Abstract
In situ polymerase chain reaction (in situ PCR), which can detect a few copies of genes within a cell by amplifying the target gene, was developed to better understand the biological functions of tissues. In this study, we optimized the protocol conditions for the detection of X chromosome-linked phosphoglycerate kinase-1 (pgk-1) gene in paraffin-embedded sections of mouse reproductive organs. The effects of various concentrations of proteinase K (PK) and PCR cycle numbers were examined. To label the amplified DNA, we used digoxigenin-dUTP (Dig), Cy-3-dUTP (Cy-3), or FluorX-dCTP (FluorX). The optimal concentration of PK was 50 µg/ml for the ovary and 10 µg/ml for the testis. Ten PCR cycles were optimal for Dig and 25 cycles were optimal for FluorX and Cy-3 in the ovary and testis. The signal-to-noise ratio of FluorX and Cy-3 for ovarian tissue was better than that of Dig. Using the above conditions, we detected 1–4 and 1–2 spots of pgk-1 in the nuclei of granulosa and germ cells, respectively. Our results indicate that in situ PCR is useful for detecting a specific gene in paraffin-embedded sections under optimized conditions of both PCR cycle number and PK concentration.
Keywords: in situ PCR, pgk-1, ovary, testis, proteinase K
I. Introduction
The polymerase chain reaction (PCR) method is suitable for detection of a few copies of a particular gene based on its extremely high sensitivity. In fact, the PCR method allows amplification of a single gene copy in a solution up a large quantity sufficient for detection by conventional gel electrophoresis and Southern blot hybridization [16]. However, the PCR method itself cannot detect the site of a target gene at the cellular level. On the other hand, in situ hybridization (ISH) is considered a powerful technique for localization of specific nucleic acid sequences at the level of individual cells with excellent detection specificity [7–10]. However, the detection sensitivity of ISH is sometimes limited. The combination of these two methods, called in situ PCR, first described by Haase et al. in 1990 [6], is an extremely highly sensitive technique to localize a single gene copy at the individual cell level [1–4, 11–13, 20, 22]. However, in situ PCR has not gained wide acceptance by the scientific community, partly because of the significant variations in the optimal conditions of the protocol, which depend on the tissues used for analysis [11, 12].
In the present study, we optimized the in situ PCR protocol for paraffin-embedded sections of the mouse reproductive organs using X chromosome-linked phosphoglycerate kinase-1 (pgk-1) gene. Pgk-1 gene within the X chromosome is ubiquitously expressed in the euchromatic X chromosome and is also used as a marker for detection of mouse sex chimeras [17, 24]. In this study, pgk-1 gene expression was analyzed in several applications as follows: (1) the effects of various concentrations of proteinase K (PK), (2) the amplification of PCR cycles, and (3) visualization dyes using digoxigenin-11-dUTP (Dig), Cy-3-dUTP (Cy-3), and FluorX-10-dCTP (FluorX). After optimization of these applications, in situ PCR was found to be excellent for detection of a specific gene in paraffin-embedded tissue sections.
II. Materials and Methods
Chemicals and biochemicals
Paraformaldehyde (PFA) was purchased from Merck (Darmstadt, Germany). PK was from Wako Pure Chemicals (Osaka, Japan). Dig was from Roche Diagnostics (Mannheim, Germany). Cy-3 and FluorX were from Amersham Biosciences Corp. (Piscataway, NJ). TaKaRa Ex Taq (Taq DNA polymerase, dNTP mixture, 10×PCR buffer, and MgCl2) was from Takara Bio Inc. (Shiga, Japan). 3,3'-diaminobenzidine-4HCl (DAB) was from Dojin Chemical Co. (Kumamoto, Japan). All other reagents used in this study were from Wako Pure Chemicals or Sigma Chemical Co. (St. Louis, MO) and were of analytical grade.
Animals and tissue preparation
The study was conducted in 6- to 8-week-old ICR mice. All experiments were conducted according to the principles and procedures outlined in the Guidelines for Animal Experimentation of Nagasaki University with the approval of the Institutional Animal Care and Use Committee. For tissue sampling, the ovaries and testes were harvested under ether anesthesia. They were cut into small pieces and divided into two groups. Tissue sections of the first group were frozen immediately in liquid nitrogen and later used for standard PCR. Those of the second group were fixed in 4% PFA in phosphate buffered saline (PBS) at room temperature overnight and embedded in paraffin. Sections (5-µm-thick) were prepared and then mounted on 3-aminopropyltriethoxysilane-coated glass slides and used for in situ PCR.
PCR method
Genomic DNA was prepared from the testis or ovary of ICR mouse with AquaPure Genomic DNA Kits (Bio-Rad, Richmond, CA) according to the instruction manual provided by the manufacturer. When the PCR conditions were optimized, the band intensity corresponding to the increased amount of DNA correlated linearly with the PCR cycle number. The bands were quantitated by densitometry (NIH Image J software, version 1.08i, http://rsb.info.nih.gov/ij/) and each value was expressed relative to that of mouse glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Primers (BEX Co., Japan) corresponding to mouse pgk1 were 5'-CACGCTTCAAAAGCGCACGTCT-3' and 5'-CTTGAGGGCAGCAGTACGGAAT-3', while those for mouse GAPDH were 5'-ACCACAGTCCATGCCATCAC-3' and 5'-TCCACCACCCTGTTGCTGTA-3'. The Gene Amp PCR System 9400 (PE Applied Biosystems, Foster City, CA) was used for gene amplification. PCR was carried out in a 20-µl final volume of reaction mixture, containing 0.5 units of TaKaRa ExTaq polymerase, 0.2 mM of each dNTP, ExTaq buffer (1×), 1 µM of each specific primer described above and 100 ng of testis or ovary DNA as a template. Each cycle of PCR included 20 sec of denaturation at 94°C, 20 sec of annealing at 55°C, and 30 sec of extension at 72°C. After 28 reaction cycles, the PCR products were separated on a 1.5% TAE-agarose gel and stained with ethidium bromide. The amplified PCR products with pgk-1 primers were loaded onto a 3% agarose gel, and the DNA bands of the predicted size were cut, purified with glass powder and integrated into pGEM-easy (Promega, Madison, WI). BigDye terminator cycle sequencing was performed against the pgk-1 plasmid and the sequence was determined with ABI PRISM 310 Genetic Analyzer (PE Applied Biosystems) using the instructions provided by the supplier.
In situ PCR method
The oligo-DNAs that can amplify part (170 bp) of the mouse X chromosome-linked pgk-1gene were used as primers as described above. The tissue sections were deparaffinized and rehydrated using standard procedures. These sections were digested with 0–100 µg/ml of PK (37°C, 15 min). After post-fixation with 4% PFA in PBS (5 min), the sections were washed in PBS and immersed in 50% formamide in 2×SSC (1×SSC=0.15 M sodium chloride and 0.015 M sodium citrate, pH 7.0) overnight at 4°C. After washing with double-distilled water (DDW, 5 min, 3 times), the sections were completely desiccated. For each slide, the amplification mixture was prepared in a final volume of 100 µl, containing 1×PCR buffer, 1.0 µg/ml forward primer, 1.0 µg/ml reverse primer, 0.2 mM dNTP, 2.5 mM MgCl2, and 2.5 U/100 µl Taq DNA polymerase. To label the amplified DNA, 5.0 µM Dig, 2.0 µM Cy-3, and 0.2 µM FluorX were used. The slides were sealed with EasiSeal (Hybaid, UK), and then placed on the heated plate of the OmniSlide System (Hybaid); the DNA in the tissues then was denatured at 94°C for 3 min. As for PCR cycle studies, 5, 10, 15, 20, 25, or 30 cycles of amplification (denaturation for 15 sec at 94°C, annealing for 15 sec at 56°C, extension for 60 sec at 72°C) were run. The slides were then heated at 72°C for 5 min. After amplification, the cover slips were removed and the slides were washed with 2×SSC (37°C, 15 min, 4 times). As a negative control, in situ PCR was performed with the omission of Taq DNA polymerase and the primers.
Visualization and image analysis
The Dig-labeled signals were detected by enzyme immunohistochemistry, and the images of the Dig-labeled sections were analyzed using an Olympus BH-2 microscope connected to a Canon CCD camera, and an Olympus SP-500 Image analyzer with Image Command program 5098 as described in detail previously [26]. Cy-3 and FluorX signals were detected using a Zeiss fluorescence microscope connected to a CCD camera (AxioCam, Carl Zeiss, Jena, Germany).
Statistical analysis
All data were expressed as mean±SD. Differences between groups were examined for statistical significance using the unpaired Student’s t-test. A P value less than 0.05 denoted the presence of a statistically significant difference. All analyses were performed with a statistical software package (StatView, version 5.0; Abacus Concepts, Berkeley, CA).
III. Results
PCR products
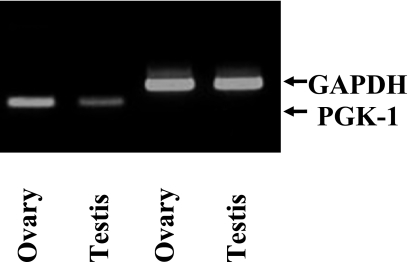
PCR products comprising 170 bp were obtained (Fig. 1) and were confirmed by DNA sequencing to be part of the pgk-1 gene. Semiquantitative analysis of these products using the NIH Image analysis system showed that the quantity of the pgk-1 gene product from the ovary was about 2-fold more than that from the testis.
Fig. 1.
PCR of pgk-1 gene in mouse ovary and testis. PCR products of pgk-1 (170 bp) were found in mouse ovary and testis.
PK concentration
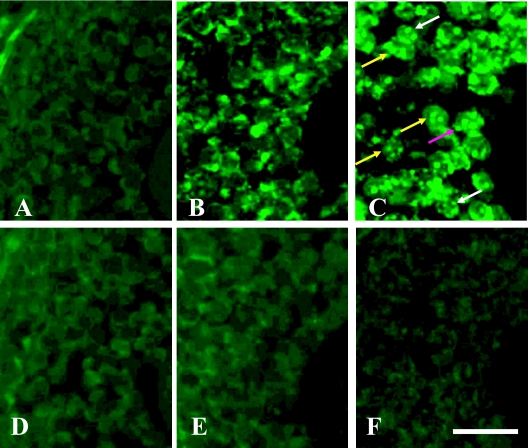
The effect of digestion by PK at concentrations from 0 to 100 µg/ml was evaluated using FluorX. The optimal concentration of PK, as evaluated by the signal-to-noise ratio, was 50 µg/ml for the ovary (Fig. 2). One to 4 pgk-1 spots were detected in the nuclei of granulosa cells (Fig. 2C). Treatment of the sections with 100 µg/ml PK resulted in diffuse spreading of the signal (data not shown). There was no difference between the use of FluorX and Cy-3 dye for in situ PCR.
Fig. 2.
Effects of various concentrations of PK in mouse ovary using FluorX. Upper panels (A, B, C) were carried out Taq polymerase and lower panels (D, E, F) were performed omitting Taq polymerase. PK was evaluated at varying concentrations from 5 µg/ml (A, D), 20 µg/ml (B, E), and 50 µg/ml (C, F). One to 4 green spots were detected in the nuclei of granulosa cells (C). Red arrow; 4 spots. Yellow arrows; 3 spots. White arrows; 2 spots. Bar=20 µm.
PCR cycle
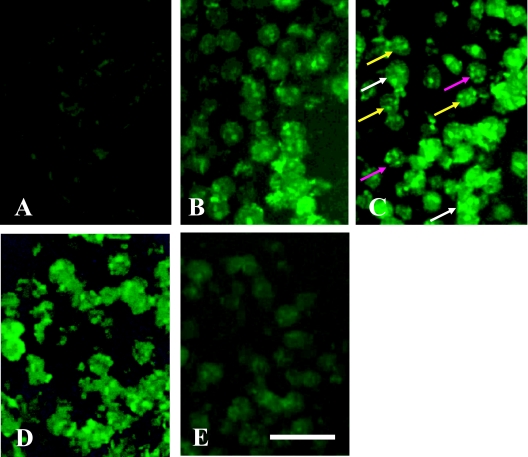
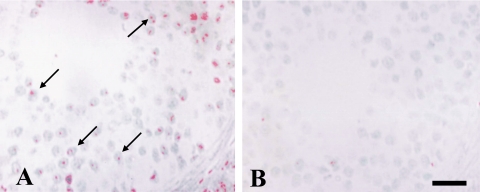
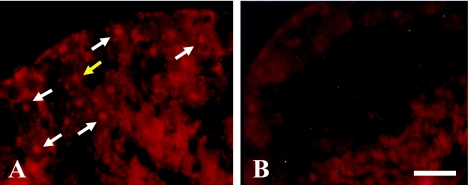
The effect of different PCR cycles (from 5 to 30 cycles) was evaluated using FluorX, Cy-3 and Dig dyes. For FluorX, 25 cycles were optimal for the detection of the pgk-1 gene in the nuclei of granulosa cells (Fig. 3C). Ten cycles was the optimum number of PCR cycles necessary to detect pgk-1 gene in the nuclei of ovarian granulosa cells using Dig (Fig. 4A). The signal-to-noise ratio of FluorX and Cy-3 for granulosa cells was better than that of Dig (Fig. 3C and Fig. 4A). Amplification over 10 cycles for Dig or over 25 cycles for Cy-3 and FluorX significantly increased the background signals due to the diffusion of PCR products into the surrounding tissues (Fig. 3D). For the testis, the concentration of PK was 10 µg/ml and 25 cycles of PCR was optimal, and 1–2 spots were detected in the nuclei of some germ cells using Cy-3 dye (Fig. 5A). Testicular sections treated with 20 µg/ml PK showed no signal for pgk-1 gene (data not shown).
Fig. 3.
Effects of different PCR cycles in mouse ovary using FluorX. Five cycles (A), 15 cycles (B), 25 cycles (C), and 30 cycles (D) of PCR were performed using Taq polymerase. One to 4 green spots were detected in the nuclei of granulosa cells (C). As a negative control, 25 cycles of PCR were performed omitting Taq polymerase (E). Red arrows; 4 spots. Yellow arrows; 3 spots. White arrows; 2 spots. Bar=20 µm.
Fig. 4.
Detection of pgk-1 in mouse ovary using Dig. One to 2 red spots were detected in the nuclei of granulosa cells using image analysis system (A). PCR cycles were 10 times, and PK concentration was 50 µg/ml. Taq polymerase was omitted as a negative control (B). Bar=20 µm.
Fig. 5.
Detection of pgk-1 in mouse testis using Cy-3. One to 2 red spots of Pgk-1 gene were detected in the nuclei of germ cells with 10 µg/ml PK concentration and 25 cycles (A). Yellow arrow; 2 spots. White arrows; 1 spot. Taq polymerase was omitted as a negative control (B). Bar=20 µm.
Number of pgk-1-positive spots in mouse granulosa cells
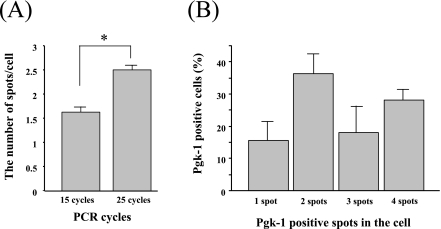
Finally, we determined the percentage of granulosa cells containing one or more pgk-1-positive spots after employing 15 cycles and 25 cycles. Five tissue samples were used in these experiments. The number of pgk-1-positive spots in granulosa cells was 1.6±0.1 spots/cell in the ovary using 15 cycles for in situ PCR and 2.5±0.1 spots/cell using 25 cycles (P<0.0001) (Fig. 6A). The percentage of pgk-1-positive granulosa cells after using 25 cycles for in situ PCR was as follows: 15.8±5.8% of the cells showed 1 spot in the nuclei, 37.2±7.1% of the granulosa cells showed 2 spots, 18.7±8.3% of the cells had 3 spots, and 27.7±4.5% had 4 spots (Fig. 6B).
Fig. 6.
Pgk-1-positive spots in mouse granulosa cells. The number of pgk-1-positive spots after employing 15 cycles and 25 cycles (A). The percentage of pgk-1-positive granulosa cells after using 25 cycles for in situ PCR (B). Data represent mean±SD. *P<0.0001.
IV. Discussion
In the present study, we assessed the utility of in situ PCR in detecting a specific gene using paraffin-embedded sections of mouse reproductive organs. Using in situ PCR, we found 1 to 4 pgk-1 spots in the nuclei of granulosa cells and 1 to 2 pgk-1 spots in the nuclei of the germ cells under optimal number of PCR cycles and PK concentration.
In practice, in situ PCR is technically difficult, and there is no standard protocol for optimal detection, especially when using paraffin-embedded tissue sections. We were able to detect 1 to 4 spots of pgk-1 in granulosa cells using our optimal conditions. However, when the PCR cycles were increased to more than 25 cycles for Cy-3 and FluorX, the signals were diffusely spread in the sections. In fact, the diffusion of the amplified products from the target cells is a major problem in this method [11, 25]. The factors that influence diffusion include excessive protease digestion [5, 18] and the use of an excessive number of PCR amplification cycles [5, 11, 21, 27]. In relation to the diffusion of the PCR products into the surrounding tissue, non-specific incorporation of labeled nucleotides into fragmented DNA, and the inhibitory effects of cross-linking of histones to DNA or PCR amplification are problematic. Long et al. [13] reported some false positive results, especially when using tissue sections, which were related to non-specific incorporation of labeled nucleotides into cellular DNA through internal priming and DNA repair. This was due, at least in part, to the exonuclease activity of Taq DNA polymerase. In our experiments, however, the diffusion of PCR products could be minimized by reducing the number of PCR cycles and the use of appropriate concentrations of PK.
Recently, in situ reverse transcription-PCR (in situ RT-PCR) has been used to detect low levels of mRNA within cells and tissue sections [2, 14, 15, 20, 23]. However, this method has certain limitations in detecting specific mRNAs. This is because detection of very few copies of mRNA in tissues or cells by this method requires the PCR reaction to be performed after carrying out reverse transcription of the specific mRNA to cDNA using reverse transcriptase. Then, it is necessary to determine whether this reaction truly occurs within the individual cell. Moreover, it is also important to establish that the RT-PCR reaction is of genomic DNA origin, and that the products truly originated from the target mRNA. For these reasons, we considered the in situ RT-PCR method to be unsuitable for evaluating the experimental results obtained by the in situ PCR method.
For in situ PCR, two main protocols, direct and indirect in situ PCR, have been described [11, 13, 25]. In the direct detection approach, the labeled nucleotides are directly incorporated into the PCR products, while the indirect method consists of an initial PCR reaction without labeled nucleotides, followed by in situ hybridization using labeled probes to the PCR products. The direct in situ PCR has been reported to increase the turn-around time and ease of in situ PCR studies due to hot-start modifications of in situ PCR without jeopardizing specificity [19].
In conclusion, in situ PCR is a potentially useful method for detecting a specific gene at cellular and subcellular levels, and may prove suitable for evaluation of the functional organization of various genes in paraffin tissue sections when used with critical PK concentration and PCR cycle number.
V. Acknowledgments
This study was supported in part by a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (No. 18390060 to T.K.).
VI. References
- 1.Asan E. Progress in focus: recent advances in histochemistry and cell biology. Histochem. Cell Biol. 2002;118:507–525. doi: 10.1007/s00418-002-0480-5. [DOI] [PubMed] [Google Scholar]
- 2.Bagasra O., Patel D., Bobroski L., Abbasi J. A., Bagasra A. U., Baidouri H., Harris T., El-Roeiy A., Lengvarszky Z., Farzadegan H., Wood C. Localization of human herpesvirus type 8 in human sperms by in situ PCR. J. Mol. Histol. 2005;36:401–412. doi: 10.1007/s10735-005-9010-9. [DOI] [PubMed] [Google Scholar]
- 3.Catzavelos C., Ruedy C., Stewart A. K., Dube I. A novel method for the direct quantification of gene transfer into cells using PCR in situ. Gene Therapy. 1998;5:755–760. doi: 10.1038/sj.gt.3300663. [DOI] [PubMed] [Google Scholar]
- 4.Chen P. C., Pan C., Yang A., Wang L., Chiang H. Detection of Epstein-Bar virus genome within thymic epithelial tumours in Taiwanese patients by nested PCR, PCR in situ hybridization, and RNA in situ hybridization. J. Pathol. 2002;197:684–688. doi: 10.1002/path.1141. [DOI] [PubMed] [Google Scholar]
- 5.Chen R. H., Fuggle S. V. In-situ cDNA polymerase chain reaction. A novel technique for detecting mRNA expression. Am. J. Pathol. 1993;143:1527–1534. [PMC free article] [PubMed] [Google Scholar]
- 6.Haase A. T., Retzel E. F., Staskus K. A. Amplification and detection of lentiviral DNA inside cells. Proc. Natl. Acad. Sci. U S A. 1990;87:4971–4975. doi: 10.1073/pnas.87.13.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koji T., Brenner R. M. Localization of estrogen receptor messenger ribonucleic acid in rhesus monkey uterus by nonradioactive in situ hybridization with digoxigenin-labeled oligodeoxynucleotides. Endocrinology. 1993;132:382–392. doi: 10.1210/endo.132.1.8419136. [DOI] [PubMed] [Google Scholar]
- 8.Koji T., Nakane P. K. Recent advances in molecular histochemical techniques: in situ hybridization and southwestern histochemistry. J. Electron Microsc. 1996;45:119–127. doi: 10.1093/oxfordjournals.jmicro.a023420. [DOI] [PubMed] [Google Scholar]
- 9.Koji T., Kanemitsu Y., Hoshino A., Nakane P. K. A novel amplification method of nonradioactive in situ hybridization signal for specific RNA with biotinylated tyramine. Acta Histochem. Cytochem. 1997;30:401–406. [Google Scholar]
- 10.Koji T. Springer Lab Manuals. Springer-Verlag; Tokyo: 2000. Molecular Histochemical Techniques. [Google Scholar]
- 11.Komminoth P., Long A. A. In-situ polymerase chain reaction. Virchows Archiv. B. 1993;64:67–73. [PubMed] [Google Scholar]
- 12.Komminoth P., Long A. A. In situ polymerase chain reaction and its applications to the study of endocrine disease. Endocr. Pathol. 1995;6:167–171. [Google Scholar]
- 13.Long A. A., Komminoth P., Lee E., Wolfe H. J. Comparison of indirect and direct in-situ polymerase chain reaction in cell preparations and tissue sections. Histochemistry. 1993;99:151–162. doi: 10.1007/BF00571876. [DOI] [PubMed] [Google Scholar]
- 14.Martinez A., Miller M., Quinn K., Unsworth E. J., Ebina M., Cuttitta F. Non-radioactive localization of nucleic acids by direct in situ PCR and in situ RT-PCR in paraffin-embedded sections. J. Histochem. Cytochem. 1995;43:739–747. doi: 10.1177/43.8.7542678. [DOI] [PubMed] [Google Scholar]
- 15.Mee A. P., Denton J., Hoyland J. A., Davies M., Mawer E. B. Quantification of vitamin D receptor mRNA in tissue sections demonstrates the relative limitations of in situ-reverse transcriptase-polymerase chain reaction. J. Pathol. 1997;182:22–28. doi: 10.1002/(SICI)1096-9896(199705)182:1<22::AID-PATH809>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 16.Mullis K. B., Falcona F. A. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 1987;155:335–350. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- 17.Nakayama H., Kuroda H., Ru X. M., Fujita J., Kitamura Y. Fast and easy detection of mouse sex chimeras using electrophoretic polymorphism of phosphoglycerate kinase-1, an X chromosome-linked enzyme. Biol. Reprod. 1988;39:923–927. doi: 10.1095/biolreprod39.4.923. [DOI] [PubMed] [Google Scholar]
- 18.Nuovo G. J., Gallery F., Macconnell P., Bloch W. Importance of different variables for enhancing in situ detection of PCR-amplified DNA. PCR Meth. Appl. 1993;2:305–312. doi: 10.1101/gr.2.4.305. [DOI] [PubMed] [Google Scholar]
- 19.Nuovo G. J., Hohman R. J., Nardone G. A., Nazarenko I. A. In situ amplification using universal energy transfer-labeled primers. J. Histochem. Cytochem. 1999;47:273–279. doi: 10.1177/002215549904700301. [DOI] [PubMed] [Google Scholar]
- 20.Nuovo G. J. The utility of in situ-based methodologies including in situ polymerase chain reaction for the diagnosis and study of viral infections. Hum. Pathol. 2007;38:1123–1136. doi: 10.1016/j.humpath.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 21.Patel V. G., Shum-Siu A., Heniford B. W., Wieman T. J., Hendler F. J. Detection of epidermal growth factor receptor mRNA in tissue sections from biopsy specimens using in-situ polymerase chain reaction. Am. J. Pathol. 1994;144:7–14. [PMC free article] [PubMed] [Google Scholar]
- 22.Pulimood A. B., Peter S., Rook G. W., Donoghue H. D. In situ PCR for Mycobacterium tuberculosis in endoscopic mucosal biopsy specimens of intestinal tuberculosis and Crohn disease. Am. J. Clin. Pathol. 2008;129:846–851. doi: 10.1309/DKKECWQWMG4J23E3. [DOI] [PubMed] [Google Scholar]
- 23.Sanno N., Jin L., Qian X., Osamura R. Y., Scheithauer B. W., Kovacs K., Lloyd R. V. Gonadotropin releasing hormone mRNA and gonadotropin releasing hormone receptor mRNA expression in pituitaries and pituitary adenomas. J. Clin. Endocrinol. Metab. 1997;82:1974–1982. doi: 10.1210/jcem.82.6.3976. [DOI] [PubMed] [Google Scholar]
- 24.Singer-Sam J., Chapman V., LeBon J. M., Riggs A. D. Parental imprinting studied by allele-specific primer extension after PCR: Paternal X chromosome-linked genes are transcribed prior to preferential paternal X chromosome inactivation. Proc. Natl. Acad. Sci. U S A. 1992;89:10469–10473. doi: 10.1073/pnas.89.21.10469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teo I. A., Shaunak S. Polymerase chain reaction in situ: an appraisal of an emerging technique. Histochem. J. 1995;27:647–659. [PubMed] [Google Scholar]
- 26.Tsukasaki S., Miyazaki M., Koji T., Abe K., Furusu A., Shin M., Suzuki D., Harada T., Ozono Y., Sakai H., Kohno S. Semi-quantitative non-radioactive in situ hybridization and its clinical application. Acta Histochem. Cytochem. 2000;33:39–47. [Google Scholar]
- 27.Zaki S. R., Heneire W., Lotheld L. M., Greer P. W., Sinha S. D., Folks T. M. In-situ polymerase chain reaction amplification: applications and current limitations. AIDS. 1994;8:1186–1187. [PubMed] [Google Scholar]