Abstract
Wogon-Sugi has been reported as a cytoplasmically inherited virescent mutant selected from a horticultural variety of Cryptomeria japonica. Although previous studies of plastid structure and inheritance indicated that at least some mutations are encoded by the chloroplast genome, the causative gene responsible for the primary chlorophyll deficiency in Wogon-Sugi, has not been identified. In this study, we identified this gene by genomic sequencing of chloroplast DNA and genetic analysis. Chloroplast DNA sequencing of 16 wild-type and 16 Wogon-Sugi plants showed a 19-bp insertional sequence in the matK coding region in the Wogon-Sugi. This insertion disrupted the matK reading frame. Although an indel mutation in the ycf1 and ycf2 coding region was detected in Wogon-Sugi, sequence variations similar to that of Wogon-Sugi were also detected in several wild-type lines, and they maintained the reading frame. Genetic analysis of the 19 bp insertional mutation in the matK coding region showed that it was found only in the chlorophyll-deficient sector of 125 full-sibling seedlings. Therefore, the 19-bp insertion in the matK coding region is the most likely candidate at present for a mutation underlying the Wogon-Sugi phenotype.
Electronic supplementary material
The online version of this article (doi:10.1007/s00294-009-0247-9) contains supplementary material, which is available to authorized users.
Keywords: Cryptomeria japonica, Wogon-Sugi, Virescent mutant, matK gene, Frameshift mutation
Introduction
In higher plants, chlorophyll-deficient mutants such as albino, variegated and virescent mutants have been selected and utilized to elucidate the development and function of the chloroplast. The virescent forms of these mutants are phenotypically characterized as having a lag in chlorophyll accumulation in young leaves (Archer and Bonnett 1987; Archer et al. 1987) and have been isolated in a wide range of flowering plants including rice (Iba et al. 1991), barley (Jain 1966), maize (Hopkins and Elfman 1984), cotton (Benedict and Kohel 1970), tobacco (Archer and Bonnett 1987; Archer et al. 1987), and peanut (Benedict and Ketring 1972).
The causative genes or alleles responsible for virescent mutations have recently been described in several plants. For example, virescent or delayed-greening mutants (cue3, cue6, cue8, and clpR1-1) of Arabidopsis are involved in the positive regulation of nuclear gene expression (López-Juez et al. 1998; Vinti et al. 2005; Koussevitzky et al. 2007). Plastid genome (plastome) mutants have been isolated in Hordeum (Rios et al. 2003; Landau et al. 2007), and the corresponding mutations have been characterized at the DNA sequence level. In the case of plastome mutants, virescence is inherited in a non-Mendelian fashion, i.e., maternally in the majority of angiosperms and occasionally biparentally.
Cryptomeria japonica (Sugi) is a coniferous tree species that belongs to the group Cupressaceae sensu lato (Kusumi et al. 2000). In C. japonica, a number of mutant varieties show traits such as dwarfism, variegation, and morphological variation in needles and shoots. Wogon-Sugi is a virescent mutant, whose new shoots change color from yellowish-white in spring to normal green in late summer. Ohba et al. (1971) used reciprocal crosses between Wogon-Sugi and wild-type Sugi to demonstrate that the yellowish-white trait of Wogon-Sugi is inherited in a non-Mendelian fashion. In crosses using Wogon-Sugi as the male parent (pollen donor) and wild-type Sugi as the female parent, Wogon-Sugi occurred at a rate of 89.6%, green and white sectors at 7.5%, and the wild-type phenotype at 2.9%. In contrast, controlled crosses using Wogon-Sugi as the female and wild-type Sugi as the male parent resulted in 99% wild-type and 1% Wogon-Sugi and chimeric seedlings. This non-Mendelian transmission was maintained even in self-pollinated or backcrossed F1 hybrids derived from the reciprocal crosses. These data suggested paternal inheritance of the plastid genome in C. japonica, and provided the initial experimental evidence that at least some types of mutations are encoded by the plastid genome.
Genomic sequencing data provide detailed information on variations and mutations at the nucleotide level. We have already determined the complete nucleotide sequence of the C. japonica chloroplast genome using the shotgun sequencing method, and determined that 116 genes are encoded in the genome (Hirao et al. 2008, GenBank accession no. AP009377). Therefore, comparison with wild-type will determine whether there is a specific mutation in the Wogon-Sugi chloroplast genome that is responsible for its phenotype.
The objective of this study was to identify the gene that is mutated in the primary chlorophyll-deficient C. japonica virescent mutant, Wogon-Sugi. To identify candidate genes, we completed nucleotide sequencing of Wogon-Sugi and Yaku-Sugi by PCR-based genome walking. For wild-type we selected Yaku-Sugi, a local race of C. japonica in Japan. To identify the specific mutation of the Wogon-Sugi chloroplast genome, comparative analyses were conducted between Wogon-Sugi and wild-type chloroplast genomes and the mutations detected were examined in 16 Wogon-Sugi and 16 wild-type plants. Finally, we examined the relationship between phenotype and genotype using chloroplast DNA markers developed based on the specific mutation of the Wogon-Sugi chloroplast genome.
Materials and methods
Chloroplast DNA sequencing, sequence assembly, and gene annotation
The virescent mutant of Cryptomeria japonica (Wogon-Sugi) is preserved in 16 individuals in the Forest Products Research Institute, Forest Tree Breeding Center (FFPRI-FTBC) in Ibaraki, Japan. To determine the complete sequence of this mutant’s chloroplast genome, a Wogon-Sugi sample, W-77, was arbitrarily selected from among 16 individual plants. In addition, one wild-type Yaku-Sugi plant was employed as a comparative sample to detect mutations in the Wogon-Sugi chloroplast genome. The wild-type was collected from a natural C. japonica forest ion Yaku Island (30°20′N and 130°30′E) and has been planted in FFPRI-FTBC.
Total cellular DNA of both plant types was prepared by the method of Shiraishi and Watanabe (1995). Approximately 100 mg of leaves was frozen in liquid nitrogen and ground in a homogenizer. The homogenized sample was mixed with 1 ml of CTAB buffer (100 mM Tris–HCl, pH 9.0, 20 mM EDTA, 2% CTAB (hexadecyltrimethylammonium bromide)) with 0.1% beta-mercaptoethanol added just prior to use. The mixture was incubated at 65°C for 60 min and centrifuged for 10 min at 12,000×g, then 600 μl of the supernatant was transferred to a 1.5 ml microcentrifuge tube. The supernatant was mixed twice with phenol/chloroform/isoamyl alcohol (25:24:1) and centrifuged for 10 min at 12,000×g. DNA was precipitated from the aqueous phase by adding 0.1 volume of 3 M sodium acetate and 2.5 volumes of ethanol. The precipitate was washed twice with 70% ethanol and dissolved in water. Extracted DNA was further purified using the DNeasy Plant Mini kit (Qiagen).
Complete nucleotide sequencing of the Wogon-Sugi and Yaku-Sugi chloroplast genomes was performed using 345 PCR genome walking primers (http://labglt.nftbc.affrc.go.jp/DNA_analysis_resource/sugi/sugi_cp_primer.html). DNA amplification reactions were carried out in a GeneAmp® PCR System 9700 (Applied Biosystems) programmed for touchdown PCR from 62 to 57°C for each primer. The PCR conditions were as follows: after initial melting at 94°C for 1 min, 30 s of denaturation at 94°C, 1 min of annealing at 62°C, and 1 min of extension at 72°C for ten cycles of amplification. At each cycle, the annealing temperature was reduced 0.5°C, followed by 30 s of denaturation at 94°C, 1 min of annealing at 57°C, and 1 min of extension at 72°C for 20 cycles, followed by a final extension at 72°C for 10 min. PCR was performed in a volume of 10 μl containing 10 mM Tris–HCl, pH 9.0, 50 mM KCl, 1.5 mM MgCl2, 0.2 mM each dNTP, 0.2 μM each primer, 100 ng genomic DNA and 0.1 U of Taq DNA polymerase (Invitrogen). Amplified PCR products were treated with exonuclease and shrimp alkaline phosphatase to remove excess dNTPs and primers. The exonuclease/alkaline phosphatase treatment was performed by mixing 5 μl PCR product with 0.2 μl exonuclease I (10 U/μl; TAKARA), 2.0 μl shrimp alkaline phosphatase (1 U/μl; Amersham), 1.0 μl SAP 10× buffer and 1.8 μl deionized water, and then incubating at 37°C for 30 min followed by 75°C for 15 min to inactivate the exonuclease and alkaline phosphatase. Cycle sequencing was performed according to the manufacturer’s instructions using BigDye® 2.0 Terminator Cycle Sequencing kit (Applied Biosystems). The sequencing primer (3.2 pmol, the same as the PCR primer), 1.0 μl ABI Dye Terminator Ready-Reaction sequencing premix and 1.5 μl 5× sequence buffer were added to the template. After a 2-min denaturation step at 96°C, dye-terminator reactions were incubated at 96°C for 15 s, 50°C for 1 s and 60°C for 4 min for 25 cycles. Excess dye terminators were removed by ethanol precipitation. The extension products were evaporated to dryness under vacuum, resuspended in Hi-DiTM formamide (Applied Biosystems), heated for 2 min at 94°C and loaded onto an ABI PRISM® model 3100 DNA sequencer (Applied Biosystems) according to the manufacturer’s directions. For sequence analysis and assembly, we used Sequencher® 3.1 software (Gene Codes Corporation). The determined sequence was annotated using DOGMA (Dual Organellar GenoMe Annotator) software (Wyman et al. 2004) after a FASTA-formatted file of the complete chloroplast genome was uploaded to the program’s server. The fully annotated chloroplast genome of Wogon-Sugi and one Yaku-Sugi plant was submitted to DDBJ GenBank with the following accession numbers: Wogon-Sugi chloroplast genome, AP010966; Yaku-Sugi chloroplast genome, AP010967. The complete nucleotide sequences of the Wogon-Sugi and Yaku-Sugi chloroplast genomes were aligned using GeneDoc software (Nicholas et al. 1997), and sequence variations between the two genomes were examined for single nucleotide polymorphisms (SNPs), simple sequence repeats (SSRs), and insertions or deletions (indels).
Verification of the mutation in the coding region by DNA sequencing
Mutations in the gene-coding regions, detected from sequence alignment, were examined in 16 wild-type plants and 16 Wogon-Sugi individuals by DNA sequencing. The 16 wild-type plants were 11 local races, Ajigasawa-Sugi, Ooshuku-Sugi, Toudou-Sugi, Makinosaki-Sugi, Honna-Sugi, Mura-Sugi, Kuma-Sugi, Tateyama-Sugi, Itoshiro-Sugi, Ashuu-Sugi, Hachirou-Sugi, plus three individuals from a natural population on Yaku Island and two plus-trees (Nagano 1, Iiyama 16) of Japan. Total cellular DNA of these 16 wild-type plants and 16 Wogon-Sugi individuals was prepared by the method of Shiraishi and Watanabe (1995). Extracted DNA was further purified using the Mag-extractor (Toyobo). DNA sequencing was performed according to the above-described PCR genome walking methods. The sequences determined were aligned using GeneDoc software (Nicholas et al. 1997). Mutations in the nucleotide sequence of the coding region were translated to predict the amino acid sequence, and then the predicted primary structures were compared.
Genetic analysis of the chloroplast genome
A controlled artificial cross was made between a wild-type plant (Yoshiki 1) as the maternal parent and W-77 Wogon-Sugi as the paternal parent. Prior to pollination, the female cones of the wild-type plant were completely enclosed in crossing bags to eliminate foreign pollen contaminants. Mature pollen was collected from Wogon-Sugi male cones for pollination. After pollination, 131 mature F1 seeds were collected, germinated, and the seedlings grown for 1 year in a greenhouse. The phenotype of all 131 seedlings was evaluated for the 20th through the 25th leaves, which were sophomoric leaves and new shoots.
The chloroplast genomes of the 131 seedlings that were characterized for phenotype were examined using a chloroplast DNA marker developed based on a mutation in the Wogon-Sugi chloroplast genome. Total cellular DNA was prepared from each individual by the method of Shiraishi and Watanabe (1995). Extracted DNA was further purified using the Mag-extractor (Toyobo). Fragment analysis was performed by sizing the PCR products. The forward primer used was 5′-AGGTTATTCTTGGGTCCGGGGGTTT-3′ and the reverse primer was 5′-ACTAAAATTTCCGTTGGTTCGGAG-3′; these were developed to detect the specific mutation in the matK coding region of the Wogon-Sugi chloroplast genome. The forward primer was labeled with HEX fluorescent dye. DNA amplification was carried out in a PTC-200 thermocycler (MJ Research). The PCR conditions were as follows: after initial melting at 94°C for 1 min, 30 s of denaturation at 94°C, 30 s of annealing at 60°C, and 1 min 30 s of extension at 72°C for 30 cycles, followed by a final extension at 72°C for 5 min. PCR was performed on samples of the same composition and volume described earlier for PCR genome walking. Amplified PCR products were separated using an ABI PRISM® model 3100 automated DNA sequencer. Labeled fragments were detected and sized using GeneScan® 350 ROX™ size standards (Applied Biosystems). All genotypes were determined using Genotyper® fragment analysis software version 3.7 genotyping software (Applied Biosystems).
Results
General characteristics of Wogon-Sugi and wild-type chloroplast genomes
The size of the Wogon-Sugi chloroplast genome was determined to be 131,804 bp, which is slightly larger than the 131,781 bp of wild-type Yaku-Sugi. Both chloroplast genomes encoded a total of 116 genes, 112 of which were single copy and two (trnI-CAU and trnQ-UUG) were duplicated as inverted repeat sequences. Of the 116 genes, there were four ribosomal RNA genes (3.5%), 30 individual transfer RNA genes (25.9%), 21 genes encoding large and small ribosomal subunits (18.1%), four genes encoding DNA-dependent RNA polymerases (3.5%), 48 genes encoding photosynthesis-related proteins (41.4%), and nine genes encoding other proteins, including those with unknown functions (7.8%). Of the 112 single copy genes, 17 contained introns, and three (clpP, trnT-GGU, and ycf68) were identified as pseudogenes. In addition, stop codons were found in the matK (maturase) coding region of the Wogon-Sugi chloroplast genome.
Sequence variations between the two chloroplast genomes
A total of 21 sequence variations (eight SNPs, nine SSRs, and four indels) were identified from the sequence alignment of the Wogon-Sugi and Yaku-Sugi chloroplast genomes (Table 1). The eight SNPs, five of the SSRs, and one indel were identified in intergenic spacer regions. The remaining four SSRs were identified in intron regions. The remaining three indels were identified in three gene coding regions: matK (maturase), ycf2 (hypothetical protein RF2) and ycf1 (hypothetical protein RF1).
Table 1.
Sequence variations between Wogon-Sugi (mutant) and Yaku-Sugi (wild-type plant) chloroplast genomes
| Location in Wogon-Sugi | Region | Mutation type | Wogon-Sugi | Yaku-Sugi |
|---|---|---|---|---|
| 10876 | rps11-rpl36 spacer | SNP | T | G |
| 16344 | rpl2 intron | SSR | (TA)9 | (TA)8 |
| 20693 | psaJ-trnP spacer | SSR | (T)19 | (T)20 |
| 21018 | trnP-trnW spacer | SNP | A | G |
| 23754 | psbJ-ψ clpP spacer | SSR | (T)10 | (T)11 |
| 23912 | psbJ-ψ clpP spacer | SNP | G | T |
| 24189 | psbJ-ψ clpP spacer | SSR | (A)11 | (A)12 |
| 39584 | chlL-trnH spacer | SSR | (T)11 | (T)10 |
| 43948 | matK coding region | Indel | 19 bp insertion | – |
| 78669 | atpI-atpH spacer | SSR | (TA)7 | (TA)6 |
| 82805 | trnG intron | SSR | (A)12 | (A)11 |
| 86642 | trnL-trnF spacer | SSR | (TA)22 | (TA)12 |
| 92241 | ndhA intron | SSR | (T)13 | (T)14 |
| 105857 | ψycf68-rps12 spacer | SNP | T | G |
| 105858 | ψycf68-rps12 spacer | SNP | C | A |
| 115914 | ycf2 coding region | Indel | 33 bp insertion | – |
| 121743 | ycf2-ycf1 spacer | SNP | A | G |
| 121981 | ycf2-ycf1 spacer | SNP | A | G |
| 128800 | ycf1 coding region | Indel | 66 bp deletion | – |
| 130055 | trnL-ccsA spacer | Indel | 19 bp deletion | – |
| 131579 | ccsA-petA spacer | SNP | G | T |
The locations and regions of polymorphic sites between Wogon-Sugi and Yaku-Sugi were documented according to the nucleotide and gene order of the annotated Wogon-Sugi chloroplast genome sequence. Mutations were classified into three types: single nucleotide polymorphism (SNP), simple sequence repeat (SSR), and insertion or deletion (indel). SSR mutations of each chloroplast genome are indicated by repeat motif and repeat number
An insertional mutation in the matK gene of the Wogon-Sugi chloroplast genome causes a frameshift mutation
The insertion or deletion mutations in coding regions matK, ycf2, and ycf1 were examined in 16 wild-type and 16 Wogon-Sugi plants by DNA sequencing. All three indels of the coding region consisted of duplicated repetitive sequences. The 19-bp insertion in the matK coding region was found only in the 16 Wogon-Sugi individuals. This insertion resulted in a reading frameshift after the amino acid residue phenylalanine (F) at the 24th amino acid residue, introducing a stop codon just after the serine (S) residue at the 39th amino acid residue (Fig. 1). The ycf2 coding region of all 32 plants was classified into four types based on the 33-bp repetitive sequence unit, which is equivalent to an insertion of 11 amino acids (Fig. 2). Four indel variants were identified among the 16 wild-type plants, which are referred to as Types I–IV in Fig. 2. Two wild-type plants had the DNA sequence depicted as Type I, with five copies of the 33-bp sequence repeat. The Type II variants had four copies (two wild-type plants), Type III had three copies (10 plants), and Type IV had two copies (two plants). All 16 Wogon-Sugi plants had the Type I indel. Despite the length variation in the ycf2 coding region, none of the variants caused a change in the reading frame. Similarly, the ycf1 coding region was classified into four types based on the 66-bp repetitive sequence unit found in all 32 plants investigated (Fig. 3): Six plants were classified as Type I, which contained five copies of the 66-bp repeat; six plants were Type II with four copies; three were Type III with three copies, and one plant was Type IV with two copies of the repeat. All Wogon-Sugi plants belonged to Type II. The indel mutations of the ycf2 and ycf1 coding regions do not shift the reading frames, but each additional repeat in the ycf2 gene is equivalent to an insertion of 11 amino acids and each additional repeat in the ycf1 gene is equivalent to an insertion of 22 amino acids.
Fig. 1.
Reading frameshift mutation in the matK coding region of the Wogon-Sugi chloroplast genome. The nucleotide and deduced amino acid sequences of part of the matK gene of Wogon-Sugi and wild-type chloroplast genomes are shown. Blue nucleotides show the insertion sequence in Wogon-Sugi, and red nucleotides show the introduced stop codon
Fig. 2.
Repetitive indel mutations in the ycf2 coding region of Wogon-Sugi and four wild-type chloroplast genomes. The nucleotide and deduced amino acid sequences of part of the ycf2 gene of Types I–IV, and of Wogon-Sugi, showing variation in the number of 66 bp insertions. Colored nucleotides show the 33-bp repetitive units
Fig. 3.
Repetitive indel mutations in the ycf1 coding region of Wogon-Sugi and four wild-type chloroplast genomes. The nucleotide and deduced amino acid sequences of part of the ycf1 gene of Types I–IV, and of Wogon-Sugi, showing variation in the number of 198 bp insertions. Colored nucleotides show the 66-bp repetitive units
The frameshift mutation of the matK coding region of Wogon-Sugi is associated with the primary chlorophyll deficiency of offspring
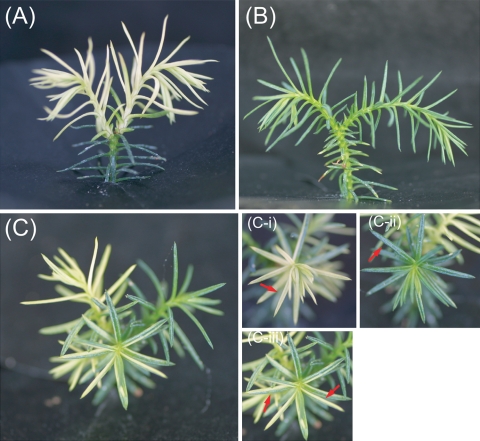
Of the 131 Wogon-Sugi seedlings produced through controlled crossing, 114 (87.0%) showed yellowish-white leaves (Fig. 4a), six (4.6%) showed normal green leaves (Fig. 4b), and 11 (8.4%) showed chimeric leaves (Fig. 4c). The chimeric seedlings exhibited yellowish-white sectors (Fig. 4c-i), normal green sectors (Fig. 4c-ii), and variegated sectors (Fig. 4c-iii).
Fig. 4.
The phenotypes obtained from an artificial cross between Wogon-Sugi (paternal parent) and wild-type (maternal parent). aWogon-Sugi type with yellowish-white leaves. b Wild-type with normal green leaves. c Chimeric type with yellowish-white sectors and normal green sectors. In the chimeric type, the yellowish-white sector, the green sector, and the variegated sectors are shown as c-i, c-ii, and c-iii, respectively. Red arrows show the sample used for genetic analysis
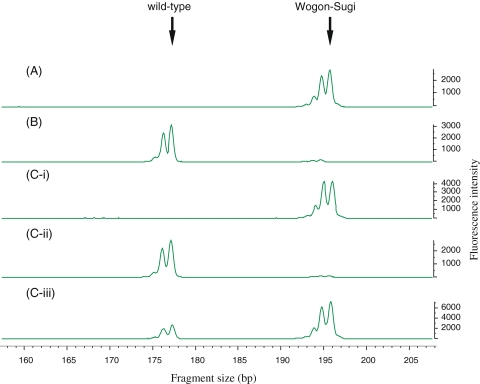
The results of genetic analysis using chloroplast DNA markers are presented in Table 2. The 114 seedlings that showed yellowish-white leaves had the genotype of the Wogon-Sugi chloroplast genome, namely the 196-bp PCR product (Fig. 5a). The six seedlings that showed normal green leaves had the genotype of the maternal wild-type chloroplast genome, namely the 177-bp PCR product (Fig. 5b). The 11 seedlings that showed chimeric leaves had the genotypes of both the paternal Wogon-Sugi and the maternal wild-type: both the 196- and 177-bp PCR products were present (Fig. 5c-i, ii, iii). For details, in each sector of chimeric seedlings, the yellowish-white sector showed the 196 bp PCR product (Fig. 5c-i), the green sector showed 177 bp PCR product (Fig. 5c-ii), and the variegated sectors showed both 196 and 177 bp PCR products (Fig. 5c-iii). That is, yellowish-white leaves that has the 196-bp PCR product is paternal inheritance of chloroplast genomes, normal green leaves that has the 177-bp PCR product is maternal inheritance, and the variegated sectors that has both 196 and 177 bp PCR products is biparental inheritance.
Table 2.
Results of genetic analysis in 131 offspring using a chloroplast DNA marker
| Inheritance patterna | ||||
|---|---|---|---|---|
| Wogon-Sugi type (Paternal) | Wild-type type (Maternal) | Chimeric type (Biparental) | Total | |
| Number of observed seedlingsb | 114 | 6 | 11 | 131 |
| Rate (%) | 87.0 | 4.6 | 8.4 | 100 |
aThe 131 offspring were obtained by an artificial cross between wild-type (Yoshiki 1; maternal parent) and Wogon-Sugi (paternal parent)
bThe chloroplast genomes of each of the 131 offspring were genotyped based on the length variation of PCR product (Fig. 5)
Fig. 5.
Fluorescently labeled PCR products from the three seedling types of Fig. 4. The expected PCR product size of Wogon-Sugi type and wild-type (Yoshiki 1) is 196 bp and 177 bp, respectively. a Yellowish-white leaves with the Wogon-Sugi type matK indel. b Normal green leaves with the wild-type matK indel. c-i Yellowish-white sector of chimeric seedling with the Wogon-Sugi type matK indel; c-ii normal green sector of chimeric seedling with the wild-type matK indel; c-iii variegated sector of chimeric seedling with the Wogon-Sugi type matK indel and the wild-type matK indel. The rightmost peak of each group corresponds to the expected PCR product (196 bp and 177 bp). On the left, peaks corresponding to slippage of the Taq polymerase are visible (195 bp and 176 bp)
Discussion
Previous study of the virescent mutation in Wogon-Sugi was restricted to observation of chloroplast structure and evaluation of the mode of inheritance of the phenotype (Ohba et al. 1971). Although these data provided the initial experimental evidence that at least some types of mutations are encoded by the chloroplast genome, the causative gene responsible for the yellowish-white (chlorophyll-deficient) leaves of Wogon-Sugi was not identified. The present results, which used the chloroplast genomic sequence and genetic analysis using chloroplast DNA markers, indicated that a frameshift mutation in the matK coding region is associated with the primary chlorophyll-deficiency traits of Wogon-Sugi.
Although comparative analysis of Wogon-Sugi and Yaku-Sugi identified 21 sequence polymorphisms (including eight SNPs, nine SSRs, and four indels), only three of the polymorphisms were located in the coding region; these three polymorphisms were indels. Therefore, these variations in the matK, ycf2, and ycf1 coding regions were considered candidates for the mutation responsible for the Wogon-Sugi phenotype. In Wogon-Sugi individuals, a 19-bp insertional mutation was found in the matK coding region. This insertion disrupts the matK reading frame. On the other hand, the indels of ycf2 and ycf1, which were respectively 33 and 66 bp repetitive sequence units, were found in both Wogon-Sugi and wild-type chloroplast genomes. Therefore, the insertional sequence in the matK coding region must be the mutation specific to the Wogon-Sugi chloroplast genome and the one responsible for the developmental chlorophyll deficiency, because the 19 bp insertion in the matK gene leads to a reading frameshift after the 24th residue, phenylalanine, and results in a stop codon after the 39th residue in the Wogon-Sugi mutant.
The type of frameshift mutation in the matK coding region has not been reported in other plant species. On the other hand, a similar indel mutation within ycf1 and ycf2 has been reported in intraspecies comparisons of Oenothera hookeri (Blasko et al. 1988) and between plants from subsection Oenothera (Nimzyk et al. 1993; Greiner et al. 2008), which were also changes in a repetitive sequence without a reading frameshift. One point to consider is the possibility that the Wogon-Sugi chloroplast genome has point mutations that prevent proper annealing of primers in a PCR reaction, and a nuclear paralog with a better fit is amplified instead. Consequently, we conducted the PCR-RFLP analysis as a supplementary analysis, namely detection of longer chloroplast regions, including matK from Wogon-Sugi total DNA with two different primer combinations. The results showed a clear size differentiation between Wogon-Sugi and wild-type (supplementary data S1); the longer regions including matK (with the insertional mutation) were only observed in Wogon-Sugi, and not for the nuclear paralog.
The matK gene, which is encoded in the trnK intron of the chloroplast genome, is utilized in systematic studies because of its high mutation rate and resolution (Shaw et al. 2005). Furthermore, indels are frequent in matK, though they primarily occur in multiples of three, maintaining the reading frame (Barthet and Hilu 2007). We conducted an additional comparative analysis with other five plant species (three angiosperm species and two gymnosperm species) to examine the conservation of the matK amino-acid sequence in C. japonica, especially in the location of the insertional mutation of Wogon-Sugi. The results indicated that the mutation point of Wogon-Sugi after the amino acid residue phenylalanine (F) at the 24th amino acid residue is highly conserved between C. japonica generally and other five plants (supplementary data S2). Thus, the location of the insertional mutation of Wogon-Sugi is likely to affect some kind of functional domain in MatK, though it is not clear whether it is an important functional domain such as a reverse-transcriptase (RT) domain, domain X (the proposed maturase functional domain), and a zinc-finger-like domain (Mohr et al. 1993).
A crossing test carried out to identify segregation confirmed that the yellowish-white trait of Wogon-Sugi is inherited in a non-Mendelian fashion (Table 2). Of the 131 F1 full-sibling offspring, 87% were Wogon-Sugi type, 8.4% were chimeric, and 4.6% were wild-type. These results were very similar to those reported by Ohba et al. (1971), suggesting that segregation of the phenotype is significantly different from the theoretical ratio of 3:1 for a Mendelian trait, and therefore one recessive nuclear gene is not responsible for this trait.
The trait of yellowish-white sectors was tightly linked with the genotype of the 19-bp frameshift mutation of the matK gene in the Wogon-Sugi chloroplast genome. A plastid DNA marker targeting the 19-bp insertion of the Wogon-Sugi matK region was useful to objectively evaluate the genotype of the Wogon-Sugi plastid genome. The yellowish-white sectors were dissected from 125 virescent offspring, including 11 chimeric offspring, and all had the genotype of the Wogon-Sugi chloroplast genome, as shown in Fig. 5. Green sectors were harvested from six green offspring and 11 chimeric offspring, and all lacked the matK insertion in the maternal chloroplast genome. These results indicate that the 19-bp insertion of the matK coding region is the most likely candidate for a polymorphism underlying the Wogon-Sugi phenotype.
The matK gene is assumed to be the splicing factor for group II introns in the chloroplast genome (Neuhaus and Link 1987). Although the maturase function of MatK is not clearly understood, in white barley its maturase-like function is indirectly associated with the mutant albostrians, which has a chloroplast ribosome deficiency that results in the loss of all chloroplast-encoded proteins including MatK (for a review, see Hess et al. 1994a; Schmitz-Linneweber and Barkan 2007). The group II intron-containing precursor transcripts of trnK, trnA, trnI, rps12, rpl2, and atpF remain unspliced in albostrians plastids (Hess et al. 1994b; Hübschmann et al. 1996; Vogel et al. 1997, 1999). Barthet and Hilu (2007) suggested that MatK has an essential function as a posttranscriptional splicing factor at a particular developmental stage, and thus its function indirectly contributes to photosynthetic competency of the chloroplast.
In the primary yellowish-white sector of new shoots in Wogon-Sugi plants, there is the possibility that matK of the mutated version lacks or has insufficient function as a result of the frameshift mutation, and that genes containing group II introns might not be spliced. However, there are several unclear aspects about the frameshift mutation of matK in the Wogon-Sugi plastid genome. For example, it is not a lethal mutation, and the yellowish-white traits of Wogon-Sugi often change to normal green in late summer (supplementary data S3). Furthermore, once the yellowish-white leaves change to green, their color is stable. C. japonica is a perennial plant and the virescence of new shoots in Wogon-Sugi is observed only early in development, even though later the shoot plastids still have the Wogon-Sugi genotype.
Regarding the behavior of the virescent mutation in Wogon-Sugi, one possibility is that a nuclear gene might be associated with the splicing of genes having the group II intron in the plastid genome. Leon et al. (1998) reviewed the range of mutants known to have altered chloroplast development and concluded that almost every step of plastid development depends on the direct action of nuclear-encoded genes. In fact, in maize (Zea mays) chloroplasts, genetic analyses have shown that nuclear-encoded proteins are associated with the splicing of at least 10 of the 17 group II introns (Jenkins et al. 1997; Jenkins and Barkan 2001; Till et al. 2001; Ostheimer et al. 2003; Ostersetzer et al. 2005). Therefore, we hypothesize that a nuclear-encoded splicing factor is recruited as an alternative splicing factor for the chloroplast during the greening of virescent Wogon-Sugi shoots. To test this hypothesis, more detailed analysis is required at the transcriptional and posttranscriptional levels to establish whether the Wogon-Sugi behavior originates with the frameshift mutation of the matK gene, and to determine its association with a nuclear-encoded splicing factor.
Electronic Supplementary Material
S1 PCR-RFLP analysis to detect only the longer region of matK from Wogon-Sugi total DNA with two different primer combinations. The forward primer of first primer set used was 5′-GGATCATAAATTGATCTTGGCAAT-3′ and the reverse primer was 5’-CCTCATGACGAATGAAGATCTATG-3’. The forward primer of the second primer set used was 5′-AGTGGGATGCCGCCTCTC-3′ and the reverse primer was 5′-GTCCTAACTAGATCGCACCATGTA-3′. PCR was performed with the same reaction composition and volume described in the Methods section for PCR genome walking. The expected fragment size of the first primer pair is 1,725 bp. The expected fragment size of the second primer is 1,945 bp. Five units of EcoR I was added to 10 µL of PCR product and the mixture was incubated overnight at 37°C. The digested products were electrophorezed on a 3% MetaPhor Agarose gel (TAKARA). The gel was run at 80 V for 4 h and then visualized by staining with SYBR Safe DNA Gel Stain (Invitrogen) (EPS 5891 kb)
S2 Amino acid sequence alignment of the matK coding region among seven chloroplast genomes including the C. japonica (Cupressaceae, AP009377) chloroplast genome using ClustalX; the position of the frameshift mutation in the Wogon-Sugi matK gene coding region is indicated. The five chloroplast genomes compared were: three angiosperms, Arabidopsis thaliana (Brassicaceae, AP000423); Oryza sativa (Poaceae, X15901); Nicotiana tabacum (Nicotianeae, Z00044); and two gymnosperms, Pinus thunbergii (Pinaceae, D17510) and Cycas taitungensis (Cycadaceae, AP009339). The histogram below the sequences represents the degree of similarity. Peaks indicate positions of high similarity and valleys indicate positions of low similarity. Conserved blocks V, VI and VII of the reverse-transcriptase (RT), and domain X (the proposed maturase functional domain) are indicated according to Mohr et al. (1993) (EPS 2407 kb)
S3 Trait of Wogon-Sugi seedlings. The color of new shoots changes from yellowish-white in spring to normal green in late summer (EPS 68848 kb)
Acknowledgments
We thank Dr. Shohab Youssefian (Akita Prefectural University) for helpful discussions, comments and advice, and Dr. Barry Jaquish (BC Ministry of Forests and Range, Victoria) for helpful comments and advice.
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s00294-009-0247-9) contains supplementary material, which is available to authorized users.
Contributor Information
Tomonori Hirao, Email: hiratomo@affrc.go.jp.
Atsushi Watanabe, Phone: +81-294-397046, FAX: +81-294-397306, Email: nabeatsu@affrc.go.jp.
References
- Archer EK, Bonnett HT (1987) Characterization of a virescent chloroplast mutant of tobacco. Plant Physiol 83:920–925 [DOI] [PMC free article] [PubMed]
- Archer EK, Hakansson G, Bonnett HT (1987) The phenotype of a virescent chloroplast mutation in Tobacco is associated with the absence of a 37.5 kD thylakoid polypeptide. Plant Physiol 83:926–932 [DOI] [PMC free article] [PubMed]
- Barthet MM, Hilu KW (2007) Expression of matK: functional and evolutionary implications. Am J Bot 94:1402–1412 [DOI] [PubMed]
- Benedict CR, Ketring DL (1972) Nuclear gene affecting greening in virescent peanut leaves. Plant Physiol 49:972–976 [DOI] [PMC free article] [PubMed]
- Benedict CR, Kohel RJ (1970) Photosynthetic rate of a virescent cotton mutant lacking chloroplast grana. Plant Physiol 45:519–521 [DOI] [PMC free article] [PubMed]
- Blasko K, Kaplan SA, Higgins KG, Wolfson R, Sears BB (1988) Variation in copy number of a 24-base pair tandem repeat in the chloroplast DNA of Oenothera hookeri strain Johansen. Curr Genet 14:287–292 [DOI] [PubMed]
- Greiner S, Wang X, Rauwolf U, Silber MV, Mayer K, Meurer J, Haberer G, Herrmann RG (2008) The complete nucleotide sequences of the five genetically distinct plastid genomes of Oenothera, subsection Oenothera: I. Sequence evaluation and plastome evolution. Nucleic Acids Res 36:2366–2378 [DOI] [PMC free article] [PubMed]
- Hess WR, Hübschmann T, Börner T (1994a) Ribosome-deficient plastids of albostrians barley: extreme representatives of non-photosynthetic plastid. Endocytobios Cell Res 10:65–80
- Hess WR, Hoch B, Zeltz P, Hübschmann T, Kössel H, Börner T (1994b) Inefficient rpl2 splicing in barley mutants with ribosome-deficient plastids. Plant Cell 6:1455–1465 [DOI] [PMC free article] [PubMed]
- Hirao T, Watanabe A, Kurita M, Kondo T, Takata K (2008) Complete nucleotide sequence of the Cryptomeria japonica D. Don. chloroplast genome and comparative chloroplast genomics: diversified genomic structure of coniferous species. BMC Plant Biol 8:70 [DOI] [PMC free article] [PubMed]
- Hopkins WG, Elfman B (1984) Temperature-induced chloroplast ribosome deficiency in virescent maize. J Hered 75:207–211
- Hübschmann T, Hess WR, Börner T (1996) Impaired splicing of the rps12 transcript in ribosome-deficient plastids. Plant Mol Biol 30:109–123 [DOI] [PubMed]
- Iba K, Takamiya K, Toh Y, Satoh H, Nishimura M (1991) Formation of functionally active chloroplasts is determined at a limited stage of leaf development in virescent mutants of rice. Dev Genet 12:342–348 [DOI]
- Jain ML (1966) Biochemical definition of yellow-virescent and light-green suppressor mutations in barley. Genetics 54:813–818 [DOI] [PMC free article] [PubMed]
- Jenkins B, Barkan A (2001) Recruitment of a peptidyl-tRNA hydrolase as a facilitator of group II intron splicing in chloroplasts. EMBO J 20:872–879 [DOI] [PMC free article] [PubMed]
- Jenkins B, Kulhanek D, Barkan A (1997) Nuclear mutations that block group II RNA splicing in maize chloroplasts reveal several intron classes with distinct requirements for splicing factors. Plant Cell 9:283–296 [DOI] [PMC free article] [PubMed]
- Koussevitzky S, Stanne TM, Peto CA, Giap T, Sjögren LLE, Zhao Y, Clarke AK, Chory J (2007) An Arabidopsis thaliana virescent mutant reveals a role for ClpR1 in plastid development. Plant Mol Biol 63:85–96 [DOI] [PubMed]
- Kusumi J, Tsumura Y, Yoshimaru H, Tachida H (2000) Phylogenetic relationships in Taxodiaceae and Cupressaceae based on matK, chlL, trnL-trnF IGS region and trnL intron sequences. Am J Bot 87:1480–1488 [DOI] [PubMed]
- Landau A, Paleo AD, Civitillo R, Jaureguialzo M, Prina AR (2007) Two infA gene mutations independently originated from a mutator genotype in barley. J Hered 98:272–276 [DOI] [PubMed]
- Leon P, Arroyo A, Mackenzie S (1998) Nuclear control of plastid and mitochondrial development in higher plants. Ann Rev Plant Physiol Plant Mol Biol 49:453–480 [DOI] [PubMed]
- López-Juez E, Jarvis RP, Takeuchi A, Page AM, Chory J (1998) New Arabidopsis cue mutants suggest a close connection between plastid and phytochrome regulation of nuclear gene expression. Plant Physiol 118:803–815 [DOI] [PMC free article] [PubMed]
- Mohr G, Perlman PS, Lambowitz AM (1993) Evolutionary relationships among group II intron-encoded proteins and identification of a conserved domain that may be related to maturase function. Nucleic Acids Res 21:4991–4997 [DOI] [PMC free article] [PubMed]
- Neuhaus H, Link G (1987) The chloroplast tRNALys (UUU) gene from mustard (Sinapsis alba) contains a class II intron potentially coding for a maturase-related polypeptide. Curr Genet 11:251–257 [DOI] [PubMed]
- Nicholas KB, Nicholas HBJ, Deerfield DWI (1997) Gene-Doc: analysis and visualization of genetic variation. EMBNEW NEWS 4:14
- Nimzyk R, Schöndorf T, Hachtel W (1993) In-frame length mutations associated with short tandem repeats are located in unassigned open reading frames of Oenothera chloroplast DNA. Curr Genet 23:265–270 [DOI] [PubMed]
- Ohba K, Iwakawa M, Okada Y, Murai M (1971) Paternal transmission of a plastid anomaly in some reciprocal crosses of Sugi, Cryptomeria japonica D. Don. Silvae Genet 20:102–107
- Ostersetzer O, Cooke AM, Watkins KP, Barkan A (2005) CRS1, a chloroplast group II intron splicing factor, promotes intron folding through specific interactions with two intron domains. Plant Cell 17:241–255 [DOI] [PMC free article] [PubMed]
- Ostheimer G, Williams-Carrier R, Belcher S, Osborne E, Gierke J, Barkan A (2003) Group II intron splicing factors derived by diversification of an ancient RNA binding module. EMBO J 22:3919–3929 [DOI] [PMC free article] [PubMed]
- Rios RD, Saione H, Robredo C, Acevedo A, Colombo N, Prina AR (2003) Isolation and molecular characterization of atrazine tolerant barley mutants. Theor Appl Genet 106:696–702 [DOI] [PubMed]
- Schmitz-Linneweber C, Barkan A (2007) RNA splicing and RNA editing in chloroplasts. In: Cell and molecular biology of plastid. Springer, Berlin, pp 213–248
- Shaw J, Lickey EB, Beck JT, Farmer SB, Liu W, Miller J, Siripun KC, Winder CT, Schilling EE, Small RL (2005) The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am J Bot 92:142–166 [DOI] [PubMed]
- Shiraishi S, Watanabe A (1995) Identification of chloroplast genome between Pinus densiflora SIEB. et Zucc. And P. thunbergia PARL. Based on the polymorphism in rbcL gene (in Japanese). J Jpn For Sor 77:429–436
- Till B, Schmitz-Linneweber C, Williams-Carrier R, Barkan A (2001) CRS1 is novel group II intron splicing factor that was derived from a domain of ancient origin. RNA 7:1227–1238 [DOI] [PMC free article] [PubMed]
- Vinti G, Fourrier N, Bowyer JR, López-Juez E (2005) Arabidopsis cue mutants with defective plastids are impaired primarily in the photocontrol of expression of photosynthesis-associated nuclear genes. Plant Mol Biol 57:343–357 [DOI] [PubMed]
- Vogel J, Hübschmann T, Börner T, Hess WR (1997) Splicing and intron-internal RNA editing of trnK-matK transcripts in barley plastids: support for MatK as an essential splice factor. J Mol Biol 270:179–187 [DOI] [PubMed]
- Vogel J, Börner T, Hess WR (1999) Comparative analysis of splicing of the complete set of chloroplast group II introns in three higher plant mutants. Nucleic Acids Res 27:3866–3874 [DOI] [PMC free article] [PubMed]
- Wyman SK, Jansen RK, Boore JL (2004) Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20:3252–3255 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S1 PCR-RFLP analysis to detect only the longer region of matK from Wogon-Sugi total DNA with two different primer combinations. The forward primer of first primer set used was 5′-GGATCATAAATTGATCTTGGCAAT-3′ and the reverse primer was 5’-CCTCATGACGAATGAAGATCTATG-3’. The forward primer of the second primer set used was 5′-AGTGGGATGCCGCCTCTC-3′ and the reverse primer was 5′-GTCCTAACTAGATCGCACCATGTA-3′. PCR was performed with the same reaction composition and volume described in the Methods section for PCR genome walking. The expected fragment size of the first primer pair is 1,725 bp. The expected fragment size of the second primer is 1,945 bp. Five units of EcoR I was added to 10 µL of PCR product and the mixture was incubated overnight at 37°C. The digested products were electrophorezed on a 3% MetaPhor Agarose gel (TAKARA). The gel was run at 80 V for 4 h and then visualized by staining with SYBR Safe DNA Gel Stain (Invitrogen) (EPS 5891 kb)
S2 Amino acid sequence alignment of the matK coding region among seven chloroplast genomes including the C. japonica (Cupressaceae, AP009377) chloroplast genome using ClustalX; the position of the frameshift mutation in the Wogon-Sugi matK gene coding region is indicated. The five chloroplast genomes compared were: three angiosperms, Arabidopsis thaliana (Brassicaceae, AP000423); Oryza sativa (Poaceae, X15901); Nicotiana tabacum (Nicotianeae, Z00044); and two gymnosperms, Pinus thunbergii (Pinaceae, D17510) and Cycas taitungensis (Cycadaceae, AP009339). The histogram below the sequences represents the degree of similarity. Peaks indicate positions of high similarity and valleys indicate positions of low similarity. Conserved blocks V, VI and VII of the reverse-transcriptase (RT), and domain X (the proposed maturase functional domain) are indicated according to Mohr et al. (1993) (EPS 2407 kb)
S3 Trait of Wogon-Sugi seedlings. The color of new shoots changes from yellowish-white in spring to normal green in late summer (EPS 68848 kb)