Abstract
Mitochondrial F1-ATPase contains a hexamer of alternating α and β subunits. The assembly of this structure requires two specialized chaperones, Atp11p and Atp12p, that bind transiently to β and α. In the absence of Atp11p and Atp12p, the hexamer is not formed, and α and β precipitate as large insoluble aggregates. An early model for the mechanism of chaperone-mediated F1 assembly (Wang, Z. G., Sheluho, D., Gatti, D. L., and Ackerman, S. H. (2000) EMBO J. 19, 1486–1493) hypothesized that the chaperones themselves look very much like the α and β subunits, and proposed an exchange of Atp11p for α and of Atp12p for β; the driving force for the exchange was expected to be a higher affinity of α and β for each other than for the respective chaperone partners. One important feature of this model was the prediction that as long as Atp11p is bound to β and Atp12p is bound to α, the two F1 subunits cannot interact at either the catalytic site or the noncatalytic site interface. Here we present the structures of Atp11p from Candida glabrata and Atp12p from Paracoccus denitrificans, and we show that some features of the Wang model are correct, namely that binding of the chaperones to α and β prevents further interactions between these F1 subunits. However, Atp11p and Atp12p do not resemble α or β, and it is instead the F1 γ subunit that initiates the release of the chaperones from α and β and their further assembly into the mature complex.
Mitochondrial F1-ATPase consists of three α and three β subunits occupying alternate positions in a hexamer that surrounds a rod-like element containing one each of γ, δ, and ϵ subunits (1–3). Three nucleotide-binding catalytic sites (CS)4 and three noncatalytic sites (NCS) alternate at the six α/β interfaces. Early work with respiratory-deficient strains of Saccharomyces cerevisiae (4) revealed that two additional mitochondrial proteins, Atp11p and Atp12p, which are not integral subunits of the enzyme, are nonetheless necessary for the assembly of F1-ATPase. Besides their failure to assemble F1, a particularly interesting feature of atp11 and atp12 mutants is that they accumulate α and β subunits as high molecular weight aggregates (4) that can be recognized as densely stained inclusion bodies in the mitochondrial matrix (5). Subsequent studies in yeast have shown that Atp12p binds to F1 α (6) and that Atp11p binds to β (7); these interactions include binding determinants in the nucleotide binding domains (NBD) of the two F1 subunits. On this basis, it is now recognized that Atp11p and Atp12p are members of two new families of molecular chaperones, pfam06644 and pfam07542 (8), which are required for the assembly of mitochondrial ATP synthase in all eukaryotes. In fact, the first nuclear genetic lesion associated to a defect of mitochondrial ATP synthase in humans was identified in the locus ATPAF2 for Atp12p and was responsible for the death of a 14-month-old infant (9). Atp12p is also present in the α subdivision of Proteobacteria, consistent with the proposed origin of mitochondria from this ancestral line (10).
The nature of the interactions between the F1 subunits and Atp11p and Atp12p has remained elusive because of the lack of structural information for these chaperones. As α and β aggregate in the absence of Atp11p and Atp12p, it is usually assumed that the F1 subunits are themselves poorly soluble, and that the two chaperones maintain them in a dispersed state until they are incorporated in the mature enzyme. Based on the analysis of the distribution of hydrophilic and hydrophobic areas on the surface of the α and β subunits of F1, and on the interaction energies between these subunits at the interfaces that provide the CS and NCS sites, Wang et al. (6) have proposed a model of F1 assembly in which Atp11p binds at the region of the β subunit that contributes to the CS site, and Atp12p binds at the region of the α subunit that contributes to the NCS site. One consequence of this particular binding of Atp11p and Atp12p to the F1 subunits is that as long as Atp11p is bound to β and Atp12p is bound to α, the two F1 subunits cannot interact at either the CS or the NCS interface. Since no other modulators of chaperone release are known, the Wang model requires an exchange of Atp11p for α and of Atp12p for β. Implied in this model is that the chaperones must themselves look very much like the α and β subunits, and that the driving force for the exchange must simply be a higher affinity of α and β for each other than for the respective chaperone partners. Here we present the structures of Atp11p from Candida glabrata and Atp12p from Paracoccus denitrificans, and we show that some features of the Wang model are correct, namely that binding of the chaperones to α and β prevents further interactions between these F1 subunits. However, Atp11p and Atp12p do not resemble α or β, and it is instead the F1 γ subunit that initiates the release of the chaperones from α and β and their further assembly into mature complex.
EXPERIMENTAL PROCEDURES
Protein Purification and Structure Determination
Mature (leaderless) C. glabrata Atp11p was produced in Escherichia coli Rosetta (DE3) cells at 37 °C. The 5′-end of the recombinant gene (Table 1) encodes the sequence Met-Gly-Gly-Ser-(His)6-Ser-Ser-Gly in front of residues 24–309 of Atp11p, such that His tag-Atp11p is produced without the 23-amino acid targeting peptide that is removed when the yeast protein is imported into mitochondria. The protein was purified via Talon Cobalt affinity column (Clontech). A selenomethionine derivative was produced in modified LeMaster media containing selenomethionine in place of Met. Both native and selenomethionine C. glabrata Atp11p crystallize in 30% PEG 400, 100 mm sodium acetate, 100 mm MES, pH 6.5, over 2–4 weeks at room temperature by sitting, hanging, or sandwich drops. The Atp11p structure was determined at 1.8 Å by single wavelength anomalous dispersion phasing with SOLVE/RESOLVE (11) at the selenium absorption peak with data collected at the APS beamline 22ID (SER-CAT). The resolution was extended to 1.5 Å with new data from the same beamline, and the structure was refined with SHELX (12) (Table 2).
TABLE 1.
Yeast strains and plasmids
| Yeast | Genotype |
|---|---|
| W303-1Aa | MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 |
| Δatp11b | MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 atp11 ::HIS3 |
| Δatp12c | MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 atp12 ::LEU2 |
| Δγd | MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 atp3 ::HIS3 |
| Plasmid | Description |
|---|---|
| pET28-cgatp11 | E. coli expression plasmid pET28a. Encodes C. glabrata Atp11p without the mitochondrial leader peptide and with a His tag at the N terminus |
| pET28-pdatp12 | E. coli expression plasmid pET28a. Encodes P. denitrificans Atp12p with a His tag and thrombin cleavage site at the N terminus |
| pET28-pdatp12(D202K) | Identical to pET28-pdatp12 with the exception of a D202K mutation that was created by PCR mutagenesis using the QuikChange protocol |
| YEp-cgatp11 | Yeast/E. coli shuttle plasmid YEp352 (2 μ) with URA3 marker gene. Encodes full-length C. glabrata Atp11p |
| YEp-pdatp12 | Yeast/E. coli shuttle plasmid YEp352 (2 μ) with URA3 marker gene. Encodes P. denitrificans Atp12p fused to the C terminus of the S. cerevisiae Atp11p mitochondrial leader peptide |
| pRS316-pdatp12 | Yeast/E. coli shuttle plasmid pRS316 (CEN) with URA3 marker gene. Encodes P. denitrificans Atp12p fused to the C terminus of the S. cerevisiae Atp11p mitochondrial leader peptide |
| pαFLAG | Yeast/E. coli shuttle plasmid YEp352 (2 μ) with URA3 marker gene. Encodes S. cerevisiae F1 α subunit with FLAG epitope fused to the C terminus |
| pβHIS6e | Yeast/E. coli shuttle plasmid pRS314 (CEN) with TRP1 marker gene. Encodes a chimeric protein composed of the S. cerevisiae OSCP mitochondrial leader peptide (targeting signal) followed by the S. cerevisiae F1 β subunit that bears a His tag at the N terminus |
TABLE 2.
X-ray data collection and refinement statistics
Each data set was collected from a single crystal at 100 K and processed with HKL2000 (41) or XDS (42, 43). Rmerge = ΣhΣi|I(h)i − 〈I(h)〉|/ΣhΣiI(h)i, where I(h)i is the ith measurement. Rcryst = Σ|Fobs − Fcalc|/Σ|Fobs|. Rfree was calculated on 10% (CNS) or 5% (SHELX) of the data omitted from refinement. In all the structures values in parentheses refer to the highest resolution shell.
| Atp11p | Atp12p | Atp12p | D202K Atp12p | |
|---|---|---|---|---|
| Data collection/reduction | SER-CAT/HKL2000 | SER-CAT/HKL2000 | LS-CAT/XDS | LS-CAT/HKL2000 |
| Space group | I41 | P1 | P212121 | P1 |
| Cell dimensions | a = b = 63.187 Å | a = 42.85 Å | a = 43.55 Å | a = 43.160 Å |
| c = 153.577 Å | b = 50.632 Å | b = 55.75 Å | b = 51.047 Å | |
| c = 67.581 Å | c = 96.62 Å | c = 67.117 Å | ||
| α = 73.87° | α = 102.91° | |||
| β = 74.19° | β = 105.45° | |||
| γ = 89.78° | γ = 90.07° | |||
| Resolution | 50-1.5 Å (1.55-1.5 Å) | 50-1.9 Å (2.02-1.9 Å) | 50-1.0 Å (1.06-1.0 Å) | 50-1.8 Å (1.83-1.8 Å) |
| Unique reflections | 44,482 (2,450) | 38,168 (4,842) | 127,208 (12,549) | 48,230 (2,304) |
| 〈Redundancy〉 | 10.3 (3.6) | 3.8 (3.2) | 14.5 (14.3) | 3.8 (3.6) |
| Completeness | 92.8% (51.2%) | 97.3% (92.4%) | 99.8% (99.9%) | 97.5% (95.6%) |
| 〈I〉/σ〈I〉 | 30.5 (3.1) | 23.3 (1.7) | 25.4 (4.0) | 15.3 (2.3) |
| Rmerge | 5.5% (34.6%) | 6.0% (53.2%) | 6.2% (74.5%) | 8.2% (55.5%) |
| Refinement | SHELX | CNS | SHELX | CNS |
| PDB entry | 2P4F | 2P4X | 2R31 | 2ZD2 |
| Rcryst | 14.1% (26.8%) | 18.7% (29.6%) | 11.9% (21.8%) | 22.5% (29.0%) |
| Rfree | 17.5% (NA)a | 23.0% (36.9%) | 14.2% (NA) | 27.9 (34.3) |
| Amino acids | 204 | 468 | 235 | 468 |
| Waters | 170 | 349 | 527 | 534 |
| 〈B〉 | 30.2 Å2 | 41.2 Å2 | 16.7 Å2 | 28.8 Å2 |
| B from Wilson plot | 18.8 Å2 | 22.6 Å2 | 5.9 Å2 | 20.70 Å2 |
| Coordinate error | 0.08 Å | 0.22 Å | 0.05 Å | 0.24 Å |
| r.m.s.d. bond length | 0.013 Å | 0.009 Å | 0.014 Å | 0.009 Å |
| r.m.s.d. angles | 2.4 | 1.4 | 2.4 | 1.4 |
| r.m.s.d. dihedral | 22.3 | 20.9 | 22.1 | 20.70 |
| r.m.s.d. improper | 1.27 | 0.99 | 1.96 | 1.01 |
| Ramachandran | ||||
| Preferred | 97.33% | 96.76% | 97.50% | 95.69% |
| Allowed | 2.14% | 2.16% | 1.88% | 3.02% |
| Outliers | 0.53% | 1.08% | 0.62% | 1.29% |
a Free R value in the high resolution shell is not available in SHELX. NA means not applicable.
Full-length P. denitrificans Atp12p was produced in E. coli from a plasmid that encodes an upstream His6 tag sequence and a thrombin recognition site (Table 1) and purified via Talon affinity chromatography. A selenomethionine derivative of this protein was produced as described for C. glabrata Atp11p. Crystals grow from around 5–20% PEG 4000, 50–250 mm sodium acetate, 100 mm HEPES, pH 7.5, at 4 °C. The Atp12p structure was determined at 1.9 Å by single wavelength anomalous dispersion phasing with SOLVE/RESOLVE (11) at the selenium absorption peak with data collected at the APS beamline 22ID (SER-CAT). A different crystal form was obtained from 28 to 30% PEG 4000, 50 mm MgCl2, 100 mm Tris-Cl, pH 8.5, in sitting drops at 4 °C. A data set at the resolution of 1.0 Å was collected at the APS beamline 21ID-F (LS-CAT). The structure was determined at 1.8 Å by Molecular Replacement with CCP4/Molrep (13), and the resolution was extended to 1.0 Å with SHELX (Table 2). Crystals of D202K Atp12p grow around 20% PEG 4000, 50–250 mm sodium acetate, 100 mm HEPES, pH 7.5, at 4 °C with 20 mm spermidine as additive. A data set at 1.8 Å was collected at the APS beamline 21ID-G (LS-CAT), and the structure refined with CNS version 1.2 (14) (Table 2).
Electrostatic Potentials
Electrostatic potentials at the solvent-accessible surface of proteins were calculated with PDB2PQR/APBS (15) using a nonlinear Poisson-Boltzmann continuum model with protein dielectric constant ϵp = 2, solvent dielectric constant ϵs = 78. Protein charges were assigned on the basis of the PARSE force field (16), assuming a pH of 8.0 for the mitochondrial matrix (17, 18), and by calculating the pKa values of all ionizable residues with the program PROPKA (19). Calculations were carried out on a uniform grid of ∼0.5 Å spacing.
Coprecipitation Experiments
These experiments utilized the yeast mutant, Δγ/pαFLAG+pβHIS6, which harbors a disrupted allele (atp3::HIS3) for F1 γ in the chromosome and carries two episomal plasmids, pαFLAG (URA3-linked) and pβHIS6 (TRP1-linked), for the simultaneous production of, respectively, F1 α that is tagged at the C terminus with the FLAG epitope (DYKDDDDK) and F1 β that is tagged at the N terminus with 6 histidine residues (see Table 1). Plasmid pβHIS6 was a gift from Dr. David Mueller (Rosalind Franklin University of Medicine and Science, Department of Biochemistry and Molecular Biology, The Chicago Medical School, North Chicago, IL). To make pαFLAG, the QuikChange method was used to insert the DNA coding for the FLAG epitope immediately in front of the TAA stop codon of yeast ATP1. The plasmid template for mutagenesis (pG50/ST2) carries the entire ATP1 coding sequence for yeast F1 α cloned in the BamHI and EcoRI sites of YEp352 (20). The sequences of the complementary primers that were used for mutagenesis are as follows: 5′- GCTACTGAATCATTTGTTGCCACTTTTGATTATAAAGATGATGATGATAAATAAtgtgagctcaaaaaataaaaa-3′ (sense strand) and 5′-tttttattttttgagctcacattaTTTATCATCATCATCTTTATAATCAAAAGTGGCAACAAATGATTCAGTAGC-3′ (antisense strand). Coding and noncoding sequences are given in uppercase and lowercase letters, respectively. ATP1 codons are in roman type and FLAG epitope codons are in boldface type. A SacI site (underlined) was created to facilitate screening clones by changing two nucleotides (double underlined) in the 3′-flanking genomic sequence of ATP1.
Mitochondria were prepared from Δγ/pαFLAG+pβHIS6, sonically irradiated, and centrifuged to produce a soluble extract of matrix proteins as described (21). For experiments with TALON metal affinity resin, 75 μl of soluble mitochondrial extract was combined with 30 μl of resin (prepared as a 50% slurry) in a final volume of 500 μl of buffer that contained 50 mm sodium phosphate, pH 7.0, 300 mm NaCl, 0.5% Triton X-100, 4 mm ATP, and 5% (w/v) sucrose. Experimental incubations with anti-FLAG M2 affinity gel were identical except that the pH was increased to 7.4 and NaCl was reduced to 150 mm. Samples were incubated end-over-end at 4 °C overnight and subsequently centrifuged for 5 min at 13,000 × g to sediment the affinity beads. The initial (post-bead) supernatant was collected, and the beads were washed three times by centrifugation with 500 μl of the appropriate buffer. Total protein was collected from the post-bead supernatant and first wash fractions by precipitating the samples (500 μl) with a 1:4 mixture chloroform:methanol. Chloroform:methanol and affinity bead precipitates were suspended in 45 μl of SDS gel loading buffer, boiled for 5 min, centrifuged, and loaded (10 μl) on a 12% SDS-polyacrylamide gel.
Miscellaneous Methods
Yeast transformations employed the lithium acetate procedure (22). The detection in Western blots of F1 α, F1 β, Atp11p, and Atp12p and assays for molecular weight standards (hemoglobin and lipoamide dehydrogenase) used in sucrose gradients were performed as described previously (6, 7).
RESULTS
Crystals of C. glabrata Atp11p and P. denitrificans Atp12p were obtained after screening several homologous proteins, including those from Homo sapiens, Mus musculus, S. cerevisiae, Candida albicans, Kluveromyces lactis, Rhodobacter capsulatus, and Arabidopsis thaliana. C. glabrata Atp11p and P. denitrificans Atp12p are 79.9 and 44.2% similar and 56.3% and 23.4% identical to the corresponding proteins in S. cerevisiae (Fig. 1). When produced from episomal plasmids C. glabrata Atp11p and P. denitrificans Atp12p rescue the respiratory defect of S. cerevisiae atp11 or atp12 deletion strains (Table 1; Fig. 2), suggesting that the mechanism by which the chaperones facilitate the assembly of F1 is maintained across evolutionary lines.
FIGURE 1.
Pairwise alignment of recombinant protein sequences with S. cerevisiae homologs. Upper, pairwise alignment of C. glabrata and S. cerevisiae Atp11p. The disordered regions of the C. glabrata structure (residues 1–93 and 163–176) are shown in orange, the N-terminal helical domain (residues 94–129) in blue, the central α/β taco (residues 130–262) in green, and the C-terminal helical domain (residues 263–298) in red. Lower, pairwise alignment of P. denitrificans and S. cerevisiae Atp12p. The smaller N-terminal domain (residues 3–83) and the larger C-terminal domain (residues 84–238) are shown in blue and red, respectively. Trp-57 and Asp-202, corresponding, respectively, to Trp-103 and Glu-289 of the yeast protein, are highlighted in blue and red boxes. C. gla, C. glabrata; P. den, P. denitrificans; S. cer., S. cerevisiae.
FIGURE 2.
Growth of S. cerevisiae mutants on nonfermentable and fermentable carbons. S. cerevisiae mutants disrupted at the genetic locus for Atp11p (Δatp11) or Atp12p (Δatp12) were transformed with a plasmid producing C. glabrata Atp11p or P. denitrificans Atp12p, and evaluated for growth on glucose and nonfermentable ethanol-glycerol (EG) plates. Wild type W303, the deletion strains Δatp11 and Δatp12, and the transformants Δatp11/YEp-cgatp11 and Δatp12/pRS316-pdatp12 were grown overnight in liquid YPD (2% glucose, 2% peptone, 1% yeast extract). The next day the cultures were adjusted to A600 = 1.0, and then serially diluted by a factor of 2. Five μl of each dilution were applied to a YPD and an EG plate (2% ethanol, 3% glycerol, 2% peptone, 1% yeast extract), and the plates were incubated at 30 °C. After 48 h, the deletion strain Δatp12 does not grow at all on EG, whereas Δatp11 displays a leaky phenotype. The transformant Δatp11/YEp-cgatp11, which produces from a multicopy plasmid C. glabrata Atp11p (56% identity with S. cerevisiae) with its own mitochondrial targeting sequence, grows on EG almost as well as the wild type. The transformant Δatp12/pRS316-pdatp12, which produces a chimeric protein in which P. denitrificans Atp12p (23% identity with S. cerevisiae) is fused to the mitochondrial targeting sequence of S. cerevisiae Atp11p, shows growth on EG, despite the plasmid being single copy; transformants of Δatp12 with the same chimeric allele expressed from a multicopy plasmid grow almost as well as the wild type (data not shown).
Structure of Atp11p
The crystal structure of C. glabrata Atp11p was initially determined at the resolution of 1.8 Å by single wavelength anomalous dispersion phasing of diffraction data from a single crystal of selenomethionine derivatized recombinant Atp11p. The resolution was subsequently extended to 1.5 Å with diffraction data from a crystal of native Atp11p (Fig. 3A; Table 2). Although the full-length protein produced in E. coli can be recovered from the crystals, the first 93 residues (out of 298, PDB numbering) are not visible in the crystal structure. However, there is an empty cavity in the density map that is large enough to harbor the disordered domain. The remaining residues 94–298 correspond to the part of S. cerevisiae Atp11p that was shown by deletion studies to retain full chaperone activity (23). The elements of sequence identity between the C. glabrata and the S. cerevisiae protein (Fig. 1) are distributed evenly throughout this region (Fig. 3B). A loop (residues 163–176) that is poorly conserved or absent in Atp11p from different sources is also disordered. The overall fold of Atp11p resembles an α/β “taco” (an incomplete antiparallel β-barrel whose concave face is filled by two α-helices) flanked by two helical domains. Only one chain of Atp11p is present in the asymmetric unit, and a survey of all symmetry mates with PISA (24) does not reveal a dimer or a higher order assembly that might be stable in solution. This conclusion is consistent with earlier sedimentation analysis of purified recombinant yeast Atp11p and of native yeast Atp11p in mitochondrial extracts, which concluded that Atp11p is a monomer (25). Structural homology searches against the entire PDB with the VAST (26) and MATRAS (27) engines do not reveal relatives for the entire protein. Searches with the individual domains reveal poor structural matches of the first helical domain (residues 94–130) with the 3-helix bundle headpiece of villin (PDB code 1WY3), of the α/β taco (residues 131–262) with the C-terminal domain of phosphoglucomutase (residues 423–547 in PDB code 3PMG), and of the C-terminal helical domain (residues 263–298) with the two C-terminal helices of E. coli HSP90 (PDB code 1SF8). At this point it does not appear that any of these weak structural homologies might be of mechanistic importance.
FIGURE 3.
Structures of C. glabrata Atp11p and P. denitrificans Atp12p. A, N-terminal helical domain (residues 94–129), the central α/β taco (residues 130–262), and the C-terminal helical domain (residues 263–298) of C. glabrata Atp11p are shown as a cartoon colored in blue, green, and red, respectively. Loops are shown as lighter colors. B, solvent-accessible surface of Atp11p, rotated by ∼90° with respect to A, is colored in transparent light blue with the regions of identity with the S. cerevisiae protein shown in a darker shade; the internal structure is shown as a cartoon colored as in A. C, characteristic “boxing glove” shape of P. denitrificans Atp12p. The N-terminal “wrist” domain (residues 3–83), and the C-terminal “hand” domain (residues 84–238) are shown as a cartoon colored in shades of blue/light-blue and red/magenta, respectively. The first 37 residues of the P. denitrificans protein, corresponding to a deletion of the S. cerevisiae protein that severely affects stability, are shown in light blue. This region corresponds approximately to an N-terminal β-hairpin. The last 42 residues of the P. denitrificans protein, corresponding to a deletion of the S. cerevisiae protein that completely eliminates the chaperone activity, are shown in magenta. The surfaces of Trp-57 and Asp-202, corresponding to Trp-103 and Glu-289 of the yeast protein, are shown in yellow and green, respectively; mutations of these residues also inactivate the protein. D–F, palm (D) and dorso (E) faces of wild type Atp12p, and the palm face of D202K (F) Atp12p from P. denitrificans are shown colored according to the electrostatic potentials (−1 kT/e, red; 1 kT/e, blue) at their solvent-accessible surface. Figures were generated with PyMOL (36).
Structure of Atp12p
The structure of P. denitrificans Atp12p was determined from two crystal forms at the resolution of 1.9 Å (sg P1, crystal form I) and 1.0 Å (sg P212121, crystal form II) (Table 2). In the asymmetric unit of crystal form I, there are two chains of Atp12p related by a local 2-fold axis, but they are loosely packed and unlikely to represent a functional dimer. A single chain occupies the asymmetric unit of crystal form II, with no neighbors in the unit cell in a position suitable to form a dimer with a large buried surface. Differences between the two crystal forms of Atp12p are distributed throughout the structure (C-α r.m.s.d. 3–238 = 0.782 Å; r.m.s.d. 3–83 = 0.685 Å; r.m.s.d. 84–238 = 0.653 Å) but are also associated to a small rotation of the N- and C-domains with respect to each other (see below). Analysis of both structures with PISA also indicates that P. denitrificans Atp12p is a monomer, in agreement with earlier sedimentation, cross-linking, and two-hybrid studies of yeast and human Atp12p (28, 29). The shape of Atp12p resembles a boxing glove, with the wrist and hand sections provided by two domains encompassing residues 3–83 (N-domain) and 84–238 (C-domain) (Fig. 3C). The hand domain of Atp12p displays a concave face, the “palm,” and a convex face, the “dorso.” Looking at the palm face of the Atp12p, there is a sharp difference in surface charge between the C-domain, which is uniformly negative, and the N-domain, which is uniformly positive (Fig. 3D). In contrast, the dorso surface is almost uniformly negatively charged (Fig. 3E).
A protein from Agrobacterium tumefaciens (PDB code 2R6I) very similar to P. denitrificans Atp12p was identified by the Midwest Center for Structural Genomics but was not characterized. Based on our study, 2R6I can now be assigned as the Atp12p chaperone of that bacterium. A structural homology search of Atp12p against the entire PDB bank identified at low threshold 2 additional neighbors for the N-domain and 67 additional neighbors for the C-domain; superposition of the best scoring neighbors onto each domain of Atp12p did not reveal a level of similarity sufficient to draw mechanistic information from the search. However, it is of interest that the core of the C-domain consists of a central helix surrounded by six other helices (Fig. 4A). This topology is reminiscent of the fold of the membrane domains of colicins, pore-forming toxins, and Bcl-2 apoptotic proteins, all of which are thought to become variably associated with biological membranes upon some conformational change (30–32). Despite this similarity, there is no evidence at this time that in vivo Atp12p might be associated with the mitochondrial inner membrane or the bacterial cell membrane.
FIGURE 4.
Details of the Atp12p structure. A, stereo view of the C-domain (from the top) showing a central helix surrounded by six other helices. B, relative motions of Atp12p N- and C-domains. Stereo view of four different structures of wild type Atp12p (shown as red, blue, yellow, and green ribbons) and of D202K Atp12p (shown as a cyan ribbon) superimposed through their N-domains to highlight the relative motion of the C-domain. C, detail of the 1.0 Å structure of wild type Atp12p around Asp-202 (visible in two conformations). D, detail of the 1.8 Å structure of D202K Atp12p around Lys-202. |2Fo − Fc| σA maps are contoured at 1σ.
Deletion of the sequence of S. cerevisiae Atp12p corresponding to the first 37 residues of the P. denitrificans sequence (Fig. 3C, light blue shading) produces a protein that supports ∼80% of the wild type level of F1 assembly in vivo but is barely detectable by Western blot in isolated mitochondria (29). This finding suggests that the N-domain is primarily necessary for stability. A W94R mutation was identified in a recessive allele of the ATPAF2 locus that caused the death of a 14-month-old homozygous patient (9). Trp-94 (corresponding to Trp-57 in P. denitrificans and to a Phe or Trp in all known sequences of Atp12p) is localized in the N-domain (Fig. 3C, yellow surface), and thus it is conceivable that the human syndrome originates from enhanced degradation of Atp12p rather than inactivation.
Deletion of the last 42 residues of the sequence of S. cerevisiae Atp12p, corresponding to the last two helices of the P. denitrificans protein (shown in magenta in Fig. 3C), completely eliminates the chaperone activity and prevents F1 assembly (29). Interestingly, in the 1.0 Å structure of Atp12p (Table 2), the last helix is clearly visible in two positions corresponding to an ∼3.0 Å shift of the helix axis (Fig. 4B). It is also worth noting that a negatively charged residue (Asp-202, shown as a green surface in Fig. 3C) of P. denitrificans Atp12p is situated immediately after a sharp kink in the penultimate helix of the protein, almost at the beginning of the C-terminal helical region whose deletion completely suppresses the chaperone activity. Substitution with Lys of Glu-289 in the C-domain of S. cerevisiae Atp12p (corresponding to Asp-202 in P. denitrificans) completely eliminates F1 assembly in vivo without decreasing the amount of Atp12p detected in isolated mitochondria (29). Other substitutions at this position, E289Q and E289A, are tolerated to the extent that there is enough ATP synthase assembled to support the growth of the Δatp12 transformants that produce the mutant proteins. However, in both cases the level of F1F0 in mitochondria is significantly decreased. Instead, the phenotype of strains that produce the yeast E289D variant is indistinguishable from wild type, which is in accord with the fact that some members of the Atp12p family, like P. denitrificans Atp12p, have an aspartic acid at this position. The effect of this mutation has also been studied in the context of purified human Atp12p (E240K in this case), and it was found to interfere with its chaperone activity in assays with the model substrate citrate synthase (28). We have determined the crystal structure of the D202K mutant form of P. denitrificans Atp12p at the resolution of 1.8 Å (Table 2); the overall protein architecture is unchanged in D202K Atp12p (C-α r.m.s.d. 5–238 = 0.339 Å). The most notable difference with the wild type is the long lysine side chain at position 202 (Fig. 4, C and D), which, by replacing the negative charge of aspartic acid with a positive charge, strikingly disrupts the uniformly negatively charged surface of the C-domain in the palm-facing view (Fig. 3F). The D202K mutation also affects the last two helices, which in different structures of the wild type and in this mutant structure are in slightly different positions (Fig. 4B). These position shifts appear also among the nine lowest frequency normal modes of Atp12p (see supplemental material) and therefore reflect intrinsic vibrational properties of the protein. Altogether, the effects of the C-terminal deletion and of the E289K mutation in S. cerevisiae Atp12p suggest that the region of Atp12p encompassing the last two helices is directly involved in the chaperone activity.
Intermediates of F1 Assembly
Earlier hypotheses on the role of Atp11p and Atp12p were based on the assumption that the regions of F1 α and β subunits most likely to promote aggregation are hydrophobic patches sequestered at the subunit interfaces in the α3β3 structure, and that Atp11p and Atp12p mimic, respectively, the α and β subunit, temporarily masking these hydrophobic regions (6). However, side-by-side analysis of the structures of both F1 and the two chaperones shows no similarity of shape between Atp11p and α and Atp12p and β. This observation is inconsistent with an assembly mechanism in which the chaperone is released (e.g. Atp11p from β) in exchange for the incoming partner subunit (e.g. α) (6), simply by virtue of a phenomenon of molecular mimicry. The likelihood that the exchange step of F1 assembly may require a specific “trigger” event was studied in yeast cells in which the nuclear gene encoding the F1 γ subunit was disrupted (Table 1), as we had observed that these cells also fail to assemble F1. Despite α and β being present in a normal amount, there is little or no hexameric F1 in soluble mitochondrial extracts from Δγ cells (Fig. 5, upper panel). Instead, most of α and β accumulate as soluble protein complexes of ∼100 kDa (Fig. 5). It appears that these complexes are stable intermediates along the path to F1 assembly, and that the γ subunit is necessary for their final progression into mature F1. When the complexes are analyzed by means of a high resolution sedimentation gradient, α and β do not comigrate; furthermore, the centers of the distributions of Atp12p and Atp11p in the gradient fractions correspond well to the centers of the distributions of α and β, respectively (Fig. 5, lower panel). As α and β have molecular masses of ∼55 and ∼52 kDa, and Atp12p and Atp11p have molecular masses of ∼34 and ∼31 kDa, the comigrations of α with Atp12p (at a position corresponding to a higher molecular weight) and of β with Atp11p (at a position corresponding to a lower molecular weight) suggest that there are two distinct populations of complexes represented, respectively, by α:Atp12p (∼89 kDa) and β:Atp11p (∼83 kDa) heterodimers.
FIGURE 5.
Western blots of sucrose gradient fractions to detect F1-ATPase assembly intermediates. Upper, purified yeast F1 (30 μg) or soluble extracts prepared by sonic irradiation of mitochondria (10 mg/ml) from wild type (WT) or Δγ isogenic yeast were mixed with molecular weight marker proteins (hemoglobin, 64 kDa; lipoamide dehydrogenase, 100 kDa) in 200 μl of buffer (20 mm Tris-HCl, pH 7.5, 4 mm ATP, 2 mm EDTA) and centrifuged through a 4.8-ml sucrose solution comprised of a linear 5–20% gradient (4.5 ml) formed over 80% sucrose (0.3 ml); the boundary between 20 and 80% sucrose is depicted by the vertical white dotted line in the schematic representation of the sucrose gradient at the top of the figure. The gradients were centrifuged in an SW55Ti rotor at 55,000 rpm for 2 h and 10 min at room temperature. Fractions (250 μl) collected from the bottom of the tubes were probed in Western blots using a mixture of polyclonal antibodies against α and β proteins and analyzed for the position of the marker proteins as described under “Experimental Procedures.” Arrowheads show the positions of hemoglobin (∼64 kDa), lipoamide dehydrogenase (∼100 kDa), and the F1 oligomer (∼360 kDa). Lower, to increase the resolution of F1 protein intermediate complexes observed in soluble extracts from Δγ yeast mitochondria (red dashed lines), the sample (100 μl) was centrifuged at room temperature through an expanded discontinuous gradient of sucrose (5–15% (4.6 ml) overlaying 80% (0.3 ml)) in an SW55Ti rotor at 28,200 rpm for 20 h. Fractions (250 μl) were analyzed in separate Western blots for the F1 α and β subunits, for Atp11p and for Atp12p. For ease of comparison, the three Western blots are shown here superimposed with their backgrounds digitally changed to three different colors as follows: yellow for α and β, green for Atp11p, and blue for Atp12p. The correspondence of peaks for Atp12p and F1 α, and for Atp11p and F1 β, is highlighted with the blue and green vertical lines, respectively.
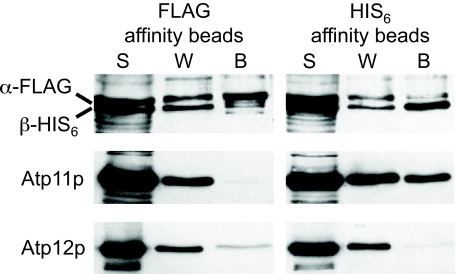
Experiments in which affinity resins were used to precipitate tagged α or β subunits from soluble mitochondrial extracts of the Δγ mutant also provided evidence that α:Atp12p and β:Atp11p complexes, but not α:β hetero-oligomers, are stable intermediates in the assembly pathway (Fig. 6). For this work, Δγ yeast was doubly transformed with two autonomously replicating plasmids, bearing different genetic markers, for the coproduction of FLAG-tagged α (α-FLAG) and His-tagged β (β-HIS6) in the mutant cells. Soluble mitochondrial extracts were prepared from doubly transformed Δγ and mixed end-over-end with either TALON metal affinity resin (HIS6 affinity beads) or with anti-FLAG M2 affinity gel (FLAG affinity beads) overnight at 4 °C. The affinity beads (Fig. 6, lane B) were collected by centrifugation, washed, and analyzed in Western blots, along with samples of the post-bead supernatants (lane S) and first wash solutions (lane W), with polyclonal antibodies against F1 α, F1 β, Atp11p, or Atp12p (Fig. 6). Although all four proteins were detected in the post-bead supernatant and first wash samples in both experiments, Western blots of the precipitated samples showed practically no evidence of F1 β-His6 coadsorbed with α-FLAG to the FLAG affinity beads or of F1 α-FLAG coadsorbed with β-His6 to the His6 affinity beads. These findings were confirmed in separate blots that were probed using commercial antibodies against either His6 or FLAG epitope (data not shown). Instead, Atp12p and Atp11p were each detected above background levels as coprecipitating proteins with α-FLAG and β-HIS6, respectively. Control experiments (data not shown) verified that the Western profiles of the bead fractions were exclusively due to the selective actions of the affinity resins as opposed to fortuitous precipitation of proteins (indicated by omitting beads in the experiment) or nonspecific binding (indicated by including 150 mm imidazole in the TALON bead experiment). Hence, in the absence of the γ subunit, α and β remain bound to their cognate chaperones and cannot interact with each other. This result is consistent with the prediction by Wang et al. (6) that Atp11p binds to a region of β that contributes to the CS site and Atp12p to a region of α that contributes to the NCS site (6), such that binding of the F1 subunits to each other or to the chaperones is mutually exclusive. Cumulatively, the evidence suggests that the γ subunit is the initial trigger for the release of the chaperones from the α:Atp12p and β:Atp11p heterodimers.
FIGURE 6.
Coprecipitation of proteins with F1 α-FLAG and with F1 β-HIS6. Western blots of proteins precipitated with affinity resins are shown (left, anti-FLAG M2 affinity gel (FLAG affinity beads); right, TALON metal affinity resin (HIS6 affinity beads)). Aliquots of the proteins present in the initial post-bead supernatants (S), in the first wash solutions (W), and in the collected bead fractions (B) were probed in separate Western blots using a mixture of polyclonal antibodies against F1 α and β or Atp11p or Atp12p antiserum. Total protein was precipitated with chloroform:MeOH from the supernatant and wash samples and suspended to the same volume of the recovered beads. Equivalent volumes (10 μl) of each fraction were loaded on the gels.
DISCUSSION
Atp11p and Atp12p are present in the mitochondrial matrix in small amounts, comparable with those of unassembled α and β subunits. In the absence of one of these subunits, as F1 cannot be assembled, the concentration of the other subunit exceeds that of the cognate chaperone and the subunit aggregates. In S. cerevisiae strains harboring a disruption of the nuclear gene encoding either the α or the β subunit, insoluble aggregates are formed even in the presence of normal amounts of functioning Atp11p and Atp12p (4). During F1 assembly, the formation of Atp11p:β and Atp12p:α intermediates may not promote directly the formation of the physiological α:β dimers, but rather act as decoys preventing the formation of nonphysiological α:α, β:β, and α:β complexes. Although this action would be critical to prevent aggregation, it also requires an initial trigger to form at least one physiological α:β dimer devoid of chaperones. After this initial event, it is conceivable that successive bindings of an Atp11p-β or Atp12p-α complex to an α:β dimer might produce a conformational change that decreases the affinity of the chaperones for their target proteins. Analysis of the F1 assembly intermediates in yeast cells, in which the nuclear gene encoding the F1 γ subunit was disrupted (), suggests that the γ subunit is the trigger factor necessary for the initial chaperone release. In the absence of structures of the Atp11p:β and Atp12p:α intermediates, it is not possible to know exactly how the γ subunit interacts with these complexes. However, a side-by-side visual inspection of Atp12p and γ reveals that the most peripheral section of the long coiled-coil of the γ subunit is remarkably similar to the C-terminal helical fragment of Atp12p (Fig. 7A), which was shown to be essential for activity. If the C-terminal helices of Atp12p are superimposed to the coiled-coil tail of the yeast γ subunit bound to α, then the surface of Atp12p complements very well the surface of the yeast α subunit (Fig. 7B), providing a reasonable guess of how the two proteins interact. The finding of a possible molecular mimicry between the C-terminal region of Atp12p and the coiled-coil tail of γ raises the possibility that F1 assembly may start with the γ subunit displacing Atp12p from the Atp12p:α heterodimer.
FIGURE 7.
A possible interaction of Atp12p with F1 α. A, superposition of the yeast F1 γ subunit (cyan) to P. denitrificans Atp12p (yellow). The C-terminal helices of Atp12p are colored in red. B, predicted interaction of P. denitrificans Atp12p with yeast F1 α (green), based on the relative position of the α and γ subunits in yeast F1. (PDB code 2hld (Mol 1)).
It had been assumed that the main reason for the existence of Atp11p and Atp12p is to protect hydrophobic regions of the surface of F1 α and β that otherwise would drive the aggregation of these subunits. However, there are no dramatic differences in the extent and distribution of hydrophobic patches between the regions of F1 α and β subunits exposed to solvent, and those buried at the subunit interface (supplemental Fig. 1). Furthermore, hydrophobic areas account for less than 50% of the total contact surface between these subunits. Thus, other forces, besides hydrophobic interactions, play an important role in F1 assembly. When electrostatic potentials are calculated for the entire F1, the oligomer surface appears almost uniformly negative, with small positive regions (data not shown). However, when electrostatic potentials are computed separately for each individual subunit of F1, the regions of the α subunits that contact the β subunits appear mostly positively charged, and the corresponding regions of the β subunits appear mostly negatively charged (supplemental Fig. 1). These observations suggest that electrostatic attraction plays a major role in the interaction of F1 α and β with their cognate chaperones and in the formation of the correct interfaces in F1. In humans, one fatal syndrome has been attributed to a defect in an F1 chaperone (9), and intermediate phenotypes with partially impaired assembly of F1 are likely to exist, in which a metabolic defect will appear only under stress. Other unrelated pathologic conditions are also characterized by the formation of nonphysiological aggregates. For example, it has been suggested that coulombic interactions play a key role in the formation of amyloid Aβ fibrils, which is the central event in the development of Alzheimer disease (33), and at least two distinct chaperones have been implicated in preventing the formation of these fibrils (34, 35). Thus, the mechanism by which Atp11p and Atp12p prevent the chaotic aggregation of F1 may be representative of a more general mechanism shared with other dedicated chaperones that also act primarily by masking electrostatic interactions.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants GM41857 (to S. H. A.) and GM69840 (to D. L. G.) from USPHS.
 The on-line version of this article (available at http://www.jbc.org) contains supplemental data.
The on-line version of this article (available at http://www.jbc.org) contains supplemental data.
The atomic coordinates and structure factors (codes 2P4F, 2P4X, 2R31, and 2ZD2) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- CS
- catalytic site
- NCS
- noncatalytic site
- PEG
- polyethylene glycol
- PDB
- Protein Data Bank
- r.m.s.d.
- root mean square deviation
- MES
- 4-morpholineethanesulfonic acid.
REFERENCES
- 1.Abrahams J. P., Leslie A. G., Lutter R., Walker J. E. (1994) Nature 370, 621–628 [DOI] [PubMed] [Google Scholar]
- 2.Bianchet M. A., Hullihen J., Pedersen P. L., Amzel L. M. (1998) Proc. Natl. Acad. Sci. U. S. A. 95, 11065–11070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kabaleeswaran V., Puri N., Walker J. E., Leslie A. G., Mueller D. M. (2006) EMBO J. 25, 5433–5442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ackerman S. H., Tzagoloff A. (1990) Proc. Natl. Acad. Sci. U. S. A. 87, 4986–4990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lefebvre-Legendre L., Salin B., Schaëffer J., Brèthes D., Dautant A., Ackerman S. H., di Rago J. P. (2005) J. Biol. Chem. 280, 18386–18392 [DOI] [PubMed] [Google Scholar]
- 6.Wang Z. G., Sheluho D., Gatti D. L., Ackerman S. H. (2000) EMBO J. 19, 1486–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Z. G., Ackerman S. H. (2000) J. Biol. Chem. 275, 5767–5772 [DOI] [PubMed] [Google Scholar]
- 8.Marchler-Bauer A., Anderson J. B., DeWeese-Scott C., Fedorova N. D., Geer L. Y., He S., Hurwitz D. I., Jackson J. D., Jacobs A. R., Lanczycki C. J., Liebert C. A., Liu C., Madej T., Marchler G. H., Mazumder R., Nikolskaya A. N., Panchenko A. R., Rao B. S., Shoemaker B. A., Simonyan V., Song J. S., Thiessen P. A., Vasudevan S., Wang Y., Yamashita R. A., Yin J. J., Bryant S. H. (2003) Nucleic Acids Res. 31, 383–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Meirleir L., Seneca S., Lissens W., De Clercq I., Eyskens F., Gerlo E., Smet J., Van Coster R. (2004) J. Med. Genet. 41, 120–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lang B. F., Gray M. W., Burger G. (1999) Annu Rev. Genet. 33, 351–397 [DOI] [PubMed] [Google Scholar]
- 11.Terwilliger T. C., Berendzen J. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sheldrick G. M. (2008) Acta Crystallogr. A 64, 112–122 [DOI] [PubMed] [Google Scholar]
- 13.CCP4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 14.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 15.Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Proc. Natl. Acad. Sci. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sitkoff D., Sharp K. A., Honig B. (1994) J. Phys. Chem. 98, 1978–1988 [Google Scholar]
- 17.Metoki K., Hommes F. A. (1984) J. Inherited Metab. Dis. 7, 9–11 [DOI] [PubMed] [Google Scholar]
- 18.Llopis J., McCaffery J. M., Miyawaki A., Farquhar M. G., Tsien R. Y. (1998) Proc. Natl. Acad. Sci. U. S. A. 95, 6803–6808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H., Robertson A. D., Jensen J. H. (2005) Proteins 61, 704–721 [DOI] [PubMed] [Google Scholar]
- 20.Hill J. E., Myers A. M., Koerner T. J., Tzagoloff A. (1986) Yeast 2, 163–167 [DOI] [PubMed] [Google Scholar]
- 21.Ackerman S. H., Tzagoloff A. (2007) Methods Mol. Biol. 372, 363–377 [DOI] [PubMed] [Google Scholar]
- 22.Schiestl R. H., Gietz R. D. (1989) Curr. Genet. 16, 339–346 [DOI] [PubMed] [Google Scholar]
- 23.Wang Z. G., Ackerman S. H. (1996) J. Biol. Chem. 271, 4887–4894 [DOI] [PubMed] [Google Scholar]
- 24.Krissinel E., Henrick K. (2007) J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 25.White M., Ackerman S. H. (1995) Arch. Biochem. Biophys. 319, 299–304 [DOI] [PubMed] [Google Scholar]
- 26.Gibrat J. F., Madej T., Bryant S. H. (1996) Curr. Opin. Struct. Biol. 6, 377–385 [DOI] [PubMed] [Google Scholar]
- 27.Kawabata T. (2003) Nucleic Acids Res. 31, 3367–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hinton A., Gatti D. L., Ackerman S. H. (2004) J. Biol. Chem. 279, 9016–9022 [DOI] [PubMed] [Google Scholar]
- 29.Wang Z. G., Ackerman S. H. (1998) J. Biol. Chem. 273, 2993–3002 [DOI] [PubMed] [Google Scholar]
- 30.Cascales E., Buchanan S. K., Duché D., Kleanthous C., Lloubès R., Postle K., Riley M., Slatin S., Cavard D. (2007) Microbiol Mol. Biol. Rev. 71, 158–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alouf J. E. (2003) Folia Microbiol. 48, 5–16 [DOI] [PubMed] [Google Scholar]
- 32.Chao D. T., Korsmeyer S. J. (1998) Annu. Rev. Immunol. 16, 395–419 [DOI] [PubMed] [Google Scholar]
- 33.Lazo N. D., Grant M. A., Condron M. C., Rigby A. C., Teplow D. B. (2005) Protein Sci. 14, 1581–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanekiyo T., Ban T., Aritake K., Huang Z. L., Qu W. M., Okazaki I., Mohri I., Murayama S., Ozono K., Taniike M., Goto Y., Urade Y. (2007) Proc. Natl. Acad. Sci. U. S. A. 104, 6412–6417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kallhoff V., Peethumnongsin E., Zheng H. (2007) Mol. Neurodegener. 2, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DeLano W. L. (2007) The PyMOL Molecular Graphics System , DeLano Scientific LLC, Palo Alto, CA [Google Scholar]
- 37.Ackerman S. H., Martin J., Tzagoloff A. (1992) J. Biol. Chem. 267, 7386–7394 [PubMed] [Google Scholar]
- 38.Bowman S., Ackerman S. H., Griffiths D. E., Tzagoloff A. (1991) J. Biol. Chem. 266, 7517–7523 [PubMed] [Google Scholar]
- 39.Paul M. F., Ackerman S., Yue J., Arselin G., Velours J., Tzagolof A., Ackerman S. (1994) J. Biol. Chem. 269, 26158–26164 [PubMed] [Google Scholar]
- 40.Mueller D. M., Puri N., Kabaleeswaran V., Terry C., Leslie A. G., Walker J. E. (2004) Protein Expr. Purif. 37, 479–485 [DOI] [PubMed] [Google Scholar]
- 41.Otwinowski Z., Minor W. (1997) in Methods in Enzymology ( Carter C. W., Sweet R. M. eds) pp. 307–326, Academic Press, New York: [DOI] [PubMed] [Google Scholar]
- 42.Kabsch W. (1988) J. Appl. Crystallogr. 21, 67–72 [Google Scholar]
- 43.Kabsch W. (1993) J. Appl. Crystallogr. 26, 795–800 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.