Abstract
Purpose
This study aimed to evaluate the prognostic significance of two functional single nucleotide polymorphisms (SNP) in the p53 pathway (p53 Arg72Pro and MDM2 T309G) in patients with esophageal cancer, and to determine the importance of histologic subtype in the SNP-outcome relationships.
Experimental Design
A cohort of 371 patients with esophageal carcinoma enrolled in Boston, USA from 1999 to 2004 were genotyped for the p53 and MDM2 SNPs. Associations between genotypes and overall survival (OS; the primary outcome) and progression-free survival (PFS) were assessed using the Kaplan-Meier method. Cox proportional hazard models, adjusted for age, stage, performance status, and smoking were developed. Interaction analyses were done for histology (adenocarcinoma versus squamous cell carcinoma).
Results
At the median follow-up of 33 months, median survival (MS) and PFS were 29.1 and 15.7 months, respectively. p53 Pro/Pro was strongly associated with shorter survival in the entire cohort (MS of 11.8 versus 29.1 months, P < 0.0001; adjusted hazard ratio for death, 2.05; 95% confidence interval, 1.30–3.24; P = 0.002 for Pro/Pro versus Arg/Arg). MDM2 G/G was associated with markedly reduced survival in squamous cell carcinoma (MS of 10.3 versus 49.4 months; adjusted hazard ratio for death, 7.9; 95% confidence interval, 2.4–26.0; P = 0.0007 for G/G versus T/T) but not in adenocarcinoma (SNP-histology interaction P = 0.004).
Conclusions
In a large prospective cohort, p53 Arg72Pro Pro/Pro was associated with a 2-fold increased risk of death in all esophageal cancers, whereas MDM2 T309G G/G was associated with a 7-fold increased risk of death in squamous cell carcinoma.
The incidence of esophageal cancer is rising at an alarming rate (1). Despite attempts to improve outcomes with aggressive multimodality therapy, prognosis remains poor with a 5-year overall survival of 16%,8 and clinical variables currently used to predict outcomes are imprecise (2, 3). Thus, the identification of molecular prognostic markers might enable further risk stratification, the first step towards individualization of treatment strategies (4).
Failure of apoptosis and cell cycle control are fundamental steps in carcinogenesis and important mechanisms in resistance to chemotherapy (5). p53 and its negative regulator, MDM2 (murine double minute 2) are key regulators of these processes (6). Both genes contain functional single nucleotide polymorphisms (SNP) known to impact tumor biology, which have been implicated in the development and prognosis of several cancers (7, 8).
A common G-to-C SNP in the p53 gene (p53 Arg72Pro) results in an amino acid change from arginine (Arg72) to proline (Pro72; ref. 9). The Pro variant exhibits decreased apoptotic potential (10) and, in keeping with this functional change, has been associated with increased risk, earlier age of onset, reduced response to chemotherapy, and early recurrence in a variety of cancers (8).
MDM2 T309G is located within the MDM2 promoter: the G variant enhances transcription factor binding leading to increased MDM2 expression and reduced apoptosis in response to DNA damage (11, 12). The G/G genotype has been associated with poor prognosis in other sporadic aerodigestive cancers (13–15). In some reports, the detrimental effect has been limited to specific subsets of the patients studied (13, 14).
Given the critical importance of the p53 pathway on malignant tumor behavior, and the previously observed biological and clinical effects of p53 Arg72Pro and MDM2 T309G, we evaluated the association of these SNPs with survival and recurrence in a large cohort of patients with esophageal cancer. Despite the practice of treating esophageal adenocarcinoma and squamous cell carcinoma as a single entity, these subtypes have distinct epidemiology, pathogenesis, tumor biology, and possibly prognosis (4, 16, 17). Moreover, tumor histologic subtype has been recognized as a determinant of the effect of MDM2 T309G on outcomes in lung cancer (14). Thus, in the present study, we also investigated whether the SNP-outcome relationships were dependent on tumor histologic subtype.
Patients and Methods
Patients
Starting January 1999, a cohort of patients with histologically confirmed esophageal carcinoma (adenocarcinoma, squamous cell carcinoma, or poorly differentiated carcinoma) was enrolled from the Massachusetts General Hospital. In 2004, recruitment was extended to the Dana-Farber Cancer Institute. Patients with pathologic diagnoses other than those listed above were excluded. There were no exclusions based on other clinical variables. The study was approved by the research ethics boards of the participating institutions. Demographic and clinical variables, and blood samples for genotyping were collected from participants at the time of study entry by trained research assistants.
For this report, analysis was restricted to patients enrolled prior to a cutoff date of October 2004 to allow for adequate follow-up to occur. Of the 404 eligible patients recruited, 33 were excluded: 6 had no blood sample for genotyping, 18 lacked outcome data, 5 had incomplete treatment data, and 4 had no genotype information for the SNPs of interest. Demographic and clinical variables are summarized in Table 1.
Table 1.
Patient demographics and treatment characteristics
| Variable | All patients, n = 371 | ADC, n = 300 | SCC, n = 63 | PDC, n = 8 |
|---|---|---|---|---|
| Age in y | ||||
| Median (range) | 64 (21–91) | 64 (21–91) | 64 (31–91) | 59 (34–78) |
| Race | ||||
| Caucasian | 358 (98%) | 294 (98%) | 56 (89%) | 8 (100%) |
| Other | 13 (2%) | 7 (2%) | 7 (11%) | 0 |
| Sex | ||||
| Male | 319 (86%) | 269 (90%) | 43 (68%) | 7 (88%) |
| Female | 52 (14%) | 31 (10%) | 20 (32%) | 1 (12%) |
| ECOG performance status | ||||
| 0–1 | 320 (86%) | 259 (86%) | 53 (84%) | 8 (100%) |
| 2 | 42 (11%) | 35 (12%) | 7 (11%) | 0 |
| 3 | 9 (2%) | 6 (2%) | 3 (5%) | 0 |
| Smoking status* | ||||
| Never | 75 (20%) | 60 (20%) | 12 (19%) | 3 (38%) |
| Former | 191 (52%) | 159 (54%) | 30 (48%) | 2 (25%) |
| Current | 102 (28%) | 78 (26%) | 21 (33%) | 3 (38%) |
| Smoking pack-years | ||||
| Median | 34.1 | 34.6 | 35.7 | 7.3 |
| Range | 0.2–212 | 0.2–212 | 0.4–149 | 0.5–72 |
| Stage | ||||
| I | 26 (7%) | 24 (8%) | 2 (3%) | 0 |
| IIa | 80 (22%) | 63 (21%) | 17 (27%) | 0 |
| IIb | 71 (19%) | 55 (18%) | 14 (22%) | 2 (25%) |
| III | 103 (28%) | 82 (27%) | 20 (32%) | 1 (12%) |
| IV | 90 (24%) | 76 (25%) | 9 (14%) | 5 (62%) |
| Treatment | ||||
| Surgery alone | 64 (17%) | 56 (19%) | 8 (13%) | 0 |
| Neoadjuvant CR | 191 (51%) | 153 (51%) | 36 (57%) | 2 (25%) |
| Neoadjuvant CT | 3 (1%) | 3 (1%) | 0 | 0 |
| Adjuvant CT or CR | 24 (6%) | 24 (8%) | 0 | 0 |
| CR† | 40 (11%) | 25 (8%) | 11 (17%) | 4 (50%) |
| CT alone | 36 (10%) | 29 (10%) | 5 (8%) | 2 (25%) |
| Radiation or PDT only | 8 (2%) | 8 (3%) | 2 (3%) | 0 |
| Supportive care only | 5 (1%) | 4 (1%) | 1 (2%) | 0 |
| Outcomes | ||||
| Death | 218 (59%) | 175 (58%) | 37 (59%) | 6 (75%) |
| Relapse/progression | 34 (9%) | 28 (9%) | 6 (10%) | 0 |
| Median survival, mo (range) | 29.1 (1.8–95.3) | 28.4 (1.8–95.3) | 33.7 (2.6–89.0) | 12.7 (2.9–31.6) |
| Median PFS, mo (range)‡ | 15.7 (1.5–89.0) | 15.7 (1.8–84.0) | 19.1 (1.5–89.0) | 12.7 (2.5–15.0) |
Abbreviations: ADC, adenocarcinoma; SCC, squamous cell carcinoma; PDC, poorly differentiated carcinoma; CT, chemotherapy; CR, chemoradiation; PDT, photodynamic therapy.
Percentages may not add up to 100 due to rounding.
Smoking status missing for three patients.
15 patients were intended to receive trimodality therapy, but due to progression of disease or complications received only chemoradiotherapy.
PFS missing for five patients (four ADC, one PDC).
End points
The primary end point was overall survival (OS) taken from the date of pathologic diagnosis to the date of death, or censored at last known follow-up. Progression-free survival (PFS), defined as the time between the date of pathologic diagnosis and the earliest of death, progression, relapse, or recurrence, was considered as a secondary end point. Outcome data were collected from clinical records and the hospital cancer registries. Where possible, death was confirmed using the Social Security Death Index (18). The last date for censoring purposes was July 2007. For individuals lost to follow-up prior to that date, censoring occurred at last date known alive for OS and last date evaluated radiologically or clinically without relapse or progression for PFS.
DNA extraction and genotyping
DNA was extracted from whole blood using the Puregene DNA Isolation Kit (Gentra Systems). The p53 Arg72Pro (rs1042522) and MDM2 T309G (rs2279744) SNPs were genotyped using the TaqMan assay and a 384-well ABI 7900HT Sequence Detection System (Applied Biosystems). Primers and probe sequences were obtained from Primer Express Software (Applied Biosystems) and are available upon request. Laboratory personnel were blinded to clinical outcomes, and validation was done in a random 5% of samples. Two authors independently reviewed the blinded genotype results.
Statistical methods
Demographic and clinical variables were compared across each of the SNP genotypes and patient groups using Fisher’s exact and Wilcoxon rank sum tests, where appropriate. Associations between SNP genotypes and survival end points were estimated using the Kaplan-Meier method, and statistical significance was assessed using the log rank test. Cox proportional hazard regression models were applied to multivariate analysis using a codominant model of inheritance, with covariates selected using a stepwise procedure. The final models for calculation of adjusted hazard ratios (AHR) for death and relapse/progression included age, stage, Eastern Cooperative Oncology Group (ECOG) performance status, and number of pack-years of smoking.
Interaction analyses were done by including a polymorphism* histology term (with polymorphism dichotomized into homozygous variant versus other) in the Cox proportional hazard models. For these tests of interaction, only adenocarcinoma and squamous cell carcinoma histologies were considered.
Two-sided statistical testing was conducted at the 0.05 level using SAS software version 9.1 (SAS Institute). The Bonferroni method was used to adjust for multiple comparisons.
Results
Patients
A total of 371 patients were included in the analyses. Ninety-eight percent of patients were Caucasian, and tumor histology was adenocarcinoma in 300 (81%), squamous cell carcinoma in 63 (17%), and poorly differentiated carcinoma in 8 (2%). Most patients were staged using computed tomography and positron emission tomography scanning, in addition to endoscopic ultrasound. For five patients, PFS was unavailable, although OS data were complete. Median follow-up for all patients was 33.1 months for OS and 32.9 months for PFS.
Genotype frequencies for the wild-type, heterozygous, and homozygous variants were 51%, 39%, and 9%, respectively, for p53 Arg72Pro; and 40%, 48%, and 12%, respectively, for MDM2 T309G. These distributions do not deviate from Hardy-Weinberg equilibrium (P > 0.10), and allelic frequencies did not differ from a cancer-free control population collected from the same institutions (19). Genotyping was complete in 97.8% to 99.7% of cases. There were no significant differences between genotyped patients and the small number of patients whose samples failed genotyping.
Tumor stage distributions were similar across p53 and MDM2 genotypes, and there was no association between genotypes and age at diagnosis, sex, race, histology, ECOG performance status, or smoking status (data not shown).
Outcomes
In univariate analyses, age, stage, ECOG performance status, and pack years of smoking were statistically significantly associated with shorter OS. Unadjusted hazard ratios for death are listed in Table 2.
Table 2.
Hazard ratios for death in univariate analysis
| Hazard ratio (95% CI) | P | |
|---|---|---|
| Clinical variables | ||
| Age* | 1.018 (1.006–1.03) | 0.003 |
| Male | 0.93 (0.63–1.37) | 0.72 |
| Tumor histology | ||
| ADC | Reference | |
| SCC | 0.93 (0.65–1.32) | 0.69 |
| PDC | 1.91 (0.84–4.32) | 0.12 |
| Stage | ||
| Node negative | Reference | |
| Node positive | 1.60 (1.13–2.26) | 0.009 |
| Metastatic | 4.17 (2.80–6.20) | <0.0001 |
| ECOG performance status | ||
| 0 | Reference | |
| 1 | 2.40 (1.16–3.49) | <0.0001 |
| 2 | 4.93 (2.51–9.68) | <0.0001 |
| Smoking Status† | ||
| Never | Reference | |
| Ex-smoker | 1.08 (0.76–1.55) | 0.66 |
| Current | 1.36 (0.92–2.03) | 0.13 |
| Pack-years smoking‡ | 1.006 (1.002–1.01) | 0.002 |
| SNPs | ||
| p53 Arg72Pro | ||
| Arg/Arg | Reference | |
| Arg/Pro | 0.92 (0.68–1.22) | 0.55 |
| Pro/Pro | 2.37 (1.53–3.68) | 0.0001 |
| MDM2 T309G | ||
| T/T | Reference | |
| T/G | 0.88 (0.66–1.18) | 0.39 |
| G/G | 1.42 (0.95–2.13) | 0.09 |
Continuous variable, defined as an increase in 1 y.
Ex-smokers is defined as individuals who had stopped smoking ≥ 1 y prior to diagnosis. Current smokers included all those who had smoked within 1 y of diagnosis.
Continuous variable, defined as an increase in 1 pack-year.
SNP genotypes and outcomes
Survival data for the entire study population are shown in Table 2 (unadjusted hazard ratios) and Table 3 (median survival and AHRs). p53 Pro/Pro was significantly associated with shorter OS and PFS in the whole cohort (Table 3 and Fig. 1, Kaplan-Meier curves). The relationship between p53 Arg72Pro and survival was consistent across all stages (data not shown).
Table 3.
p53 pathway genotypes and outcomes by SNP in all patients
| Genotype | n | Overall survival
|
Progression-free survival
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Months | Log-rank P | AHR* (95% CI) | P† | Months | Log-rank P | AHR* (95% CI) | P† | ||
| p53 Arg72Pro | |||||||||
| Arg/Arg | 186 | 29.1 | <0.0001 | Reference | 17.4 | 0.0018 | Reference | ||
| Arg/Pro | 143 | 35.5 | 0.84 (0.62–1.13) | 0.25 | 17.8 | 0.99 (0.75–1.32) | 0.97 | ||
| Pro/Pro | 34 | 11.8 | 2.05 (1.30–3.24) | 0.002 | 7.9 | 2.03 (1.29–3.18) | 0.002 | ||
| MDM2 T309G | |||||||||
| T/T | 149 | 29.6 | 0.06 | Reference | 18.0 | 0.09 | Reference | ||
| T/G | 178 | 30.6 | 0.91 (0.67–1.22) | 0.52 | 15.0 | 0.93 (0.71–1.23) | 0.62 | ||
| G/G | 43 | 22.0 | 1.15 (0.76–1.74) | 0.51 | 10.5 | 1.14 (0.76–1.72) | 0.52 | ||
Abbreviations: AHR, adjusted hazard ratio for death (OS) or relapse/progression (PFS).
Adjusted for: age, stage, ECOG performance status, pack-years of smoking.
P value corresponding to Cox proportional hazards model.
Fig. 1.

Kaplan-Meier analysis of (A) OS and (B) PFS in all participants by p53 Arg72Pro genotype.
Tumor histology subgroup and interaction analyses
In adjusted models, the association between p53 Pro/Pro and both relapse/progression (AHR, 1.92; 95% confidence interval [95% CI], 1.15–3.21; P = 0.01) and death (AHR, 2.29; 95% CI, 1.36–3.86; P = 0.002) remained significant in the adenocarcinoma subgroup (n = 300). The AHR point estimates were similar in magnitude and direction at 1.78 (95% CI, 0.55–5.79) for death and 3.00 (95% CI, 0.98–9.20) for relapse/progression in the squamous cell carcinoma subgroup (n = 63), and no interaction between histology and the SNP-outcome relationships was identified.
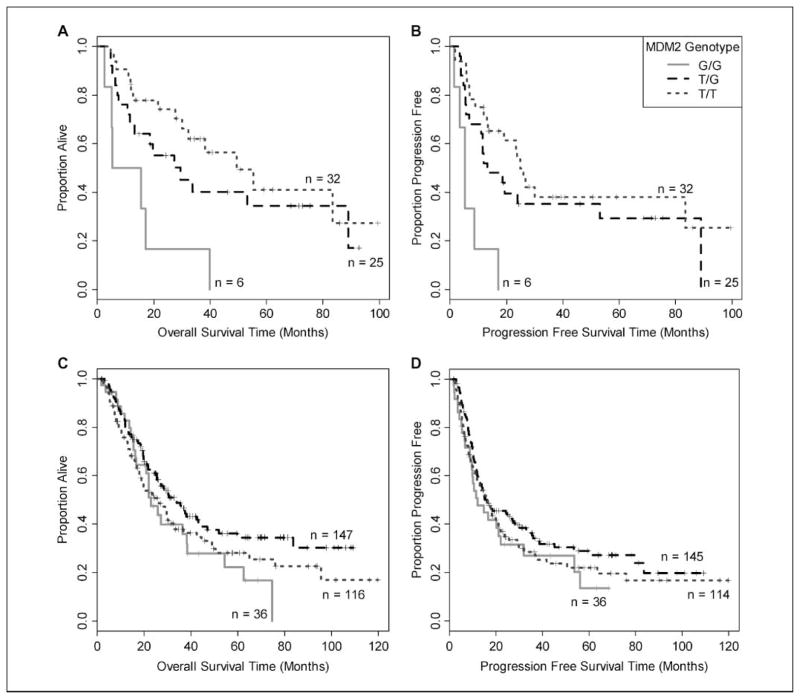
In contrast, MDM2 G/G showed a significant association with worse outcome in the squamous cell carcinoma population, but not in adenocarcinoma or the overall cohort. In patients with squamous cell carcinoma, median OS and PFS were markedly shorter in individuals with the G/G genotype (Table 4 and Fig. 2, Kaplan-Meier curves), and AHRs were large and highly statistically significant. In adenocarcinoma, the heterozygous T/G genotype was weakly associated with longer OS (but not PFS) in the adjusted model (AHR for death, 0.70; 95% CI, 0.50–0.99; P = 0.04), although this association does not remain significant after Bonferroni correction, and may represent a chance finding. In the multivariate model, there was a significant interaction between histology and MDM2 genotype (P = 0.004). This relationship between the MDM2 T309G polymorphism and survival was consistent across all stages (data not shown).
Table 4.
MDM2 T309G genotype and survival by tumor histology
| Genotype | n | Overall survival
|
Progression-free survival
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Months | Log-rank P | AHR* (95% CI) | P† | Months | Log-rank P | AHR* (95% CI) | P† | ||
| Squamous cell carcinoma | |||||||||
| T/T | 32 | 49.4 | 0.003 | Reference | 24.8 | 0.0004 | |||
| T/G | 25 | 29.5 | 1.81 (0.86–3.79) | 0.12 | 13.3 | 2.09 (1.04–4.22) | 0.18 | ||
| G/G | 6 | 10.3 | 7.89 (2.40–26.0) | 0.0007 | 5.3 | 9.79 (3.00–31.9) | 0.0002 | ||
| Adenocarcinoma | |||||||||
| T/T | 116 | 26.1 | 0.10 | Reference | 16.1 | 0.27 | |||
| T/G | 147 | 33.6 | 0.70 (0.50–0.99) | 0.04 | 16.1 | 0.79 (0.58–1.07) | 0.13 | ||
| G/G | 36 | 22.9 | 0.85 (0.53–1.36) | 0.49 | 12.1 | 0.89 (0.56–1.41) | 0.61 | ||
Adjusted for: age, stage, ECOG performance status, pack years of smoking.
P value corresponding to Cox Proportional Hazards model.
Fig. 2.
Kaplan-Meier analysis of OS and PFS by MDM2 T309G genotype in histologic subgroups. The G/G genotype was associated with significantly shorter OS and PFS in the squamous cell carcinoma subgroup (A, B), but not in the adenocarcinoma subgroup (C, D; P = 0.004 for SNP-histology interaction).
Trimodality treatment subgroup
The majority of our patients were intended to receive cisplatin-based trimodality therapy (n = 196). We explored the impact of SNP genotypes on outcomes in this intention-to-treat subgroup. A total of 181 of these individuals completed all three treatment components; the remainder had complications or disease progression that resulted in early termination of therapy. SNP genotype frequencies, histology, race, gender, ECOG performance status, and age distributions in this subgroup were similar to the overall cohort. No intention-to-treat patients had stage I disease, and a smaller proportion had stage IV disease (17% versus 24%).
In these patients, as in the overall population, the p53 Pro/Pro genotype was associated with a shorter OS (AHR for death, 2.22; 95% CI, 1.11–4.47; P = 0.02) and PFS (AHR for relapse/progression, 2.15; 95% CI, 1.06–4.35; P = 0.03). Median survival was 11.8 months for Pro/Pro homozygous individuals, compared with 30.6 months in patients with the Arg/Arg genotype (P = 0.007). As observed in the entire study cohort, the MDM2 T309G genotype was not associated with outcomes in this treatment subgroup. In the subset of patients with squamous cell carcinoma intended for cisplatin-based trimodality therapy, the number of patients was small (n = 37), and no outcome association with MDM2 T309G was detected.
Bonferroni corrections
In evaluating the statistical significance of our primary results, we considered all of the 13 primary comparisons done in this cohort of patients and reported here and elsewhere (20, 21). When adjusted for these, the association between p53 Arg72Pro and outcomes in all cases, as well as MDM2 T309G SNP-histology interaction remained statistically significant.
Discussion
In this large, predominantly Caucasian, cohort of patients with esophageal cancer and mature outcome data, we have shown that variant genotypes in two key p53 pathway SNPs, p53 Arg72Pro and MDM2 T309G, are associated with shortened survival. The p53 Pro/Pro genotype was associated with a 2-fold increased risk of death and relapse/progression in the entire population, regardless of histology, and after Bonferroni correction for multiple comparisons. The MDM2 G/G genotype was associated with a >7-fold increased risk of death in the subgroup of patients with squamous cell carcinoma, but not among those with adenocarcinoma.
The association between p53 Pro/Pro genotype and worse prognosis in our cohort is in keeping with the functional consequences of this polymorphism: the p53 Pro variant protein has been shown to have a reduced ability to induce apoptosis attributed, at least in part, to impaired mitochondrial trafficking (10), and inhibition of p73-dependent apoptosis (22, 23). This variant has been associated with worse outcome and resistance to chemotherapy in several other cancers (23–28). The only published study to assess the impact of the p53 Arg72Pro polymorphism in patients with esophageal cancer, which included a smaller number of patients (n = 210) and shorter follow-up time, did not show a statistically significant association with survival or recurrence, although the hazard ratio point estimates were similar to our results (29).
We also found that MDM2 T309G G/G is associated with markedly worse OS and PFS, although in this case the detrimental effect was limited to patients with squamous cell carcinoma, as shown by both subgroup and interaction analyses. The G variant of this SNP is known to increase promoter-binding affinity, leading to up-regulation of MDM2, and consequent inhibition and down-regulation of the p53 pathway (8). A similar histology-specific relationship between MDM2 G/G and adverse outcomes was recently shown in a cohort of patients with non–small cell lung cancer (14).
The observed histologic specificity of the effect of MDM2 T309G could be accounted for by differences in the molecular biology of adenocarcinoma and squamous cell carcinoma, which are well recognized (30). These tumor types exhibit more than 500 differentially expressed genes (31). In North American and European populations, somatic mutations in p53 occur with greater frequency in squamous cell carcinoma than in adenocarcinoma (67% versus 46%) and the type and localization of p53 mutations are different (32, 33). Although MDM2 T309G has been shown to modulate the association between alterations of p53 and outcomes in breast cancer (34), Heist et al. did not find a correlation between MDM2 T309G and p53 status or expression in non–small cell lung cancer (14),9 suggesting that a more complex or tumor-specific relationship might exist. As routine untreated tissue availability is limited in esophageal cancer and tumor samples were not available to investigate the relationship among tumor p53 alterations, SNP genotypes, and outcomes in our cohort, prospective studies including tissue collection are warranted to evaluate this potential association.
Because the SNPs assessed in this study modulate a common pathway, a joint assessment of the MDM2 and p53 SNPs would be justified. However, only seven patients carried both risk genotypes, making such an analysis infeasible.
In summary, we show, in a large cohort of patients with esophageal cancer, that p53 Arg72Pro Pro/Pro is strongly associated with shortened survival in esophageal cancer. Further, MDM2 T309G G/G is associated with shortened survival in esophageal squamous cell carcinoma. Our results show the prognostic value of SNPs in the p53 pathway and underscore the distinct differences between squamous cell and adenocarcinomas of the esophagus. Although these findings do not at present allow for the selection of a particular therapeutic strategy, they do identify poor-risk groups that may have a differential response to treatment on the basis of altered p53 pathway function. The predictive value of these SNPs can only be assessed in randomized controlled trials.
Acknowledgments
Grant support: NIH grant R01 CA074386, Doris Duke Charitable Foundation, Alan B. Brown Chair in Molecular Genomics, Posluns Family Foundation, and CIHR operating grant to Dr. G Liu.
Footnotes
SEER Cancer Statistics Review, 1975–2004, National Cancer Institute. Bethesda, MD, http://seer.cancer.gov/csr/1975_2004/, based on November 2006 SEER data submission, posted to the SEER web site, 2007.
Personal communication.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Pohl H, Welch HG. The role of overdiagnosis and reclassification in the marked increase of esophageal adenocarcinomaincidence. J Natl Cancer Inst. 2005;97:142–6. doi: 10.1093/jnci/dji024. [DOI] [PubMed] [Google Scholar]
- 2.Lagarde SM, ten Kate FJ, Reitsma JB, Busch OR, van Lanschot JJ. Prognostic factors in adenocarcinoma of the esophagus or gastroesophageal junction. J Clin Oncol. 2006;24:4347–55. doi: 10.1200/JCO.2005.04.9445. [DOI] [PubMed] [Google Scholar]
- 3.Rizk NP, Venkatraman E, Bains MS, et al. American Joint Committee on Cancer staging system does not accurately predict survival in patients receiving multimodality therapy for esophageal adenocarcinoma. J Clin Oncol. 2007;25:507–12. doi: 10.1200/JCO.2006.08.0101. [DOI] [PubMed] [Google Scholar]
- 4.Ajani JA. Carcinoma of the esophagus: is biology screaming in my deaf ears? J Clin Oncol. 2005;23:4256–8. doi: 10.1200/JCO.2005.12.911. [DOI] [PubMed] [Google Scholar]
- 5.Kirsch DG, Kastan MB. Tumor-suppressor p53: implications for tumor development and prognosis. J Clin Oncol. 1998;16:3158–68. doi: 10.1200/JCO.1998.16.9.3158. [DOI] [PubMed] [Google Scholar]
- 6.Rayburn E, Zhang R, He J, Wang H. MDM2 and human malignancies: expression, clinical pathology, prognostic markers, and implications for chemotherapy. Curr Cancer Drug Targets. 2005;5:27–41. doi: 10.2174/1568009053332636. [DOI] [PubMed] [Google Scholar]
- 7.Bond GL, Levine AJ. A single nucleotide polymorphism in the p53 pathway interacts with gender, environmental stresses and tumor genetics to influence cancer in humans. Oncogene. 2007;26:1317–23. doi: 10.1038/sj.onc.1210199. [DOI] [PubMed] [Google Scholar]
- 8.Pietsch EC, Humbey O, Murphy ME. Polymorphisms in the p53 pathway. Oncogene. 2006;25:1602–11. doi: 10.1038/sj.onc.1209367. [DOI] [PubMed] [Google Scholar]
- 9.Harris N, Brill E, Shohat O, et al. Molecular basis for heterogeneity of the human p53 protein. Mol Cell Biol. 1986;6:4650–6. doi: 10.1128/mcb.6.12.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dumont P, Leu JI, Della Pietra ACR, George DL, Murphy M. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet. 2003;33:357–65. doi: 10.1038/ng1093. [DOI] [PubMed] [Google Scholar]
- 11.Bond GL, Hu W, Bond EE, et al. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell. 2004;119:591–602. doi: 10.1016/j.cell.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 12.Bond GL, Hu W, Levine A. A single nucleotide polymorphism in the MDM2 gene: from a molecular and cellular explanation to clinical effect. Cancer Res. 2005;65:5481–4. doi: 10.1158/0008-5472.CAN-05-0825. [DOI] [PubMed] [Google Scholar]
- 13.Ohmiya N, Taguchi A, Mabuchi N, et al. MDM2 promoter polymorphism is associated with both an increased susceptibility to gastric carcinoma and poor prognosis. J Clin Oncol. 2006;24:4434–40. doi: 10.1200/JCO.2005.04.1459. [DOI] [PubMed] [Google Scholar]
- 14.Heist RS, Zhou W, Chirieac LR, et al. MDM2 polymorphism, survival, and histology in early-stage non-small-cell lung cancer. J Clin Oncol. 2007;25:2243–7. doi: 10.1200/JCO.2006.08.8914. [DOI] [PubMed] [Google Scholar]
- 15.Asomaning K, Reid AE, Zhou W, et al. MDM2 promoter polymorphism and pancreatic cancer risk and prognosis. Clin Cancer Res. 2008;14:4010–5. doi: 10.1158/1078-0432.CCR-07-4187. [DOI] [PubMed] [Google Scholar]
- 16.Rohatgi PR, Swisher SG, Correa AM, et al. Histologic subtypes as determinants of outcome in esophageal carcinoma patients with pathologic complete response after preoperative chemoradiotherapy. Cancer. 2006;106:552–8. doi: 10.1002/cncr.21601. [DOI] [PubMed] [Google Scholar]
- 17.Siewert JR, Ott K. Are squamous and adenocarcinomas of the esophagus the same disease? Semin Radiat Oncol. 2007;17:38–44. doi: 10.1016/j.semradonc.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 18.Bradbury PA, Heist RS, Kulke MH, et al. A rapid outcomes ascertainment system improves the quality of prognostic and pharmacogenetic outcomes from observational studies. Cancer Epidemiol Biomarkers Prev. 2008;17:204–11. doi: 10.1158/1055-9965.EPI-07-0470. [DOI] [PubMed] [Google Scholar]
- 19.Cescon DW, Liu G, Zhai R, et al. p53 Arg72Pro, MDM2 T309G and CCND1 G870A polymorphisms are not associated with susceptibility to esophageal adenocarcinoma. A ACR Meeting Abstracts. 2008:3866. doi: 10.1111/j.1442-2050.2009.00960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cescon DW, Bradbury PA, Asomaning K, et al. p53 Arg72Pro, MDM2 T309G, CCND1 G870A polymorphisms, histology and esophageal cancer prognosis [May 20 suppl; abstr 11033] J Clin Oncol. 2008:26. [Google Scholar]
- 21.Bradbury PA, Marshall AL, Kulke MH, et al. Prognostic significance of nuclear excision (NER) and base excision (BER) DNA repair gene polymorphisms in esophageal cancer. J Clin Oncol; 2007 ASCO Annual Meeting Proceedings Part I; Jun 20, 2007. p. 2511. Supplement. [Google Scholar]
- 22.Bergamaschi D, Gasco M, Hiller L, et al. p53 polymorphism influences response in cancer chemotherapy via modulation of p73-dependent apoptosis. Cancer Cell. 2003;3:387–402. doi: 10.1016/s1535-6108(03)00079-5. [DOI] [PubMed] [Google Scholar]
- 23.Sullivan A, Syed N, Gasco M, et al. Polymorphism in wild-type p53 modulates response to chemotherapy in vitro and in vivo. Oncogene. 2004;23:3328–37. doi: 10.1038/sj.onc.1207428. [DOI] [PubMed] [Google Scholar]
- 24.Wang YC, Chen CY, Chen SK, Chang YY, Lin P. p53 codon 72 polymorphism in Taiwanese lung cancer patients: association with lung cancer susceptibility and prognosis. Clin Cancer Res. 1999;5:129–34. [PubMed] [Google Scholar]
- 25.Starinsky S, Figer A, Ben-Asher E, et al. Genotype phenotype correlations in Israeli colorectal cancer patients. Int J Cancer. 2005;114:58–73. doi: 10.1002/ijc.20645. [DOI] [PubMed] [Google Scholar]
- 26.Tommiska J, Eerola H, Heinonen M, et al. Breast cancer patients with p53 Pro72 homozygous genotype have a poorer survival. Clin Cancer Res. 2005;11:5098–103. doi: 10.1158/1078-0432.CCR-05-0173. [DOI] [PubMed] [Google Scholar]
- 27.Toyama T, Zhang Z, Nishio M, et al. Association of TP53 codon 72 polymorphism and the outcome of adjuvant therapy in breast cancer patients. Breast Cancer Res. 2007;9:R34. doi: 10.1186/bcr1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu Y, Yao L, Ouyang T, et al. p53 Codon 72 polymorphism predicts the pathologic response to neoadjuvant chemotherapy in patients with breast cancer. Clin Cancer Res. 2005;11:7328–33. doi: 10.1158/1078-0432.CCR-05-0507. [DOI] [PubMed] [Google Scholar]
- 29.Wu X, Gu J, Wu TT, et al. Genetic variations in radiation and chemotherapy drug action pathways predict clinical outcomes in esophageal cancer. J Clin Oncol. 2006;24:3789–98. doi: 10.1200/JCO.2005.03.6640. [DOI] [PubMed] [Google Scholar]
- 30.Lin J, Beerm DG. Molecular biology of upper gastrointestinal malignancies. Semin Oncol. 2004;31:476–86. doi: 10.1053/j.seminoncol.2004.04.019. [DOI] [PubMed] [Google Scholar]
- 31.Greenawalt DM, Duong C, Smyth GK, et al. Gene expression profiling of esophageal cancer: comparative analysis of Barrett’s esophagus, adenocarcinoma, and squamous cell carcinoma. Int J Cancer. 2007;120:1914–21. doi: 10.1002/ijc.22501. [DOI] [PubMed] [Google Scholar]
- 32.Olivier M, Eeles R, Hollstein M, Khan MA, Harris CC, Hainaut P. The IARC TP53 database: new online mutation analysis and recommendations to users. Hum Mutat. 2002;19:607–14. doi: 10.1002/humu.10081. [DOI] [PubMed] [Google Scholar]
- 33.Royds JA, Iacopetta B. p53 and disease: when the guardian angel fails. Cell Death Differ. 2006;13:1017–26. doi: 10.1038/sj.cdd.4401913. [DOI] [PubMed] [Google Scholar]
- 34.Boersma BJ, Howe TM, Goodman JE, et al. Association of breast cancer outcome with status of p53 and MDM2 SNP309. J Natl Cancer Inst. 2006;98:911–9. doi: 10.1093/jnci/djj245. [DOI] [PubMed] [Google Scholar]