Abstract
Communities of ammonia-oxidizing archaea (AOA) and bacteria (AOB) in freshwater sediments and those in association with the root system of the macrophyte species Littorella uniflora, Juncus bulbosus, and Myriophyllum alterniflorum were compared for seven oligotrophic to mesotrophic softwater lakes and acidic heathland pools. Archaeal and bacterial ammonia monooxygenase alpha-subunit (amoA) gene diversity increased from oligotrophic to mesotrophic sites; the number of detected operational taxonomic units was positively correlated to ammonia availability and pH and negatively correlated to sediment C/N ratios. AOA communities could be grouped according to lake trophic status and pH; plant species-specific communities were not detected, and no grouping was apparent for AOB communities. Relative abundance, determined by quantitative PCR targeting amoA, was always low for AOB (<0.05% of all prokaryotes) and slightly higher for AOA in unvegetated sediment and AOA in association with M. alterniflorum (0.01 to 2%), while AOA accounted for up to 5% in the rhizospheres of L. uniflora and J. bulbosus. These results indicate that (i) AOA are at least as numerous as AOB in freshwater sediments, (ii) aquatic macrophytes with substantial release of oxygen and organic carbon into their rhizospheres, like L. uniflora and J. bulbosus, increase AOA abundance; and (iii) AOA community composition is generally determined by lake trophy, not by plant species-specific interactions.
Oxygen release from the roots of macrophyte species such as Littorella uniflora (L.) Asch. (shore weed), Lobelia dortmanna L. (water lobelia), and Glyceria maxima (Hartm.) Holmb. (reed sweet grass) stimulates nitrification and coupled nitrification-denitrification in the rhizosphere compared to that in unvegetated sediment (2, 36, 40). These interactions are of high ecological relevance especially in oligotrophic systems, since enhanced nitrogen loss due to rhizosphere-associated denitrification can retard natural eutrophication and succession of plant communities (1). While the microbial communities involved in coupled nitrification-denitrification have been well studied in rice paddy soils (7, 11), less information is available for natural freshwater sediments, especially those from oligotrophic lakes (2, 26).
The first key step of coupled nitrification-denitrification, the oxidation of ammonia to nitrite, is catalyzed by two groups of prokaryotes—the ammonia-oxidizing bacteria (AOB) (24) and the only recently recognized ammonia-oxidizing archaea (AOA) (22). For both groups, the gene encoding the alpha-subunit of ammonia monooxygenase (amoA) has been widely used as a functional marker to analyze their community compositions (15, 25); recent studies demonstrated the ubiquity of AOA and their predominance over AOB in a broad range of environments (32, 38). AOA, but not AOB, were also strongly enriched in the rhizosphere of the freshwater macrophyte Littorella uniflora in a mesotrophic Danish lake, suggesting that AOA were primarily responsible for increased rates of nitrification in the rhizosphere of this plant species (19). Moreover, ammonia oxidizer communities differed between rhizosphere and unvegetated sediment, indicating a plant-specific effect on AOA and AOB community composition. The objectives of this study were therefore to test whether (i) AOA generally predominate over AOB in freshwater sediments and especially in macrophyte rhizospheres and (ii) macrophytes have species-specific effects on abundance and community composition of AOA and AOB in rhizosphere sediments and on root surfaces.
To address these questions, two shallow heathland pools and five lakes in Denmark and Germany, ranging from low-pH and dystrophic sites to neutral-pH and oligotrophic and mesotrophic sites, were chosen, and three macrophyte species—Littorella uniflora, Juncus bulbosus L. (bulbous rush), and Myriophyllum alterniflorum DC. (alternate water milfoil)—were selected as model systems. These plant species differ in nitrogen nutrition, extent of radial oxygen loss, and lifestyle, presumably resulting in differential, plant species-specific effects on rhizosphere- and root-associated AOA and AOB communities. L. uniflora prefers nitrate as the nitrogen source, while J. bulbosus prefers ammonium (41, 45); oxygen release is high to moderate from the roots of L. uniflora and J. bulbosus (9, 12) but is minor from the roots of M. alterniflorum (M. Herrmann, P. Stief, and A. Schramm, unpublished results); L. uniflora and J. bulbosus remain photosynthetically active throughout the year, while only the below-ground parts of M. alterniflorum are retained during winter.
Rhizosphere sediments and roots from each plant species were sampled from three different sites per species, and unvegetated sediment was obtained from all seven sites. The comparison of samples from these different sites and compartments (rhizosphere, root surface, unvegetated sediment) allowed an evaluation of the importance of plant species relative to that of environmental conditions related to lake trophic status and pH on ammonia oxidizer communities.
MATERIALS AND METHODS
Study sites and sampling.
Unvegetated sediment and sediment from the rhizospheres of L. uniflora, J. bulbosus, and M. alterniflorum were sampled from five Danish softwater lakes (lakes Hampen, Kalgård, Grane Langsø, Søby, and Almind) and two humic, slightly to moderately acidic heathland pools in northwest Germany (nature reserve “Heiliges Meer”). For details, see Table 1.
TABLE 1.
Sampling sites, sample types, and chemical composition of sediment and sediment pore watera
| Site | Plant species | Sample type | Sample name | Sediment
|
Pore water
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| C/N ratio | N (%) | C (%) | Concn (μmol liter−1) of:
|
Free NH3 (nmol liter−1) | pH | ||||||
| NO2− | NO3− | NH4+ | |||||||||
| Lake Hampen (A) (mesotrophic) | Unvegetated sediment | AS | 8.1 ± 0.5 | 0.02 ± 0.01 | 0.17 ± 0.03 | 0.4 ± 0.1 | 1.3 ± 0.2 | 84.7 ± 63.0 | 212.7 ± 158.3 | 6.8 | |
| L. uniflora | Rhizosphere | LAR | 10.6 ± 0.2 | 0.06 ± 0.02 | 0.52 ± 0.16 | 0.5 ± 0.1 | 58.2 ± 22.8 | 20.0 ± 7.2 | 50.3 ± 18.2 | 6.8 | |
| Root surface | LARp | ||||||||||
| M. alterniflorum | Rhizosphere | MAR | 10.2 ± 2.2 | 0.02 ± 0.00 | 0.17 ± 0.02 | 0.4 ± 0.3 | 2.1 ± 1.4 | 34.9 ± 24.8 | 87.7 ± 62.2 | 6.8 | |
| Root surface | MARp | ||||||||||
| Lake Kalgård (B) (oligotrophic) | Unvegetated sediment | BS | 18.1 ± 4.6 | 0.01 ± 0.00 | 0.11 ± 0.02 | 0.4 ± 0.1 | 2.7 ± 0.5 | 6.3 ± 1.6 | 12.5 ± 3.2 | 6.7 | |
| L. uniflora | Rhizosphere | LBR | 14.6 ± 0.8 | 0.03 ± 0.01 | 0.35 ± 0.19 | 0.4 ± 0.1 | 5.9 ± 2.1 | 9.5 ± 2.1 | 15.0 ± 3.4 | 6.6 | |
| Root surfaces | LBRp | ||||||||||
| Lake Grane Langsø (C) (oligotrophic) | Unvegetated sediment | CS | 13.1 ± 2.6 | 0.03 ± 0.01 | 0.36 ± 0.19 | 0.6 ± 0.2 | 5.1 ± 0.9 | 7.8 ± 3.1 | 12.4 ± 4.8 | 6.6 | |
| L. uniflora | Rhizosphere | LCR | 10.2 ± 0.5 | 0.03 ± 0.01 | 0.30 ± 0.04 | 0.6 ± 0.1 | 64.8 ± 23.0 | 5.0 ± 3.6 | 6.3 ± 4.5 | 6.5 | |
| Root surface | LCRp | ||||||||||
| J. bulbosus | Rhizosphere | JCR | 16.3 ± 1.5 | 0.10 ± 0.03 | 1.45 ± 0.55 | 0.8 ± 0.2 | 18.7 ± 16.2 | 5.1 ± 0.5 | 6.4 ± 0.7 | 6.5 | |
| Root surface | JCRp | ||||||||||
| Lake Søby (D) (mesotrophic) | Unvegetated sediment | DS | 8.7 ± 1.2 | 0.02 ± 0.00 | 0.15 ± 0.03 | 1.4 ± 1.1 | 1.3 ± 1.2 | 15.5 ± 6.2 | 245.2 ± 99.0 | 7.6 | |
| M. alterniflorum | Rhizosphere | MDR | 10.7 ± 0.3 | 0.01 ± 0.00 | 0.11 ± 0.04 | 0.5 ± 0.2 | 6.5 ± 2.4 | 9.1 ± 3.1 | 181.7 ± 62.5 | 7.7 | |
| Root surface | MDRp | ||||||||||
| Lake Almind (F) (mesotrophic) | Unvegetated sediment | FS | 13.4 ± 1.4 | 0.01 ± 0.00 | 0.15 ± 0.02 | 0.2 ± 0.1 | 1.5 ± 0.4 | 23.6 ± 7.2 | 59.3 ± 18.1 | 6.8 | |
| M. alterniflorum | Rhizosphere | MFR | 14.4 ± 2.5 | 0.02 ± 0.01 | 0.18 ± 0.06 | 0.4 ± 0.2 | 1.5 ± 0.5 | 31.1 ± 14.0 | 78.1 ± 35.1 | 6.8 | |
| Root surface | MFRp | ||||||||||
| Heathland pool G (G) (dystrophic) | Unvegetated sediment | GS | 17.5 ± 0.3 | 0.85 ± 0.38 | 12.63 ± 5.55 | 0.3 ± 0.0 | 1.7 ± 0.3 | 30.9 ± 13.2 | 49.0 ± 20.9 | 6.6 | |
| J. bulbosus | Rhizosphere | JGR | 16.0 ± 0.8 | 1.39 ± 0.56 | 18.82 ± 6.63 | 0.5 ± 0.1 | 197.8 ± 130.8 | 34.2 ± 6.5 | 13.6 ± 2.6 | 6.0 | |
| Root surface | JGRp | ||||||||||
| Heathland pool H (H) (dystrophic) | Unvegetated sediment | HS | 28.7 ± 8.4 | 1.22 ± 0.57 | 27.23 ± 4.82 | 0.0 ± 0.0 | 10.0 ± 3.2 | 48.4 ± 21.6 | 1.2 ± 0.5 | 4.8 | |
| J. bulbosus | Rhizosphere | JHR | 23.9 ± 3.4 | 1.60 ± 0.55 | 31.72 ± 7.81 | 0.0 ± 0.0 | 8.2 ± 2.0 | 52.3 ± 2.4 | 2.1 ± 0.1 | 5.0 | |
| Root surface | JHRp | ||||||||||
Values are the average ± standard deviation of results obtained from replicate sediment cores (n = 3). Pore water extracts from replicate cores were pooled for measurement of pH. Concentrations of free NH3 were calculated based on pH and NH4+ concentrations.
Sediment cores were taken in triplicate from within monospecies stands of L. uniflora, J. bulbosus, and M. alterniflorum and from unvegetated sediment at a water depth of 5 to 60 cm in September 2005 for molecular analysis (for heathland pool H, April 2006; for heathland pool G and Lake Søby, September 2006) and in June 2006 for chemical analysis (for heathland pool H, April 2006; for heathland pool G and Lake Søby, September 2006). Unvegetated sediment samples were obtained from the upper 1.5 cm to include the oxic-anoxic interface (typically the zone of coupled nitrification-denitrification). Rhizosphere sediment samples were obtained from all over the root zone by shaking off sediment that was loosely adhering to the roots. Samples were transferred in the field to sterile 50-ml Falcon tubes. Plant roots from which rhizosphere sediment samples had been obtained were gently washed with lake water to remove the remaining sediment, separated from the green plant parts, and transferred to sterile 50-ml Falcon tubes. All samples were kept on ice during transport. In the laboratory, samples for DNA extraction and samples for chemical analyses were frozen to −80°C and to −21°C, respectively, while pore water samples were extracted within 24 h.
Sediment chemical analysis.
Pore water samples were extracted from unvegetated sediment and rhizosphere sediment by the centrifugation of about 75 g of sediment at 1,000 rpm for 5 min in combination with filtration through glass microfiber filters (Whatman) and were analyzed for pH and NO2− plus NO3− (6), NO2− (16), and NH4+ (5) concentrations. Concentrations of free NH3 were calculated based on pH and NH4+ concentrations (pKa of NH4+/NH3 is 9.4 at 20°C). For analyses of total organic C and total N contents, sediment samples were dried at 110°C for 76 h, homogenized using a mortar, treated with H2SO3 for 64 h, and dried again at 70°C for 24 h. C/N analyses were performed on an NA 1500 nitrogen analyzer (Carlo Erba Strumentazione).
DNA extraction, PCR amplification, cloning, and sequence analysis.
DNA was extracted in triplicate from 200 mg sediment by combining enzymatic and chemical cell lysis with the FastDNA Spin kit for soil (Qbiogene Inc.) (13). For the analysis of microbial communities on root surfaces, DNA was extracted from 100 mg plant roots following the same protocol. Archaeal and bacterial amoA genes were amplified using the HotStarTaq master mix kit (Qiagen) with published protocols and primer sets Arch-AmoAF/Arch-AmoAR (14) and AmoA-1F/AmoA-2R-TC (34). Bands of the correct size were excised and purified (GenElute gel extraction kit; Sigma). Purified PCR products from triplicate DNA extracts of the triplicate sediment cores were pooled for subsequent cloning (pGEM-T cloning kit; Promega). For samples of unvegetated sediment, one clone library was constructed per site for each gene. For bacterial amoA, one combined rhizosphere sediment/root surface clone library was constructed for each plant species and site, and denaturing gradient gel electrophoresis (DGGE) (34) was used to differentiate rhizosphere and root surface populations by assigning clones to specific bands in the environmental DGGE patterns. Bands not represented by cloned inserts were excised and reamplified for the direct sequencing of purified PCR products (34). For archaeal amoA, separate clone libraries were constructed for rhizosphere sediment and root samples of each plant species and each site. In total, 16 bacterial and 25 archaeal amoA clone libraries were constructed, plasmids were extracted using the GenElute plasmid preparation kit (Sigma), and inserts were sequenced by Macrogen (South Korea). Alignments, translations to amino acids, and phylogenetic analyses were done using the ARB package (30). Phylogenetic trees of bacterial and archaeal AmoA were constructed as consensus trees of neighbor-joining and parsimony analysis (1,000 replicates). Maximum likelihood analysis was also performed but with a reduced number of sequences. Operational taxonomic units (OTUs) were defined based on a 97% identity cutoff on the amino acid level using DOTUR (43), and Good's coverage was calculated for each clone library (47).
qPCR.
Relative abundances of AOA and AOB were determined by quantitative PCR (qPCR) as described previously (19). In brief, copy numbers of 16S rRNA genes and bacterial or archaeal amoA genes were determined in triplicate for each sediment sample, using universal primers 907F/1492R (27) and amoA primers Arch-AmoAF/Arch-AmoAR (14) and AmoA-1F/AmoA-2R-TC (34) with brilliant SYBR green qPCR master mix (Stratagene) on an Mx3005P instrument (Stratagene). qPCR reactions contained 25 ng environmental DNA or 25 ng salmon sperm DNA in standard reactions. Standard curves were prepared from serial dilutions of Bacteroides fragilis ATCC 25285 genomic DNA for 16S rRNA genes and serial dilutions of plasmids containing an environmental archaeal amoA sequence (clone AS_AOA_2 [GenBank accession number EU309881]) or the amoA sequence of Nitrosospira multiformis ATCC 25196, respectively. Standard curves were linear from 107 to 102 16S rRNA gene copies and from 5 × 108 to 5 × 101 amoA gene copies; i.e., the amoA genes could not be reliably quantified when there were <50 gene copies (the limit of quantification). Detection limits were five gene copies for the functional genes. Efficiencies of the qPCR reactions were 88 to 97% for amoA and 75 to 83% for 16S rRNA genes. The specificities of PCR products were confirmed by melting curve analysis and agarose gel electrophoresis. Environmental DNA templates were spiked with standard plasmid DNA to test for the presence of PCR inhibitors. The abundances of AOA and AOB were expressed relative to the number of prokaryotic cells as estimated from 16S rRNA gene copy numbers, assuming 2.5 and one amoA gene copies per AOB and AOA, respectively (29), one 16S rRNA gene copy per AOA, and 3.6 copies of the 16S rRNA gene per average prokaryotic cell (21). Relative abundances were preferred over absolute values for this study, as they are less sensitive to differences in DNA extraction efficiencies; these can be expected when working with sample types that differ substantially in their contents of humic substances.
Statistical analysis.
To compare the clone libraries between different sites, macrophyte species, and compartments, Jaccard similarity values were calculated (44). These values consider how many OTUs are shared between clone libraries relative to the total number of OTUs in these clone libraries. Similarity matrices were turned into dendrograms to visualize the clustering of AOA and AOB communities based on their similarities in OTU composition, utilizing the programs “Clustering Calculator” (http://www2.biology.ualberta.ca/jbrzusto/cluster.php) and “Interactive Tree of Life” (http://itol.embl.de). Differences between sites in the chemical parameters and relative abundances of AOA were analyzed by one-way analysis of variance (ANOVA) and Tukey's least significant difference test (P = 0.05; XLSTAT). To test if differences in compartment, plant species, or lake trophic status had a general effect on the number of detected OTUs for AOA and AOB, samples obtained from the different sites were grouped according to compartment, plant species, or lake trophic status, and groups were compared using one-way ANOVA and Tukey's least significant difference test (P = 0.05; XLSTAT). Spearman's rank correlation coefficients were calculated to analyze if relative abundances of AOA or the numbers of detected OTUs of AOA and AOB were correlated to environmental parameters (XLSTAT). Relative abundances of AOB were excluded from correlation analysis, since the copy numbers of bacterial amoA genes usually remained below the quantification or detection limit of the qPCR, and the relative abundances given represent only maximum estimates based on the quantification limit or detection limit.
Nucleotide sequence accession numbers.
Nonredundant sequences of archaeal and bacterial amoA genes obtained in this study have been deposited in the GenBank database under accession numbers EU667645 to EU667773 (for bacterial amoA) and EU667774 to EU667992 (for archaeal amoA). Most of the sequences obtained from site A have been published previously (accession numbers EU309859 to EU309918) (19).
RESULTS
Sediment analysis and site characterization.
Based on the composition of the littoral vegetation, literature data (50), and our own chemical data (Table 1), sampling sites were grouped into three categories. Oligotrophic lakes Kalgård and Grane Langsø were characterized by very sandy sediments with a low content of total N and organic C, C/N ratios between 10.2 and 18.1, low concentrations of NH4+ and free NH3 in the pore water, and pH ranging from 6.5 to 6.7; littoral vegetation was composed of isoetid species such as L. uniflora, Lobelia dortmanna, and Isoetes lacustris (31, 39). Mesotrophic lakes Hampen, Almind, and Søby had a slightly higher pH, higher concentrations of NH4+, and higher availability of free NH3 in the sediment pore water, while C/N ratios were lower; mixed stands of L. uniflora and M. alterniflorum and dense stands of Phragmites australis indicate increasingly mesotrophic conditions (17, 28). Two humic heathland pools were characterized as dystrophic (51) based on their high content of organic C and total N in the sediments, high C/N ratios, and, due to their low pH, a very low availability of NH3 despite concentrations of NH4+ in the pore water being higher than those at the oligotrophic sites; the vegetation was dominated by Juncus bulbosus and Sphagnum cuspidatum. Nitrite concentrations in the sediment pore water were low at all sites. Nitrate concentrations were generally low in the pore water of the rhizosphere of M. alterniflorum and in unvegetated sediment, except for Lake Grane Langsø and heathland pool H, but were considerably higher in the rhizospheres of L. uniflora and J. bulbosus at most of the sites, indicating active nitrification. Concentrations of NH4+ were usually lower in the rhizosphere than in unvegetated sediment.
Community composition of AOA and AOB.
Between 8 and 24 clones were sequenced per archaeal amoA clone library to achieve coverages of 82 to 100%, resulting in a total of 306 archaeal amoA sequences. Based on 97% translated amino acid identity, they were assigned to 13 OTUs, most of which included a large number of sequences from different sites; only three OTUs were represented by single sequences. The number of detected OTUs per clone library ranged from 1 to 5. For bacterial amoA, between 16 and 90 clones per library were screened by DGGE prior to sequencing, and sequences were obtained from 1 to 33 clones per library. Additionally, 1 to 10 sequences per library were obtained directly from environmental DGGE bands not represented by clones. Including sequences derived from clones and DGGE bands, library coverages were 71 to 100%. The resulting 191 bacterial AmoA sequences were assigned to 15 OTUs, with one to six OTUs per clone library.
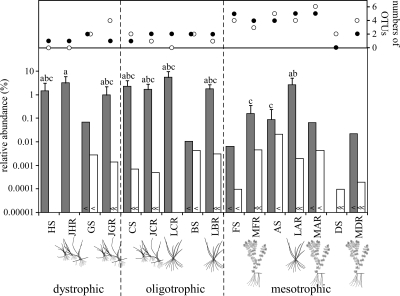
The number of detected OTUs of both AOA and AOB was highest in the rhizospheres of L. uniflora and M. alterniflorum at mesotrophic Lake Hampen (site A). The acidic heathland pool (site H) showed the lowest diversity, with only a single OTU for AOA AmoA and no bacterial amoA genes detected (Fig. 1). One-way ANOVA indicated that neither plant species nor compartment had a significant site-independent influence on the number of detected OTUs of AOA or AOB, while mesotrophic sites could significantly be distinguished from dystrophic and oligotrophic sites (P = 0.05). For archaeal and bacterial AmoAs, four and six OTUs, respectively, occurred exclusively in the rhizosphere or on root surfaces, while two and four OTUs, respectively, were restricted to unvegetated sediment. Out of these compartment-specific OTUs, only a single bacterial AmoA OTU occurred at more than one site.
FIG. 1.
Number of detected OTUs of AOA (closed circles) and AOB (open circles) and relative abundances of AOA (gray columns) and AOB (white columns) in unvegetated sediment and rhizosphere sediment. For definitions of the abbreviations for samples, see Table 1. Relative abundance values are the means ± the standard deviations of nine qPCR reactions with triplicate DNA extracts from three environmental replicates. Columns represent maximum estimates, where amoA gene copy numbers remained below the quantification limit of 50 copies (<) or the detection limit of 5 copies (≪) per reaction. Letters above columns indicate significant differences between samples.
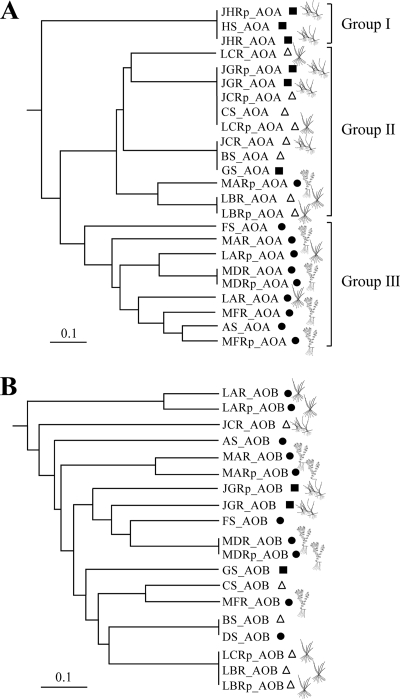
Ammonia oxidizer communities often showed high similarities between rhizosphere- and root surface-associated communities from the same plant species and site, based on the number of OTUs shared between clone libraries (Jaccard indices [Fig. 2]). However, neither AOA nor AOB communities clustered according to plant species across the sites. While AOB communities did not show any apparent clustering, AOA communities formed three major groups representing sample types (Fig. 2A). Group I contained only a single OTU, which occurred in unvegetated sediment and in the rhizosphere and root surface of J. bulbosus at heathland pool H, which is characterized by a low pH, high C/N ratios, and a very low availability of free NH3; group II included samples mostly derived from oligotrophic sites, characterized by a higher pH, lower C/N ratios, and an increasing availability of free NH3; group III included communities obtained exclusively from mesotrophic sites with higher pH values, lower C/N ratios, and higher concentrations of free NH3 than those of the sites for the other two groups.
FIG. 2.
Similarities of clone libraries of archaeal AmoA (A) and bacterial AmoA (B) among sites, plant species, and compartments, based on Jaccard values. The origins of sequences are indicated by closed circles (mesotrophic sites), open triangles (oligotrophic sites), and closed squares (dystrophic sites). For the definitions of abbreviations, see Table 1.
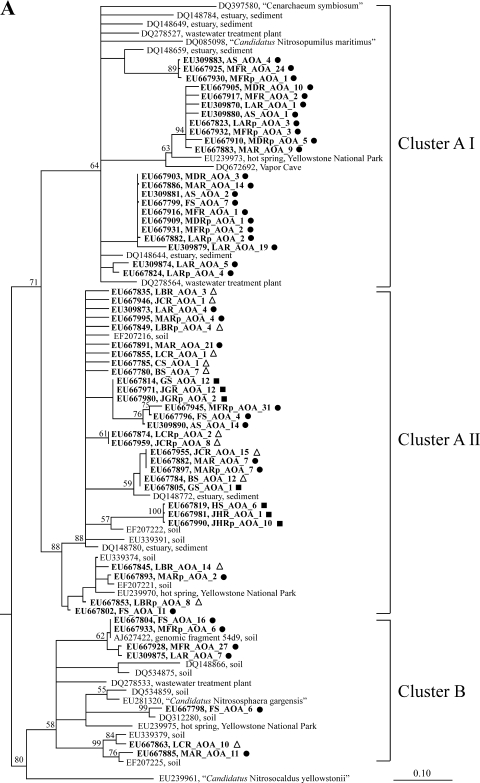
Phylogenetic affiliation of archaeal AmoA sequences.
AOA AmoA sequences affiliated with the proposed clusters A I, A II, and B (19) (Fig. 3A), where clusters A I and B harbor representatives of AOA that have been confirmed by either cultivation or metagenomic studies (cluster A I, “Candidatus Nitrosopumilus maritimus” and Cenarchaeum symbiosum; cluster B, soil metagenomic fragment 54d9 and “Candidatus Nitrososphaera gargensis”). The majority of the freshwater-derived sequences affiliated with clusters A I and A II.
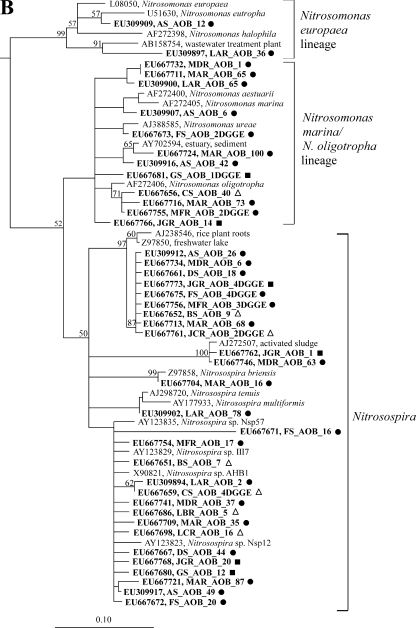
FIG. 3.
Phylogenetic affiliation of archaeal AmoA (A) and bacterial AmoA (B). Protein distance trees are displayed with bootstrap values (>50%) from parsimony analysis (1,000 replicates). Branchings not supported by both methods are drawn as multifurcations. The major lineages were also supported by maximum likelihood analysis with a reduced number of sequences. For all the OTUs obtained in this study, accession numbers and clone numbers for representative sequences from each clone library are shown in boldface type. For the definitions of abbreviations, see Table 1. The origins of sequences are indicated by closed circles (mesotrophic sites), open triangles (oligotrophic sites), and closed squares (dystrophic sites).
Cluster A II harbored sequences derived from all three different types of lake trophic statuses investigated in this study—dystrophic, oligotrophic, and mesotrophic—and these sequences were closely related to sequences obtained from soil, estuarine sediments, and hot springs. Sequences obtained from acidic heathland pool H (HS_AOA, JHR_AOA, JHRp_AOA) were distinct from all other sequences within cluster A II (Fig. 3A). All sequences affiliating with cluster A I and, with one exception, cluster B had been obtained from mesotrophic sites (A, D, and F), and for almost all samples obtained from mesotrophic sites, more OTUs affiliated with cluster A I or B than with cluster A II.
Phylogenetic affiliation of bacterial AmoA sequences.
Bacterial AmoA sequences affiliated with groups commonly found in freshwater habitats (24), i.e., the Nitrosospira, Nitrosospira marina/Nitrosomonas oligotropha, and Nitrosomonas europaea lineages (Fig. 3B). With few exceptions (GS_AOB_1_DGGE, CS_AOB_40, JGR_AOB_14), Nitrosomonas-like sequences were only obtained from mesotrophic sites (A, D, and F); only two sequences from the mesotrophic site A were related to Nitrosomonas europaea/Nitrosomonas eutropha, while all other Nitrosomonas-like sequences affiliated with the N. marina/N. oligotropha lineage. The majority of bacterial AmoA sequences was related to Nitrosospira-like sequences, particularly to a group of uncultured AOB retrieved from freshwater and rice paddy soils. Low-diversity AOB communities at oligotrophic sites, such as Lake Kalgård (site B) and Lake Grane Langsø (site C), consisted almost exclusively of Nitrosospira-related AOB.
Relative abundances of AOA and AOB.
AOA dominated numerically at most of the sites and were up to 3 orders of magnitude more abundant than were AOB. Unvegetated sediment of mesotrophic Lake Søby (site D) was the only sample in which no archaeal amoA genes were detected, while bacterial amoA genes were not detected in acidic heathland pool H (site H), in samples from the root surface of J. bulbosus and from the rhizosphere of L. uniflora from oligotrophic Lake Grane Langsø (C), or in samples from the root surface of M. alterniflorum from mesotrophic Lake Almind (F) (Fig. 1). For some of the other sites, bacterial amoA genes were not detected in all replicates of a certain sample type. Variation among replicate sediment cores was higher than variation among replicate DNA extracts for all three compartments, which indicated a high spatial heterogeneity of the distribution of the two groups of ammonia oxidizers.
With few exceptions, relative abundances of AOA were high (0.1 to 5.6% of all prokaryotes) in the dystrophic and oligotrophic sediments and in the rhizosphere/root surface system of L. uniflora and J. bulbosus. In contrast, archaeal amoA gene copy numbers were <50 per reaction in most mesotrophic sediments, including the rhizosphere/root surface system of M. alterniflorum (Fig. 1).
Bacterial amoA gene copy numbers were always below the limit of quantification (<50 copies per reaction) and, in many samples, even below the detection limit of the qPCR (five copies per reaction). However, bacterial amoA genes were still detectable in most samples by the standard PCR; therefore, 50 or 5 amoA gene copies per reaction were used to estimate the maximum relative abundance of AOB (Fig. 1). Based on these maximum estimates, relative abundances of AOB were highest at mesotrophic sites in association with both the rhizosphere and root surfaces of M. alterniflorum (maxima 0.005% and 0.055% of all prokaryotes, respectively) (Fig. 1).
AOA and AOB on root surfaces.
With a few exceptions, OTUs of AOA and AOB that occurred on the root surface of a given plant species at a given site could also be retrieved from the corresponding rhizosphere, with the number of detected OTUs on the root surface being equal to or lower than that detected in the rhizosphere. In particular, a decrease in the OTU numbers of root surface communities was observed for J. bulbosus at site C and for L. uniflora at site A (AOA and AOB) and C (AOA only). Relative abundances of AOA were, in general, lower on the root surfaces of L. uniflora and J. bulbosus than those in the respective rhizospheres, while there was no clear trend for M. alterniflorum or for the relative abundances of AOB (data not shown).
Relationship between abundance and community composition of AOA and AOB and environmental parameters.
Relative abundances of AOA were negatively correlated to ammonia availability and pH and were positively correlated to the sediment content of organic C and N, while there was no significant correlation with C/N ratios (Table 2). No such correlation analysis was performed for the relative abundances of AOB, since these data were only maximum estimates. For a range of 2 to 213 nmol liter−1 ammonia and pH 4.8 to 6.8, the number of detected OTUs of both AOA and AOB was strongly positively correlated to ammonia availability and pH. However, this correlation was no longer significant for AOA if samples from Lake Søby (site D) with the maximum ammonia availability and pH were included in the analysis (data not shown). The number of detected OTUs of AOA was negatively correlated to C/N ratios and sediment content of C and N (Table 2).
TABLE 2.
Spearman's rank correlation coefficients of environmental parameters and relative abundances of AOA or numbers of detected OTUs of AOA and AOB in unvegetated sediment and rhizosphere sedimenta
| Parameter | Correlation to sediment characteristic
|
||||
|---|---|---|---|---|---|
| NH3 availability | pH | C (%) | N (%) | C/N ratio | |
| Relative abundance of AOA | −0.671** | −0.637** | 0.685** | 0.668** | 0.163 |
| No. of OTUs of AOA | 0.801*** | 0.889*** | −0.679** | −0.647* | −0.614* |
| No. of OTUs of AOB | 0.885*** | 0.776** | −0.504 | −0.457 | −0.587* |
Samples from site D were excluded from correlation analysis between OTU richness and environmental parameters. Significance levels are indicated as follows: ***, 0.001; **, 0.01; *, 0.05.
DISCUSSION
Influence of plant species versus influence of lake trophic status on community compositions of AOA and AOB.
In the present study, we observed a general effect of the lake trophic status on the diversity of ammonia-oxidizing prokaryotes, with an increase in the number of detected OTUs from dystrophic and oligotrophic lakes to mesotrophic lakes. Plant species or compartment—unvegetated sediment, rhizosphere, and root surface—had no such effect. Previous reports of the influence of plant species on the community composition of rhizosphere-associated ammonia oxidizers are contradictory. While Nicolaisen et al. (35) and Kowalchuk et al. (26) did not find differences in AOB community composition between bulk soil and the rhizospheres of Oryza sativa and Glyceria maxima, other studies indicated a specific effect of the plant on rhizosphere-associated communities (7, 11, 19). However, none of these studies compared several plant species and sites. Competition with plant roots for reduced nitrogen compounds may select for certain types of rhizosphere-associated ammonia oxidizers (3), suggesting that plant species which preferentially utilize ammonium, such as J. bulbosus, support different ammonia oxidizers in their rhizosphere from those supported by plant species which prefer nitrate, such as L. uniflora (45). Similar distinct effects may result from differences in the extent of oxygen release between macrophyte species.
Comparison of the compositions of AOA and AOB communities from seven lakes and three plant species in this study clearly showed that plant species-specific traits, such as nitrogen source utilization, extent of oxygen release, and lifestyle, did not select for specific ammonia oxidizers that would always be associated with the root system of a particular plant species. On the contrary, environmental conditions related to lake trophic status, such as ammonia availability, pH, and contents of humic acids, probably had a stronger effect on the conditions in the rhizosphere than did the plant species itself, resulting in a direct correspondence of especially the AOA communities to lake trophic status. AOB communities did not show any obvious clustering patterns. The lack of plant species-specific associations also indicates the absence of specific symbiotic interactions between AOA or AOB and the macrophyte species investigated.
Phylogenetic affiliation of AOA and AOB.
The affiliation of sequences obtained from freshwater environments with sequences derived from marine and estuarine habitats, wastewater treatment plants, and hot springs indicates that the phylogeny of archaeal AmoA is not primarily linked to general habitat types, as was originally proposed (14). In contrast, the results obtained in this study suggest that environmental factors related to lake trophic status, such as pH and ammonia availability, may play a critical role in the phylogenetic affiliation of freshwater AOA. While cluster A II harbored sequences retrieved from all lake types investigated in this study, affiliation with clusters A I and B was largely restricted to AmoA sequence types obtained from mesotrophic lakes. Moreover, sequences obtained from unvegetated sediment and J. bulbosus rhizosphere and root surfaces at acidic heathland pool H formed a single distinct OTU within cluster A II, suggesting a specific adaptation of the corresponding AOA phylotype to extreme site conditions of low pH, high C content of the sediment, high C/N ratios, and extremely low availability of ammonia. A clear effect of pH on AOA community composition and phylogenetic affiliation of the respective phylotypes in soils was recently reported by Nicol et al. (33), with similar patterns for both archaeal amoA and crenarchaeotal 16S rRNA genes.
Nitrosospira- and Nitrosomonas oligotropha-related AOB dominated most of the AOB communities found in this study. These organisms have previously been reported from oligotrophic freshwater environments and from neutral to acidic soils (23, 49). Affiliation with Nitrosomonas lineages other than N. oligotropha was restricted to sequences obtained from mesotrophic sites at a higher availability of ammonia, which agrees with the higher substrate requirements reported for the corresponding AOB lineages (23).
Regulating factors for AOA in freshwater environments.
AOA dominated the ammonia oxidizer communities associated with all investigated plant species from all sites, which confirms and extends previous observations of archaeal predominance in the rhizospheres of L. uniflora (19) and Oryza sativa (11). AOA can contribute significantly to ammonia oxidation in certain soils and estuarine environments (8, 33, 42); since increased nitrification rates (19, 36) and increased nitrate concentrations (found in this study) in the rhizospheres of L. uniflora and J. bulbosus coincided with an increased abundance of AOA but not AOB, it is likely that AOA also contribute to nitrification in freshwater environments, at least in macrophyte rhizospheres. If so, AOA are apparently adapted to low ammonia availability and low pH, as their relative abundance was negatively correlated with pH and ammonia availability, and AOA were absent from unvegetated sediment at site D, which has the highest pH and ammonia availability. These data that point to a less important role of AOA under mesotrophic to eutrophic conditions and at higher pHs agree well with the pH-dependent, contrasting patterns of bacterial and archaeal amoA transcript numbers reported for soils (33).
Rhizosphere-associated nitrification is stimulated by oxygen release from the roots of aquatic macrophytes (36, 40), and plant species-specific differences in the extent of oxygen release are therefore likely to influence the abundance of rhizosphere-associated ammonia oxidizers. In fact, the plant species with the highest oxygen release—L. uniflora and J. bulbosus (9, 12)—harbored the highest relative abundances of AOA, while the low oxygen availability in the rhizosphere of M. alterniflorum (Herrmann et al., unpublished) resulted in relative AOA abundances only slightly higher than those in unvegetated sediment. However, since oxygen release by the three plant species was not recorded when sediment samples for molecular analyses were taken, the observed trend cannot be directly proven by statistical means. Differences in the extent of oxygen release between plant species may also be responsible for differences in the distribution patterns of AOA within the rhizosphere/root system. Relative abundances of AOA on the root surface of L. uniflora that are lower than those in the rhizosphere may be due to the sensitivity of ammonia oxidizers to very high oxygen concentrations of up to 450 μM (12). Differences between compartments were less pronounced for J. bulbosus with its moderate oxygen release (9), and no clear trend was observed for M. alterniflorum with its low oxygen release.
The positive correlation of relative abundances of AOA to sediment content of organic C and N indicates that not only root oxygen release but also root exudation of organic carbon and nitrogen compounds may play a role for the association of AOA with the rhizosphere of freshwater macrophytes. Uptake of amino acids has been shown for marine Crenarchaeota (18, 37), and utilization of root exudates has been proposed for mesophilic Crenarchaeota in the rhizosphere or mycorrhizosphere of terrestrial plants (4, 46). The C and N contents of L. uniflora and J. bulbosus rhizospheres were increased compared with those in unvegetated sediment, indicating a substantial release of organic substances via root exudation (10, 48). In contrast, the effect of M. alterniflorum on sediment chemistry was much weaker, and earlier studies did not indicate a stimulating effect for M. alterniflorum on microbial processes in the sediment (20). The abundance pattern of AOA in line with the extent of root exudation may suggest mixotrophic or heterotrophic utilization of root exudates by freshwater AOA; such a metabolism would also provide an explanation for the high relative abundances of AOA in unvegetated sediment with high contents of organic C and N (sites C and H). With the data available, no statistical evidence can be provided for the suggested relationship between root exudation and abundance of rhizosphere-associated ammonia-oxidizing archaea. Here, further studies including a more detailed characterization of root exudation and the carbon flow in the rhizosphere, i.e., by employing techniques such as stable isotope probing, are needed to gain more insight into the potential utilization of root exudates by this group of microorganisms.
Conclusion.
The community composition of AOA in oligotrophic to mesotrophic freshwater sediments is strongly determined by factors related to lake trophic status, such as pH, ammonia availability, and sediment content of organic C and N; there was no indication for species-specific effects of freshwater macrophytes on the community composition of rhizosphere-associated AOA or AOB. Where present, AOA were more abundant than AOB; the macrophytes L. uniflora and J. bulbosus, especially, supported high numbers of AOA, possibly by releasing oxygen and organic root exudates into their rhizospheres.
Acknowledgments
This study was financially supported by the Deutsche Forschungsgemeinschaft (project HE 5205/1-1) and the Danish Research Council (grant 2117-05-0027).
We thank Britta Poulsen for excellent assistance with field and laboratory work; Bente Lomstein, Rikke Holm, and Peter Stief for help with the chemical analyses; and Kilian Stoecker for sharing his AOB amoA ARB database. Kasper Urup Kjeldsen is acknowledged for his help with clone library analysis. We thank the owners of Lake Kalgård and Lake Grane Langsø as well as the “Westfälisches Museum für Naturkunde” for permission to obtain sediment samples.
Footnotes
Published ahead of print on 20 March 2009.
REFERENCES
- 1.Adema, E. B., J. Van de Koppel, H. A. J. Meijer, and A. P. Grootjans. 2005. Enhanced nitrogen loss may explain alternative stable states in dune slack succession. Oikos 109:374-386. [Google Scholar]
- 2.Bodelier, P. L. E., J. A. Libochant, C. W. P. M. Blom, and H. J. Laanbroek. 1996. Dynamics of nitrification and denitrification in root-oxygenated sediments and adaptation of ammonia-oxidizing bacteria to low oxygen or anoxic habitats. Appl. Environ. Microbiol. 62:4100-4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bollmann, A., M.-J. Bär-Gilissen, and H. J. Laanbroek. 2002. Growth at low ammonium concentrations and starvation response as potential factors involved in niche differentiation among ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 68:4751-4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bomberg, M., G. Jurgens, A. Saano, R. Sen, and S. Timonen. 2003. Nested PCR detection of Archaea in defined compartments of pine mycorrhizospheres developed in boreal forest humus microcosms. FEMS Microbiol. Ecol. 43:163-171. [DOI] [PubMed] [Google Scholar]
- 5.Bower, C. E., and T. Holm-Hansen. 1980. A salicylate-hypochlorite method for determining ammonia in seawater. Can. J. Fish. Aquat. Sci. 37:794-798. [Google Scholar]
- 6.Braman, R. S., and S. A. Hendrix. 1989. Nanogram nitrite and nitrate determination in environmental and biological materials by vananadium(III) reduction with chemiluminescence detection. Anal. Chem. 61:2715-2718. [DOI] [PubMed] [Google Scholar]
- 7.Briones, A. M., S. Okabe, Y. Umemiya, N.-B. Ramsing, W. Reichardt, and H. Okuyama. 2002. Influence of different cultivars on populations of ammonia-oxidizing bacteria in the root environment of rice. Appl. Environ. Microbiol. 68:3067-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caffrey, J. M., N. Bano, K. Kalanetra, and J. T. Hollibaugh. 2007. Ammonia oxidation and ammonia-oxidizing bacteria and archaea from estuaries with differing histories of hypoxia. ISME J. 1:660-662. [DOI] [PubMed] [Google Scholar]
- 9.Chabbi, A. 1999. Juncus bulbosus as a pioneer species in acidic lignite mining lakes: interactions, mechanisms and survival strategies. New Phytol. 144:133-142. [Google Scholar]
- 10.Chabbi, A., M. E. Hines, and C. Rumpel. 2001. The role of organic carbon excretion by bulbous rush roots and its turnover and utilization by bacteria under iron plaques in extremely acid sediments. Environ. Exp. Bot. 46:237-245. [Google Scholar]
- 11.Chen, X.-P., Y.-G. Zhu, Y. Xia, J.-P. Shen, and J.-Z. He. 2008. Ammonia-oxidizing archaea: important players in paddy rhizosphere soil? Environ. Microbiol. 10:1978-1987. [DOI] [PubMed] [Google Scholar]
- 12.Christensen, P. B., N. P. Revsbech, and K. Sand-Jensen. 1994. Microsensor analysis of oxygen in the rhizosphere of the aquatic macrophyte Littorella uniflora (L.) Ascherson. Plant Physiol. 105:847-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foesel, B. U., A. Gieseke, C. Schwermer, P. Stief, L. Koch, E. Cytryn, J. R. de la Torré, J. van Rijn, D. Minz, H. L. Drake, and A. Schramm. 2008. Nitrosomonas Nm143-like ammonia oxidizers and Nitrospira marina-like nitrite oxidizers dominate the nitrifier community in a marine aquaculture biofilm. FEMS Microbiol. Ecol. 63:192-204. [DOI] [PubMed] [Google Scholar]
- 14.Francis, C. A., K. J. Roberts, J. M. Beman, A. E. Santoro, and B. B. Oakley. 2005. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. USA 102:14683-14688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Francis, C. A., J. M. Beman, and M. M. M. Kuypers. 2007. New processes and players in the nitrogen cycle: the microbial ecology of anaerobic and archaeal ammonia oxidation. ISME J. 1:19-27. [DOI] [PubMed] [Google Scholar]
- 16.Grasshoff, K., M. Ehrhardt, and K. Kremling. 1983. Methods of seawater analysis, 2nd ed. Verlag Chemie, Weinheim, Germany.
- 17.Hansen, L., G. F. Krog, and M. Søndergaard. 1986. Decomposition of lake phytoplankton. 1. Dynamics of short-term decomposition. Oikos 46:37-44. [Google Scholar]
- 18.Herndl, G. J., T. Reinthaler, E. Teira, H. van Aken, C. Veth, A. Pernthaler, and J. Pernthaler. 2005. Contribution of Archaea to total prokaryotic production in the deep Atlantic Ocean. Appl. Environ. Microbiol. 71:2303-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herrmann, M., A. M. Saunders, and A. Schramm. 2008. Archaea dominate the ammonia-oxidizing community in the rhizosphere of the freshwater macrophyte Littorella uniflora. Appl. Environ. Microbiol. 74:3279-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karjalainen, H., G. Stefansdottir, L. Tuominen, and T. Kairesalo. 2001. Do submersed plants enhance microbial activity in the sediment? Aquat. Bot. 69:1-13. [Google Scholar]
- 21.Klappenbach, J. A., P. R. Saxman, J. R. Cole, and T. M. Schmidt. 2001. rrndb: the Ribosomal RNA Operon Copy Number Database. Nucleic Acid Res. 29:181-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Könneke, M., A. E. Bernhard, J. R. de la Torre, C. B. Walker, J. B. Waterbury, and D. A. Stahl. 2005. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543-546. [DOI] [PubMed] [Google Scholar]
- 23.Koops, H.-P., and A. Pommerening-Röser. 2001. Distribution and ecophysiology of the nitrifying bacteria emphasizing cultured species. FEMS Microbiol. Ecol. 37:1-9. [Google Scholar]
- 24.Koops, H. P., U. Purkhold, A. Pommerening-Röser, G. Timmermann, and M. Wagner. 2003. The lithoautotrophic ammonia-oxidizing bacteria, p. 778-811. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes, 3rd ed., vol. 5. Springer, New York, NY. doi: 10.1007/0-387-30745-1_36. [DOI] [Google Scholar]
- 25.Kowalchuk, G. A., and J. R. Stephen. 2001. Ammonia-oxidizing bacteria: a model for molecular microbial ecology. Annu. Rev. Microbiol. 55:485-529. [DOI] [PubMed] [Google Scholar]
- 26.Kowalchuk, G. A., P. L. E. Bodelier, G. H. J. Heilig, J. R. Stephen, and H. J. Laanbroek. 1998. Community analysis of ammonia-oxidizing bacteria, in relation to oxygen availability in soils and root-oxygenated sediments, using PCR, DGGE and oligonucleotide probe hybridization. FEMS Microbiol. Ecol. 27:339-350. [Google Scholar]
- 27.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, New York, NY.
- 28.Lauridsen, T. L., J. P. Jensen, E. Jeppesen, and M. Søndergaard. 2003. Response of submerged macrophytes in Danish lakes to nutrient loading reductions and biomanipulation. Hydrobiologia 506-509:641-649. [Google Scholar]
- 29.Leininger, S., T. Urich, M. Schloter, L. Schwark, J. Qi, G. W. Nicol, J. I. Prosser, S. C. Schuster, and C. Schleper. 2006. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442:806-809. [DOI] [PubMed] [Google Scholar]
- 30.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Madsen, T. V. 1985. A community of submerged aquatic CAM plants in lake Kalgaard, Denmark. Aquat. Bot. 23:97-108. [Google Scholar]
- 32.Nicol, G. W., and C. Schleper. 2006. Ammonia-oxidising crenarchaeota: important players in the nitrogen cycle? Trends Microbiol. 14:207-212. [DOI] [PubMed] [Google Scholar]
- 33.Nicol, G. W., S. Leininger, C. Schleper, and J. I. Prosser. 2008. The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ. Microbiol. 10:2966-2978. [DOI] [PubMed] [Google Scholar]
- 34.Nicolaisen, M. H., and N. B. Ramsing. 2002. Denaturing gel electrophoresis (DGGE) approaches to study the diversity of ammonia-oxidizing bacteria. J. Microbiol. Methods 50:189-203. [DOI] [PubMed] [Google Scholar]
- 35.Nicolaisen, M. H., N. Risgaard-Petersen, N. P. Revsbech, W. Reichardt, and N. B. Ramsing. 2004. Nitrification-denitrification dynamics and community structure of ammonia-oxidizing bacteria in a high yield irrigated Philippine rice field. FEMS Microbiol. Ecol. 49:359-369. [DOI] [PubMed] [Google Scholar]
- 36.Ottosen, L. D. M., N. Risgaard-Petersen, and L. P. Nielsen. 1999. Direct and indirect measurements of nitrification and denitrification in the rhizosphere of aquatic macrophytes. Aquat. Microb. Ecol. 19:81-91. [Google Scholar]
- 37.Ouverney, C. C., and J. A. Fuhrman. 2000. Marine planktonic Archaea take up amino acids. Appl. Environ. Microbiol. 66:4829-4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prosser, J. I., and G. W. Nicol. 2008. Relative contributions of archaea and bacteria to aerobic ammonia oxidation in the environment. Environ. Microbiol. 10:2931-2941. [DOI] [PubMed] [Google Scholar]
- 39.Riis, T., and K. Sand-Jensen. 1998. Development of vegetation and environmental conditions in an oligotrophic Danish lake over 40 years. Freshw. Biol. 40:123-134. [Google Scholar]
- 40.Risgaard-Petersen, N., and K. Jensen. 1997. Nitrification and denitrification in the rhizosphere of the aquatic macrophyte Lobelia dortmanna L. Limnol. Oceanogr. 42:529-537. [Google Scholar]
- 41.Roelofs, J. G. M., T. E. Brandrud, and A. J. P. Smolders. 1994. Massive expansion of Juncus bulbosus L. after liming of acidified SW Norwegian lakes. Aquat. Bot. 48:187-202. [Google Scholar]
- 42.Schauss, K., A. Focks, S. Leininger, A. Kotzerke, H. Heuer, S. Thiele-Bruhn, S. Sharma, B.-M. Wilke, M. Matthies, K. Smalla, J. C. Munch, W. Amelung, M. Kaupenjohann, M. Schloter, and C. Schleper. 2009. Dynamics and functional relevance of ammonia-oxidizing archaea in two agricultural soils. Environ. Microbiol. 11:446-456. [DOI] [PubMed] [Google Scholar]
- 43.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schloss, P. D., and J. Handelsman. 2006. Introducing SONS, a tool for operational taxonomic unit-based comparisons of microbial community memberships and structures. Appl. Envion. Microbiol. 72:6773-6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schuurkes, J. A. A. R., C. J. Kok, and C. Den Hartog. 1986. Ammonium and nitrate uptake by aquatic plants from poorly buffered and acidified waters. Aquat. Bot. 24:131-146. [Google Scholar]
- 46.Simon, H. M., C. E. Jahn, L T. Bergerud, M. K. Sliwinski, P. J. Weimer, D. K. Willis, and R. M. Goodman. 2005. Cultivation of mesophilic soil crenarchaeotes in enrichment cultures from plant roots. Appl. Environ. Microbiol. 71:4751-4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singleton, D. R., M. A. Furlong, S. L. Rathbun, and W. B. Whitman. 2001. Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl. Environ. Microbiol. 67:4374-4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Søndergaard, M. 1983. Heterotrophic utilization and decomposition of extracellular carbon released by the aquatic angiosperm Littorella uniflora (L.) Aschers. Aquat. Bot. 16:59-73. [Google Scholar]
- 49.Speksnijder, A., G. A. Kowalchuk, K. Roest, and H. J. Laanbroek. 1998. Recovery of a Nitrosomonas-like 16S rDNA sequence group from freshwater habitats. Syst. Appl. Microbiol. 21:321-330. [DOI] [PubMed] [Google Scholar]
- 50.Vestergaard, O., and K. Sand-Jensen. 2000. Alkalinity and trophic state regulate aquatic plant distribution in Danish lakes. Aquat. Bot. 67:85-107. [Google Scholar]
- 51.Wetzel, R. G. 2001. Limnology—lake and river ecosystems. Elsevier Academic Press, San Diego, CA.