Abstract
Cardiac slow delayed rectifier (IKs) channel is composed of KCNQ1 (pore-forming) and KCNE1 (auxiliary) subunits. Although KCNE1 is an obligate IKs component that confers the uniquely slow gating kinetics, KCNE2 is also expressed in human heart. In vitro experiments suggest that KCNE2 can associate with the KCNQ1-KCNE1 complex to suppress the current amplitude without altering the slow gating kinetics. Our goal here is to test the role of KCNE2 in cardiac IKs channel function. Pulse-chase experiments in COS-7 cells show that there is a KCNE1 turnover in the KCNQ1-KCNE1 complex, supporting the possibility that KCNE1 in the IKs channel complex can be substituted by KCNE2 when the latter is available. Biotinylation experiments in COS-7 cells show that although KCNE1 relies on KCNQ1 coassembly for more efficient cell surface expression, KCNE2 can independently traffic to the cell surface, thus becoming available for substituting KCNE1 in the IKs channel complex. Injecting vesicles carrying KCNE1 or KCNE2 into KCNQ1-expressing oocytes leads to KCNQ1 modulation in the same manner as KCNQ1+KCNEx (where x = 1 or 2) cRNA coinjection. Thus, free KCNEx peptides delivered to the cell membrane can associate with existing KCNQ1 channels to modulate their function. Finally, adenovirus-mediated KCNE2 expression in adult guinea pig ventricular myocytes exhibited colocalization with native KCNQ1 protein and reduces the native IKs current density. We propose that in cardiac myocytes the IKs current amplitude is under dynamic control by the availability of KCNE2 subunits in the cell membrane.
The slow delayed rectifier (IKs) channel functions as a “repolarization reserve” in the heart (1). It provides needed outward currents to prevent excessive action potential prolongation when the β-adrenergic tone is high or when there is a blockade of the rapid delayed rectifier channel as in acquired long QT syndrome (1). The IKs channel is composed of a pore-forming component (the KCNQ1 channel, also known as Kv7.1 or KvLQT1) and an auxiliary regulatory component (the KCNE1 subunit, also known as minK or IsK) (2, 3). KCNE1 association with KCNQ1 produces the unique IKs channel phenotype: very slow activation and deactivation rates and a positive voltage range of activation. There has been a strong interest in understanding how KCNQ1 and KCNE1 interact with each other (4–12). Such knowledge is important for understanding why the IKs channel malfunctions under pathological conditions (13, 14) or when congenital mutations occur to KCNQ1 or KCNE1 (15–17). This information is also useful for the design of antiarrhythmic agents targeting malfunctioning IKs channels (18).
More KCNE subunits (KCNE2–KCNE5) have been cloned and characterized (19). In heterologous expression systems, each of these new members of the KCNE subfamily can associate with the KCNQ1 channel and confer distinct channel phenotypes (20). KCNE3, like KCNE1, increases KCNQ1 current amplitude, whereas KCNE2, KCNE4, and KCNE5 decrease or abolish KCNQ1 current. KCNE5, like KCNE1, shifts the voltage dependence of KCNQ1 activation in the positive direction, whereas KCNE2 and KCNE3 make KCNQ1 constitutively active. The KCNE2, KCNE3, and KCNE4 proteins have been detected in human heart (21–23). If human cardiac myocytes coexpress KCNQ1 with KCNE1-KCNE4, changing the pattern of KCNQ1 association with these different KCNE subunits can have a dramatic impact on the membrane electrical activity (24). What is the pattern of KCNQ1 association with these different KCNE subunits in the heart? How is this regulated?
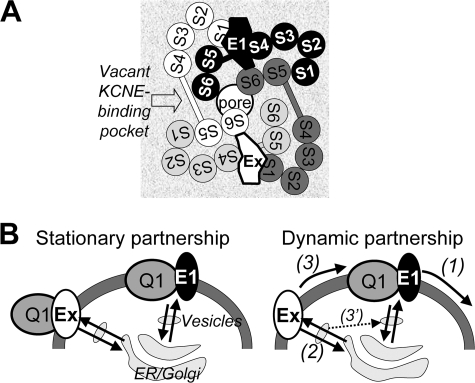
Before addressing these questions, several points need to be considered. First, KCNE1 is an obligate component of the cardiac IKs channel, because only it can confer the unique IKs gating phenotype. Second, the subunit stoichiometry in a KCNQ1-KCNE complex is likely 4:2, i.e. 2 KCNE subunits per KCNQ1 tetramer channel (4, 25). Fig. 1A depicts a schematic view of the arrangement of transmembrane helices in a KCNQ1-KCNE complex. The arrangement of KCNQ1 subunits is based on crystal structures of Kv channels (26, 27). The location of the putative “KCNE-binding pocket” (28) is based on experimental data on how KCNQ1 and KCNE1 interact with each other (5, 7, 8, 11, 12, 29). Third, a KCNQ1 channel can simultaneously bind two different KCNE subunits, and when this occurs there is a hierarchy in how the KCNQ1 gating kinetics is modulated (30, 31). For example, a ternary complex of KCNQ1-KCNE1-KCNE2 will behave like a typical IKs channel, without a constitutive current component as is seen in the KCNQ1-KCNE2 complex (31). That is, the gating modifying effects of KCNE1 override those of KCNE2. Therefore, a likely scenario in the heart is that KCNE1 serves as the major regulatory subunit that determines the IKs phenotype (i.e. KCNQ1-(KCNE1)2 or KCNQ1-(KCNE1)(KCNEx), where x = 2–4; Fig. 1A).
FIGURE 1.
A, schematic arrangement of transmembrane helices in a KCNQ1-KCNE channel complex. The view is from the extracellular perspective. The arrangement of transmembrane helices in the Q1 channel is based on the Kv channel crystal structures (Protein Data Bank codes 2A79 and 2R9R) (26, 27). The four Q1 subunits are arranged around a central pore and are color-coded as white, light gray, dark gray, and black. The voltage-sensing domain (S1–S4) of each Q1 subunit is juxtaposed to the pore domains (S5 and S6) of the adjacent Q1 subunit. Experimental data suggest that the E1 transmembrane domain resides in a KCNE-binding pocket between voltage-sensing domains of two adjacent Q1 subunits (12, 28), where E1 simultaneously interacts with S1 (12) and S4 (11) of two adjacent Q1 voltage-sensing domains and with S6 of a third Q1 subunit (5, 7, 8, 29). Furthermore, optimally two KCNE subunits simultaneously bind to a Q1 channel (4, 25), likely in diagonal positions as depicted here. In the cardiac IKs channel, E1 is an obligate component (occupying one KCNE-binding pocket), whereas E1 or another KCNE subunit (Ex) occupies the other KCNE-binding pocket. B, two possible scenarios of Q1-KCNE association. Left panel, stationary partnership where Q1 and KCNE subunits form stable complexes without dissociation or exchange of the KCNE components during transits among ER, Golgi apparatus, cytosolic vesicles, and the cell surface membrane. Right panel, dynamic partnership where E1 can dissociate from the Q1 channel (process (1)) and the vacancy is filled by another KCNE subunit, Ex, which can take place in the cell surface membrane (process (3)) after Ex traffics to the cell surface independent of Q1 (process (2)) or during transits among cytosolic vesicular compartments (process 3′).
We focused on KCNE2 because its mutations have been linked to long QT syndrome (LQT6) or familial atrial fibrillation/short QT syndrome (32, 33). This points to a role of KCNE2 in maintaining the cardiac electrical stability, although the molecular mechanism is not clear. In heterologous expression systems KCNE2 can associate with several voltage-gated cation channels that are important in shaping the cardiac action potentials: ERG1 (32), Kv4 (34), and HCN (35) (pore-forming components of the rapid delayed rectifier, transient outward, and pacemaker channels, respectively). However, the effects of KCNE2 on these channels are modest. On the other hand, KCNE2 can dramatically reduce the current amplitude when it is coexpressed with KCNQ1-KCNE1 (31). This occurs by two mechanisms: 1) forming a KCNQ1-KCNE1-KCNE2 ternary complex with reduced current amplitude and typical IKs gating kinetics (31), 2) forming a KCNQ1-(KCNE2)2 complex as low conductance background channels (31, 36). Indeed, the linkage between KCNE2 and KCNQ1 provides the most likely explanation for why a KCNE2 mutation, R27C, that relieves the current-suppressing effects on KCNQ1, can augment outward currents and increase the risk for atrial fibrillation/short QT syndrome (33).
How likely is the coassembly of KCNQ1-KCNE1-KCNE2 or KCNQ1-(KCNE2)2 complexes in the heart? To begin to tackle this question, we considered two general scenarios of KCNQ1-KCNE association (Fig. 1B). In the first scenario, KCNQ1 and KCNE subunits form stable complexes and stay together throughout their life span in cells (stationary partnership). Based on our observation that KCNQ1 and KCNE subunits can be coimmunoprecipitated when they are translated in a cell-free system in the presence of microsomal membranes (equivalent to endoplasmic reticulum (ER))3 (31), coassembly of KCNQ1-KCNE complexes likely begins in the ER. In the scenario of a stationary partnership, the pattern of KCNQ1 association with different KCNE subunits is mainly determined by the temporal and spatial relationship of transcription and translation among these subunits. In the second scenario, KCNQ1-KCNE partnership is less stable. KCNE can dissociate from the KCNE-binding pocket in the KCNQ1 channel, allowing the vacancy to be filled by other different KCNE subunits. In this case (dynamic partnership), the pattern of KCNQ1 association with KCNE subunits will be influenced by the binding affinities of different KCNQ1-KCNE combinations and the pattern of KCNE subunit trafficking among different subcellular compartments (37) (Fig. 1B).
In this study, we used COS-7 and oocyte expression systems to address three questions: 1) Can KCNE1 dissociate from the KCNQ1-KCNE1 complex? 2) Can KCNE2 achieve a substantial cell surface expression on its own? 3) Can free KCNE2 subunits in the cell membrane associate with KCNQ1 channel that have vacancies in the KCNE-binding pockets? These are labeled as Processes (1), (2), and (3) in the “dynamic partnership” scenario of Fig. 1B. We also used the adenovirus-mediated gene transfer approach to seek evidence for KCNE2 modulation of native IKs channels in adult cardiac myocytes. In the following text, Q1, E1, and E2 stand for KCNQ1, KCNE1, and KCNE2, respectively. The term “KCNE” or “Ex” is used when referring to multiple types of KCNE subunits.
EXPERIMENTAL PROCEDURES
Molecular Biology
Human Q1, E1, E2, and rat Kv4.3 cDNAs were generous gifts from Drs. M. T. Keating (Harvard University), R. Sansom (Merck Company), G. W. Abbott (Cornell Medical School), and P. Serodio (New York University). Rat KChIP2b was cloned from rat heart using the reverse transcription-PCR method. Q1 was subcloned into vector pcDNA3.1/V5-His-TOPO (Invitrogen, for COS-7 expression) or pSP64 (Promega, for oocyte expression). In the former case, in-frame V5 (GKPIPNPLLGLDST) and His6 epitope tags were attached to the C terminus of Q1. Epitope tagging did not perturb Q1 channel function or Q1-KCNE interaction (12). A c-Myc epitope tag was inserted between residues 20 and 21 in the extracellular domain of E2, which did not interfere with E2 function (34).
Pulse-Chase Experiments
COS-7 cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal calf serum (Hyclone) and penicillin/streptomycin in a moist 5% CO2 chamber at 36 °C. The cells were plated at a subconfluence level the day before transfection. COS-7 cells were transfected with Q1 and E1 or Kv4.3 and KChIP2 at a cDNA molar ratio (pore-forming subunit: auxiliary subunit) of 1:2. One day later, the cells were depleted of methionine by incubation in serum-free, methionine-free Dulbecco's modified Eagle's medium for 30 min at 36 °C. Then [35S]Met (200 μCi/ml, [Met] = ∼0.2 mm) was added, and the cells were incubated for 60 min at 36 °C to label newly translated proteins with [35S]Met. Afterward, the cells were washed and incubated in Dulbecco's modified Eagle's medium containing 2 mm unlabeled Met for different amounts of “chase” time. At the end of specified chase times, the cells were washed, lysed, and solubilized in an immunoprecipitation buffer that contained 1% Triton X-100, 50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1 mm EDTA, 1 mg/ml BSA, and protease inhibitor mixture. After centrifugation to remove debris, 5 μl of whole cell lysate (WCL) was set aside for later immunoblot experiments as “direct input.” The remaining WCL was subjected to immunoprecipitation as follows. After preclearing WCL by incubation in protein G beads, immunoprecipitating Ab was added, and the WCL/Ab mixture was incubated overnight at 4 °C. Then blocked protein G beads (nonspecific binding sites blocked by incubation in 3% BSA) were added, and the WCL/Ab/protein G beads mixture was incubated at 4 °C for 4 h with rocking. The beads were collected and washed, and the proteins were eluted by incubation in sample buffer containing 2% SDS. The original WCLs and immunoprecipitates were fractionated by SDS-PAGE. Part of the proteins was blotted to polyvinylidene difluoride membrane and probed by suitable Abs. The background-subtracted intensities of immunoreactive bands were quantified by densitometry (ChemiImager model 4400, α-Innotech). For the immunoprecipitates, the radioactivity of proteins in the gels was quantified by PhosphorImager. The following Abs were used. A V5 mouse mAb targeting the V5 epitope fused to the C terminus of Q1 was used to coimmunoprecipitate Q1 and Q1-associated E1. A Q1 goat pAb (Santa Cruz) and an E1 rabbit pAb (Alomone) were used in immunoblot quantification of protein levels. A Kv4.3 rabbit pAb (Alomone) was used to coimmunoprecipitate Kv4.3 and Kv4.3-associated KChIP2. Kv4.3 and KChIP2 mouse mAbs (NeuroMAb) were used in immunoblot quantification of protein levels.
Biotinylation Experiments
COS-7 cells were transfected with E1 or E2 cDNA alone or with Q1 cDNA (cDNA molar ratio of Ex:Q1 = 2:1). Two days later, the cells were washed with cold phosphate-buffered saline twice, followed by incubation in 0.4 mm amine-reactive, disulfide bond containing biotin derivative (EZ-link sulfo-NHS-SS-biotin; Pierce) on ice for 30 min. The biotinylation reaction was quenched by 25 mm Tris-HCl. The cells were washed with cold phosphate-buffered saline twice, digested with trypsin, and collected. The cells were lysed by sonication in 120 μl of lysis buffer (20 mm Tris-HCl, 0.2 m NaCl, 1 mm EDTA, 1% Triton X-100, with protease inhibitor mixture, pH 7.5). After centrifugation to remove debris, the supernatants (WCLs) were collected. The protein concentrations in WCLs were measured. For each sample, 10% of WCL was saved as direct input for the following immunoblot experiments. To the remaining WCL, a slurry of NeutrAvidin-agarose beads was added at 50 μl of beads/200 μg of protein, and the mixture was incubated at 4 °C overnight. The beads were collected and washed with lysis buffer six times. Biotinylated proteins were eluted by incubating beads in sample buffer containing mercaptoethanol. Biotinylated fractions, direct inputs, and negative control (NeutrAvidin-retrieved fraction from cells without biotin labeling) were loaded to 4–20% Tricine gel and fractionated by electrophoresis. After blotting the proteins to polyvinylidene difluoride membrane, the membrane was probed with E1, E2, or Q1 (V5) Ab. After ECL to quantify the E1, E2, and Q1 band intensities, the membrane was stripped and reprobed with an actin mAb. The background-subtracted band intensities were determined by densitometry.
Vesicle Injection and Oocyte Expression
The procedures were modified from those described previously (38, 39). COS-7 cells expressing E1 or c-Myc-tagged E2 were homogenized in a “vesicular buffer” (containing 50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1 mm EDTA, 1 mg/ml BSA, and protease inhibitor mixture) with a Teflon glass homogenizer. After low speed centrifugation to remove debris, the supernatant was centrifuged at 100,000 × g at 4 °C for 2 h. The pellet (membrane vesicles) was washed, aliquotted at 10 μl/vial, and stored at −80 °C till use. Protein expression of E1 and c-Myc-E2 was confirmed in an immunoblot experiment on solubilized vesicles.
The oocytes were uninjected (control oocytes) or injected with Q1 cRNA (10 ng/oocyte, Q1-oocytes). The procedures of cRNA transcription, oocyte preparation, and cRNA injection have been described before (12). Twenty-four h later, each oocyte was injected with 50 nl of vesicles. After incubating the oocytes for different amounts of time, incorporation of E1 or c-Myc-E2 to oocyte cell membrane was tested by voltage clamping or by cell surface chemiluminescence as described below.
Whole oocyte membrane currents were recorded using the “two-cushion pipette” voltage clamp method (40). Voltage clamp was done at room temperature (24–26 °C) in a low chlorine ND96 solution (96 mm NaOH, 2 mm KOH, 1.8 mm CaCl2, 2 mm MgSO4, 5 mm HEPES, 2.5 mm sodium pyruvate, titrated to pH 7.5 with methanesulfonic acid) with OC-725B or OC-725C amplifier (Warner Instruments). Voltage clamp protocol generation and data acquisition were controlled by pClamp 5.5 via a 12-bit D/A and A/D converter (DMA; Axon Instruments, CA). The current data were low pass filtered at 1 kHz (Frequency Devices) and stored on disks for off-line analysis.
Oocyte cell surface chemiluminescence was monitored as described before (41). Intact oocytes were incubated with c-Myc mAb (1 μg/ml) for 1 h at 4 °C. After washing with the ND96 solution 8 times, 5 min each, the oocytes were incubated with horseradish peroxidase-conjugated anti-mouse secondary Ab for 40 min at 4 °C. The oocytes were washed with ND96 5 times, 10 min each. Individual oocytes were placed in the wells of a 96-well plate in 50 μl of ND96. After adding 50 μl of SuperSignal ELISA femto maximum sensitivity substrate (Pierce) and allowing the reaction to proceed for 20 s at room temperature, the signals at 405 nm were integrated for 10 s using a luminator (Wallac Victor 2).
Adult Guinea Pig Ventricular Myocyte Primary Culture, Adenovirus Infection, Myocyte Patch Clamp Recording, and Detecting Immunofluorescence with Confocal Microscopy
Guinea pig ventricular myocytes were isolated as previously described (31) under sterile conditions. The cells were allowed to recover in KB for 2 h at room temperature. Then the medium was changed to nominally calcium-free normal Tyrode's (147 mm NaCl, 4 mm KCl, 5.5 mm dextrose, 5 mm HEPES, pH 7.3), and the calcium concentration was raised stepwise to 1 mm during 2 h. The cells were plated on laminin-coated coverslip and allowed to recover for ∼4 h in a serum-free medium (medium 199 (Invitrogen) supplemented with BSA, l-carnitine, creatine, taurine, and penicillin/streptomycin as previously described (42)) at 36 °C in a 5% CO2 moist incubator. Then the medium volume was reduced to 0.6 ml (per 35-mm dish), and adenovirus harboring eGFP or HA-tagged E2 (35) was added at ∼109 viral particles/dish. The cells were incubated with virus for 2 h at 36 °C in 5% CO2 incubator with occasional gentle shaking to facilitate contacts of viral particles with myocytes. Afterward, the dish was replenished with 2 ml of fresh medium described above. The cells were incubated for another 1–2 days before patch clamp recording or fixation/immunostaining for confocal microscopy.
Membrane voltage and currents were recorded using the whole cell variant of the patch clamp technique with AxoPatch200 amplifiers. Voltage clamp was controlled by Clampex of pClamp10 via Digidata 1440A. The pipette solution contained 125 mm potassium aspartate, 20 mm KCl, 10 mm potassium-ATP, 10 mm HEPES, 10 mm EGTA, and 1 mm magnesium, pH 7.3. The tip resistance ranged from 1.5 to 2.5 mΩ, and 95% of the series resistance was compensated. The myocytes were superfused with normal Tyrode's (2 mm CaCl2 added to the nominally calcium-free Tyrode's described above) at 33–34 °C. After forming whole cell recording configuration, the bath solution was switched to a sodium- and calcium-free Tyrode's (NaCl and CaCl2 substituted with equimolar choline-Cl and MgCl2). This solution removed the interference from sodium and calcium channel currents as well as Na/Ca exchanger current, facilitating the quantification of IKs.
The myocytes were fixed in 4% paraformaldehyde, permeabilized by 0.2% Triton X-100, and probed with the following primary/secondary antibody pairs: Q1 goat pAb (C-20, Santa Cruz)/Alexa568-conjugated anti-goat and HA mouse mAb/Alexa488-conjugated anti-mouse. The nuclei were stained by 4′,6-diamidino-2-phenylindole. The myocytes were viewed with a Zeiss 510 Meta confocal laser-scanning microscope. The pixel contents of Q1 immunofluorescence were quantified using ImageJ.
RESULTS
Pulse-Chase Experiments Reveal a Turnover of KCNE1 Subunit in the KCNQ1-KCNE1 Complex
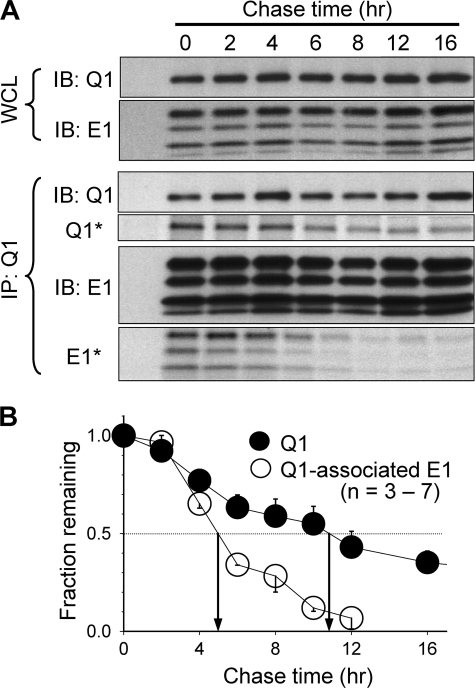
COS-7 cells coexpressing E1 and Q1 were metabolically labeled with [35S]Met for 1 h. After different amounts of chase time, the cells were lysed and incubated in a buffer containing 1% Triton X-100. This mild detergent treatment was sufficient to solubilize the cell membrane without disrupting noncovalent protein-protein associations (43). Q1 and Q1-associated E1 were coimmunoprecipitated from the WCLs. The time courses of decay of radioactivity in Q1 and Q1-associated E1 were measured from the immunoprecipitates (IPs). In these experiments, the protein levels of total Q1 and E1 in WCLs, as well as protein levels of Q1 and Q1-associated E1 in IPs, were also measured. As is shown in the top two rows of Fig. 2A (IB: Q1 and IB: E1 in WCL), the whole cell levels of Q1 and E1 proteins were quite stable. This indicates that during a chase time of up to 16 h, as labeled Q1 and E1 were degraded, unlabeled proteins were synthesized so that the whole cell pool of Q1 and E1 proteins was maintained at a steady-state. As is shown in the Q1 and E1 immunoblots of the IPs (Fig. 2A, IB: Q1 and IB: E1 in IP: Q1), the protein levels of Q1 and Q1-associated E1 were also stable during the chase times. These data confirm that we have achieved a consistent coimmunoprecipitation of Q1 and Q1-associated E1. Therefore, the decrease in radioactivity after longer chase times (Q1* and E1* in IP: Q1) was not due to a loss of proteins. Instead, the decrease in radioactivity reflected a dilution of labeled proteins by unlabeled counterparts.
FIGURE 2.
Pulse-chase experiments to test the turnover rates of KCNQ1 and KCNQ1-associated KCNE1. A, immunoblot and autoradiograph images from a representative experiment. COS-7 cells expressing Q1 and E1 were metabolically labeled with [35S]Met for 1 h and chased with unlabeled Met for up to 16 h (chase times marked at the top). WCLs were subjected to Q1 and E1 coimmunoprecipitation with V5 mAb targeting the V5 epitope engineered into the C terminus of Q1 and Q1-associated E1. Upper two rows, monitoring total Q1 and E1 protein levels in WCLs. Lower four rows, monitoring protein levels (IB: Q1 and IB: E1) and radioactivity levels (Q1* and E1*) of Q1 and Q1-associated E1 in the immunoprecipitates (IP: Q1). These data are from the same gel: immunoprecipitates were fractionated by SDS-PAGE, and part of the proteins was blotted to polyvinylidene difluoride membrane for immunoblot (IB) measurement, and the remaining proteins in the gel were used for radioactivity measurement. B, data summary. Autoradiograph band intensities at different chase times were normalized to the value at time zero (fraction remaining) and plotted against chase times. The multiple E1 bands represented differentially glycosylated forms (12), and band intensities were combined for quantification. Shown are the means ± S.E. from three to seven experiments. Arrows point to the estimated half-time (T½) of turnover for Q1 and Q1-associated E1 (11 and 5 h, respectively).
A comparison between Q1 and Q1-associated E1 in terms of the rate of decay of radioactivity will reveal the stability of their partnership. If Q1 and E1 form a stable complex that does not dissociate during the proteins' life span in the cell, they will be degraded simultaneously. In this case, we expect to see a similar rate of decay of radioactivity in Q1 and Q1-associated E1. On the other hand, if labeled E1 can dissociate from the radioactive Q1-E1 complex and the vacancy can be filled by newly synthesized (and thus unlabeled) E1 during the chase time, we expect to see that the radioactivity of Q1-associated E1 decays faster than that of Q1. Fig. 2B summarizes the time courses of decay of radioactivity pooled from 3 to 7 experiments. The estimated half-time of decay of radioactivity of Q1-associated E1 is much shorter than that of Q1 (5 and 11 h, respectively). These data indicate that labeled E1 subunits have dissociated from Q1-E1 complexes and are replaced by unlabeled E1 peptides, i.e. there is a turnover of E1 subunit in the Q1-E1 complex. Our biochemical data support a previous functional study (44): oocytes injected with Q1 and E1 cRNAs manifested typical IKs currents in initial days following cRNA injection. However, after longer incubation (up to 14 days), the currents gradually lost the E1-endowed phenotype and became more like Q1 expressed alone, as if E1 gradually dissociated from the Q1-E1 complexes.
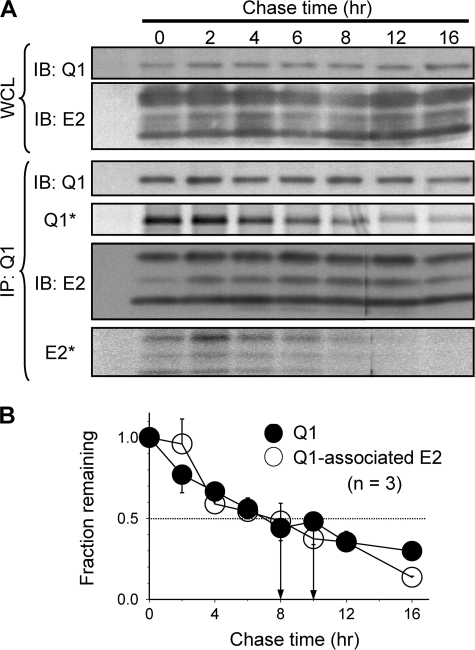
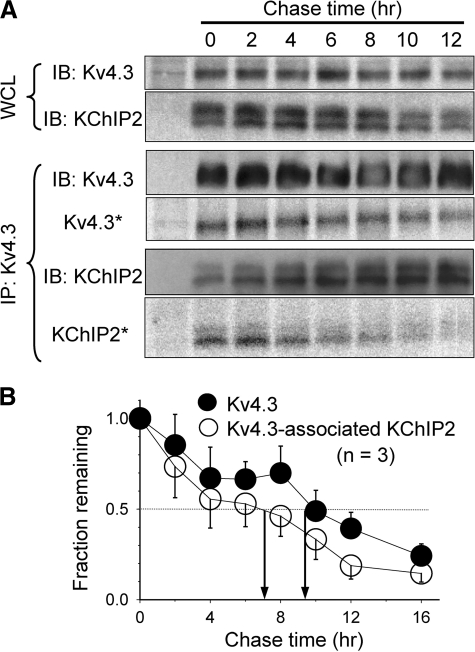
To test whether this is the general case for Kv channel α/β subunit association or whether it is a property of the Q1-E1 complex, we used the same pulse-chase approach to probe the stability of Q1-E2 and Kv4.3-KChIP2 complexes in COS-7 cells. The levels of radioactivity in Q1 and Q1-associated E2 decayed at a similar rate (estimated T½ of decay: 10 and 8 h, respectively; Fig. 3). The same was true for Kv4.3 and Kv4.3-associated KChIP2 (T½ of decay: 9 and 7 h, respectively; Fig. 4). Therefore, the partnerships of Q1-E2 and Kv4.3-KChIP2 complexes were more stable than that of Q1-E1. The data shown in Fig. 2 therefore support a dynamic partnership between Q1 and E1, i.e. process (1) in Fig. 1B occurs in cells.
FIGURE 3.
Pulse-chase experiments to test the turnover rates of KCNQ1 and KCNQ1-associated KCNE2. The experimental procedures and format of data presentation are the same as those described for Fig. 2. The multiple E2 bands represented differentially glycosylated forms (21), and band intensities were combined for quantification. The data in B were summarized from three experiments. The estimated T½ values of turnover of Q1 and Q1-associated E2 are 10 and 8 h, respectively. IB, immunoblot.
FIGURE 4.
Pulse-chase experiments to test the turnover rates of Kv4. 3 and Kv4.3-associated KChIP2. The experimental procedures and format of data presentation are the same as those described for Fig. 2. KChIP2 migrates as two closely spaced bands. Both bands were included in quantification. The data in B were summarized from three experiments. The estimated T½ values of turnover of Kv4.3 and Kv4.3-associated KChIP2 are 9 and 7 h, respectively. IB, immunoblot.
Biotinylation Experiments Reveal That KCNE2 Achieves a Higher Cell Surface Expression That Is Less Dependent on KCNQ1 than KCNE1
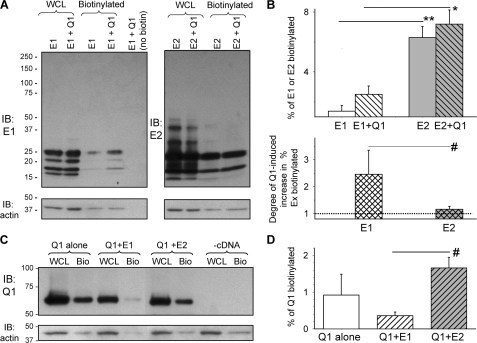
Previously, it has been shown in COS-7 and Chinese hamster ovary cells that E1 requires Q1 coassembly to reach the cell surface (45). To test whether this is the case for E2, we performed biotinylation experiments to quantify the steady-state cell surface E2 level when expressed alone or coexpressed with Q1 (n = 6). For comparison, we also quantified steady-state cell surface E1 level in the absence and presence of Q1 (n = 4). COS-7 cells were transfected with E2 or E1, alone or with Q1. Forty-eight h after transfection, the cells were incubated with sulfo-NHS-SS-biotin for 30 min on ice (to minimize internalization). This biotin derivative specifically reacted with ϵ-amines of lysine side chains exposed on the cell surface (two each on E1 and E2). The degree of cell surface expression was calculated as described in Fig. 5 legend. The degree of contamination of biotinylated fraction by cytosolic proteins (caused by unintentional cell membrane breakage or biotin internalization) was quantified by the biotinylated fraction of actin. It amounted to 2.4 ± 0.3% (Fig. 5A, lower panels) and was corrected.
FIGURE 5.
Biotinylation experiments to test cell surface KCNE1, KCNE2, and KCNQ1 expression. A and B, cell surface E1 and E2 expression and influence of Q1 coassembly. Surface proteins bearing exposed lysine residues in COS-7 cells were labeled with an amine-reactive biotin derivative on ice for 30 min. After cell lysis, 10% of the WCLs were reserved for direct immunoblotting, and the remaining WCLs were incubated with NeutrAvidin-conjugated agarose beads to retrieve biotinylated proteins. WCL, biotinylated (cell surface) fraction, and transfected cDNA(s) are marked at the top. A, immunoblot images from a representative experiment. Upper panels, E1 (left panel) and E2 (right panel) immunoblots. Lower panels, actin immunoblots from the same membranes after stripping. The rightmost lane in the left panel is NeutrAvidin-retrieved fraction from whole cell lysates without biotin label (control for NeutrAvidin specificity). Band intensities were determined by densitometry and background-subtracted. The multiple (differentially glycosylated) E1 or E2 bands were combined for quantification. The percentage of cell surface fraction was calculated as: % biotinylated = 100*[biotinylated/(biotinylated + WCL*10)], where biotinylated and WCL denote immunoblot band intensities in these two fractions. The band intensity values for WCL needed to be multiplied by 10 because only 10% of WCL was used for direct immunoblotting. B, data summary. Upper panel, percentage of total E1 or E2 that was biotinylated, expressed alone or coexpressed with Q1. Lower panel, increase in biotinylated E1 or E2 by Q1 coexpression, expressed as ratio of the percentage biotinylated Ex in “Ex+Q1” versus “Ex alone”. C and D, cell surface Q1 expression and influence of E1 or E2 coassembly. C, representative immunoblot images. Upper panel, Q1 immunoblots of WCL and biotinylated fraction (Bio). Lower panel, actin immunoblot images from the same gels. The cDNAs used for transfection are listed on top; -cDNA represents untransfected control COS-7 cells. D, percentage of total Q1 that was biotinylated when expressed alone or coexpressed with E1 or E2. The data in B and D were pooled from three to six experiments. **, p < 0.001; *, p < 0.01; #, p < 0.05.
Fig. 5A and the upper panel of Fig. 5B show that E1 alone was poorly expressed in the cell surface (1.4 ± 0.4% of total). Coexpression with Q1 increased the cell surface E1 level to 2.5 ± 0.6% of total, a factor of 2.5 ± 0.9. On the other hand, E2 expressed alone achieved a significantly higher cell surface expression (6.3 ± 0.8% of total, p = 0.001 versus E1 expressed alone). Q1 coexpression had little impact (E2+Q1, cell surface E2 expression 7.2 ± 1.0% of total, a ratio of only 1.2 ± 0.1). The difference in the increase in the percentage of cell surface level caused by Q1 coexpression between E1 and E2 was significant (Fig. 5B, lower panel). These data support the likelihood of process (2) in Fig. 1B: E2 can achieve cell surface expression independent of Q1.
With such a distinct difference between E1 and E2 in their ability to exit ER and reach the plasma membrane (37), can they influence the degree of Q1 cell surface expression when coexpressed with Q1? We quantified the Q1 cell surface level when expressed alone or coexpressed with E1 or E2. Fig. 5C depicts immunoblot images from a representative experiment. The data averaged from three experiments show that Q1 can achieve a higher cell surface expression when coexpressed with E2 than with E1 (Fig. 5D, p < 0.01).
Vesicle Injection Experiments in Oocytes Reveal That Free KCNE Subunits Delivered to Cell Membranes Can Associate with KCNQ1 Channels and Modulate Their Function
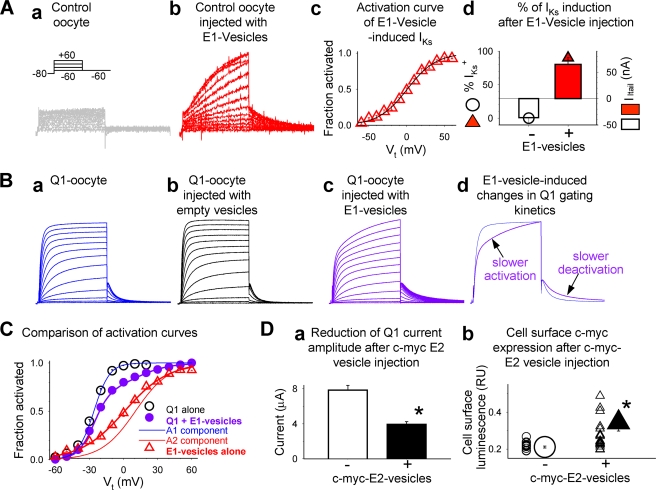
It has been shown that vesicles containing transmembrane channels or receptors injected into oocytes can fuse with oocyte cell membrane, leading to an incorporation of functional channels or receptors in the oocyte surface membrane (38, 39). Injecting vesicles prepared from E1-expressing COS-7 cells (E1 vesicles) into control oocytes (expressing endogenous xQ1 (3)) produced a typical IKs current within a few hours, and the current was stable for up to 22 h (Fig. 6A). Injecting E1 vesicles into Q1-oocytes (expressing large Q1 current encoded by injected Q1 cRNA) induced a slow phase of current activation and deactivation, similar to the time course of IKs (Fig. 6B). Analysis of the voltage dependence of activation in Q1-oocytes injected with E1 vesicles suggests that there are 2 channel populations: Q1 alone and E1-modified Q1 (Fig. 6C). On the other hand, injecting empty vesicles (prepared from untransfected COS-7 cells) into Q1-oocytes had no effect on either the gating kinetics (Fig. 6B) or the voltage dependence of activation (data not shown). These data support the efficiency of delivering KCNE subunits to the oocyte cell surface membrane by injecting KCNE-bearing vesicles. Furthermore, after the arrival at the cell surface membrane the E1 subunits can move laterally and associate with native or exogenous Q1 channels to modulate their function in the same manner as seen in Q1 + E1 coexpression experiments.
FIGURE 6.
Oocyte vesicle injection experiments to test whether KCNE peptides delivered to the cell membranes can modulate KCNQ1 channel function. The vesicles were prepared from COS-7 cells transfected with cDNA encoding E1 or c-Myc-E2 (c-Myc epitope engineered into E2 extracellular domain (34)) or without transfection (E1, c-Myc E2, and empty vesicles, respectively). The vesicles were injected into control oocytes (without cRNA injection) or Q1-oocytes (oocytes injected with Q1 cRNA, 10 ng/oocyte, 24 h before vesicle injection). All current traces were elicited by the voltage clamp protocol diagrammed in panel a of A: from a holding voltage of −80 mV, 2-s pulses to test voltage (Vt) ranging from −60 to +60 mV in 10-mV increments are applied once every 15 s. The test pulses were followed by a repolarizing step to −60 mV to monitor tail current amplitudes. A, testing the effects of injecting E1 vesicles into control oocytes. Panel a, representative current traces recorded from a control oocyte. Panel b, IKs current detected in a control oocyte ∼10 h after E1 vesicle injection. Panel c, average isochronal (2 s) activation curve of IKs current recorded from control oocytes injected with E1 vesicles. Peak amplitudes of tail currents were normalized by the estimated maximal tail current amplitude (fraction activated) and plotted against Vt. The data were fit with a single Boltzmann function: fraction activated = 1/[1 + exp ((V0.5 − Vt)/k)], where V0.5 (1.0 ± 2.2 mV) and k (19.1 ± 0.6 mV) are half-maximal activation voltage and slope factor (n = 11). Panel d, percentage of E1 vesicle injected oocytes that manifested IKs (% IKs+, symbols, left coordinate) and the mean IKs current amplitude in these oocytes (measured from peak amplitude of tail currents after Vt +60 mV, histogram bars, right coordinate). The data were collected from 14 oocytes without E1 vesicle injection, and from 22 oocytes 8–22 h after E1 vesicle injection. B, testing the effects of injecting E1 or empty vesicles into Q1-oocytes. Panel a, current traces from a Q1-oocyte without vesicle injection. Panel b, current traces from a Q1-oocyte ∼20 h after empty vesicle injection. Panel c, current traces from a Q1-oocyte ∼10 h after E1 vesicle injection. Panel d, superimposed current traces from two Q1-oocytes, one without (blue) and the other with (purple) E1 vesicle injection. The arrows point to the slow activation and deactivation phases induced by E1 vesicles. C, average isochronal activation curves from three groups of oocytes: 1) Q1-oocytes without vesicle injection (Q1 alone, black open circles), 2) Q1-oocytes injected with E1 vesicles (purple closed circles), and 3) control oocytes injected with E1 vesicles (E1 vesicles alone, same data as in panel c of A). Group 1 data were fit with a single Boltzmann function as described above (V0.5 = −27.0 ± 0.6 mV, k = 7.6 ± 0.2 mV, n = 5). Group 2 data were fit with a double Boltzmann function: fraction activated = A1/[1 + exp ((V1 − Vt)/k1)] + A2/[1 + exp ((V2 − Vt)/k2)], where A1 (0.70 ± 0.04) and A2 (0.30 ± 0.04) represent the fractions of current activated in the negative and positive voltage ranges, V1 (−26.0 ± 0.6 mV), k1 (6.9 ± 0.3 mV), V2 (10.4 ± 3.8 mV), and k2 (14.9 ± 0.7 mV) are the half-maximum voltages and slope factors of the two components (n = 13). The thin curves are activation curves calculated for the A1 (blue) and A2 (red) components. D, injecting c-Myc E2 vesicles led to a decrease in Q1 current amplitude and oocyte cell surface expression of c-Myc epitope. Panel a, mean current amplitudes measured as time-dependent currents during a 2-s step to +40 mV in Q1-oocytes injected with empty or c-Myc E2 vesicles (open and closed histogram bars, respectively). Panel b, magnitudes of cell surface luminescence in Q1-oocytes without or with c-Myc E2 vesicle injection (3–22 h after vesicle injection). The data from individual oocytes are shown as small symbols, and the mean data are shown as large symbols. *, p < 0.01.
After validating the vesicle injection technique, we tested the effects of delivering E2 to oocyte cell membrane on Q1 channel function. Injecting c-Myc E2 vesicles into Q1-oocytes caused a significant decrease in the current amplitude (Fig. 6D, panel a). This is consistent with the current-suppressing effect of E2 on Q1 (31). This likely resulted from incorporation of E2 subunits into the oocyte cell membrane, because we detected cell surface c-Myc epitope (fused to the E2 extracellular domain (34)) by immunoluminescence in intact oocytes (41) (Fig. 6D, panel b). These data support process (3) in Fig. 1B: free E2 subunits present in the cell surface membrane can associate with Q1 channels and modulate their function.
We further tested whether injecting E2 vesicles into oocytes that had been injected with Q1 and E1 cRNAs could cause a decrease in the IKs current amplitude without altering its gating kinetics, as was expected based on our previous observations (31). We made repeated attempts, with alterations in the quantity of Q1 + E1 cRNAs injected (10/3 and 5/1.5 ng/oocyte) and the timing of vesicle injection after cRNA injection (ranging from a few hrs to 48 h) but could not detect any sign of E2 suppression of Q1-E1 current amplitude in these oocytes. We suspect that the amount of E2 peptides delivered in 50 nl of vesicle suspension was limited and could not effectively compete with E1 in the Q1-E1 complexes.
Adenovirus-mediated KCNE2 Expression in Adult Guinea Pig Ventricular Myocytes Causes a Decrease in the Native IKs Current Density
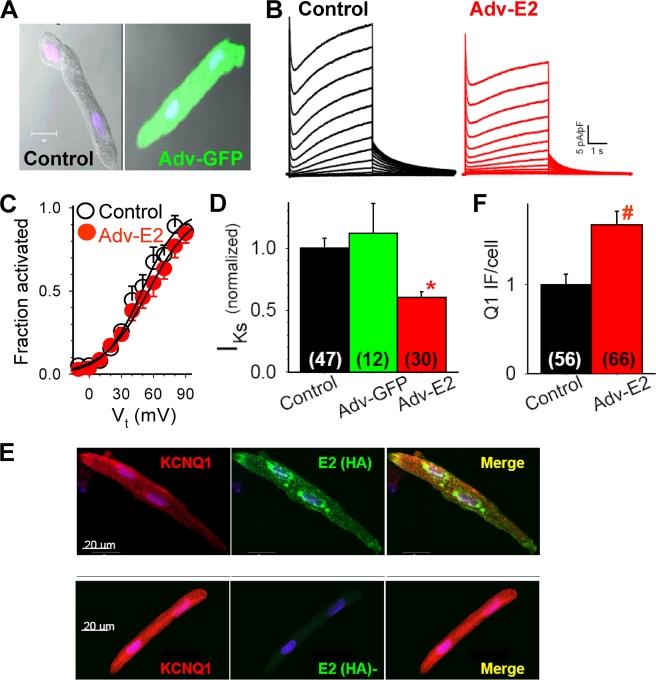
We set up the adult myocyte primary culture technique (42). Ventricular myocytes isolated from young adult guinea pigs (2–3 month) could be maintained in a serum-free medium for 4–5 days while maintaining an elongated morphology and clear striations (Fig. 7A). We used adenovirus carrying the eGFP gene (Adv-GFP) to test the efficiency of gene transfer into these adult myocytes. Incubating myocytes with ∼ 2 × 1010 viral particles/ml for 2 h, followed by 24–36 h of culture, led to 98% GFP+ myocytes (Fig. 7A). Importantly, adenovirus-mediated eGFP gene transfer did not affect the IKs current density relative to time control myocytes (myocytes incubated under the same conditions for a similar amount of time without adenovirus infection).
FIGURE 7.
Adenovirus-mediated KCNE2 expression in adult guinea pig ventricular myocytes to test whether KCNE2 can associate with native KCNQ1-KCNE1 to modulate IKs current density. Adult (2–3 months old) guinea pig ventricular myocytes were maintained in serum-free medium for 4 days. The myocytes were infected with adenoviruses harboring eGPF gene (Adv-GFP) or KCNE2 (Adv-E2, with an HA tag engineered into the C terminus) (35) at ∼ 2 × 1010 particles/ml for 2 h. Whole cell currents were recorded on days 3 and 4. A, top panel, superimposed differential interference contrast and fluorescence images of two myocytes, the one on the right infected with Adv-GFP and the one on the left as time control. B, Adv-E2 infection reduced IKs without altering gating kinetics. C, Adv-E2 infection did not change the voltage dependence of IKs activation. D, IKs current density in time control, Adv-GFP, and Adv-E2 infected guinea pig ventricular myocytes (normalized by the mean value in control myocytes). E, confocal images of immunofluorescence from two cultured guinea pig ventricular myocytes, stained for Q1 (Q1 goat Ab/Alexa568 anti-goat, left panel), and HA tag of E2 (HA mouse Ab/Alexa488 anti-mouse, middle panel). Right panel, merge of Alexa568 and Alexa488 signals. F, pixel contents of Q1 immunofluorescence (Q1 IF) per cell in control and Adv-E2-infected myocytes (normalized to the mean value from control myocytes). In D and F, the numbers of cells studied are listed in parentheses. *, p < 0.01.
After confirming the efficiency of adenovirus-mediated gene transfer and the lack of effects on IKs by this process per se, we proceeded to test the effects of adenovirus-mediated E2 (with an HA tag fused to the C terminus) (35) expression on native IKs. Relative to time controls, myocytes infected with adenovirus-E2 (HA) manifested a significantly reduced mean IKs current density without changes in the gating kinetics or voltage dependence of activation (Fig. 7, B–D). To test whether the reduction in IKs current density was secondary to a decrease in the Q1 protein level, we used immunofluorescence/confocal microscopy to compare the pixel contents of Q1 immunofluorescence in myocytes expressing adenovirus-transfected E2 (monitored by the HA immunofluorescence) with those of myocytes without exogenous E2 expression. The myocytes were processed under the same conditions. Confocal images were obtained on the same day with identical setting in laser intensity and detector gain. The numerical aperture was adjusted to include fluorescence signals from an optical section of 10 μm (i.e. covering approximately the whole cell depth). Pixel contents of Q1 immunofluorescence were measured from 56 control (HA−) and 66 E2-expressing (HA+) myocytes. There was a modest increase in Q1 pixel contents in E2-expressing myocytes (Fig. 7F).
The HA immunofluorescence manifested the expected subcellular distribution pattern based on that of native E2 in cardiac myocytes (21, 31): t-tubules and cell surface (Fig. 7E). However, there were often strong HA immunofluorescence signals in the perinuclear region, reflecting a high level of E2-HA biosynthesis activity in the ER-Golgi apparatus. Importantly, the signals of HA immunofluorescence and Q1 immunofluorescence overlapped in the t-tubules and cell surface (Merge in Fig. 7E), supporting a colocalization of these two proteins in the myocytes. These data are consistent with observations from heterologous experiments: E2 can associate with the IKs (Q1-E1) channel complex to reduce the current amplitude without affecting the slow gating kinetics typical of the cardiac IKs channels (31).
DISCUSSION
Our major findings and data interpretation can be summarized as follows. First, pulse-chase experiments showed that there was a faster decay of 35S radioactivity in Q1-associated E1 than that of Q1, indicating a time-dependent dilution of 35S-labeled E1 in the Q1-E1 complex by unlabeled E1, i.e. E1 can dissociate from Q1 and be replaced by new E1. Second, when expressed alone in COS-7 cells, the steady-state cell surface E2 level was much higher than that of E1. Q1 coexpression increased E1 cell surface level but did not markedly impact on the cell surface E2 level. These data suggest that E1 requires Q1 coassembly for cell surface expression, whereas E2 can traffic to the cell surface by itself. Third, E1 vesicle injection experiments in oocytes validated the efficiency of delivering functional KCNE subunits to the oocyte cell surface that could then associate with Q1 channels to modulate their function. Injecting c-Myc-tagged E2 vesicles into oocytes caused a decrease in Q1 current amplitude and an appearance of cell surface c-Myc epitope, supporting the notion that functional E2 can associate with Q1 channels that have vacancies in the KCNE-binding pockets and reduces the current amplitude. Finally, adenovirus-mediated E2 (with HA tag) expression in adult guinea pig ventricular myocytes caused a decrease in the native IKs current density, without altering its voltage dependence or kinetics of gating. Because the pixel contents of Q1 immunofluorescence were modestly increased by adenovirus-mediated E2 expression, the decrease in IKs current density could not be explained by a reduction in the Q1 protein level. Furthermore, there was a colocalization between HA immunofluorescence (reflecting exogenous E2) and native Q1. These data are consistent with our previous observations from heterologous expression experiments that E2 can associate with the IKs (Q1-E1) complex to suppress its current amplitude without altering its unique gating kinetics. Together, our data support the likelihood of processes (1), (2), and (3) outlined in Fig. 1B. We propose that E2 can function as a kinetically silent suppressor of native IKs current amplitude in cardiac myocytes.
Structural Basis for the Dynamic Partnership between KCNQ1 and KCNE1 Subunits
Because a Q1 channel optimally binds two KCNE subunits (4, 25), the asymmetric structure will always leave vacancies in the KCNE-binding pockets (Fig. 1A). This will allow a competition between free KCNE subunits and Q1-bound KCNE subunits for binding to the pockets. On the other hand, KChIP2 binds to the cytoplasmic N-terminal domain of Kv4.3 channels in a 4:4 stoichiometry (46). This symmetric structure leaves no room for further KChIP2 binding, contributing to a more stable partnership.
The E2 turnover time in the Q1-E2 complex was similar to that of Q1, indicating a more stable Q1-E2 partnership than that of Q1-E1. KCNE association with the Q1 channel was mediated mainly by the KCNE transmembrane domain (47, 48). It has been suggested that the lower the transmembrane domain hydrophobicity, the higher the binding affinity of KCNE subunits to their partner α-subunit channels (48). This is not likely to be the reason for a more stable Q1-E2 partnership than Q1-E1, because the E2 transmembrane domain is modestly more hydrophobic than that of E1; the grand average of hydrophobicity (GRAVY) values (based on calculation using the Prot Param software at the ExPASy web site) are 2.378 and 2.083 for the transmembrane domains of E2 and E1, respectively. Recent data suggest Q1-KCNE interactions also occur in the cytoplasmic region (49). Extramembranous interactions between Q1 and E2 may contribute to a stable partnership between the two.
KCNQ1-KCNE Association, Trafficking, and IKs Current Amplitude in the Heart
In cardiac myocytes that coexpress Q1 with multiple KCNE subunits, the regulation of Q1-KCNE complex formation likely takes place at several levels. First, the expression levels of different KCNE subunits, and thus their availability, may influence the degree of their association with Q1 by mass action. Genes encoding these KCNE subunits have different core promoter elements (50). Their transcription is under differential regulation, depending on the cell type as well as physiological or pathological conditions of the heart (51–53). Second, coassembly of Q1 with KCNE subunits likely begins in the ER (31). Thus, the temporal and spatial relationship between Q1 and different KCNE subunits in terms of their transcription and translation will influence the pattern of their association. Third, the trafficking pattern of KCNE subunits can influence their association with Q1. A recent report showed that artificially forcing E2 to shuttle between ER and Golgi could increase the amount of E2 associated with hERG, another E2 partner α-subunit channel (37). It is suggested that E2 and hERG are mainly coassembled in the Golgi, and forcing E2 to shuttle between ER and Golgi can increase the chance of their encounter (37). An equivalent process is depicted in Fig. 1B (process 3′): E2 may exchange with E1 in the Q1-E1 complex during transits through the cytosolic vesicular compartments. However, under normal conditions, most E2 should traffic to the cell surface with short transit times in the cytosolic compartments (37). Therefore, the competition between E2 and E1 for binding to their partner α-subunit channels is likely to take place mainly in the cell surface membrane (process 3 in Fig. 1B).
Recent studies have revealed multi-tiered regulation of Q1 channel folding and trafficking in cells (49). The distal C termini of Q1subunits form a coiled-coil structure that mediates Q1-specific tetramerization (54). Furthermore, calmodulin binding to a proximal C-terminal domain is critical for Q1 folding and trafficking to the cell surface (55, 56). Our data suggest further influence on Q1 trafficking by coassembly with KCNE subunits: Q1 coexpressed with E2 exhibited a higher cell surface expression than Q1 coexpressed with E1 (Fig. 5, C and D). How does this reconcile with the observations that E1 coexpression increases Q1 current amplitude, whereas E2 coexpression exerts the opposite effect? The answer may lie in the effects of these two KCNE subunits on the single channel conductance of their partner α-subunit channels. E1 coassembly with Q1 increased the single channel conductance of Q1 (57, 58). Our previous observations suggested that E2 coassembly may reduce the Q1 single channel conductance (59): E2 coassembly altered ion selectivity of the Q1 channel pore, and a single point mutation in the E2 transmembrane domain (I63C) switched the E2 phenotype from suppressing Q1 current to enhancing Q1 current, i.e. Q1-(E2-I63C) became Q1-(E1-like) (59). There is also a precedent: E2 coassembly with hERG reduces the hERG single channel conductance (32).
Physiological Implications
IKs functions as a repolarization reserve in the heart (1). Previous studies have shown that stressful conditions in the heart can increase the IKs amplitude by two mechanisms involving Q1. First, an elevation in β-adrenergic tone will cause protein kinase A-mediated phosphorylation of serine at position 27 in Q1. This leads to a negative shift in the voltage dependence of IKs activation and an increase in IKs open probability within the plateau voltage range of cardiac action potentials (60). Second, the release of stress hormones such as cortisol can induce the serum- and glucocorticoid-inducible kinase 1, which enhances the trafficking of Q1-carrying vesicles to the cell surface and thus increases the IKs current amplitude (61). The findings reported here suggest that E2 may be a “silent” IKs suppressor. Thus, a shift in the balance between E1 and E2 expression levels may allow a dynamic control of IKs current amplitude. Indeed, it has been shown that heart failure is linked to an increase in E1 transcription but a decrease in E2 transcription (52). This may be a third mechanism by which stressful conditions in the heart cause an increase in the IKs current amplitude.
This work was supported, in whole or in part, by National Institutes of Health Grants HL-46451 (to G.-N. T.) and HL-28958.
- ER
- endoplasmic reticulum
- IP
- immunoprecipitate(s)
- BSA
- bovine serum albumin
- WCL
- whole cell lysate
- Ab
- antibody
- mAb
- monoclonal antibody
- pAb
- polyclonal antibody
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- GFP
- green fluorescent protein
- HA
- hemagglutinin.
REFERENCES
- 1.Jost M., Virág L., Bitay M., Takács J., Lengyel C., Biliczki P., Nagy Z., Bogáts G., Lathrop D. A., Papp J. G., Varró A. ( 2005) Circulation 112, 1392– 1399 [DOI] [PubMed] [Google Scholar]
- 2.Barhanin J., Lesage F., Guillemare E., Fink M., Lazdunski M., Romey G. ( 1996) Nature 384, 78– 80 [DOI] [PubMed] [Google Scholar]
- 3.Sanguinetti M. C., Curran M. E., Zou A., Shen J., Spector P. S., Atkinson D. L., Keating M. T. ( 1996) Nature 384, 80– 83 [DOI] [PubMed] [Google Scholar]
- 4.Chen H., Kim L. A., Rajan S., Xu S., Goldstein S. A. N. ( 2003) Neuron 40, 15– 23 [DOI] [PubMed] [Google Scholar]
- 5.Tapper A. R., George A. L., Jr. ( 2001) J. Biol. Chem. 276, 38249– 38254 [DOI] [PubMed] [Google Scholar]
- 6.Kurokawa J., Motoike H. K., Kass R. S. ( 2001) J. Gen. Physiol. 117, 43– 52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Melman Y. F., Um S. Y., Krumerman A., Kagan A., McDonald T. V. ( 2004) Neuron 42, 927– 937 [DOI] [PubMed] [Google Scholar]
- 8.Panaghie G., Tai K. K., Abbott G. W. ( 2006) J. Physiol. 570, 455– 467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seebohm G., Strutz-Seebohm N., Ureche O., Baltaev R., Lampert A., Kornichuk G., Kamiya K., Wuttke T. V., Lerche H., Sanguinetti M. C., Lang F. ( 2006) Biophys. J. 90, 2235– 2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rocheleau J. M., Kobertz W. R. ( 2008) J. Gen. Physiol. 131, 59– 68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakajo K., Kubo Y. ( 2007) J. Gen. Physiol. 130, 269– 281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu X., Jiang M., Hsu K. L., Zhang M., Tseng G. N. ( 2008) J. Gen. Physiol. 131, 589– 603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang M., Cabo C., Yao J., Boyden P. A., Tseng G. N. ( 2000) Cardiovasc. Res. 48, 34– 43 [DOI] [PubMed] [Google Scholar]
- 14.Liu X. S., Jiang M., Zhang M., Tang D., Clemo H. F., Higgins R. S., Tseng G. N. ( 2007) Am. J. Physiol. Heart Circ. Physiol. 292, H560– 571 [DOI] [PubMed] [Google Scholar]
- 15.Splawski I., Shen J., Timothy K. W., Lehmann M. H., Priori S., Robinson J. L., Moss A. J., Schwartz P. J., Towbin J. A., Vincent G. M., Keating M. T. ( 2000) Circulation 102, 1178– 1185 [DOI] [PubMed] [Google Scholar]
- 16.Chen Y. H., Xu S. J., Bendahhou S., Wang X. L., Wang Y., Xu W. Y., Jin H. W., Sun H., Su X. Y., Zhuang Q. N., Yang Y. Q., Li Y. B., Liu Y., Xu H. J., Li X. F., Ma N., Mou C. P., Chen Z., Barhanin J., Huang W. ( 2003) Science 299, 251– 254 [DOI] [PubMed] [Google Scholar]
- 17.Hong K., Piper D. R., Diaz-Valdecantos A., Brugada J., Oliva A., Burashnikov E., Santos-de-Soto J., Grueso-Montero J., Diaz-Enfante E., Brugada P. F., Sanguinetti M. C., Brugada R. ( 2005) Cardiovasc. Res. 68, 433– 440 [DOI] [PubMed] [Google Scholar]
- 18.Ruan Y., Liu N., Napolitano C., Priori S. G. ( 2008) Circ. Arrhythmia Electrophysiol. 1, 290– 297 [DOI] [PubMed] [Google Scholar]
- 19.McCrossan Z. A., Abbott G. W. ( 2004) Neuropharmacology 47, 787– 821 [DOI] [PubMed] [Google Scholar]
- 20.Bendahhou S., Marionneau C., Haurogne K., Larroque M. M., Derand R., Szuts V., Escande D., Demolombe S., Barhanin J. ( 2005) Cardiovasc. Res. 67, 529– 538 [DOI] [PubMed] [Google Scholar]
- 21.Jiang M., Zhang M., Tang D. G., Clemo H. F., Liu J., Holwitt D., Kasirajan V., Pond A. L., Wettwer E., Tseng G.-N. ( 2004) Circulation 109, 1783– 1788 [DOI] [PubMed] [Google Scholar]
- 22.Delpon E., Cordeiro J. M., Nunez L., Thomsen P. E. B., Guerchicoff A., Pollevick G. D., Wu Y., Kanters J. K., Larsen C. T., Burashnikov E., Christiansen M., Antzelevitch C. ( 2008) Circ. Arrhythmia Electrophysiol. 1, 209– 218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manderfield L. J., George A. L., Jr. ( 2008) FEBS J. 275, 1336– 1349 [DOI] [PubMed] [Google Scholar]
- 24.Tseng G. N. ( 2007) Heart Rhythm 4, 1542– 1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morin T. J., Kobertz W. R. ( 2008) Proc. Natl. Acad. Sci. U. S. A. 105, 1478– 1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Long S. B., Campbell E. B., MacKinnon R. ( 2005) Science 309, 897– 903 [DOI] [PubMed] [Google Scholar]
- 27.Long S. B., Tao X., Campbell E. B., MacKinnon R. ( 2007) Nature 450, 376– 382 [DOI] [PubMed] [Google Scholar]
- 28.Kang C., Tian C., Sönnichsen F. D., Smith J. A., Meiler J., George A. L., Jr., Vanoye C. G., Kim H. J., Sanders C. R. ( 2008) Biochemistry 47, 7999– 8006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang K. W., Tai K. K., Goldstein S. A. N. ( 1996) Neuron 16, 571– 577 [DOI] [PubMed] [Google Scholar]
- 30.Morin T. J., Kobertz W. R. ( 2007) ACS Chem. Biol. 2, 469– 473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu D. M., Jiang M., Zhang M., Liu X. S., Korolkova Y. V., Tseng G. N. ( 2006) Heart Rhythm 3, 1469– 1480 [DOI] [PubMed] [Google Scholar]
- 32.Abbott G. W., Sesti F., Splawski I., Buck M. E., Lehmann M. H., Timothy K. W., Keating M. T., Goldstein S. A. N. ( 1999) Cell 97, 175– 187 [DOI] [PubMed] [Google Scholar]
- 33.Yang Y., Xia M., Jin Q., Bendahhou S., Shi J., Chen Y., Liang B., Lin J., Liu Y., Liu B., Zhou Q., Zhang D., Wang R., Ma N., Su X., Niu K., Pei Y., Xu W., Chen Z., Wan H., Cui J., Barhanin J., Chen Y. ( 2004) Am. J. Hum. Genet. 75, 899– 905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang M., Jiang M., Tseng G. N. ( 2001) Circ. Res. 88, 1012– 1019 [DOI] [PubMed] [Google Scholar]
- 35.Qu J., Kryukova Y., Potapova I. A., Doronin S. V., Larsen M., Krishnamurthy G., Cohen I. S., Robinson R. B. ( 2004) J. Biol. Chem. 279, 43497– 43502 [DOI] [PubMed] [Google Scholar]
- 36.Tinel N., Diochot S., Borsotto M., Lazdunski M., Barhanin J. ( 2000) EMBO J. 19, 6326– 6330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Um S. Y., McDonald T. V. ( 2007) PLoS-One 2, e933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marsal J., Tigyi G., Miledi R. ( 1995) Proc. Natl. Acad. Sci. U. S. A. 92, 5224– 5228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tian C., Vanoye C. G., Kang C., Welch R. C., Kim H. J., George A. L., Jr., Sanders C. R. ( 2007) Biochemistry 46, 11459– 11472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schreibmayer W., Lester H. A., Dascal N. ( 1994) Pflugers Arch. 426, 453– 458 [DOI] [PubMed] [Google Scholar]
- 41.Rocheleau J. M., Gage S. D., Kobertz W. R. ( 2006) J. Gen. Physiol. 128, 721– 729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ellingsen O., Davidoff A. J., Prasad S. K., Berger H. J., Springhorn J. P., Marsh J. D., Kelly R. A., Smith T. W. ( 1993) Am. J. Physiol. Heart Circ. Physiol. 265, H747– 754 [DOI] [PubMed] [Google Scholar]
- 43.Nakahira K., Shi G., Rhodes K. J., Trimmer J. S. ( 1996) J. Biol. Chem. 271, 7084– 7089 [DOI] [PubMed] [Google Scholar]
- 44.Poulsen A. N., Klaerke D. A. ( 2007) Biochem. Biophys. Res. Commun. 363, 133– 139 [DOI] [PubMed] [Google Scholar]
- 45.Chandrasekhar K. D., Bas T., Kobertz W. R. ( 2006) J. Biol. Chem. 281, 40015– 40023 [DOI] [PubMed] [Google Scholar]
- 46.Wang H., Yan Y., Liu Q., Huang Y., Shen Y., Chen L., Chen Y., Yang Q., Hao Q., Wang K., Chai J. ( 2007) Nat. Neurosci. 10, 32– 39 [DOI] [PubMed] [Google Scholar]
- 47.Tapper A. R., George A. L., Jr. ( 2000) J. Gen. Physiol. 116, 379– 390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y., Sesti F. ( 2007) Biophys. J. 93, 3083– 3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haitin Y., Attali B. ( 2008) J. Physiol. 586, 1803– 1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lundquist A. L., Turner C. L., Ballester L. Y., George A. L., Jr. ( 2006) Genomics 87, 119– 128 [DOI] [PubMed] [Google Scholar]
- 51.Gaborit N., Steenman M., Lamirault G., Le Meur N., Le Bouter S., Lande G., Léger J., Charpentier F., Christ T., Dobrev D., Escande D., Nattel S., Demolombe S. ( 2005) Circulation 112, 471– 481 [DOI] [PubMed] [Google Scholar]
- 52.Lundquist A. L., Manderfield L. J., Vanoye C. G., Rogers C. S., Donahue B. S., Chang P. A., Drinkwater D. C., Murray K. T., George A. L., Jr. ( 2005) J. Mol. Cell Cardiol. 38, 277– 287 [DOI] [PubMed] [Google Scholar]
- 53.Radicke S., Cotella D., Graf E. M., Banse U., Jost N., Varró A., Tseng G. N., Ravens U., Wettwer E. ( 2006) Cardiovasc. Res. 71, 695– 703 [DOI] [PubMed] [Google Scholar]
- 54.Wiener R., Haitin Y., Shamgar L., Fernández-Alonso M. C., Martos A., Chomsky-Hecht O., Rivas G., Attali B., Hirsch J. A. ( 2008) J. Biol. Chem. 283, 5815– 5830 [DOI] [PubMed] [Google Scholar]
- 55.Ghosh S., Nunziato D. A., Pitt G. S. ( 2006) Circ. Res. 98, 1048– 1054 [DOI] [PubMed] [Google Scholar]
- 56.Shamgar L., Ma L., Schmitt N., Haitin Y., Peretz A., Wiener R., Hirsch J., Pongs O., Attali B. ( 2006) Circ. Res. 98, 1055– 1063 [DOI] [PubMed] [Google Scholar]
- 57.Sesti F., Goldstein S. A. N. ( 1998) J. Gen. Physiol. 112, 651– 663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang Y., Sigworth F. J. ( 1998) J. Gen. Physiol. 112, 665– 678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu X. S., Zhang M., Jiang M., Wu D. M., Tseng G. N. ( 2007) J. Membr. Biol. 216, 117– 127 [DOI] [PubMed] [Google Scholar]
- 60.Marx S. O., Kurokawa J., Reiken S., Motoike H., D'Armiento J., Marks A. R., Kass R. S. ( 2002) Science 295, 496– 499 [DOI] [PubMed] [Google Scholar]
- 61.Seebohm G., Strutz-Seebohm N., Birkin R., Dell G., Bucci C., Spinosa M. R., Baltaev R., Mack A. F., Korniychuk G., Choudhury A., Marks D., Pagano R. E., Attali B., Pfeufer A., Kass R. S., Sanguinetti M. C., Tavare J. M., Lang F. ( 2007) Circ. Res. 100, 686– 692 [DOI] [PubMed] [Google Scholar]