptible Ureaplasma clinical isolate, and no mutations were found in the coding region that would explain its failure to mediate tetracycline resistance. An amino acid substitution (D82N) was found in the ParC subunit of the ciprofloxacin-resistant isolate, adjacent to the S83L mutation reported by other investigators in many ciprofloxacin-resistant Ureaplasma isolates. It is now possible to detect antibiotic resistance in Ureaplasma within 48 h of positive culture without prior knowledge of bacterial load, identifying them for further molecular analysis.
Ureaplasmas and mycoplasmas are eubacteria belonging to the class Mollicutes. These unique organisms are the smallest self-replicating cells that lack a cell wall. Lack of a rigid cell wall prevents reactivity with Gram staining and makes them insusceptible to antibiotics that target bacterial cell walls (i.e., β-lactams and glycopeptides), while imparting a frailty that mostly limits them to a parasitic existence in association with the eukaryotic cells of their host (39). Shepard first described ureaplasmas, or T-mycoplasmas, in the 1950s, following isolation from a male patient with nongonococcal urethritis (31). This remains the most widely recognized patient group; however, they are also commonly found as commensals in the genital tract in as many as 80% of women of child-bearing age (30). Although the factors leading to intrauterine infection are unclear, detection of Ureaplasma in the amniotic fluid is frequently associated with chorioamnionitis, spontaneous abortion, and premature birth (13, 14, 40). It has been suggested that the rate of vertical transmission is inversely proportional to gestational age at time of delivery (1). Furthermore, the presence of Ureaplasma in the lungs of very premature neonates has been associated with the development of chronic lung disease (or bronchopulmonary dysplasia) and long-term hospitalization (7, 19, 29, 30).
Ureaplasma spp. are susceptible to bacteriostatic agents such as protein synthesis-inhibiting tetracyclines and macrolides as well as bactericidal agents, including fluoroquinolones. For premature neonatal patients, however, the removal of a microbe must be balanced against the potential (and often unknown) toxicity of the antibiotics in these patients (38). For this reason, physicians routinely favor macrolide antibiotics such as erythromycin when deciding to treat Ureaplasma infections in premature neonates.
Only a limited number of reports have been made regarding the trends in resistance among Ureaplasma isolates from neonates (18, 34, 37), and all of these comment on the lack of a standardized methodology, which hinders comparison of results. Factors such as inoculum size, pH of media, and time of incubation are known to dramatically alter the MIC, and standardized means of detecting resistance are needed.
Here, we describe a modified breakpoint analysis in a 96-well broth microdilution format that enables a concurrent determination of bacterial load in a sample simultaneously with the determination of resistance without prior knowledge of bacterial load. For isolates showing resistance within the breakpoint concentration, full MICs can be determined using a similar full-plate methodology. The breakpoint method was used to screen 15 strains isolated during 2006 and 2007 from bronchoalveolar lavage samples from neonates of various gestational ages at the University Hospital of Wales (UHW), Cardiff, United Kingdom, in addition to 46 frozen isolates from a reference repository of samples submitted to the Health Protection Agency at Colindale (London, United Kingdom) from 2003 to present. Within these 61 isolates, we identified one erythromycin-/clarithromycin-resistant, one ciprofloxacin-resistant, and one tetracycline-resistant Ureaplasma strain. Further PCR and sequence analyses were performed to determine the mechanism of resistance.
MATERIALS AND METHODS
USM and culture conditions.
Ureaplasmas were grown in commercially available ureaplasma selective medium (USM) purchased from Mycoplasma Experience Ltd. (Surrey, United Kingdom). While the exact formula of the commercially obtained medium is proprietary, the simple broth base medium was supplemented with yeast extract, 1 g/liter urea, and 10% porcine serum, with phenol red as the pH indicator (starting pH, 6.65) and with 2.5 μg/ml amphotericin B and 0.25 mg/ml ampicillin (neither of which inhibit the growth of ureaplasmas). The adapted microbroth technique was carried out in flat-bottom 96-well plates covered with adhesive sealing tape (Elkay, Basingstoke, United Kingdom) in a humidified tissue culture incubator set at an ambient CO2 concentration at 37°C. The sealing tape does not allow gas exchange, and the humidified incubator ensures that the integrity of the outermost wells is the same as those that are more central to the plate.
Sample collection.
Bronchoalveolar lavage (BAL) samples were collected from 20 preterm and term neonates requiring mechanical ventilation on the neonatal ward at the UHW (Cardiff, United Kingdom). Samples were initially screened for Ureaplasma infection by addition of 25 μl BAL fluid to 2 ml USM and incubation at 37°C for 1 week. Samples that turned a clear red color (indicating Ureaplasma growth in the absence of contaminating bacteria) following incubation were confirmed by PCR using the Ureaplasma-specific primers U4 and U5, which selectively amplify the urease gene (see below). Positive samples were diluted 1:100 and 1:1,000 in USM, incubated overnight, divided into aliquots, and frozen at −80°C for later batch analysis. Fifteen clinical Ureaplasma isolates were successfully archived at −80°C in USM and used for later susceptibility testing. An additional 46 archived Ureaplasma isolates were revived from a potential 77 samples submitted for testing to the Health Protection Agency (HPA) at Colindale. The HPA clinical laboratory confirmed that no samples used were contaminated with mycoplasma using specific culture methods. Known serovar (SV) isolates used as reference strains were also obtained from HPA, although SV1 (DKF-1) and SV9 (Vancouver isolate) were originally obtained from the Institute of Medical Microbiology, University of Aarhus, Denmark.
Screening of HPA isolates using a modified breakpoint method.
Erythromycin was purchased from Sigma-Aldrich (Dorset, United Kingdom) as a 1-mg/ml stock solution, while azithromycin, clarithromycin, ciprofloxacin, doxycycline, and tetracycline (also purchased from Sigma-Aldrich) were prepared as 1-mg/ml stocks dissolved in dimethyl sulfoxide or ethanol as directed by the manufacturer. Working stocks of antibiotics consisted of freshly diluted antibiotics at concentrations of 64 mg/liter in USM. An adapted breakpoint analysis was used to screen for the presence of resistant mutants within the 61 isolates. Although no official breakpoint values are available for Ureaplasma, we determined the following values based on the normal ranges of MICs reported for Ureaplasma, as stated in Cumitech 34 (36). As seen in Fig. 1, the bottom row of wells contained 2 mg/liter tetracycline, the next row up contained 4 mg/liter ciprofloxacin, the second row contained 4 mg/liter erythromycin, and the top row did not contain an antibiotic for a growth control. Two milliliters of USM was inoculated with either growing or frozen Ureaplasma spp. the night before titration was to be performed. Twenty microliters of this culture of unknown color-changing units (CCU) was then added to each well from A1 to A4, and serial 10-fold dilutions were performed from columns A to H. In this manner, the 96-well plate was organized to investigate the susceptibility of three different strains to three antibiotics per plate by repeating the layout for wells A5 to H8 and A9 to H12 as described above, but using different test strains of Ureaplasma. Plates were sealed and incubated at 37°C in a humidified cell culture incubator with ambient CO2 for 48 h, at which time color change within the growth control rows had ceased. From the titration of the antibiotic-free row, it was possible to calculate back through the titration from 1 CCU, the final well in which color change occurred, to the recommended 104 CCUs. Any strain which showed growth in antibiotic-containing media at this level was subsequently fully investigated using the full-plate method (see below), with additional MIC determinations for the macrolides clarithromycin and azithromycin for suspected erythromycin-resistant isolates and doxycycline for suspected tetracycline-resistant isolates. SV9 (Vancouver strain) was used as a positive control for tetracycline resistance; however, no known characterized resistant strains were available to use as controls for erythromycin and ciprofloxacin resistance. USM (with and without antibiotics) was also incubated in the absence of added Ureaplasma spp. to serve as a negative color-changing control. Validation of this technique was performed by determining the antibiotic MICs for ATCC (American Type Culture Collection, Bethesda, MD) isolates of Staphylococcus aureus (ATCC 29213), Enterococcus faecalis (ATCC 29212), and Escherichia coli (ATCC 25922). MICs determined in Mueller-Hinton medium for erythromycin, ciprofloxacin, and tetracycline were within the accepted MIC ranges for these bacteria.
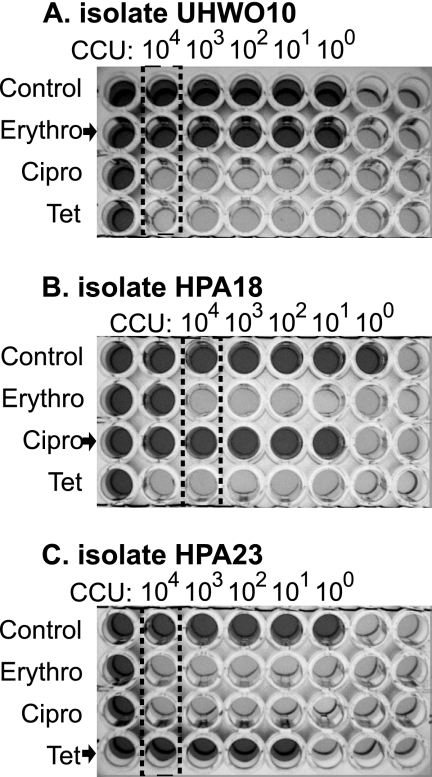
FIG. 1.
Photographs of antibiotic breakpoint investigation showing an erythromycin (Erythro)-resistant strain (UHWO10) (A), a ciprofloxacin (Cipro)-resistant strain (HPA18) (B), and a tetracycline (Tet)-resistant strain (HPA23) (C). Dark red wells indicating growth of ureaplasma appear dark gray, while orange-yellow wells representing no ureaplasma growth appear light gray in the grayscale photograph. Columns containing 104 CCU are identified by a dotted box, and comparison of growth in the absence of antibiotics (control) was used to determine resistance to 4 mg/liter erythromycin, 4 mg/liter ciprofloxacin, or 2 mg/liter tetracycline.
Modified broth microdilution technique for determination of antibiotic MICs.
In a 96-well plate, wells A1 to H1 received 360 μl of 64 mg/liter antibiotic in USM, and 180 μl sterile USM was added to the remaining wells (A2 to H12) of the plate. Using a multichannel pipette, rows of doubling dilutions were made by transferring 180 μl from row A1 to H11 (and an excess of 180 μl was discarded from row A11 to H11; see the instructional diagram in Fig. S1 in the supplemental material). Rows A12 to H12 remained free of antibiotic for unrestricted growth comparison. Thus, an antibiotic gradient was created from 64 mg/liter to 0.0625 mg/liter for the antibiotic in question. Twenty microliters of Ureaplasma from the overnight culture of unknown CCU was added to each well in the columns A1 to A12 (1:10 dilution). A 10-fold dilution curve of bacteria was then titrated at 90 degrees across the antibiotic gradient (i.e., A1 to -12 to H1 to -12). Plates were sealed and incubated at 37°C in a humidified cell culture incubator with ambient CO2 for 48 h, at which time color change within the growth control had ceased (see Fig. 2). The MIC was defined as the lowest concentration of antibiotic that prevented a color change after 48 h when read at 104 CCU (relative to growth in the antibiotic-free medium). USM (with and without antibiotics) was also incubated in the absence of added Ureaplasma isolates to serve as a negative color-changing control.
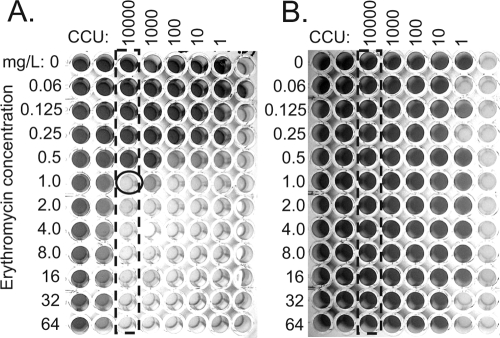
FIG. 2.
Full-plate determination of MIC for erythromycin for a susceptible SV1 isolate (A) and a resistant (UHWO10) isolate (B) in plates containing a gradient of antibiotic from 64 mg/liter to 0.06 mg/liter. Dark red wells indicating growth of ureaplasma appear dark gray, while orange-yellow wells representing no ureaplasma growth appear light gray in the grayscale photograph. Columns representing 104 CCU growth in the absence of antibiotic are shown by the dotted box, and for the susceptible strain, a circle shows the first concentration of erythromycin to inhibit growth (MIC = 1 mg/liter). Since the resistant isolate (UHWO10) grew even in the presence of 64 mg/liter erythromycin, the MIC is determined to be >64 mg/liter.
Speciation of Ureaplasma isolates.
Confirmation of Ureaplasma was determined by amplification of the Ureaplasma-specific urease gene (a 430-bp DNA product) using Blanchard and coworkers' (5) U4 and U5 primers (Table 1). Clinical isolates were further separated into species based on PCR primers which amplify a region of the multiple-banded antigen (MBA), as published by Teng et al. (35), also listed in Table 1. Primer set UM-1 for Ureaplasma parvum yielded a 403-bp product, with a 448-bp product for Ureaplasma urealyticum. For antibiotic-resistant U. parvum strains, SVs were determined by sequencing the UM-1 primer set amplicon and comparing SV-specific base compositions at nucleotides −54 to −56 and −82 to −84 (17, 35). Bacterial DNA from a 1.5-ml overnight culture was released by boiling lysis (95°C for 10 min) following centrifugation at 13,000 × g for 10 min, removal of all USM, and resuspension in 50 μl sterile water. A 20-μl PCR (containing 1× GoTaq Flexi buffer [Promega], 1.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates, 0.25 μM each primer, and 1.25 units of GoTaq DNA polymerase) was performed on 1 μl (as well as 1 μl of a 1:100 dilution to lessen possible polymerase inhibitor concentrations) for 35 cycles using the annealing temperatures listed in Table 1. To determine the DNA sequence, five 20-μl PCRs were pooled and purified from other reaction components using a Novagen SpinPrep PCR clean-up kit (Merck Chemicals Ltd., Nottingham, United Kingdom) per the manufacturer's instructions and then sequenced using the BigDye Terminator v3.1 cycle sequencing kit and analyzed with an ABI Prism 3130xl genetic analyzer (Applied Biosystems, Warrington, United Kingdom).
TABLE 1.
Primers and conditionsa
| Primers used for PCR amplification | Primer sequence | Annealing temp (°C) | Size (bp) | Reference |
|---|---|---|---|---|
| Ureaplasma-specific urease gene primers | ||||
| U4 | 5′ ACGACGTCCATAAGCAACT 3′ | 54 | 430 | 5 |
| U5 | 5′ CAATCTGCTCGTGAAGTATTAC 3′ | |||
| UM-1 | ||||
| UMS-125 | 5′ GTATTTGCAATCTTTATATGTTTTCG 3′ | 52 | 403 (U. p) | 35 |
| UMA226 | 5′ CAGCTGATGTAAGTGCAGCATTAAATTC 3 | 448 (U. u) | ||
| Erythromycin resistance primers | ||||
| 23S OP1 domain V | ||||
| MH23S-11 | 5′ TAACTATAACGGTCCTAAGG 3′ | 56 | 1,339 | 22 |
| UP23S-OP1 | 5′ ACCACCATTCAATGTTTGAC 3′ | |||
| 23S OP2 domain V | ||||
| MH23S-11 | 5′ TAACTATAACGGTCCTAAGG 3′ | 56 | 1,427 | 22 |
| UP23S-OP2R2 | 5′ CGTATACTTTGCCATAGTGTTGCC 3′ | |||
| L4 | ||||
| UPL4-U | 5′ TCTATTGATGGTAACTTCGC 3′ | 60 | 392 | 22 |
| UPL4-R | 5′ GTTGAAGGTGTTTCTAAATCGC 3′ | |||
| L22 | ||||
| UPL22-U | 5′ TTCGCACCGTAAAGCTTCTC 3′ | 60 | 458 | 22 |
| UPL22-R | 5′ GTTCTGGATCAACGTTTTCG 3′ | |||
| Tetracycline resistance primers | ||||
| TetM primers for screening | ||||
| TetMF | 5′ TTATCAACGGTTTATCAGG 3′ | 48 | 397 | 4 |
| TetMR | 5′ CGTATATATGCAAGACG 3′ | |||
| TetM primers for sequencing | ||||
| TetMF-78 | 5′ GTATACCTATGGTTATGC 3′ | 48 | 901 | |
| TetMR | 5′ CGT ATA TAT GCA AGA CG 3′* | |||
| TetMF | 5′ TTATCAACGGTTTATCAGG 3′ | 54 | 1,715 | |
| TetMR 2123 | 5′ GCATTTCGGACAATAGAGGGGG 3′* | |||
| Ciprofloxacin resistance primers | ||||
| GyrA | ||||
| gyrA-1 | 5′ TTGCTGCTTTCGAAAACGG 3′ | 50 | 336 | 3 |
| gyrA-2 | 5′ CTGATGGTAAAACACTTGG 3′ | |||
| GyrB | ||||
| gyrB-3 | 5′ CCTGGTAAATTAGCTGACTG 3′ | 55 | 310 | 3 |
| gyrB-4 | 5′ TTCGAATATGACTGCCATC 3′ | |||
| ParC | ||||
| parC-5 | 5′ ACGCAATGAGTGAATTAGG 3′ | 55 | 309 | 3 |
| parC-6 | 5′ CACTATCATCAAAGTTTGGAC 3′ | |||
| ParE | ||||
| parE-7 | 5′ ATGGGCGGAAAATTAACGC 3′ | 55 | 313 | 3 |
| parE-8 | 5′ CTTGGATGTGACTACCATCG 3′ |
PCR primers used for the amplification and sequencing of genes associated with resistance to respective antibiotics. Annealing temperatures and predicated product sizes are indicated. U. p, U. parvum; U. u, U. urealyticum; *, primers which were designed for this study.
Amplification and sequence determination of putative resistance targets.
For erythromycin- and ciprofloxacin-resistant isolates, mutations in bacterial genes previously associated with resistance (3, 22) were investigated.
DNA encoding bacterial 23S rRNA gene, L22, and L4 proteins was amplified and sequenced for macrolide-resistant strains; gyrase subunits and topoisomerase subunits were sequenced for ciprofloxacin-resistant strains, and tetM genes from both tetracycline-susceptible and -resistant isolates were sequenced. PCR primers and annealing temperatures are outlined in Table 1. Amplicons were purified and sequenced as detailed above.
RESULTS
Species distribution among Ureaplasma isolates tested.
Seventy-five percent (15/20) of the archived UHW samples were successfully revived from −80°C storage, while only 59.7% (46/77) of archived HPA samples were successfully revived. This may reflect the relatively short storage time for the former samples (less than 18 months) compared to that of the latter samples (up to 4 years), although differences in isolate preparation prior to freezing them may have equally contributed. For all 61 revived samples, 49 were found to belong to U. parvum (80%) and 12 to U. urealyticum (20%). A minor difference in species distribution was identified between HPA and UHW collections, but this was not deemed significant by Fisher's exact test analysis (78% U. parvum versus 87% U. parvum, respectively).
Examples of breakpoint and detailed MIC determination.
Ninety-six-well plates were set out as detailed in Materials and Methods for breakpoint screening of antibiotic resistance. An example showing strains resistant to erythromycin, ciprofloxacin, and tetracycline is shown in Fig. 1. Nonturbid, dark red wells (which appear dark gray in the figure) are indicative of positive Ureaplasma growth in the selective medium. If the last well showing growth in the 10-fold dilution (in the top row, lacking antibiotics) is considered to have 1 CCU, then four columns to the left represents 103 CCU, and five columns to the left represents 104 CCU (isolated by the dotted box). Following previously published recommendations that Ureaplasma resistance needs to be assessed using an input of 104 CCU (36), resistance-to-threshold antibiotic concentrations (4 mg/liter erythromycin, 4 mg/liter ciprofloxacin, or 2 mg/liter tetracycline) were identified by comparison of wells within the dotted box. Figure 1 shows breakpoint identification of an erythromycin-resistant clinical SV1 isolate (UHWO10; i.e., 10th sample from UHW patient O) (Fig. 1, top), a ciprofloxacin-resistant clinical SV1 isolate (HPA18; i.e., 18th HPA sample) (middle), and a tetracycline-resistant clinical SV6 isolate (HPA23) (bottom). While we have used the 104 CCU column to assess antibiotic resistance, using 103 CCU would be equally valid, and no difference in our MICs was found when reading either the 103 or 104 CCU column.
For those isolates identified by breakpoint analysis, MICs for a single antibiotic were determined using a range of concentrations with a full-plate single antibiotic assay (Fig. 2). Using the same method used for determining which column contained 104 CCU (isolated by dotted box), examples of an erythromycin-susceptible clinical isolate (HPA18; MIC, 1 mg/liter) (Fig. 2A) and the highly resistant clinical isolate (UHWO10; MIC, >64 mg/liter) (Fig. 2B) are shown.
Summary of MICs among resistant isolates.
All isolates were examined for resistance to erythromycin, ciprofloxacin, and tetracycline with the resistant isolates summarized in Table 2. Six isolates were identified to have an MIC greater than the normal MIC range previously published in Cumitech 34 (0.02 to 4 mg/liter) (36), giving the isolates a prevalence of 9.8%, although only one of these isolates (UHWO10) is likely to represent true antibiotic resistance with an MIC of >64 mg/liter. Among the erythromycin-resistant isolates, azithromycin and clarithromycin MICs were comparable to those of susceptible controls, with the exception of the highly resistant UHWO10, which was additionally resistant to clarithromycin (MIC = 4 mg/liter) but susceptible to azithromycin (normal MIC ranges, 0.5 to 4 mg/liter for azithromycin and <0.004 to 2 mg/liter for clarithromycin) (36). A single tetracycline-resistant clinical isolate, HPA23, was identified with tetracycline and doxycycline MICs of 64 and 16 mg/liter, respectively (1.6% prevalence). A single ciprofloxacin-resistant clinical isolate, HPA18, was also identified with an MIC of 8 mg/liter (1.6% prevalence).
TABLE 2.
MICs for resistant and control U. parvum and U. urealyticum strains
| Isolate | Species (SV) | MICs with indicated antibioticsa
|
|||||
|---|---|---|---|---|---|---|---|
| Ery | Azi | Cla | Tet | Dox | Cip | ||
| HPA3 | U. urealyticum | 4 | 0.5 | <0.125 | 2 | 0.5 | 4 |
| HPA6 | U. urealyticum | 8 | <0.25 | <0.125 | 2 | 0.5 | 4 |
| HPA12 | U. urealyticum | 8 | <0.25 | <0.125 | 2 | 0.5 | 4 |
| HPA17 | U. urealyticum | 8 | 0.5 | <0.125 | 1 | 0.25 | 4 |
| HPA18 | U. parvum (SV1) | 4 | <0.25 | <0.125 | 2 | 0.25 | 8 |
| HPA20 | U. urealyticum | 8 | <0.25 | <0.125 | 2 | 0.5 | 4 |
| HPA23 | U. parvum (SV6) | 4 | <0.25 | <0.125 | 64 | 16 | <2 |
| HPA32 | U. parvum (SV14) | 8 | <0.25 | <0.125 | 1 | 0.25 | <2 |
| UHWJM | U. parvum | 4 | <0.25 | <0.125 | 2 | 0.5 | 4 |
| UHWO10 | U. parvum (SV1) | >64 | 2 | 4 | 1 | 0.25 | 4 |
| UHWP2 | U. parvum | 4 | <0.25 | <0.125 | 2 | 0.5 | 4 |
| UHWQ3 | U. parvum (SV1) | 4 | <0.25 | <0.125 | 1 | 0.25 | 4 |
MIC results from a selection of resistant isolates (in boldface) and susceptible isolates, as determined by growth comparison at 104 CCU. Ery, erythromycin; Azi, azithromycin; Cla, clarithromycin; Tet, tetracycline; Dox, doxycycline; Cip, ciprofloxacin.
Molecular characterization of macrolide resistance.
Mutations in the 23S rRNA genes (two separate operons) and associated L4 or L22 protein from the prokaryotic ribosomal complex have previously been reported to be associated with macrolide resistance (22). These genes were sequenced from the highly resistant clinical isolate UHWO10 as well as the five isolates with an erythromycin MIC of 8 mg/liter and a selection of the susceptible isolates. No mutations were found in either the 23S rRNA operon or the L22-encoding gene of any of the isolates; however, a 6-bp deletion was identified in the L4 protein gene from the highly resistant UHWO10. This in-frame deletion resulted in the loss of arginine and glutamine residues at residue numbers 66 and 67, respectively (ΔR66Q67) (Table 3). This region was sequenced in clinical isolates representing susceptible SV1 strains (representing an SV control) and other U. parvum strains. All were identical to the published SV3 genome sequence (accession number AF222894) (12). Furthermore, this region was also conserved in the SV8 reference strain and a U. urealyticum clinical strain (Table 3). Comparison of all sequences found three species-specific conserved nucleotide polymorphisms (all silent) in the L4 protein gene (U. parvum T309, G357, and C373 compared to U. urealyticum C309, A357, and T373). While 80% of all isolates were found to belong to U. parvum, four out of five of the isolates with MICs of 8 mg/liter belonged to U. urealyticum. Therefore, 33% of the all U. urealyticum isolates identified were found to be less susceptible to erythromycin (but susceptible to clarithromycin and azithromycin) (Table 2).
TABLE 3.
Partial protein sequence alignments of antibiotic-resistant and -susceptible isolates
| Isolate tested with indicated antibiotica | Sequenceb |
|---|---|
| Erythromycin | |
| L4 isolates | |
| SV3s | 63KPWRQKHT70 |
| UHWO10r | 63KPW--KHT70 |
| Tetracycline | |
| TetM isolates | |
| SV9 Seattler | 209HNCSLFPVYHGSAKNNIGID228 |
| HPA23r | 209HNCSLFPVYHGSAKNNIGID228 |
| HPA6s | 209QNCSLFPLYHGSAKSNIGID228 |
| SV9 Vancouverr | 209QNCSLFPLYHGSAKSNIGID228 |
| Fluoroquinolone | |
| ParC isolates | |
| Consensuss | 78HPHGDSSIYE87 |
| HPA18r | 78HPHGNSSIYE87 |
| Bebear UUg2-5r | 78HPHGDLSIYE87 |
| Bebear UUcr | 78HPHGDSSIYK87 |
Superscript letters (r and s) indicate resistance and susceptibility, respectively, to the indicated antibiotics.
Boldfaced amino acids identify areas of sequence divergence within a variable region of the protein. The sequence of TetM of the Vancouver strain was determined at the UHW. Underlined amino acids identify areas of sequence divergence relative to the sequences of susceptible strains. Previous S83L amino acid substitutions arising from mutation and E87K substitutions associated with resistance as described by Bebear et al. (2) are also shown. -, deletion of amino acids R and Q.
Molecular characterization of tetracycline resistance.
Tetracycline-resistant bacterial strains are often found to have the transferable tetM genetic element encoded in their genome. The SV9 (Vancouver) reference strain, known to contain the tetM gene (25), demonstrated a tetracycline-resistant (Tetr) phenotype, with tetracycline and doxycycline MICs of >64 mg/liter and 64 mg/liter, respectively. Only one U. parvum clinical isolate (HPA23) was found to be Tetr and doxycycline resistant (Table 2), coincident with the identification of the tetM gene by PCR (data not shown). PCR screening of all 61 isolates identified a tetM-positive U. urealyticum clinical isolate (HPA6) that was susceptible to both tetracycline and doxycycline (Table 2). The entire tetM gene was sequenced and compared to resistant strains SV9 (Vancouver) and HPA23 and susceptible strain HPA6 (Table 3). No mutations in the coding region of the tetM gene were found for HPA6 that could account for the inability of a resultant protein to mediate tetracycline resistance, nor was any alteration to the endogenous tetM gene promoter found that may have stopped TetM protein expression. Therefore, we have no explanation for the susceptibility of this tetM-positive isolate. Interestingly, three amino acid polymorphisms (H209Q, V216L, N223S) were found in HPA23 that did not affect function and were identical to those originally reported for a Tetr strain isolated in Seattle, Washington, in 1984 (Table 3) (28).
Molecular characterization of ciprofloxacin resistance.
A single ciprofloxacin-resistant (Cipr) clinical isolate, HPA18, was identified with an MIC of 8 mg/liter. Mutations in the subunits of the bacterial gyrase (gyrA and gyrB) and topoisomerase IV (parC and parE) genes, known as the quinolone resistance-determining regions, have previously been associated with resistance to other fluoroquinolone antibiotics (2, 3, 10, 11, 41, 42). The sequences of these genes were determined from the Cipr HPA18 isolate and several susceptible clinical isolates and compared to both known SV reference strains and the published SV3 sequence (12). A single base pair change at position 244 (G244A) in the parC gene was identified (Table 3), resulting in a codon change and subsequent substitution of aspartic acid for asparagine (D82N) relative to those of other U. parvum sequences from susceptible controls from both species.
DISCUSSION
Summary of mutations found.
This study describes a modified breakpoint analysis used to screen for antibiotic resistance among a large number of clinical Ureaplasma samples of unknown initial CCU titration. U. parvum strains resistant to ciprofloxacin and tetracycline as well as one highly erythromycin-resistant strain were identified. The species of all clinical isolates were determined by PCR analysis of the gene encoding the MBA protein, and in many cases, the SV was determined through analysis of the amplicon sequence in this region. Potential underlying molecular mechanisms of antibiotic resistance were investigated, based on target genes identified by previous investigators. A unique deletion of ΔR66Q67 was identified in the L4 protein of the bacterial 50S ribosomal complex in an isolate with high erythromycin resistance, a unique point mutation of D82N in the ParC subunit of bacterial topoisomerase in the ciprofloxacin-resistant isolate, and the presence of the tetM element in the tetracycline-resistant isolate.
Advantages of the breakpoint method.
The primary advantage to using the modified breakpoint methodology described here is the simultaneous determination of input inoculum for determination of susceptibility at the 104 CCU inoculum. Using this methodology, the investigator also has the option of determining the amount of CCU reduction at a defined antibiotic concentration. By defining the final well in which color change occurred in the growth control as 1 CCU, comparison of growth in the column representing an initial inoculum of 104 CCU (as indicated in Fig. 1) identifies resistant organisms to be further interrogated by the full-plate, single-antibiotic method. As seen in Fig. 1, the presence of a resistant isolate is readily noticeable: 10-fold reduction (or less) in growth was observed at a 2 or 4 mg/liter concentration of antibiotic. Further investigation with the full-plate method found that less than 10-fold reduction in growth was seen for UHWO10 when grown in up to 64 mg/liter erythromycin (Fig. 2).
Ciprofloxacin resistance.
Ciprofloxacin was used as a representative of the bactericidal fluoroquinolone antibiotic family that targets two bacterial topoisomerases, namely topoisomerase IV and DNA gyrase. Numerous mechanisms have been described by which bacteria can mediate resistance to fluoroquinolones, including mutation of the topoisomerase genes, decreased membrane permeability, active drug efflux, modification by a fluoroquinolone-inactivating enzyme, or the presence of a Qnr protein (27). We have identified a point mutation leading to an amino acid substitution which was two amino acids downstream from the proposed active site of the ParC protein (20). Six separate groups have investigated a total of 32 resistant isolates and reported mutations in Ureaplasma genes associated with fluoroquinolone resistance (2, 3, 10, 11, 41, 42). Nineteen of 32 fluoroquinolone-resistant strains investigated by these groups have the same mutation in the parC gene, S83L (or S80L, if using homology to the E. coli position to identify the location) (Table 3); furthermore, similar mutations in ParC have been found in fluoroquinolone-resistant S. aureus and Streptococcus pneumoniae, which are reported to have MICs similar to those of resistant Ureaplasma strains (15). The mutation in the Cipr isolate D82N is adjacent to this region, and this isolate has a ciprofloxacin MIC similar to that of isolate UUc identified by Bebear et al. (2), who also found an E87K mutation in the near vicinity, indicating this is a dominant target for fluoroquinolone resistance. (Table 3).
Erythromycin resistance.
This investigation also identified one highly resistant erythromycin resistant isolate (MIC, >64 mg/liter); however, five additional isolates each consistently showed an MIC of 8 mg/liter, which is higher than the range of 0.02 to 4 mg/liter listed in the Cumitech 34 reference manual (36). Interestingly, 4/5 of the less susceptible isolates were U. urealyticum, which represented 33% of the total U. urealyticum isolates screened, suggesting a possible inherently increased tolerance of U. urealyticum isolates to erythromycin relative to that of U. parvum isolates. There is much less information available about the mechanism of erythromycin resistance in Ureaplasma spp. In fact, this is the first full molecular characterization of a clinical Ureaplasma isolate with erythromycin resistance. The physiological mechanism of erythromycin resistance was first indicated by Palu et al. (21) almost 20 years ago, who demonstrated reduced radiolabeled erythromycin binding to ribosomes of resistant Ureaplasma compared to that of a susceptible strain. However, technical advances since that report have greatly enhanced the ability to investigate molecular mechanisms. Investigation of other macrolide-resistant bacteria has identified mutations that inhibit erythromycin binding to the bacterial 50S ribosomal complex, where it exhibits bacteriostatic activity through inhibition of protein synthesis. Pereyre et al. serially passaged a U. parvum reference strain 45 to 50 times in increasing amounts of erythromycin and sequenced associated genes: two 23S rRNA operons and L4 and L22 ribosome-associated proteins (22). They consistently found mutation of the erythromycin-binding site around nucleotide position 2067 (2058 numbering based on homology to E. coli) near the peptidyl transferase loop in domain V of one of the 23S rRNA operons and occasionally found additional mutation of the associated L4 or L22 protein. Dongya et al. identified a number of point mutations within the 23S rRNA of 18 clinical isolates with various degrees of resistance to the macrolides josamycin, clarithromycin, roxithromycin, and azithromycin; however, they did not investigate resistance to erythromycin (9). Interestingly, none of the mutations described were in the region of nucleotide 2067. Our investigation did not find mutations in either the 23S rRNA operon or the L22 protein but did detect a deletion of two adjacent amino acids in the L4 protein for the highly resistant strain (MIC, >64 mg/liter). No mutations were found in the isolates with an erythromycin MIC of 8 mg/liter, yet compensating mutations outside of the investigated regions, DNA methylation, expression of bacterial proteins that modify macrolides, or increased drug efflux via ion channels may have been responsible for the relative increase in tolerance to erythromycin for these strains. Kenny and Cartwright previously showed that susceptibility to erythromycin was reduced in acidic growth medium; however, medium pH had to fall below 6.5 before an effect was observed (16). As our USM had a pH of 6.65, the MICs were determined relative to each other in the same batch of USM, and the findings were consistent upon repeated testing. As a result, we do not feel that the reduced susceptibility of these five isolates was due to a pH artifact for the USM used.
Mutations within the highly conserved region of L4 have previously been noted in both laboratory-derived and clinically isolated macrolide-resistant pneumococci and E. coli strains (6, 32, 33). Both a substitution mutation (G69C) and an insertion mutation (6-bp insertion between the codons encoding Q67 and K68) were found in two resistant pneumococcus strains (32, 33). Clinical isolates found by the same group made up a distinct clonal cluster which contained a substitution from 69GTG71 to 69TPS71 as well as one isolate which contained a 6-amino-acid insertion within the same region (33). A similar mutation (K63E) within L4 of E. coli has also been found to be associated with resistance to erythromycin (6) and was located adjacent to the deletion described here. The resultant three-dimensional alteration to the entire ribosomal complex as a result of the structural L4 mutation must prevent the interaction of the C5 sugar of the macrolide with the residues around A2067 (A2058 E. coli numbering) and abrogate the ability of erythromycin to inhibit protein synthesis in the resistant strain (23).
Tetracycline resistance.
Currently, the only known mechanism of tetracycline resistance for mollicutes is the presence of the tetM-transferable genetic element. It was first described in Ureaplasma by Roberts and Kenny in 1986 (24) and has been the focus of investigations by several groups since then. Unlike the studies by Robertson et al. (26), who found that all 26 of their Tetr isolates were also resistant to erythromycin at or above 2 mg/liter, our control SV9 Tetr and clinical Tetr isolates were susceptible to erythromycin (MIC, 4 mg/liter). Blanchard et al. (4) have previously shown that screening Ureaplasma isolates by PCR readily identified Tetr strains. However, we found a tetracycline-susceptible strain (HPA6) that screened tetM positive with these strains. Recently, Degrange et al. identified two tetM-positive Mycoplasma hominis isolates that were tetracycline susceptible (8). One of these M. hominis isolates had a 1,260-bp insertion in the leader peptide sequence (likely preventing successful transcription), while no mutations were found within the tetM gene or promoter region of the second isolate. Like the latter case for our Tets tetM+ HPA6 strain, we could find no explanation for the susceptibility when comparing the sequence to that of the control Tetr SV9 (Vancouver) strain. We also found that our Tetr strains were resistant to both tetracycline and doxycycline, which is in contrast with the findings of Blanchard et al. (4). They identified 21 clinical Tetr Ureaplasma isolates and found that 8 were resistant to doxycycline, 2 were intermediate, and 11 were susceptible. It has been proposed that doxycycline is less of an inducer of tetM transcription in some organisms than tetracycline. In this study, we demonstrated that our clinical tetracycline-resistant isolate HPA23, which harbored a functional TetM protein, was additionally resistant to doxycycline.
Conclusion.
In conclusion, we have developed a reliable method to determine antimicrobial susceptibility of Ureaplasma isolates without prior knowledge of inoculum size. Although this is the second molecular investigation of macrolide resistance in clinical Ureaplasma isolates, more erythromycin-resistant strains are required to determine the prevalence of mutations in the relevant genes associated with resistance. Screening for the tetM gene could identify tetracycline-resistant strains; however, we found one false-positive strain, which raises a note of caution for the use of PCR screening for antibiotic resistance. It appears that mutation within an 8- amino-acid region of topoisomerase IV parC is commonly found in fluoroquinolone-resistant Ureaplasma, with S83L being the dominant mutation, but molecular screening methods would have to employ more-complex methods, such as capillary electrophoresis-based single-strand conformation polymorphism, to screen for all potential mutations.
Supplementary Material
Acknowledgments
We thank Phil Davies and Sister Dianne Nuttall for their help in acquiring neonatal BAL samples from neonates at the UHW. We also thank the parents, babies, and staff of the neonatal unit at the UHW for their participation in this study.
NCM is funded by a project grant from the Wellcome Trust.
Footnotes
Published ahead of print on 9 March 2009.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Abele-Horn, M., J. Peters, O. Genzel-Boroviczeny, C. Wolff, A. Zimmermann, and W. Gottschling. 1997. Vaginal Ureaplasma urealyticum colonization: influence on pregnancy outcome and neonatal morbidity. Infection 25:286-291. [DOI] [PubMed] [Google Scholar]
- 2.Bebear, C. M., H. Renaudin, A. Charron, M. Clerc, S. Pereyre, and C. Bebear. 2003. DNA gyrase and topoisomerase IV mutations in clinical isolates of Ureaplasma spp. and Mycoplasma hominis resistant to fluoroquinolones. Antimicrob. Agents Chemother. 47:3323-3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bebear, C. M., H. Renaudin, A. Charron, D. Gruson, M. Lefrancois, and C. Bebear. 2000. In vitro activity of trovafloxacin compared to those of five antimicrobials against mycoplasmas including Mycoplasma hominis and Ureaplasma urealyticum fluoroquinolone-resistant isolates that have been genetically characterized. Antimicrob. Agents Chemother. 44:2557-2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanchard, A., D. M. Crabb, K. Dybvig, L. B. Duffy, and G. H. Cassell. 1992. Rapid detection of tetM in Mycoplasma hominis and Ureaplasma urealyticum by PCR: tetM confers resistance to tetracycline but not necessarily to doxycycline. FEMS Microbiol. Lett. 74:277-281. [DOI] [PubMed] [Google Scholar]
- 5.Blanchard, A., J. Hentschel, L. Duffy, K. Baldus, and G. H. Cassell. 1993. Detection of Ureaplasma urealyticum by polymerase chain reaction in the urogenital tract of adults, in amniotic fluid, and in the respiratory tract of newborns. Clin. Infect. Dis. 17(Suppl. 1):S148-S153. [DOI] [PubMed] [Google Scholar]
- 6.Chittum, H. S., and W. S. Champney. 1994. Ribosomal protein gene sequence changes in erythromycin-resistant mutants of Escherichia coli. J. Bacteriol. 176:6192-6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davies, P. L., N. C. Maxwell, and S. Kotecha. 2006. The role of inflammation and infection in the development of chronic lung disease of prematurity. Adv. Exp. Med. Biol. 582:101-110. [DOI] [PubMed] [Google Scholar]
- 8.Degrange, S., H. Renaudin, A. Charron, C. Bebear, and C. M. Bebear. 2008. Tetracycline resistance in Ureaplasma spp. and Mycoplasma hominis: prevalence in Bordeaux, France, from 1999 to 2002 and description of two tet(M)-positive isolates of M. hominis susceptible to tetracyclines. Antimicrob. Agents Chemother. 52:742-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dongya, M., X. Wencheng, M. Xiaobo, and W. Lu. 2008. Transition mutations in 23S rRNA account for acquired resistance to macrolides in Ureaplasma urealyticum. Microb. Drug Resist. 14:183-186. [DOI] [PubMed] [Google Scholar]
- 10.Duffy, L., J. Glass, G. Hall, R. Avery, R. Rackley, S. Peterson, and K. Waites. 2006. Fluoroquinolone resistance in Ureaplasma parvum in the United States. J. Clin. Microbiol. 44:1590-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geiβdörfer, W., G. Sandner, S. John, A. Gessner, C. Schoerner, and K. Schröppel. 2008. Ureaplasma urealyticum meningitis in an adult patient. J. Clin. Microbiol. 46:1141-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glass, J. I., E. J. Lefkowitz, J. S. Glass, C. R. Heiner, E. Y. Chen, and G. H. Cassell. 2000. The complete sequence of the mucosal pathogen Ureaplasma urealyticum. Nature 407:757-762. [DOI] [PubMed] [Google Scholar]
- 13.Gray, D. J., H. B. Robinson, J. Malone, and R. B. Thomson, Jr. 1992. Adverse outcome in pregnancy following amniotic fluid isolation of Ureaplasma urealyticum. Prenat. Diagn. 12:111-117. [DOI] [PubMed] [Google Scholar]
- 14.Horowitz, S., M. Mazor, R. Romero, J. Horowitz, and M. Glezerman. 1995. Infection of the amniotic cavity with Ureaplasma urealyticum in the midtrimester of pregnancy. J. Reprod. Med. 40:375-379. [PubMed] [Google Scholar]
- 15.Ince, D., and D. C. Hooper. 2000. Mechanisms and frequency of resistance to premafloxacin in Staphylococcus aureus: novel mutations suggest novel drug-target interactions. Antimicrob. Agents Chemother. 44:3344-3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kenny, G. E., and F. D. Cartwright. 1993. Effect of pH, inoculum size, and incubation time on the susceptibility of Ureaplasma urealyticum to erythromycin in vitro. Clin. Infect. Dis. 17(Suppl. 1):S215-S218. [DOI] [PubMed] [Google Scholar]
- 17.Kong, F., X. Zhu, W. Wang, X. Zhou, S. Gordon, and G. L. Gilbert. 1999. Comparative analysis and serovar-specific identification of multiple-banded antigen genes of Ureaplasma urealyticum biovar 1. J. Clin. Microbiol. 37:538-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matlow, A., C. Th'ng, D. Kovach, P. Quinn, M. Dunn, and E. Wang. 1998. Susceptibilities of neonatal respiratory isolates of Ureaplasma urealyticum to antimicrobial agents. Antimicrob. Agents Chemother. 42:1290-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maxwell, N. C., P. L. Davies, and S. Kotecha. 2006. Antenatal infection and inflammation: what's new? Curr. Opin. Infect. Dis. 19:253-258. [DOI] [PubMed] [Google Scholar]
- 20.Morais Cabral, J. H., A. P. Jackson, C. V. Smith, N. Shikotra, A. Maxwell, and R. C. Liddington. 1997. Crystal structure of the breakage-reunion domain of DNA gyrase. Nature 388:903-906. [DOI] [PubMed] [Google Scholar]
- 21.Palu, G., S. Valisena, M. F. Barile, and G. A. Meloni. 1989. Mechanisms of macrolide resistance in Ureaplasma urealyticum: a study on collection and clinical strains. Eur. J. Epidemiol. 5:146-153. [DOI] [PubMed] [Google Scholar]
- 22.Pereyre, S., M. Metifiot, C. Cazanave, H. Renaudin, A. Charron, C. Bebear, and C. M. Bebear. 2007. Characterisation of in vitro-selected mutants of Ureaplasma parvum resistant to macrolides and related antibiotics. Int. J. Antimicrob. Agents 29:207-211. [DOI] [PubMed] [Google Scholar]
- 23.Poehlsgaard, J., and S. Douthwaite. 2002. The macrolide binding site on the bacterial ribosome. Curr. Drug Targets Infect. Disord. 2:67-78. [DOI] [PubMed] [Google Scholar]
- 24.Roberts, M. C., and G. E. Kenny. 1986. Dissemination of the tetM tetracycline resistance determinant to Ureaplasma urealyticum. Antimicrob. Agents Chemother. 29:350-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roberts, M. C., and G. E. Kenny. 1986. TetM tetracycline-resistant determinants in Ureaplasma urealyticum. Pediatr. Infect. Dis. 5:S338-S340. [DOI] [PubMed] [Google Scholar]
- 26.Robertson, J. A., G. W. Stemke, S. G. Maclellan, and D. E. Taylor. 1988. Characterization of tetracycline-resistant strains of Ureaplasma urealyticum. J. Antimicrob. Chemother. 21:319-332. [DOI] [PubMed] [Google Scholar]
- 27.Robicsek, A., J. Strahilevitz, G. A. Jacoby, M. Macielag, D. Abbanat, C. H. Park, K. Bush, and D. C. Hooper. 2006. Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 12:83-88. [DOI] [PubMed] [Google Scholar]
- 28.Sanchez-Pescador, R., J. T. Brown, M. Roberts, and M. S. Urdea. 1988. The nucleotide sequence of the tetracycline resistance determinant tetM from Ureaplasma urealyticum. Nucleic Acids Res. 16:1216-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schelonka, R. L., B. Katz, K. B. Waites, and D. K. Benjamin, Jr. 2005. Critical appraisal of the role of Ureaplasma in the development of bronchopulmonary dysplasia with metaanalytic techniques. Pediatr. Infect. Dis. J. 24:1033-1039. [DOI] [PubMed] [Google Scholar]
- 30.Schelonka, R. L., and K. B. Waites. 2007. Ureaplasma infection and neonatal lung disease. Semin. Perinatol. 31:2-9. [DOI] [PubMed] [Google Scholar]
- 31.Shepard, M. C. 1954. The recovery of pleuropneumonia-like organisms from Negro men with and without nongonococcal urethritis. Am. J. Syph. Gonorrhea Vener. Dis. 38:113-124. [PubMed] [Google Scholar]
- 32.Tait-Kamradt, A., T. Davies, P. C. Appelbaum, F. Depardieu, P. Courvalin, J. Petitpas, L. Wondrack, A. Walker, M. R. Jacobs, and J. Sutcliffe. 2000. Two new mechanisms of macrolide resistance in clinical strains of Streptococcus pneumoniae from Eastern Europe and North America. Antimicrob. Agents Chemother. 44:3395-3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tait-Kamradt, A., T. Davies, M. Cronan, M. R. Jacobs, P. C. Appelbaum, and J. Sutcliffe. 2000. Mutations in 23S rRNA and ribosomal protein L4 account for resistance in pneumococcal strains selected in vitro by macrolide passage. Antimicrob. Agents Chemother. 44:2118-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tay, S. T., N. Y. Boo, T. B. Khoo, A. S. Koay, and M. Y. Rohani. 1997. Prevalence and antibiotic susceptibility of Ureaplasma urealyticum in Malaysian neonates with respiratory distress. Med. J. Malaysia 52:409-411. [PubMed] [Google Scholar]
- 35.Teng, L. J., X. Zheng, J. I. Glass, H. L. Watson, J. Tsai, and G. H. Cassell. 1994. Ureaplasma urealyticum biovar specificity and diversity are encoded in multiple-banded antigen gene. J. Clin. Microbiol. 32:1464-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waites, K., C. Bebear, J. Robertson, D. Talkington, and G. Kenny. 2000. Cumitech 34: laboratory diagnosis of mycoplasmal infections. ASM Press, Washington, DC.
- 37.Waites, K. B., D. T. Crouse, and G. H. Cassell. 1992. Antibiotic susceptibilities and therapeutic options for Ureaplasma urealyticum infections in neonates. Pediatr. Infect. Dis. J. 11:23-29. [DOI] [PubMed] [Google Scholar]
- 38.Waites, K. B., D. T. Crouse, and G. H. Cassell. 1993. Therapeutic considerations for Ureaplasma urealyticum infections in neonates. Clin. Infect. Dis. 17(Suppl. 1):S208-S214. [DOI] [PubMed] [Google Scholar]
- 39.Waites, K. B., B. Katz, and R. L. Schelonka. 2005. Mycoplasmas and ureaplasmas as neonatal pathogens. Clin. Microbiol. Rev. 18:757-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Witt, A., A. Berger, C. J. Gruber, L. Petricevic, P. Apfalter, C. Worda, and P. Husslein. 2005. Increased intrauterine frequency of Ureaplasma urealyticum in women with preterm labor and preterm premature rupture of the membranes and subsequent cesarean delivery. Am. J. Obstet. Gynecol. 193:1663-1669. [DOI] [PubMed] [Google Scholar]
- 41.Xie, X., and J. Zhang. 2006. Trends in the rates of resistance of Ureaplasma urealyticum to antibiotics and identification of the mutation site in the quinolone resistance-determining region in Chinese patients. FEMS Microbiol. Lett. 259:181-186. [DOI] [PubMed] [Google Scholar]
- 42.Zhang, W., Y. Wu, W. Yin, and M. Yu. 2002. Study of isolation of fluoroquinolone-resistant Ureaplasma urealyticum and identification of mutant sites. Chin. Med. J. (Engl. Ed.) 115:1573-1575. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.