Abstract
Polaromonas sp. strain JS666 is the only bacterial isolate capable of using cis-dichloroethene (cDCE) as a sole carbon and energy source. Studies of cDCE degradation in this novel organism are of interest because of potential bioremediation and biocatalysis applications. The primary cellular responses of JS666 to growth on cDCE were explored using proteomics and transcriptomics to identify the genes upregulated by cDCE. Two-dimensional gel electrophoresis revealed upregulation of genes annotated as encoding glutathione S-transferase, cyclohexanone monooxygenase, and haloacid dehalogenase. DNA microarray experiments confirmed the proteomics findings that the genes indicated above were among the most highly upregulated by cDCE. The upregulation of genes with antioxidant functions and the inhibition of cDCE degradation by elevated oxygen levels suggest that cDCE induces an oxidative stress response. Furthermore, the upregulation of a predicted ABC transporter and two sodium/solute symporters suggests that transport is important in cDCE degradation. The omics data were integrated with data from compound-specific isotope analysis (CSIA) and biochemical experiments to develop a hypothesis for cDCE degradation pathways in JS666. The CSIA results indicate that the measured isotope enrichment factors for aerobic cDCE degradation ranged from −17.4 to −22.4‰. Evidence suggests that cDCE degradation via monooxygenase-catalyzed epoxidation (C=C cleavage) may be only a minor degradation pathway under the conditions of these experiments and that the major degradation pathway involves carbon-chloride cleavage as the initial step, a novel mechanism. The results provide a significant step toward elucidation of cDCE degradation pathways and enhanced understanding of cDCE degradation in JS666.
The prevalence of chlorinated solvents, such as tetrachloroethene and trichloroethene, as groundwater contaminants is due largely to their common use as dry-cleaning solvents and metal degreasers and improper disposal (62, 67). Under anaerobic conditions, some organisms are capable of reductive dechlorination of tetrachloroethene to ethene (44, 45), but such organisms are not ubiquitous, and the accumulation of the suspected carcinogen cis-dichloroethene (cDCE) often limits chloroethene degradation in the field (15). Polaromonas sp. strain JS666 is the only isolate capable of growth-coupled, aerobic cDCE oxidation (7). JS666 may be useful for the bioremediation of cDCE-contaminated sites where this common groundwater contaminant has migrated into aerobic zones. Investigations of cDCE degradation in JS666 are also of interest because of the potential for discovery of novel degradation mechanisms that may be useful for biocatalysis.
The cDCE metabolic pathways in JS666 have yet to be elucidated. Genomic analysis of JS666 indicated the presence of many genes potentially involved in cDCE degradation that are distributed among the chromosome and two large plasmids, but it did not reveal the presence of an obvious cDCE degradation operon (43). Mattes et al. suggested that cDCE degradation pathways in JS666 may have evolved recently from horizontal gene transfer because of the presence of numerous transposable elements and duplication of potential degradative genes on the chromosome and plasmids (43). Gene transfer is often important in the evolution of novel catabolic pathways (30), and biodegradative pathways likely assembled from multiple sources have been noted for other xenobiotic compounds (35). A newly evolved cDCE degradation pathway in JS666 assembled by recruitment of degradative enzymes involved in other functions would complicate prediction of cDCE pathways based solely on genomics since the pathway may not be organized into a single operon and degradative genes may be surrounded by genes not involved in degradation.
Coleman et al. established that JS666 cells harbor monooxygenase activity by detection of the corresponding epoxides produced by cells incubated with ethene and propene (7). Their results were consistent with the known mechanism of chloroethene degradation in aerobic bacteria, which is thought to occur primarily via monooxygenase-catalyzed epoxidation (62). However, the gene responsible for the observed monooxygenase activity in JS666 is not known. Epoxides are highly reactive compounds that cause damage to cells by forming adducts with macromolecules (65). Many organisms utilize epoxide-transforming enzymes, including glutathione S-transferases (GSTs) (63, 64), epoxide hydrolases (66), epoxide dehydrogenases (21), and epoxide carboxylases (1), to degrade and detoxify the epoxides.
The GSTs are a diverse family of proteins that can catalyze the conjugation of glutathione (GSH) to a number of different xenobiotic substrates (68). Mammalian GSTs have been extensively characterized, although much less is known about their bacterial counterparts (3, 22). In mammalian systems, there are two mechanisms by which GSH conjugation of haloalkenes can occur: (i) addition and elimination, in which the sulfur nucleophile from GSH is added to the haloalkene, resulting in removal of the halide, and (ii) addition reactions in which the sulfur nucleophile from GSH is added to the double bond of the haloalkene (3). Most of the few bacterial GSTs for which a function is known appear to be required for growth of organisms on xenobiotic compounds (69). Some GSTs function as dehalogenases, as demonstrated by the GSH-dependent enzyme dichloromethane dehalogenase, which catalyzes the conversion of dichloromethane to formaldehyde and inorganic chloride in Methylobacterium dichloromethanicum DM4 (formerly Hyphomicrobium sp. strain DM2) (38).
In this study, we integrated proteomic and transcriptomic data to reveal genes upregulated by growth on cDCE in JS666. We correlated the omics data with data from compound-specific isotope analysis (CSIA), enzyme assays, and metabolite analysis to delineate the primary cellular responses of JS666 to cDCE and formulate a hypothesis for cDCE degradation pathways.
MATERIALS AND METHODS
Chemicals and media.
cDCE (>99% pure), stabilized with hydroquinone monomethyl ether (MEHQ), was obtained from TCI America. JS666 was grown on minimal salts medium (MSM) as described by Coleman et al. (7), and 1/4× Trypticase soy agar plates were routinely used to evaluate culture purity.
Culture techniques.
Cultures of JS666 were maintained in 160-ml serum bottles fitted with Teflon-coated butyl rubber stoppers and containing 100 ml of MSM and 60 ml of air. cDCE was added to cultures in 4-μl increments to obtain a nominal concentration of 0.52 mM. The actual aqueous concentration was 0.48 mM when partitioning between phases was considered. cDCE starvation and cDCE concentrations above 1 mM were avoided because of increased lag phases and inhibition of degradation, respectively. After the degradation of two to four additions of cDCE, 5 ml of culture was transferred to fresh medium. Cultures were incubated at room temperature (22°C) in the dark with constant shaking (150 rpm).
Cells used for omics experiments were prepared from cultures of JS666 grown on MSM supplemented with acetate, glycolate, or cDCE. cDCE-grown cultures were used as inocula for all cultures, but cultures were maintained on glycolate or acetate as the sole carbon source (10 mM) for multiple transfers to ensure that no residual cDCE remained. Cells were harvested from cultures in mid-exponential phase by centrifugation at 4°C. cDCE cultures were harvested after the degradation of 8 μl of cDCE to minimize the concentration of MEHQ preservative at the time of harvest. Cell pellets containing 800 to 1,600 μg of protein were stored at −70°C until they were used to preserve proteins and RNA for omics experiments.
Analytical methods.
cDCE was analyzed by injection of headspace samples (100 μl) into a gas chromatograph equipped with a flame ionization detector and a 1% SP-1000 column (60/80 Carbopack B; Supelco) that was operated isothermally at 175°C. cDCE was quantified (μmol/bottle) using standards prepared in water with the same liquid-to-headspace gas ratio as experimental bottles.
To enable calculation of optimal protein loading for two-dimensional (2D) gels, the protein concentration was quantified using a modified Lowry protein assay (Pierce Biotechnology, Inc.) according to the manufacturer's instructions.
Proteomics.
2D gel electrophoresis (2D-GE) was conducted essentially as described by Sauer and Camper (50), using the principles described by Gorg et al. (18). For preparation of crude protein extracts, frozen cell pellets (800 μg) were suspended in 1 ml of Tris-EDTA buffer (10 mM Tris-Cl, 1 mM EDTA [pH 8.0]) containing 0.5 mg of phenylmethylsulfonyl fluoride and 0.67 mg of sodium azide and disrupted by sonication (six times for 2 s, 20 Hz, 4°C) in a Tekmar sonic disruptor. Proteins were precipitated from crude extracts with 100 μl of a trichloroacetic acid-acetone mixture (10%) and resolubilized in 700 μl of buffer containing urea, thiourea, dithiothreitol, 3-[(3-cholamidopropyl)-dimethylammonio]propanesulfonate (CHAPS), and ampholytes (Amersham 3-10 NL IPG buffer or 3-10 Pharmalytes). The soluble protein solution (350 μl) was applied to 18-cm Immobiline 3-10 NL or 3-11 NL IPG strips by in-gel rehydration. Isoelectric focusing at 17°C was used to separate the proteins in the first dimension. After isoelectric focusing, proteins were equilibrated and separated in the second dimension by gel electrophoresis using a Hoefer Dalt vertical system with 12% acrylamide gels at 17°C. 2D gels were prepared in triplicate. Gels were stained with Coomassie brilliant blue, which was followed by destaining in a 30% ethanol-10% acetic acid mixture. Gels were imaged with an Epson Expression 1680 scanner equipped with a transparency adaptor. The molecular weights of proteins in the gels were estimated based on comparison of the vertical distances migrated to the vertical distances that Precision Plus unstained protein standards (Bio-Rad) migrated. The isoelectric point was estimated from the horizontal distances that the spots migrated in the gel compared to pH gradients in the IPG strips supplied by the manufacturer.
Twenty-eight proteins from proteomic experiments performed with acetate or glycolate as the reference substrate were digested with trypsin and analyzed using liquid chromatography coupled to tandem mass spectrometry (LC/MS-MS) and/or matrix-assisted laser desorption ionization-tandem time of flight MS (MALDI-TOF/TOF MS). LC/MS-MS analyses were conducted at the Mass Spectrometry Laboratory at Montana State University or at the Mass Spectrometry Core at Cornell University. At Montana State University, peptides were separated on a microfluidic ChipCube interface and detected with an ESI-Trap XCT Ultra instrument (Agilent). At Cornell, peptides were separated by a LC Packings Ultimate integrated capillary high-performance liquid chromatography (HPLC) system (Dionex) and detected with a hybrid triple quadrupole linear ion trap mass spectrometer (4000 Q Trap; Applied Biosystems). MALDI-TOF/TOF MS was conducted at the Mass Spectrometry Core at the University of Texas Medical Branch with a 4800 MALDI-TOF/TOF proteomic analyzer (Applied Biosystems). The MASCOT search engine was used to compare peptide masses determined by MS to masses of sequences in the NCBInr bacterial database. Acceptable protein identifications required expectation values of 0.01 and 0.001 for LC-MS/MS and MALDI-TOF/TOF MS, respectively. The significance of a protein match was further verified by comparing the molecular weight or isoelectric point based on the distance that the protein spot migrated in the gel to values in the database.
Transcriptomics.
Total RNA was isolated from frozen cell pellets using a hot phenol-chloroform extraction method similar to that described by Cury and Koo (9). During RNA extraction and purification, RNaseOUT (Invitrogen) was added to samples to prevent RNA degradation. The extracted RNA was precipitated overnight in ethanol, which was followed by removal of contaminating DNA using a Turbo DNA-free DNA kit (Ambion). RNA was enriched in mRNA using an RNeasy mini clean-up kit (Qiagen). Enriched RNA was further purified by a second ethanol precipitation. cDNA was synthesized from the total RNA using an Invitrogen SuperScript double-stranded cDNA synthesis kit according to NimbleGen's instructions (47). The purity and quantity of the RNA and cDNA were confirmed using a NanoDrop ND-1000 spectrophotometer. The quality of the RNA and cDNA was verified using a FlashGel system (Cambrex). All hybridizations and labeling were performed by NimbleGen Systems Inc. as described by Ulijasz et al. (60).
Custom, whole-genome expression arrays for Polaromonas sp. strain JS666 were synthesized by NimbleGen Systems, Inc. Arrays were designed to obtain full coverage of open reading frames (ORFs) and a 50-bp tiling of the intergenic regions. Frequency filtering of the intergenic regions was carried out to remove low-complexity regions and repeats. Long oligonucleotides (60-mers) were synthesized on ultra-high-density, one-plex arrays containing 385,000 features. The arrays contained five replicate probes for each of the target sequences.
Microarray data were analyzed with the Genespring software (Agilent Technologies) at the Genomics Core at Montana State University. Data were normalized with a robust multichip analysis scheme with per-chip and per-gene median polishing. Samples were filtered to identify differential expression (>1.5-fold change on cDCE compared to glycolate). A threshold of 100 for the baseline raw signal intensity in at least one replicate transcript was required to reduce noise due to low expression. The significance of the differential expression was evaluated using a one-way analysis of variance Welch t test with a Benjamini-Hochberg multiple-test correction with a false discovery rate (FDR) of 5% of the genes identified (P < 0.05). Hierarchical clustering of transcripts using a standard correlation similarity measure was used to group triplicate samples (containing technical and biological replicates) and transcripts based on the similarity of gene expression patterns. Biological replicates were obtained from separate cultures, and technical replicates were extracted from aliquots of the same culture and processed separately.
Enzyme assays.
GST activity was assayed using 1-chloro-2,4-dinitrobenzene (CDNB) and cDCE as substrates. GST activity with CDNB was determined spectrophotometrically at 340 nm by conjugation of reduced GSH to CDNB as described by Habig and Jakoby (19). The standard reaction mixture (1 ml) contained 50 mM potassium phosphate buffer (pH 8.5), 5 mM GSH, 1.0 mM CDNB, and 2.5 to 25 μg of protein from cell extract. GST activity with cDCE as a substrate was assayed by monitoring the disappearance of cDCE in the headspace by using gas chromatography. The reaction mixture (1 ml) contained 50 mM phosphate buffer at pH 6.5, 7.5, or 8.5, 5 mM GSH, 18 mM cDCE, and 100 to 200 μg protein from the cell extract. Reactions were performed in duplicate, and cDCE was monitored at time zero and after 3 h. Other enzymes, including pyridoxamine 5-phosphate oxidase (PNPox) (72), haloacid dehalogenase (HAD) (31), chloroacetaldehyde dehydrogenase (CAD) (10), and cyclohexanone monooxygenase (CMO) (20), were assayed using previously described methods.
Metabolic profiling.
Cell extracts for metabolite detection were prepared by suspension of frozen (−70°C) cell pellets containing ∼1,600 μg of protein in 500 μl of water, followed by sonication to disrupt cells and then filtration through 0.2-μm cellulose membrane filters (Spartan 13; Whatman). An HPLC-MS method was developed for detection of GSH based on a method described by Simmonds et al. (55). Metabolites were separated with an Agilent series 1100 HPLC equipped with a Supelcosil LC-8 guard column and an Intersil ODS-3 column (C18; 250 nm by 4.6 nm; particle size, 5 μm). The buffer system consisted of 0.1% formic acid in water (buffer A) and 0.1% formic acid in methanol (buffer B). The initial mobile phase was 1% buffer B for 4 min, and this was followed by ramping up to 25% buffer B by 27 min, ramping down to 1% buffer B in 1 min, and equilibration with 1% buffer B for 4 min. A flow rate of 0.4 ml/min was used for the column.
Positive-mode electrospray ionization was performed using a series 6300 Agilent SL ion trap mass spectrometer for detection of compounds with mass-to-charge ratios (m/z) ranging from 50 to 700. The MS and flow conditions were optimized for detection of GSH standards (m/z 308) prepared in water. GSH was detected with an MS detector at a retention time of 14 min.
CSIA.
Cultures used for isotope analysis were prepared in triplicate (biological replicates) by adding ∼30 μmol of cDCE to 100 ml of MSM in 260-ml bottles fitted with mininert screw caps. After the degradation of cDCE, additional cDCE was added in 30-μmol increments. The third addition of cDCE occurred after a starvation period of 9 days to assess the effects of cDCE starvation on isotope enrichment factors. cDCE concentrations and isotope values for cultures and controls (nonbiological and killed) were determined in triplicate (technical replicates) throughout the course of degradation. CSIA was performed as described previously by Chartrand et al. (5). The total analytical uncertainty, including both accuracy and reproducibility, is ±0.5‰ (52).
CSIA measures the ratio of heavy elements to light elements (i.e., 13C/12C) in a sample compared to that in a standard (V-PDB in the case of carbon). The results are expressed as delta values in units (‰) per mille obtained using the following equation:
 |
(1) |
where Rsample and Rstd are the 13C/12C ratios for the sample and the standard, respectively. Stable isotope fractionation (i.e., a change in the 13C/12C ratio) during biodegradation occurs due to differences in the reaction rates and activation energies of heavy and light atoms present at a reacting bond, with light-isotope bonds in general reacting more quickly (16, 46). The degree of fractionation depends strongly on the type of bond being cleaved and, therefore, the mechanism of the initial step in degradation (5, 11, 24, 25). During biodegradation of many chlorinated ethenes and chlorinated ethanes, it has been found that the preferential breakage of the bond containing the lighter isotope (resulting in enrichment of the heavy isotope in the remaining substrate) can be described by the Rayleigh equation (4, 25, 27, 53, 57):
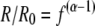 |
(2) |
where R is the isotope composition (13C/12C) of a substrate at any given time, R0 is the initial isotope composition of the substrate, f is the fraction of the substrate remaining, and α is the fractionation factor, which is a measure of the difference in the reaction rates of heavy- and light-isotope molecules and is a constant value throughout the conversion (42). The fractionation factor is determined by plotting ln f versus ln (R/R0) and determining the slope (m) of the linear regression, where m = (α − 1). The fractionation can also be described by the isotope enrichment factor (ɛ), where:
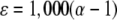 |
(3) |
Microarray accession numbers.
Microarray data have been deposited in the Gene Expression Omnibus database (www.ncbi.hlm.nih.gov/projects/geo) under the following accession numbers: normalized data, GSE15204; platform, GPL8287; and raw data files, GSM379684 to GSM379689.
RESULTS AND DISCUSSION
cDCE induction experiments for selection of reference substrates.
For differential expression studies, it is important that reference substrates for proteomics and transcriptomics do not induce cDCE degradation. Ethanol, acetate, and glycolate were screened as potential reference substrates. Cells grown on ethanol that were washed and suspended in medium containing cDCE as the sole carbon source began degrading cDCE rapidly, similar to controls that began consuming ethanol and oxygen rapidly after resuspension (Fig. 1A and B). We interpreted this to mean that ethanol may induce cDCE degradation and is not an appropriate reference substrate for 2D-GE. In contrast, cells grown on acetate that were washed and suspended in medium containing cDCE exhibited a lag phase of 25 to 30 days before cDCE degradation began, unlike controls that began consuming acetate and oxygen rapidly after resuspension (Fig. 1C and D). Glycolate-grown cells treated similarly exhibited a lag phase of 4 to 14 days before cDCE degradation began (data not shown). The reason for the extended lag phase before cDCE degradation by cells grown on acetate or glycolate is not known, but we speculate that it may be associated with issues of gene regulation or with solvent toxicity caused by transferring cells from a preferred substrate to a solvent, such as cDCE. The results indicate that acetate and glycolate do not induce cDCE degradation and are appropriate reference substrates for 2D-GE. Acetate was used as a reference substrate for the initial 2D-GE experiments.
FIG. 1.
Effects of growth substrate on subsequent cDCE degradation. Ethanol-grown cultures were washed and suspended in medium containing cDCE (A) or ethanol (B), and acetate-grown cultures were washed and suspended in medium containing cDCE (C) or acetate (D). Ethanol and acetate concentrations were monitored indirectly by determining the decrease in the partial pressure of oxygen (pO2). All cultures had approximately the same initial level of biomass (i.e., an optical density at 600 nm of 0.05). The data are the results of duplicate experiments (filled and open symbols).
Proteins upregulated by cDCE.
2D-GE was used to identify proteins upregulated by cDCE compared to the results obtained with the reference substrate acetate. Fifteen differentially expressed proteins extracted from a cDCE-grown culture were excised and used for subsequent MS analysis using LC/MS-MS (Fig. 2A). The results indicated that cDCE upregulated proteins associated with the glyoxylate cycle, including the putative glyoxylate carboligase (Bpro4561), 2-hydroxy-3-oxopropionate reductase (Bpro4563), and hydroxypyruvate isomerase (Bpro4562) (Table 1). Other interesting upregulated proteins include a GST (Bpro0645), two HADs (Bpro5186 and Bpro0530), and a PNPox (Bpro0646). The upregulated HAD genes are paralogs whose products share 99% amino acid identity.
FIG. 2.
Overlays of 2D gel images comparing proteins extracted from cDCE-grown (blue) cells to proteins extracted from acetate-grown (red) cells (A) and proteins extracted from cDCE-grown (blue) cells to proteins extracted from glycolate-grown (red) cells (B). Differentially expressed spots in cDCE gels that were subsequently analyzed by MS are circled.
TABLE 1.
Proteins upregulated in cells grown on cDCE compared to cells grown on acetatea
| Sample | Locus tag | Putative enzyme functionb | Protein scorec | Peptide countd |
|---|---|---|---|---|
| 1 | Bpro0645 | GST-like protein | 115 | 1 |
| 2 | Bpro0645 | GST-like protein | 181 | 1 |
| 2 | Bpro3647 | 3-Oxoacyl-(acyl carrier protein) reductase | 143 | 1 |
| 3 | Bpro4563 | 2-Hydroxy-3-oxopropionate reductase | 547 | 4 |
| 4 | Bpro4562 | Hydroxypyruvate isomerase | 435 | 5 |
| 4 | Bpro2567 | Short-chain dehydrogenase/reductase | 144 | 2 |
| 4 | Bpro4841 | Peroxidase | 126 | 1 |
| 5 | Bpro5186 | HAD, type II | 110 | 1 |
| 6 | Bpro0646 | PNPox-related protein | 85 | 1 |
| 10 | Bpro0650 | DEAD/DEAH box helicase-like protein | 256 | 2 |
| 12 | Bpro4561 | Glyoxylate carboligase | 614 | 6 |
| 13 | Bpro4561 | Glyoxylate carboligase | 564 | 4 |
There were no significant JS666 hits for samples 7 to 9, 11, 14, and 15.
Proteins upregulated by both reference substrates are indicated by bold type. Annotations were derived from the DOE Joint Genome Institute Integrated Microbial Genomes database.
The protein score (or Mowse score) is −10 × log (P), where P is the probability that the observed match is a random event.
Number of peptides with an expectation value of <0.01 as identified using LC-MS/MS. The expectation value corresponds to the number of matches that are expected to occur by chance alone.
Glycolate, which feeds directly into the glyoxylate cycle, was selected as a reference substrate in the next 2D gel experiment to avoid differential expression of well-characterized glyoxylate cycle proteins. Thirteen differentially expressed proteins from a cDCE-grown culture were analyzed using LC/MS-MS and/or MALDI-TOF/TOF MS (Fig. 2B). Identification of proteins, including a GST, HAD, and PNPox, as significantly upregulated in cDCE-grown cells confirmed previous 2D-GE findings (Table 2). In addition to the previously detected proteins, a putative CMO was upregulated by cDCE. It is important to note that although MALDI-TOF/TOF MS provides better mass resolution than LC/MS-MS, it is not effective for identifying multiple proteins in one spot because of the lack of a chromatography step to separate proteins prior to analysis.
TABLE 2.
Proteins upregulated in cells grown on cDCE compared to cells grown on glycolate
| Sample | Locus tag | Putative enzyme functiona | Protein scoreb | Peptide countc |
|---|---|---|---|---|
| 1 | Bpro0645 | GST-like protein | 325 | 8 |
| 2 | Bpro2715 | 2-C-methyl-d-erythritol 2,4-cyclodiphosphate synthase | 109 | 2 |
| 3 | Bpro5186 | HAD, type II (plasmid 1) | 663 | 11 |
| 3 | Bpro0530 | HAD, type II (chromosome) | 638 | 9 |
| 4 | Bpro5186 | HAD, type II (plasmid 1) | 582 | 11 |
| 4 | Bpro0530 | HAD, type II (chromosome) | 534 | 9 |
| 5 | Bpro5186 | HAD, type II (plasmid 1) | 721 | 14 |
| 5 | Bpro0530 | HAD, type II (chromosome) | 559 | 11 |
| 6 | Bpro0646 | PNPox-related protein | 280 | 7 |
| 7 | Bpro2322 | 2-Isopropylmalate synthase | 146 | 2 |
| 8 | Bpro5565 | CMO | 186 | 3 |
| 8 | Bpro2322 | 2-Isopropylmalate synthase | 123 | 2 |
| 8 | Bpro0759 | Chaperonin GroEL | 70 | 1 |
| 9 | Bpro2029 | Trigger factor | 404 | 9 |
| 9 | Bpro3157 | Protein of unknown function DUF404 | 265 | 5 |
| 9 | Bpro5565 | CMO | 70 | 1 |
| 9 | Bpro1807 | Glutamine synthetase, type I | 68 | 1 |
| 11 | Bpro0646 | PNPox-related protein | 207 | 6 |
| 13 DCEd | Bpro4562 | Hydroxypyruvate isomerase | 790 | 16 |
| 13 GLYd | Bpro4562 | Hydroxypyruvate isomerase | 722 | 16 |
Proteins upregulated by both reference substrates are indicated by bold type. Annotations were derived from the DOE Joint Genome Institute Integrated Microbial Genomes database.
Samples 2 and 7 to 9 were identified using LC-MS/MS. All other samples were identified using MALDI-TOF/TOF MS. Spot 10 had 12 protein matches, which are not listed.
Number of significant peptides with an expectation value of <0.001 for MALDI-TOF/TOF MS samples and an expectation value of <0.01 for LC-MS/MS samples.
All proteins were from 2D gel spots that were present only in cDCE gels, with the exception of samples 13 DCE and 13 GLY, which were highly upregulated in the cDCE gel.
Genome-wide transcriptional response to cDCE.
Transcriptomic experiments using DNA microarrays were carried out to complement proteomic experiments. We identified 856 transcripts that were differentially expressed at least 1.5-fold with an FDR of 5% for cDCE-grown cells and glycolate-grown cells. The transcriptional profile indicated that 140 ORFs and 77 regions annotated as intergenic regions were upregulated by cDCE at least 1.5-fold (FDR, 5%). Transcriptomic analysis indicated that genes annotated as encoding GSH, HAD, CMO, and PNPox were among the most highly upregulated transcripts, confirming the proteomic findings (Table 3). Other interesting transcripts upregulated by cDCE were annotated as transcripts encoding (i) transport-related proteins, including two sodium/solute symporters and ABC transporter components, (ii) a carbon monoxide dehydrogenase (CO DHase), and (iii) an α/β-hydrolase fold (Hlase), whose gene is located in the putative CMO operon. The putative Hlase was upregulated 7.3-fold, but it did not meet the 5% FDR significance threshold with a P value of 0.056.
TABLE 3.
Selected transcripts upregulated in cells grown on cDCE compared to cells grown on glycolate
| Sample | Locus taga | P valueb | Change (fold)c | Region or putative enzyme function |
|---|---|---|---|---|
| 1 | ctg94:3530095-ctg94:3530164 | 0.015 | 122.9 | Intergenic region |
| 2 | Bpro3336 | 0.005 | 111.0 | ABC transporter, extracellular ligand-binding receptor |
| 3 | Bpro0645d | 0.012 | 99.8 | GST-like protein |
| 4 | Bpro0646 | 0.012 | 87.5 | PNPox-related protein |
| 5 | Bpro3335 | 0.009 | 70.0 | ABC transporter, inner membrane translocator |
| 6 | Bpro0530 | 0.011 | 53.3 | HAD, type II |
| 7 | Bpro5186 | 0.013 | 51.5 | HAD, type II |
| 8 | Bpro0532 | 0.007 | 42.7 | Hypothetical protein |
| 9 | ctg94:3611882-ctg94:3611943 | 0.011 | 41.7 | Intergenic region |
| 10 | Bpro0531 | 0.015 | 40.9 | Sodium/solute symporter |
| 11 | Bpro5185 | 0.017 | 39.2 | Sodium/solute symporter |
| 12 | Bpro3334 | 0.010 | 30.8 | ABC transporter, inner membrane translocator |
| 13 | Bpro3333 | 0.013 | 27.1 | ABC transporter, ATPase component |
| 14 | Bpro0529 | 0.013 | 24.3 | Conserved hypothetical protein |
| 15 | ctg94:581640-ctg94:581753 | 0.033 | 21.7 | Intergenic region |
| 16 | ctg94:2876701-ctg94:2876845 | 0.011 | 20.8 | Intergenic region |
| 17 | ctg94:529265-ctg94:529416 | 0.014 | 20.0 | Intergenic region |
| 18 | Bpro5184 | 0.004 | 19.0 | Predicted membrane protein |
| 19 | Bpro3332 | 0.005 | 18.3 | ABC transporter, ATPase component |
| 20 | Bpro2731 | 0.016 | 17.9 | Conserved hypothetical protein |
| 23 | Bpro2396 | 0.017 | 14.8 | Heme peroxidase |
| 30 | Bpro0577 | 0.025 | 10.4 | CO DHase |
| 31 | Bpro5565 | 0.024 | 10.1 | CMO |
| 40 | Bpro5566 | 0.056 | 7.3 | Hlase |
| 44 | Bpro5564 | 0.010 | 6.3 | Alcohol dehydrogenase |
| 75 | Bpro3866 | 0.027 | 3.6 | Universal stress protein (UspA) |
| 81 | Bpro5301 | 0.010 | 3.5 | Cytochrome P450 |
| 83 | Bpro2732 | 0.018 | 3.4 | Transposase |
| 92 | Bpro4792 | 0.030 | 3.2 | Transposase |
| 95 | Bpro4575 | 0.024 | 3.1 | Transposase |
| 100 | Bpro3227 | 0.040 | 3.0 | Universal stress protein (UspA) |
Intergenic regions, which do not have locus tags, are indicated by their positions in the contig.
P values are based on a one-way analysis of variance Welch t test with Benjamini-Hochberg multiple-test correction and an FDR of 5% for the genes identified.
Genes upregulated at least 1.5-fold in cDCE-grown cells compared to glycolate-grown cells with an FDR of 5% (P < 0.05). The P value for Bpro5566 was 0.056.
Genes and the corresponding proteins that were identified as upregulated by growth on cDCE in microarray and proteomic experiments are indicated by bold type.
Transcripts significantly upregulated by cDCE were distributed in 20 cluster of orthologous group (COG) functional categories (Fig. 3). Aside from the genes for general function prediction (31 genes) and genes with unknown functions (16 genes), the most abundant genes upregulated by cDCE were associated with amino acid transport and metabolism (26 genes). An enrichment analysis using Fisher's exact test revealed that four of the COGs were significantly overrepresented (P < 0.10) in the set of genes upregulated by cDCE, including COGs for (i) cell wall, membrane, and envelope biogenesis, (ii) intracellular trafficking, secretion, and vesicular transport, (iii) transcription, and (iv) translation and ribosomal structure and biogenesis. The results indicate that the functions of many genes remain to be determined and that intracellular transport and cell membrane biogenesis may be important in cDCE degradation in JS666. The enrichment of membrane biogenesis genes among the genes upregulated by cDCE is interesting given that the toxicity of solvents such as cDCE to organisms is hypothesized to occur by permeabilization of cell membranes (29, 54). One adaptive mechanism that some organisms utilize to counteract the debilitating effects of solvent exposure includes biogenesis to repair and rebuild damaged membranes (29, 54).
FIG. 3.
Distribution of genes upregulated by cDCE based on COG functional categories. Statistically enriched COGs (P < 0.10, Fisher's exact test) are indicated by light gray bars.
In a hierarchical clustering of transcripts, the upregulated GST and CMO transcripts grouped separately (Fig. 4). The levels of expression of GST were similar to those of the upregulated HAD and PNPox enzymes. The transcripts whose expression was similar to that of the upregulated CMO transcript included the transcript of a putative upregulated CO DHase. Genes with similar expression levels (but not necessarily up- or downregulation) are sometimes involved in related functions or metabolic pathways or are coregulated (26).
FIG. 4.
Hierarchical clustering of transcripts. The rows represent selected transcripts that were upregulated by growth on cDCE (>1.5-fold change with an FDR of 5%). The columns represent individual microarrays, and the samples in columns 1 and 3 are technical replicates that were extracted separately from aliquots of the same biological culture. The samples in columns 1 and 2 and the samples in columns 2 and 3 are biological replicate pairs. Darker gray indicates high levels of expression, while lighter gray indicates low levels. Clusters of genes with expression similar to that of the GST or CMO gene are indicated by a gray background. GLY, glycolate.
It is interesting that over 35% of the 217 upregulated transcripts were in intergenic regions. In fact, the most highly upregulated transcript (123-fold change) determined using microarrays was annotated as an intergenic region in the predicted ABC transporter operon. The upregulated transcripts could represent noncoding RNAs that may be involved in gene regulation (51).
The transcriptomic experiments revealed upregulation of 14 genes in the COG functional category for posttranslational modification, protein turnover, and chaperones. 2D gels revealed the presence of spot chains (i.e., multiple spots with slightly different isoelectric points) for several upregulated proteins, including GST, HAD, PNPox, and CMO, suggesting that the enzymes were likely posttranslationally modified (70). Collectively, the results indicate that posttranslational modifications of proteins may be an important factor in cDCE degradation in JS666.
Numerous transcripts, including 450 ORFs and 189 intergenic regions, were downregulated at least 1.5-fold (FDR, 5%) by cDCE compared to the results with glycolate. Many of these downregulated genes, such as genes encoding ATPases and oxidases, are predicted to be involved in energy production and conversion (65 genes; P = 0.05, Fisher's exact test). The slower growth on cDCE than on glycolate could explain the downregulation of energy generation genes.
Confirmation of enzyme function using biochemical experiments.
The results of transcriptomic and proteomic analyses presented here provide the basis for future work to formulate and test hypotheses by using enzyme assays, knockouts, and heterologous expression. Such experiments are beyond the scope of the present work, but it is relevant to mention that preliminary enzyme assays have failed to reveal the mechanism of the initial attack on cDCE (R. B. Payne and J. C. Spain, unpublished data). HAD (31), PNPox (72), and GST (19) enzyme activities seem to be induced during growth on cDCE, whereas CAD activity (10) seems to be constitutive. HAD catalyzes the conversion of chloroacetic acid to glycolate and the conversion of dichloroacetic acid to chloroglycolate. Other researchers confirmed that when Bpro0530 was expressed in Escherichia coli and purified, it was active with chloroacetic acid (71). Adjacent to the upregulated GST gene on the genome is a putative gene encoding PNPox, which has been implicated in the conversion of vitamin B6 to its active form (72). The addition of pyridoxamine HCl (final concentration, 100 μl/liter), one of the forms of vitamin B6, did not improve the cDCE degradation activity in JS666. GST enzyme activity was detected when Bpro0645 was overexpressed in E. coli by measuring the conjugation of GSH to the substrate CDNB (19); however, attempts to detect CMO (20) or GST activity with cDCE as a substrate were unsuccessful (R. B. Payne and J. C. Spain, unpublished data). The failure to detect enzyme activities could reflect inappropriate assay conditions, especially since monooxygenase activity was present in whole cells, as indicated by detection of epoxides produced from ethene and propene (7). CAD catalyzes the conversion of chloroacetaldehyde to chloroacetic acid (61). It is unclear which gene on the long list of constitutively expressed dehydrogenase genes was responsible for the observed CAD enzyme activity in JS666.
The fact that GST activity was detected in cell extracts led us to assay for the presence of GSH in cell extracts. HPLC-MS analysis revealed that GSH was present in extracts prepared from cDCE-grown cells (data not shown). GSH was also detected in extracts prepared from glycolate-grown cells at slightly lower levels, although the difference was not statistically significant (P = 0.087, unpaired t test). GSH, which is present at concentrations up to 10 mM in many cells, has multiple cellular roles as a cofactor for biochemical reactions and a primary defense against oxidative stress. The results indicate that GSH, a necessary cofactor for the upregulated GST, is present in JS666 cells.
cDCE upregulates enzymes with antioxidant functions, and degradation is inhibited by elevated oxygen levels.
Transcriptomic analysis revealed that cDCE caused upregulation of genes predicted to be involved in (i) a general stress response, including universal stress proteins and transposases, which can be induced by stress, and (ii) an oxidative stress response, including a GST, PNPox, and peroxidase. Oxidative stress can be caused by the production of reactive intermediates or reactive oxygen species (ROS), which are continuously produced during aerobic respiration (22). In other organisms, it has been shown that GST and its cofactor reduced GSH can function in a coordinated adaptive response to oxidative stress since the expression of GSTs was shown to be upregulated by exposure to prooxidants (22) and strains with knockouts of GST-like proteins were more susceptible to hydrogen peroxide than the parent strains (36). Reduced GSH is known to play a central role as an intracellular antioxidant that scavenges free radicals and reduces hydrogen peroxide (23). The results suggest that in JS666 GST and GSH may help alleviate the effects of oxidative stress produced by growth on cDCE.
PNPox is the primary enzyme involved in the conversion of vitamin B6 to the biologically active form (48). PNPox enzyme activity was detected in extracts of cDCE-grown cells. Vitamin B6 can alleviate the effects of oxidative stress by scavenging ROS (2, 37, 49, 58). Vitamin B6 is also involved in maintaining GSH levels in the cell since the synthesis of GSH depends on the availability of cysteine, which is synthesized by vitamin B6-dependent enzymes (58). While further work is needed to elucidate the function of the PNPox in cDCE degradation in JS666, this enzyme appears to be linked with GST in JS666 and other organisms. Peroxidases are known to participate in an oxidative stress response by eliminating hydrogen peroxide (8, 39). A putative peroxidase gene was among the genes upregulated by cDCE, but enzyme activity was not confirmed in JS666. The results suggest that the upregulation of the PNPox and peroxidase in JS666 may be an adaptive response to oxidative stress.
JS666 cultures that were challenged with higher concentrations of oxygen (partial pressures of oxygen, 0.34 and 0.77 atm) degraded less cDCE than cultures exposed to ambient or subambient levels (33). A high oxygen level did not appear to inhibit glycolate or succinate degradation. These results, along with those described above, suggest that growth on cDCE generates an oxidative stress response causing upregulation of enzymes with antioxidant functions. The inhibition is not likely due to ROS that are continually produced during aerobic respiration since enzymes associated with oxidative stress were upregulated by growth on cDCE, but it might be due to reactive intermediates, such as GSH conjugates or epoxides, that cause increased damage to cellular constituents in the presence of high oxygen levels (22, 65). Although it is possible that the MEHQ used as a cDCE preservative was responsible for some of the observed differential expression, this is unlikely due to the low concentrations. Leelakrinangsak et al. observed that effects were elicited by 2-methylhydroquinone (41), but the concentrations were 7 orders of magnitude higher than the concentration of MEHQ used here.
CSIA.
CSIA was conducted to investigate the mechanism of the initial step in cDCE oxidation in JS666. The measured isotope enrichment factors for aerobic cDCE degradation by JS666 ranged from −17.4 to −22.4‰. The data used to calculate enrichment factors fit a Rayleigh model with coefficients of determination for the Rayleigh linear regression (R2) of >0.97, suggesting that fractionation due to cDCE assimilation by JS666 is due to a single rate-limiting step. Using the approach outlined by Elsner et al. (13), measured enrichment factors calculated by a Rayleigh regression using CSIA data can be converted into kinetic isotope effects (KIE) (12k/13k). In the case of molecules that have only one carbon atom, kinetic isotope effects can be directly related to isotope enrichment factors by the following equation:
 |
(4) |
where ɛ is the experimentally determined isotope enrichment factor (in ‰) and 12k/13k is the primary kinetic isotope effect of the reaction. cDCE molecules contain two carbon atoms, and because of the low natural abundance of 13C, generally only one of them at most is 13C. Since the reaction occurs only at one of the two carbon atoms, there is competition between the 13C position and the 12C position in a single cDCE molecule. To correct for this competition, a factor of 2 must be introduced into equation 4:
 |
(5) |
This correction is described in standard textbooks (46), and these principles have been applied in many recent studies in order to identify specific reaction mechanisms that function during degradation of organic contaminants.
The large fractionation values associated with aerobic cDCE degradation in JS666 (−17.4 to −22.4‰) correspond to KIE values of 1.03 to 1.04. These KIE values are consistent both with theoretical predictions for C-Cl bond cleavage (17) and with the results of previous experiments investigating C-Cl bond cleavage for chlorinated ethenes (4, 28, 40, 57) and ethanes (12, 25). In contrast, previous studies describing fractionation associated with aerobic biodegradation of vinyl chloride (VC) via epoxidation (5, 6) reported fractionation values in the range from −8.2‰ ± 0.1‰ to −7.0‰ ± 0.3‰ for aerobic VC assimilation. These values correspond to KIE values of 1.007 to 1.008, which are very close to the expected value for epoxidation (1.011) (56). For cDCE, the KIE values in this study differ significantly from previously reported values for cometabolic oxidation in studies that reported nonresolvable fractionation (6) or very little fractionation (−7.1 to −9.8‰) (59). The latter values correspond to KIE values of 1.007 to 1.010, again consistent with fractionation observed during biodegradation of VC via epoxidation and confirmed by abiotic experiments (56).
While previous studies showed small fractionation effects consistent with the cleavage of the carbon-carbon double bond (C=C) by an alkene monooxygenase, the results of this study suggest that this might not be the major cDCE transformation pathway. KIE values of 1.03 to 1.04 indicate that the major transformation pathway proceeds via C-Cl cleavage. To the best of our knowledge, an aerobic haloalkene degradation mechanism involving a C-Cl cleavage in the first step has never been demonstrated in a microbial system, suggesting that cDCE degradation in JS666 may involve a novel pathway.
Proposed cDCE degradation pathways.
cDCE degradation pathways are proposed based on several lines of evidence, including the (i) identification of upregulated enzymes by proteomic and transcriptomic analyses, (ii) bioinformatic analysis of upregulated enzymes to predict functions, (iii) verification of enzyme functions by enzyme assays and other biochemical experiments, (iv) CSIA, and (v) observations of degradation behavior in culturing experiments. Based on the evidence presented above, the following are likely candidates for enzymes involved in cDCE degradation: HADs (Bpro0530 and Bpro5186), GST (Bpro0645), CMO (Bpro5565), Hlase (Bpro5566), and CO DHase (Bpro0577).
The results of a comparative genomic analysis of JS666 and Polaromonas napthalenivorans CJ2 support the hypothesis that the genes encoding the enzymes mentioned above are important in cDCE degradation in JS666. CJ2, whose 16S rRNA exhibits 97% nucleotide identity with the 16S rRNA of JS666, is capable of degrading naphthalene but not capable of cDCE degradation (34). Thus, genes present in JS666 but not in CJ2 may be more likely to be involved in cDCE degradation. Comparative genomic analysis indicated that homologs of the putative Hlase, CMO, GST, HADs, CO DHase, and PNPox are not likely present in CJ2 (data not shown).
The oxidation of halogenated alkenes, such as cDCE, is thought to occur primarily by monooxygenase-catalyzed epoxidations (14, 62). The detection of epoxides produced from ethene and propene (7) supports the view that a monooxygenase is involved in the first step in cDCE degradation in JS666. There are several possible mechanisms by which the DCE epoxide could be transformed in JS666 that are in agreement with the omics data, including (i) GST-catalyzed formation of glutathione conjugates, (ii) Hlase-catalyzed formation of 1,2-dichloroethane-1,2-diol, or (iii) spontaneous decomposition of the DCE epoxide to dichloroacetaldehyde.
Evidence from hierarchical clustering of transcripts indicates that the CMO and GST group separately, suggesting that they may be involved in separate degradation pathways, with one pathway involving a GST and HAD and another pathway involving a CMO, Hlase, and CO DHase (Fig. 5). Two possible cDCE degradation pathways are consistent with the hierarchical clustering results, including (i) GST-catalyzed dehalogenation followed by CAD- and HAD-catalyzed formation of glycolate and (ii) monooxygenase-catalyzed epoxidation followed by hydrolase-catalyzed formation of a diol and dehydrogenase-catalyzed formation of glyoxylate (Fig. 6). The higher levels of expression of enzymes predicted to be involved in the GST-catalyzed dehalogenation pathway than of the enzymes predicted to be involved in the monooxygenase-catalyzed epoxidation pathway (Fig. 5) suggest that the GST-catalyzed dehalogenation is the primary cDCE degradation pathway in JS666 and that monooxygenase-catalyzed epoxidation is a less important transformation pathway. This interpretation is also supported by the results of carbon isotope analysis.
FIG. 5.
Expression of selected transcripts in cDCE-grown cells, expressed in arbitrary fluorescence units. The levels of expression of corresponding transcripts in glycolate-grown cells were low (data not shown). The following enzymes were examined: HAD 1 (Bpro5186), HAD 2 (Bpro0530), GST (Bpro0645), CMO (Bpro5565), CO DHase (Bpro0577), and Hlase (Bpro5566).
FIG. 6.
Proposed cDCE degradation pathways.
The large carbon isotope fractionation effect associated with cDCE degradation in JS666 suggests that the major cDCE degradation pathway involves carbon-chlorine bond cleavage in the initial step, which is consistent with GST-catalyzed dehalogenation but is not consistent with the carbon-carbon double-bond cleavage that occurs during monoxygenase-catalyzed epoxidation. When degradation proceeds via two distinct pathways in one organism, the measured carbon isotope enrichment factor represents a weighted average of the two pathways that is strongly dependent on how much each pathway contributes (11). When one pathway is dominant, however, especially if the larger fractionation effect is associated with that pathway, fractionation associated with the minor pathway is unlikely to be observed. In other words, the large fractionation effect seen in the samples indicates that C-Cl cleavage is the dominant transformation pathway but does not rule out the possibility that there is a small contribution from the monoxygenase pathway. GSTs have been shown to act as dehalogenases in dichloromethane metabolism in M. dichloromethanicum DM4 (38), although an analogous reaction has never been observed for two carbon compounds in bacteria. In mammalian systems, GST-catalyzed dehalogenation of chlorinated ethenes occurs primarily by an addition-elimination reaction mechanism in which the sulfur nucleophile from GSH is added to the haloalkene, resulting in removal of the halide (3).
The presence of two cDCE degradation pathways may also explain the observation of two degradation phenotypes in JS666 depending on the culture technique employed (32). One degradation phenotype is characterized by growth-coupled cDCE degradation and has high maximum specific degradation rates, whereas the other is characterized by apparent cometabolic degradation in which biomass decreases over time and the maximum degradation rates are approximately 1/10 the published values.
Transport and cDCE degradation.
A high degree of upregulation of transport-related genes, including genes encoding a putative ABC transporter and two sodium/solute symporters, is associated with cDCE degradation. In JS666, growth on cDCE resulted in upregulation of all of the putative protein components necessary to form an ABC transporter, including two ATPases (Bpro3332 and Bpro3333), two inner membrane translocators (Bpro3334 and Bpro3335), and one extracellular ligand-binding receptor (Bpro3336). The genes encoding two putative sodium/solute symporters (Bpro0531 and Bpro5185), which share 100% amino acid identity and are upregulated 40-fold by cDCE, are located adjacent to the genes encoding upregulated HADs in JS666 on the chromosome and plasmid. Furthermore, an enrichment analysis revealed that genes in the intracellular trafficking, secretion, and vesicular transport COG were statistically overrepresented (P = 0.095, Fisher's exact test) in the set of genes upregulated by cDCE. The absence of proteins with high sequence similarity in the naphthalene-degrading strain CJ2 further supports this hypothesis.
Conclusions.
An integrated omics approach revealed genes upregulated by cDCE in JS666. The results of this analysis, in conjunction with the results of CSIA and enzyme assays, suggest that oxidative stress and transport are correlated with cDCE degradation and establish a basis for prediction of cDCE degradation pathways. A number of genes that might be important in cDCE degradation were identified, including the genes responsible for the observed HAD and GST enzyme activities, as well as genes likely to be responsible for the observed monooxygenase and PNPox activities. The results of both molecular techniques and CSIA suggest that cDCE degradation via monooxygenase-catalyzed epoxidation may be only a minor pathway and that the initial step in the major cDCE degradation pathway involves carbon-chloride bond cleavage. We hypothesize that the C-Cl cleavage may be due to a GST-catalyzed dehalogenation reaction. Further research is needed to determine whether cDCE degradation proceeds via this novel mechanism. Experiments are under way to confirm the functional activity of upregulated enzymes and to identify their roles in the cDCE degradation pathways of JS666.
Acknowledgments
This research was supported in part by contracts with Environmental Security Technology Certification Program and Strategic Environmental Research and Development Program. L.K.J. was the recipient of an NSF graduate research fellowship.
This work was made possible by collaborations between the Center for Biofilm Engineering at Montana State University and Cornell University. We thank John Neuman, Robin Gerlach, Elinor Pulcini, Stewart Clark, Pat Secor, Al Cunningham, and Peg Dirckx at the Center for Biofilm Engineering for their technical assistance and use of their labs. We thank Ray B. Payne for sharing his unpublished results of enzyme assays. We also acknowledge Kate McInnerney of the Montana State University Functional Genomics Core; Tony Haag of the Mass Spectrometry Core at the University of Texas Medical Branch; Sheng Zhang of Proteomics and Mass Spectrometry Core at Cornell University; Rachel Neal of the Montana State University Mass Spectrometry Laboratory; Elizabeth Edwards, Alison Waller, and Sarah Hirschorn of the University of Toronto; and Shirley Nishino of the Georgia Institute of Technology.
Footnotes
Published ahead of print on 10 April 2009.
REFERENCES
- 1.Allen, J. R., D. D. Clark, J. G. Krum, and S. A. Ensign. 1999. A role for coenzyme M (2-mercaptoethanesulfonic acid) in a bacterial pathway of aliphatic epoxide carboxylation. Proc. Natl. Acad. Sci. USA 96:8432-8437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anand, S. S. 2005. Protective effect of vitamin B6 in chromium-induced oxidative stress in liver. J. Appl. Toxicol. 25:440-443. [DOI] [PubMed] [Google Scholar]
- 3.Anders, M. W., and W. Dekant. 1998. Glutathione-dependent bioactivation of haloalkenes. Annu. Rev. Pharmacol. Toxicol. 38:501-537. [DOI] [PubMed] [Google Scholar]
- 4.Bloom, Y., R. Aravena, D. Hunkeler, E. Edwards, and S. K. Frape. 2000. Carbon isotope fractionation during microbial dechlorination of trichloroethene, cis-1,2-dichloroethene, and vinyl chloride: implications for assessment of natural attenuation. Environ. Sci. Technol. 34:2768-2772. [Google Scholar]
- 5.Chartrand, M. M. G., A. Waller, T. E. Mattes, M. Elsner, G. Lacrampe-Couloume, J. M. Gossett, E. A. Edwards, and B. Sherwood Lollar. 2005. Carbon isotopic fractionation during aerobic vinyl chloride degradation. Environ. Sci. Technol. 34:1064-1070. [DOI] [PubMed] [Google Scholar]
- 6.Chu, K. H., S. Mahendra, D. L. Song, M. E. Conrad, and L. Alvarez-Cohen. 2004. Stable carbon isotope fractionation during aerobic biodegradation of chlorinated ethenes. Environ. Sci. Technol. 38:3126-3130. [DOI] [PubMed] [Google Scholar]
- 7.Coleman, N. V., T. E. Mattes, J. M. Gossett, and J. C. Spain. 2002. Biodegradation of cis-dichloroethene as the sole carbon source by a β-proteobacterium. Appl. Environ. Microbiol. 68:2726-2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Comtois, S. L., M. D. Gidley, and D. J. Kelly. 2003. Role of the thioredoxin system and the thiol-peroxidases Tpx and Bcp in mediating resistance to oxidative and nitrosative stress in Helicobacter pylori. Microbiology 149:121-129. [DOI] [PubMed] [Google Scholar]
- 9.Cury, J. A., and H. Koo. 2007. Extraction and purification of total RNA from Streptococcus mutans biofilms. Anal. Biochem. 365:208-214. [DOI] [PubMed] [Google Scholar]
- 10.Dijk, J. A., J. Gerritse, G. Schraa, and A. J. M. Stams. 2004. Degradation pathway of 2-chloroethanol in Pseudomonas stutzeri strain JJ under denitrifying conditions. Arch. Microbiol. 182:514-519. [DOI] [PubMed] [Google Scholar]
- 11.Elsner, M., M. Chartrand, N. Vanstone, G. L. Couloume, and B. Sherwood Lollar. 2008. Identifying abiotic chlorinated ethene degradation: characteristic isotope patterns in reaction products with nanoscale zero-valent iron. Environ. Sci. Technol. 42:5963-5970. [DOI] [PubMed] [Google Scholar]
- 12.Elsner, M., D. M. Cwiertny, A. L. Roberts, and B. Sherwood Lollar. 2007. 1,1,2,2-Tetrachloroethane reactions with OH−, Cr(II), granular iron, and a copper-iron bimetal: insights from product formation and associated carbon isotope fractionation. Environ. Sci. Technol. 41:4111-4117. [DOI] [PubMed] [Google Scholar]
- 13.Elsner, M., L. Zwank, D. Hunkeler, and R. P. Schwarzenbach. 2005. A new concept linking observable stable isotope fractionation to transformation pathways of organic pollutants. Environ. Sci. Technol. 39:6896-6916. [DOI] [PubMed] [Google Scholar]
- 14.Ensign, S. A. 2001. Microbial metabolism of aliphatic alkenes. Biochemistry 40:5845-5853. [DOI] [PubMed] [Google Scholar]
- 15.Fennell, D. E., A. B. Carroll, J. M. Gossett, and S. H. Zinder. 2001. Assessment of indigenous reductive dechlorinating potential at a TCE-contaminated site using microcosms, polymerase chain reaction analysis, and site data. Environ. Sci. Technol. 35:1830-1839. [DOI] [PubMed] [Google Scholar]
- 16.Galimov, E. M. (ed.). 1985. The biological fractionation of isotopes. Academic Press, Orlando, FL.
- 17.Glod, G., W. Angst, C. Holliger, and R. P. Schwarzenbach. 1997. Corrinoid-mediated reduction of tetrachloroethene, trichloroethene, and trichlorofluoroethene in homogeneous aqueous solution: reaction kinetics and reaction mechanisms. Environ. Sci. Technol. 31:253-260. [Google Scholar]
- 18.Gorg, A., C. Obermaier, G. Boguth, A. Harder, B. Scheibe, R. Wildgruber, and W. Weiss. 2000. The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis 21:1037-1053. [DOI] [PubMed] [Google Scholar]
- 19.Habig, W. H., and W. B. Jakoby. 1981. Assays for differentiation of glutathione S-transferase. Methods Enzymol. 77:398-405. [DOI] [PubMed] [Google Scholar]
- 20.Hage, J. C., and S. Hartmans. 1999. Monooxygenase-mediated 1,2-dichloroethane degradation by Pseudomonas sp. strain DCA1. Appl. Environ. Microbiol. 65:2466-2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartmans, S., F. J. Weber, D. P. M. Somhorst, and J. A. M. Debont. 1991. Alkene monooxygenase from Mycobacterium—a multicomponent enzyme. J. Gen. Microbiol. 137:2555-2560. [DOI] [PubMed] [Google Scholar]
- 22.Hayes, J. D., J. U. Flanagan, and I. R. Jowsey. 2005. Glutathione transferases. Annu. Rev. Pharmacol. Toxicol. 45:51-88. [DOI] [PubMed] [Google Scholar]
- 23.Hayes, J. D., and L. I. McLellan. 1999. Glutathione and glutathione-dependent enzymes represent a co-ordinately regulated defence against oxidative stress. Free Radic. Res. 31:273-300. [DOI] [PubMed] [Google Scholar]
- 24.Hirschorn, S. K., M. J. Dinglasan-Panlilio, E. A. Edwards, G. Lacrampe-Couloume, and B. Sherwood Lollar. 2007. Isotope analysis as a natural reaction probe to determine mechanisms of biodegradation of 1,2-dichloroethane. Environ. Microbiol. 9:1651-1657. [DOI] [PubMed] [Google Scholar]
- 25.Hirschorn, S. K., M. J. Dinglasan, M. Elsner, S. A. Mancini, G. Lacrampe-Couloume, E. A. Edwards, and B. Sherwood Lollar. 2004. Pathway dependent isotopic fractionation during aerobic biodegradation of 1,2-dichloroethane. Environ. Sci. Technol. 38:4775-4781. [DOI] [PubMed] [Google Scholar]
- 26.Huang, R., A. Wallqvist, and D. G. Covell. 2006. Comprehensive analysis of pathway or functionally related gene expression in the National Cancer Institute's anticancer screen. Genomics 87:315-328. [DOI] [PubMed] [Google Scholar]
- 27.Hunkeler, D., R. Aravena, and B. J. Butler. 1999. Monitoring microbial dechlorination of tetrachloroethene (PCE) in groundwater using compound-specific stable carbon isotope ratios: microcosm and field studies. Environ. Sci. Technol. 33:2733-2738. [Google Scholar]
- 28.Hunkeler, D., R. Aravena, and E. Cox. 2002. Carbon isotopes as a tool to evaluate the origin and fate of vinyl chloride: laboratory experiments and modeling of isotope evolution. Environ. Sci. Technol. 36:3378-3384. [DOI] [PubMed] [Google Scholar]
- 29.Isken, S., and J. A. M. de Bont. 1998. Bacteria tolerant to organic solvents. Extremophiles 2:229-238. [DOI] [PubMed] [Google Scholar]
- 30.Janssen, D. B., J. E. Oppentocht, and G. J. Poelarends. 2001. Microbial dehalogenation. Curr. Opin. Biotechnol. 12:254-258. [DOI] [PubMed] [Google Scholar]
- 31.Janssen, D. B., A. Scheper, L. Dijkhuizen, and B. Witholt. 1985. Degradation of halogenated aliphatic compounds by Xanthobacter autotrophicus GJ10. Appl. Environ. Microbiol. 49:673-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jennings, L. K. 2005. Culturing and enumeration of Polaromonas sp. strain JS666 for its use as a bioaugmentation agent in the remediation of cis-dichloroethene-contaminated sites. M.S. thesis. Cornell University, Ithaca, NY.
- 33.Jennings, L. K. 2009. Proteomic and transcriptomic analyses reveal genes upregulated by cis-dichloroethene in Polaromonas JS666. Ph.D. dissertation. Cornell University, Ithaca, NY. [DOI] [PMC free article] [PubMed]
- 34.Jeon, C. O., M. Park, H.-S. Ro, W. Park, and E. L. Madsen. 2006. The naphthalene catabolic (nag) genes of Polaromonas naphthalenivorans CJ2: evolutionary implications for two gene clusters and novel regulatory control. Appl. Environ. Microbiol. 72:1086-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson, G. R., R. K. Jain, and J. C. Spain. 2002. Origins of the 2,4-dinitrotoluene pathway. J. Bacteriol. 184:4219-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanai, T., K. Takahashi, and H. Inoue. 2006. Three distinct-type glutathione S-transferases from Escherichia coli important for defense against oxidative stress. J. Biochem. (Tokyo) 140:703-711. [DOI] [PubMed] [Google Scholar]
- 37.Kannan, K., and S. K. Jain. 2004. Effect of vitamin B-6 on oxygen radicals, mitochondrial membrane potential, and lipid peroxidation in H2O2-treated U937 monocytes. Free Radic. Biol. Med. 36:423-428. [DOI] [PubMed] [Google Scholar]
- 38.Kohler-Staub, D., and T. Leisinger. 1985. Dichloromethane dehalogenase of Hyphomicrobium sp. strain DM2. J. Bacteriol. 162:676-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.La Carbona, S., N. Sauvageot, J. C. Giard, A. Benachour, B. Posteraro, Y. Auffray, M. Sanguinetti, and A. Hartke. 2007. Comparative study of the physiological roles of three peroxidases (NADH peroxidase, alkyl hydroperoxide reductase and thiol peroxidase) in oxidative stress response, survival inside macrophages and virulence of Enterococcus faecalis. Mol. Microbiol. 66:1148-1163. [DOI] [PubMed] [Google Scholar]
- 40.Lee, P. K. H., M. E. Conrad, and L. Alvarez-Cohen. 2007. Stable carbon isotope fractionation of chloroethenes by dehalorespiring isolates. Environ. Sci. Technol. 41:4277-4285. [DOI] [PubMed] [Google Scholar]
- 41.Leelakriangsak, M., N. T. T. Huyen, S. Towe, N. van Duy, D. Becher, M. Hecker, H. Antelmann, and P. Zuber. 2008. Regulation of quinone detoxification by the thiol stress sensing DUF24/MarR-like repressor, YodB in Bacillus subtilis. Mol. Microbiol. 67:1108-1124. [DOI] [PubMed] [Google Scholar]
- 42.Mariotti, A., J. C. Germon, P. Hubert, P. Kaiser, R. Letolle, A. Tardieux, and P. Tardieux. 1981. Experimental-determination of nitrogen kinetic isotope fractionation—some principles—illustration for the denitrification and nitrification processes. Plant Soil 62:413-430. [Google Scholar]
- 43.Mattes, T. E., A. K. Alexander, P. M. Richardson, A. C. Munk, C. S. Han, P. Stothard, and N. V. Coleman. 2008. The genome of Polaromonas sp. strain JS666: insights into the evolution of a hydrocarbon- and xenobiotic-degrading bacterium, and features of relevance to biotechnology. Appl. Environ. Microbiol. 74:6405-6416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maymó-Gatell, X., T. Anguish, and S. H. Zinder. 1999. Reductive dechlorination of chlorinated ethenes and 1,2-dichloroethane by “Dehalococcoides ethenogenes” strain 195. Appl. Environ. Microbiol. 65:3108-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maymó-Gatell, X., Y. T. Chien, J. M. Gossett, and S. H. Zinder. 1997. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science 276:1568-1571. [DOI] [PubMed] [Google Scholar]
- 46.Melander, L., and W. H. Saunders, Jr. (ed.). 1980. Reaction rates of isotopic molecules. John Wiley & Sons, New York, NY.
- 47.NimbleGen 2006, posting date. Protocols for synthesis of double-stranded cDNA. http://www.nimblegen.com/protocols/NGS_cDNA_Synthesis2006_09_28.pdf.
- 48.Pedelacq, J. D., B. S. Rho, C. Y. Kim, G. S. Waldo, T. P. Lekin, B. W. Segelke, B. Rupp, L. W. Hung, S. I. Kim, and T. C. Terwilliger. 2006. Crystal structure of a putative pyridoxine 5′-phosphate oxidase (Rv2607) from Mycobacterium tuberculosis. Proteins Struct. Funct. Bioinform. 62:563-569. [DOI] [PubMed] [Google Scholar]
- 49.Sang, Y. Y., J. M. Barbosa, H. Z. Wu, R. D. Locy, and N. K. Singh. 2007. Identification of a pyridoxine (pyridoxamine) 5′-phosphate oxidase from Arabidopsis thaliana. FEBS Lett. 581:344-348. [DOI] [PubMed] [Google Scholar]
- 50.Sauer, K., and A. K. Camper. 2001. Characterization of phenotypic changes in Pseudomonas putida in response to surface-associated growth. J. Bacteriol. 183:6579-6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Serganov, A., and D. J. Patel. 2007. Ribozymes, riboswitches and beyond: regulation of gene expression without proteins. Nat. Rev. Genet. 8:776-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sherwood Lollar, B., S. K. Hirschorn, M. M. G. Chartrand, and G. Lacrampe-Couloume. 2007. An approach for assessing total instrumental uncertainty in compound-specific carbon isotope analysis: implications for environmental remediation studies. Anal. Chem. 79:3469-3475. [DOI] [PubMed] [Google Scholar]
- 53.Sherwood Lollar, B. S., G. F. Slater, J. Ahad, B. Sleep, J. Spivack, M. Brennan, and P. MacKenzie. 1999. Contrasting carbon isotope fractionation during biodegradation of trichloroethylene and toluene: implications for intrinsic bioremediation. Org. Geochem. 30:813-820. [Google Scholar]
- 54.Sikkema, J., J. A. M. de Bont, and B. Poolman. 1995. Mechanisms of membrane toxicity of hydrocarbons. Microbiol. Rev. 59:201-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Simmonds, A. C., C. A. Reilly, R. M. Baldwin, B. I. Ghanayem, D. L. Lanza, G. S. Yost, K. S. Collins, and P. G. Forkert. 2004. Bioactivation of 1,1-dichloroethylene to its epoxide by CYP2E1 and CYP2F enzymes. Drug Metab. Dispos. 32:1032-1039. [PubMed] [Google Scholar]
- 56.Singleton, D. A., S. R. Merrigan, J. Liu, and K. N. Houk. 1997. Experimental geometry of the epoxidation transition state. J. Am. Chem. Soc. 119:3385-3386. [Google Scholar]
- 57.Slater, G. F., B. Sherwood Lollar, B. E. Sleep, and E. A. Edwards. 2001. Variability in carbon isotopic fractionation during biodegradation of chlorinated ethenes: implications for field applications. Environ. Sci. Technol. 35:901-907. [DOI] [PubMed] [Google Scholar]
- 58.Taysi, S. 2005. Oxidant/antioxidant status in liver tissue of vitamin B-6 deficient rats. Clin. Nutr. 24:385-389. [DOI] [PubMed] [Google Scholar]
- 59.Tiehm, A., K. R. Schmidt, B. Pfeifer, M. Heidinger, and S. Ertl. 2008. Growth kinetics and stable carbon isotope fractionation during aerobic degradation of cis-1,2-dichloroethene and vinyl chloride. Water Res. 42:2431-2438. [DOI] [PubMed] [Google Scholar]
- 60.Ulijasz, A. T., D. R. Andes, J. D. Glasner, and B. Weisblum. 2004. Regulation of iron transport in Streptococcus pneumoniae by RitR, an orphan response regulator. J. Bacteriol. 186:8123-8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van der Ploeg, J., M. P. Smidt, A. S. Landa, and D. B. Janssen. 1994. Identification of chloroacetaldehyde dehydrogenase involved in 2,2-dichloroethane degradation. Appl. Environ. Microbiol. 60:1599-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Hylckama Vlieg, J. E. T., and D. B. Janssen. 2001. Formation and detoxification of reactive intermediates in the metabolism of chlorinated ethenes. J. Biotechnol. 85:81-102. [DOI] [PubMed] [Google Scholar]
- 63.van Hylckama Vlieg, J. E. T., J. Kingma, W. Kruizinga, and D. B. Janssen. 1999. Purification of a glutathione S-transferase and a glutathione conjugate-specific dehydrogenase involved in isoprene metabolism in Rhodococcus sp. strain AD45. J. Bacteriol. 181:2094-2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van Hylckama Vlieg, J. E. T., J. Kingma, A. J. van den Wijngaard, and D. B. Janssen. 1998. A glutathione S-transferase with activity towards cis-1,2-dichloroepoxyethane is involved in isoprene utilization by Rhodococcus sp. strain AD45. Appl. Environ. Microbiol. 64:2800-2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van Hylckama Vlieg, J. E. T., G. J. Poelarends, A. E. Mars, and D. B. Janssen. 2000. Detoxification of reactive intermediates during microbial metabolism of halogenated compounds. Curr. Opin. Microbiol. 3:257-262. [DOI] [PubMed] [Google Scholar]
- 66.van Loo, B., J. Kingma, M. Arand, M. G. Wubbolts, and D. B. Janssen. 2006. Diversity and biocatalytic potential of epoxide hydrolases identified by genome analysis. Appl. Environ. Microbiol. 72:2905-2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vogel, T. M., C. S. Criddle, and P. L. McCarty. 1987. Transformations of halogenated aliphatic compounds. Environ. Sci. Technol. 21:722-736. [DOI] [PubMed] [Google Scholar]
- 68.Vuilleumier, S. 1997. Bacterial glutathione S-transferases: what are they good for? J. Bacteriol. 179:1431-1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vuilleumier, S., and M. Pagni. 2002. The elusive roles of bacterial glutathione S-transferases: new lessons from genomes. Appl. Microbiol. Biotechnol. 58:138-146. [DOI] [PubMed] [Google Scholar]
- 70.Wang, J. J., T. Y. Ying, H. L. Wang, Z. X. Shi, M. Z. Li, K. He, E. L. Feng, J. Wang, J. Yuan, T. Li, K. H. Wei, G. F. Su, H. C. Zhu, X. M. Zhang, P. T. Huang, and L. Y. Huang. 2005. 2-D reference map of Bacillus anthracis vaccine strain A16R proteins. Proteomics 5:4488-4495. [DOI] [PubMed] [Google Scholar]
- 71.Wong, M. 2008. Discovery and characterization of hydrolytic dehalogenases from genomic data. M.S. thesis. University of Toronto, Toronto, Ontario, Canada.
- 72.Zhao, G. S., and M. E. Winkler. 1995. Kinetic limitation and cellular amount of pyridoxine (pyridoxamine) 5′-phosphate oxidase of Escherichia coli K-12. J. Bacteriol. 177:883-891. [DOI] [PMC free article] [PubMed] [Google Scholar]








