Abstract
Rapid progress in genome research creates a wealth of information on the functional annotation of mammalian genome sequences. However, as we accumulate large amounts of scientific information we are facing problems of how to integrate and relate the data produced by various genomic approaches. Here, we propose the novel concept of an organ atlas where diverse data from expression maps to histological findings to mutant phenotypes can be queried, compared and visualized in the context of a three-dimensional reconstruction of the organ. We will seek proof of concept for the organ atlas by elucidating genetic pathways involved in development and pathophysiology of the kidney. Such a kidney atlas may provide a paradigm for a new systems-biology approach in functional genome research aimed at understanding the genetic bases of organ development, physiology and disease.
Key Words: EuReGene, kidney, genome, development, pathophysiology, genetics
Introduction
Elucidation of the human and other vertebrate genomes heralds a new era in biomedical research that offers unprecedented opportunities to understand disease processes and to identify strategies to improve health. Deciphering the three billion base pairs of the mammalian genome provides us with a blueprint of the genetic map that defines the activities of cells and tissues, organs and entire organisms. However, before being able to fully exploit the wealth of information presented in genomes, we need to assign functions to individual genes and we need to understand their interaction in intricate complex networks that control all biological processes.
According to current estimates, mammalian genomes encode 20,000 to 30,000 genes (www.ensemble.org). Taking into account that, by differential splicing or RNA editing, multiple transcripts may originate from a single gene, mammalian tissues are expected to express a minimum of 40,000 different proteins.1 Based on previous analysis of the human genome,2 approximately one third of all mammalian genes can be grouped into known functional categories such as transcription factors, receptors and ligands, or structural proteins. Another third exhibits sequence similarity to known genes, providing at least an educated guess about their possible functions. This leaves us with no less than 30% of the mammalian genome (or 13.000 genes) that needs de novo functional annotation—a tremendous challenge in current genome research.
Application of transgenic technologies in model organisms has arguably been the single most successful approach to uncover the previously unknown function of new genes. In a process known as reverse genetics, we can manipulate the expression of individual genes and study the (patho)physiological consequences of altered gene expression in cells, tissues or intact organisms. Aided by major technological advances in molecular biology, we can today easily inactivate the expression of genes in living organisms using, for example, gene targeting or silencing by small interference RNAs and antisense oligonucleotide knock-down. We can ectopically express or overexpress genes, using pronuclear injection of gene constructs or somatic cell gene transfer, and we can evaluate the effects of such manipulations using a variety of sophisticated methods including gene expression profiling by micro arrays, in vivo imaging, and high-throughput in situ hybridizations. With respect to kidney development and disease, transgenic approaches, for example, have elucidated the crucial role of Wt1 and Wnt4 as key genes in nephrogenesis3–6 and helped to uncover the importance of chloride channels ClC-K and ClC-5 in ion handling of the kidney and in the occurrence of renal salt wasting syndromes in humans.7–13
Understanding the Complexity of Genetic Systems
While the remarkable success of reverse genetics is well appreciated, we have to keep in mind that studying one gene at a time may pose conceptual problems when trying to understand the complexity of biological systems. Much like the famous “six blind men from Indostan”, in the poem by John Godfrey Saxe (1816–1887), who fail to accurately describe the appearance of an elephant because each of them is fooled by the selective impression of touching trunk, tail or ear of the animal, we have to be aware that studying individual genes or gene pathways inevitably falls short of revealing the complete picture of a physiological process. For example, while monogenic approaches in kidney research have uncovered major gene pathways controlling volume and electrolyte homeostasis,14,15 the majority of patients with end-stage renal failure are not affected by single gene mutations in such master genes but rather by a complex interplay of environmental and genetic risk factors influencing susceptibility to the most prevalent renal diseases such as hypertension-induced kidney damage, diabetic nephropathy and glomerulonephritis.
Thus, novel concepts are emerging in which the study of an organism is viewed as an interacting network of genes, proteins and biochemical reactions-interactions that are ultimately responsible for an organism's form and (mal)functions. In this new discipline, commonly referred to as Systems Biology, the individual activities and collective interaction of genes, proteins and other components in an organism are characterized as an integrated network. For Systems Biology to work, novel tools from theoretical disciplines such as bioinformatics and applied mathematics have to be developed and combined with classical wet-lab experiments of molecular biology, biochemistry or physiology.
Towards a new systems approach in kidney research, the European Renal Genome Project (EuReGene) will pursue combining the effectiveness of monogenic strategies in model organisms with novel bioinformatics programs to develop an organ atlas that describes the spatial and temporal relationship of major gene pathways in kidney development and disease. It will integrate available information from many sources into the framework of a four-dimensional atlas where novel functional relationships can be uncovered in an environment that is not constrained by current hypotheses.
Why the Kidney?
A prerequisite for an organ atlas is a target tissue with a well-defined structure-function relationship. Before outlining the concept of a kidney atlas in detail, we have to consider why the kidney may be an appropriate tissue to develop the concept of an organ atlas.
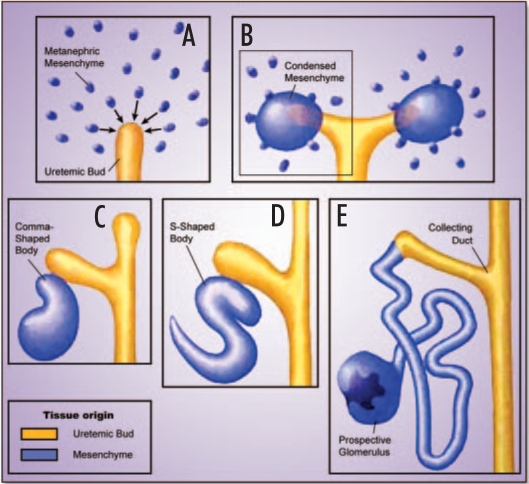
In terms of developmental programs, nephrogenesis recapitulates major steps in organ formation, including the differentiation of primordial cells into an array of highly specialized cell types and the instructive interaction of different germ layers to form a complex organ structure. Development of the metanephros, the permanent kidney of mammals, proceeds in three steps, each of which is essential for the formation of a functional excretory organ.16,17 It encompasses the induction, growth and branching of the ureter from the mesonephric or Wolffian duct (Fig. 1A), the initiation of nephrogenesis as a result of reciprocal interactions between the ureter and the metanephric mesenchyme (Fig. 1B), and finally the patterning and terminal differentiation of the nephron (Fig. 1C–E). The emerging mature kidney consists of a complex three-dimensional assembly of more than twenty different cell types capable of executing complex physiological functions.
Figure 1.
Development of the metanephros. This figure highlights the major steps in nephrogenesis starting with the formation, growth and branching of the ureteric bud from the Wolffian duct elicited by inductive signals from the metanephric blastema (A). In turn, signals originating from the tip of the ureter induce the metanephric mesenchyme to condense and undergo nephrogenesis (B). Finally, metanephric development proceeds through a complex series of morphogenic events including comma- and S-shaped body structures to give rise to the nephron and its specific components (C).
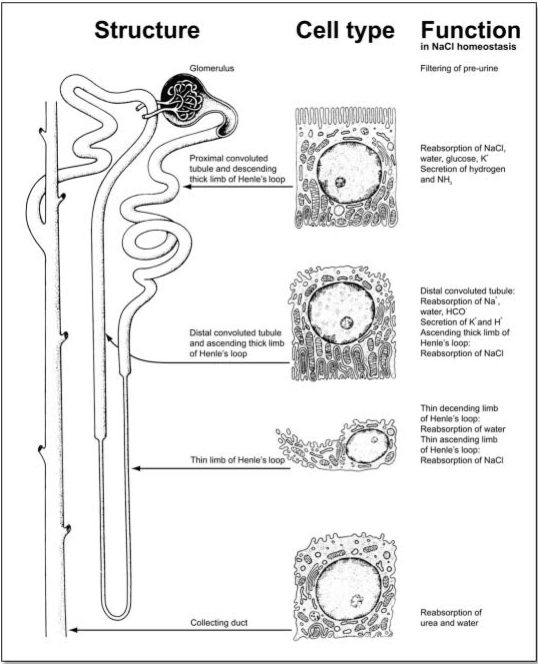
As well as representing a paradigm for organogenesis, the kidney also proves an attractive model for elucidating the complex interaction of many specialized cell types in a mature organ. This notion is particularly supported by the fact that its well-defined tissue architecture is closely aligned with biochemical function (Fig. 2). The adult human kidney consists of approximately one million nephrons, functional units that are composed of differentiated structures and cell types. The glomerulus acts as selective filter that retains the bulk of plasma proteins but permits passage of water and solutes, which, for the major part, are subsequently reabsorbed along specialized nephron segments. Podocytes are specialized cells with interdigitated foot processes in the glomerulus. They synthesize the slit diaphragm proteins, nephrin and podocin, which play an essential role in determining glomerular permeability. Mutations in these genes and injury to podocytes lead to proteinuria, a hallmark of most glomerular diseases. Renal vessels, including glomerular capillaries are lined by endothelial cells. They are key actors in vasculitis, triggered by immune processes or high blood pressure causing tissue damage and renal failure. Following passage through the glomerulus, the ultra filtrate enters the proximal tubule. Here, epithelial cells endocytose many low molecular weight proteins and potentially toxic drug metabolites that are freely filtered. The bulk of ion and water reabsorption also takes place in this nephron segment whereas finer adjustments are made in more distal segments such as the distal convoluted tubules and the collecting ducts. These highly regulated processes involve transporters that use the energy stored in transmembrane ionic gradients, as well as ion-specific channels and ion-motive ATPases. In addition to the specialized NaCl-reabsorbing function, which drives fluid reabsorption and urinary concentration, renal ion transport is essential for renal cell volume adjustment, intracellular pH regulation, and acidification of intracellular vesicles. Finally, passive paracellular pathways, driven by electrochemical gradients, are required for renal handling of divalent ions such as magnesium and calcium. This ion transport route is governed by claudins, a family of tight junction proteins that enable ion specific reabsorption.18,19 Mutations affecting the function of proteins involved in transcellular or paracellular transport are the cause of electrolyte disorders, proteinuria and tubular acidosis.
Figure 2.
Structural and functional organization of the adult nephron. The nephron is organized in distinct structural elements (left panel) that are characterized by unique specialized cell types (middle panel), each of which performs specific tasks in volume and electrolyte homeostasis (right panel). For reasons of simplification, described activities of renal cell types focus on water and sodium chloride handling along the nephron segments, an activity that is central to the regulation of body volume and electrolyte balance, and to blood pressure regulation. (Figure adapted from, The Urinary System. In: Junqueira LC, Carneiro J, Kelley RO. Eds. Basic Histology. 8th edition. Connecticut: Appleton & Lange,1995:369).
The Kidney Atlas Project
The aim of the kidney atlas is to provide a unified graphical environment in which to visualize, query, and explore data generated from a variety of experimental approaches in renal development and (patho)physiology. It will enable an integrated access to gene function information whereby diverse data can be explored as part of a global view of mechanisms and concepts in nephrology. Such an atlas presents a novel systems approach for studying developmental and disease processes in an intact organ that may provide a new paradigm in functional genome research. In the following, we will discuss the conceptual framework of the organ atlas in detail and we will highlight the major scientific projects in renal development and disease that will be incorporated in this atlas.
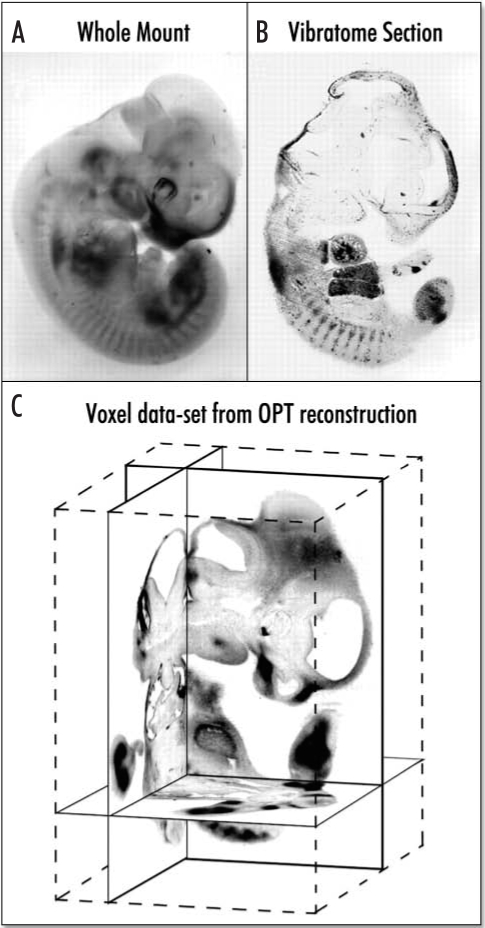
The core of the atlas will be a series of three-dimensional (3D) reconstructions of the whole kidney. Initially, we will reconstruct mouse kidneys at embryonic day E17.5 and at 12 weeks of age, prototypes for the embryonic and the adult organs, respectively. 3D models of the kidney have greatly facilitated studies of development and disease, and confocal microscopy and magnetic resonance imaging technologies have played an important part in studying the kidney structure. However, both techniques have their limitations and are not ideal for structures the size of the mouse kidney. Of several alternative techniques applicable to this size range, the most promising is optical projection tomography (OPT) invented by James Sharpe.20,21 OPT produces 3D images of structures several hundred micrometers to 1.5 cm in size with a resolution of 10 µm—without the need to cut sections, a requirement for other techniques such as confocal microscopy (Fig. 3). An overriding advantage of OPT is that it captures images of specimens stained by conventional methods for in situ hybridization (ISH) and immunohistochemistry. Resulting images can be studied as 3D objects or as digital sections (Fig. 3). With respect to the kidney atlas, our initial task is to adapt OPT technology to renal specimens and to describe the tissue architecture and molecular anatomy of the normal developing and adult mouse kidney at two levels of resolution. Low-resolution, (approx 10 µm) models will be built from OPT scans of kidneys stained with antibodies and fluorescent reporters that mark nephrons, the vasculature and branching collecting system. High-resolution models will be reconstructed from serial plastic sections (approx. 3 µm).
Figure 3.
Optical projection tomography. Optical projection tomography (OPT) has been developed for 3D reconstruction of whole mount in situ or immunohistological analysis of early mouse embryos. (A). An E11.5 mouse embryo whole-mount stained for expression of the Sox9 gene (in which the staining is clearly visible, but determining exactly which tissues are expressing is not possible). (B) A 50-µm thick vibratome section cut through a similar whole-mount stained embryo, in which expression can be seen in several tissues. (C) The 3D OPT reconstruction of the embryo shown in (A). Virtual sections in three orthogonal planes are shown, within the context of the full 3D block of voxel data.
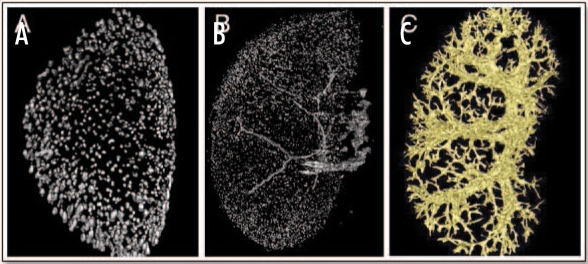
In pilot experiments, we have developed protocols that enabled us to visualize several renal structures in 3D using OPT of the adult mouse kidney. For detection of glomeruli, we have taken advantage of a transgenic mouse line expressing β-galactosidase in all glomerular podocytes.22 LacZ staining of the glomeruli can be readily detected using OPT and used to depict the total number of glomeruli present in the adult organ (Fig. 4A). As an alternative method, we infused mouse kidneys with beads that attach to glomeruli and also enabled us to visualize this nephron segment (Fig. 4B). For detection of the vasculature, we can infuse kidneys with molten latex and use OPT to reconstruct the entire renal vessel tree (Fig. 4C).
Figure 4.
Three-dimensional reconstruction of glomeruli and vasculature in adult mouse kidneys. (A and B) OPT was used to reconstruct the 3D architecture of the glomeruli in the adult mouse kidney using lacZ staining in mice expressing the β-galactosidase gene in podocytes (A) or infusion of beads that are trapped in the glomeruli of normal mice (B). (C) OPT reconstruction of the vascular tree in the adult mouse kidney infused with molten latex. (Pictures courtesy of Jane Armstrong, University of Edinburgh and Annemieke IJpenberg, MRC, Edinburgh).
The Databases
In the kidney atlas, 3D reconstructions of the mouse kidney will be integrated with a database system that holds data from across the EuReGene project. These scientific data will be mapped onto the organ model in such a way that specific data can be related to a distinct position and developmental time point within the kidney where and when these data are relevant. The kinds of data represented in the atlas project range from ISH patterns, to global gene expression profiles, to description of mutant phenotypes. Most of the information relates to the mouse kidney, but the atlas will also represent the evolutionary dimension by including models and data from Xenopus. These projects cover topics in three important areas of current research activities in nephrology, namely renal development, renal pathophysiology, and complex genetics of renal diseases. To ascertain comparability of data across all projects, the consortium will use common standards for experimental procedures such as a unified ontology for annotation of renal structures, and compliance to MIAME standards (http://www.mged.org/Workgroups/MIAME/miame.html) for gene expression profiling. Concerning the laboratory animals, wherever possible studies will be performed in male mice of the C57BL/6 strain at E17.5 for the embryonic and 12 weeks of age for the adult stage.
In renal development (Table 1A).
Table 1.
Overview of scientific projects in three areas of kidney research that will be implemented in the kidney atlas
| Area of research | Project | Approach |
| A. Renal Development | ||
| Systematic approaches | Renal expression maps of major gene families (solute carriers, G-protein coupled receptors, transcription factors) | Automated high throughput ISH on embryonic and adult mouse kidney sections, and Xenopus embryos |
| Global renal gene expression profiling | Oligonucleotide micro arrays on E9.0, E10.5, E12.5, E14.5 and E17.5, and adult mouse kidneys | |
| Monogenic approaches | Ureter induction, growth and branching | Developmental studies in Wt1-, Wnt11-, Sox8, Sox9, vHNF1-deficient mice |
| Inductive signals in nephrogenesis provided by Wnt4 | Studies on inductive signals in nephrogenesis | |
| Patterning and terminal differentiation | Studies on differentiation of renal cell types by siRNA knockdown approaches in cell lines and organ cultures as well as morpholino knockdowns in Xenopus embryos | |
| B. Renal (Patho)physiology | ||
| Monogenic approaches in renal physiology | Ion transport | Mouse models with defects in claudins, chloride channels and sodium transporters |
| Acid-base and phosphate transport | Mouse models of functional deficiency for NaPi-IIa and accessory factors | |
| Protein transport | Mouse models of endocytosis defects in proximal tubules (megalin, cubilin, ClC-5, Dab2) | |
| Monogenic approaches in renal pathology | Glomerulosclerosis | Studies in Wt1- and podocin-deficient mice, and in animals with induced glomerular expression of Pax2 |
| Inflammation | Mouse and rat models of hypertension-induced renal damage and vasculitis | |
| Nephrotoxicity | Studies on nephrotoxicity induced by drugs in rat and mouse models | |
| C. Complex Genetics | ||
| Mapping of modifier genes in established renal disease models | Proteinuric kidney damage | Mapping studies in Munich Wistar Fromter rat model of proteinuria |
| Diabetic nephropathy | Mapping studies in ENU mouse models of types 2 diabetes | |
| Renal stone disease | Mapping studies in ENU mouse models of hypercalciuric kidney stone disease | |
| Glomerulosclerosis | Mapping in mouse model of focal segmental glomerulosclerosis | |
| Mapping of new disease genes | Volume and electrolyte disorders | Screen for volume and electrolyte anomalies in ENU mice |
Details of these projects can be found at www.euregene.org.
Major efforts will be directed at elucidating the basis of congenital renal and urogenital tract anomalies as a major cause of morbidity and mortality in children. Uncovering the developmental programming of the kidney is crucial to understanding the resulting diseases as well as the repair processes whereby the kidney responds to insults (as in diabetes and hypertension). Ultimately, this may lead to strategies in stem cell technology for renal replacement therapy. Important questions in this respect include what is the inductive signal from the ureteric tip that induces nephrogenesis, what drives nephron patterning, and how is vasculogenesis during kidney development regulated?
In renal pathophysiology (Table 1B).
Our studies aim at understanding the genetic origins of volume, electrolyte and blood pressure disorders caused by abnormalities in renal cell physiology. Chief among recent advances in this field are the identification of key genes in renal development (e.g., transcription factors, signaling molecules), renal structure (e.g., podocyte proteins) and renal function (e.g., transporters, ion channels, endocytic receptors). However, presently we know little about the regulatory pathways in which they act. Thus, we aim at developing new mouse and rat models of renal pathophysiology for defining disease processes through comparative genomic analysis. We will apply improved cell (tubular, endothelial, glomerular, interstitial) and organ systems, and functional genomic tools (transcriptome analysis, imaging techniques) to define regulatory maps of cell growth and differentiation, homeostasis and cell-cell interaction. Construction of regulatory maps will be imperative to understand disease processes, and, more importantly, it will identify future targets for therapeutic intervention.
In complex genetics (Table 1C).
We focus on the identification of modifiers and susceptibility factors that contribute to complex traits underlying the majority of human renal diseases (e.g., diabetic nephropathy, proteinuria, glomerulosclerosis). To do so, we will explore established animal models of complex renal diseases to characterize modifiers in expression and in linkage studies, and we will apply the results of large-scale ENU screens to identify novel mouse models of renal disorders.
Perspective
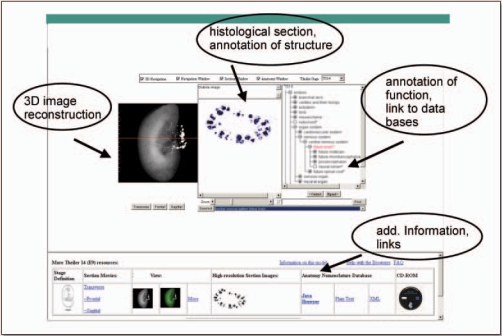
By the end of the EuReGene project period (2005–2008), the full kidney atlas will be made available as a public resource. The atlas will provide an intuitively easy way to browse and explore diverse data (Fig. 5). Users will be able to browse the data spatially, moving the mouse cursor across images of kidney sections and pulling up the different kinds of data that relate to each position. They will be able to query data textually, where this has been annotated. A query interface will enable users to ask where, and when during development, particular genes are expressed or to ask which genes are expressed in particular locations. To the extent that data have been spatially mapped to the models, it will be possible to use, for example, the coexpression domain of two genes to ask which other genes are expressed in this region, or enable us to link ISH data to micro array expression data not only by gene name but by consistent textual and spatial annotations of in situ patterns and the origin of micro array samples. In another example, we plan to link data on mutant kidney phenotypes to gene expression patterns by cross-referencing through gene name and the anatomy or spatial localization of the affected parts. Initially, the data bases incorporated in the kidney atlas will mainly consist of data produced within the consortium (Table 1). Eventually, other genome resources can be included through a common bioinformatics interface forming the basis for a large integrative resource for genome research into the genetics of kidney development and disease.
Figure 5.
EuReGene kidney organ atlas. This figure represents an outline of the kidney organ atlas providing a unified graphical environment of the kidney architecture from 3D reconstructions and histological sections in which to visualize, query, and explore data generated from a variety of experimental approaches in renal development and (patho)physiology.
Acknowledgements
EuReGene (www.euregene.org) is an Integrated Project funded by the European Community as part of the Framework program 6 (FP6 005085). For more information contact Iwan Meij, EuReGene management, Max-Delbrueck-Center for Molecular Medicine, Berlin, Germany (i.meij@mdc-berlin.de).
Footnotes
Previously published online as an Organogenesis E-publication: http://www.landesbioscience.com/journals/organogenesis/abstract.php?id=2118
References
- 1.Larsson TP, Murray CG, Hill T, Fredriksson R, Schioth HB. Comparison of the current RefSeq, Ensembl and EST databases for counting genes and gene discovery. FEBS Lett. 2005;579:690–698. doi: 10.1016/j.febslet.2004.12.046. [DOI] [PubMed] [Google Scholar]
- 2.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 3.Hammes A, Guo JK, Lutsch G, Leheste JR, Landrock D, Ziegler U, Gubler MC, Schedl A. Two splice variants of the Wilms' tumor 1 gene have distinct functions during sex determination and nephron formation. Cell. 2001;106:319–329. doi: 10.1016/s0092-8674(01)00453-6. [DOI] [PubMed] [Google Scholar]
- 4.Stark K, Vainio S, Vassileva G, McMahon AP. Epithelial transformation of metanephric mesenchyme in the developing kidney regulated by Wnt-4. Nature. 1994;372:679–683. doi: 10.1038/372679a0. [DOI] [PubMed] [Google Scholar]
- 5.Kreidberg JA, Sariola H, Loring JM, Maeda M, Pelletier J, Housman D, Jaenisch R. WT-1 is required for early kidney development. Cell. 1993;74:679–691. doi: 10.1016/0092-8674(93)90515-r. [DOI] [PubMed] [Google Scholar]
- 6.Saulnier DM, Ghanbari H, Brandli AW. Essential function of Wnt-4 for tubulogenesis in the Xenopus pronephric kidney. Dev Biol. 2002;248:13–28. doi: 10.1006/dbio.2002.0712. [DOI] [PubMed] [Google Scholar]
- 7.Silva IV, Cebotaru V, Wang H, Wang XT, Wang SS, Guo G, Devuyst O, Thakker RV, Guggino WB, Guggino SE. The ClC-5 knockout mouse model of Dent's disease has renal hypercalciuria and increased bone turnover. J Bone Miner Res. 2003;18:615–623. doi: 10.1359/jbmr.2003.18.4.615. [DOI] [PubMed] [Google Scholar]
- 8.Wang SS, Devuyst O, Courtoy PJ, Wang XT, Wang H, Wang Y, Thakker RV, Guggino S, Guggino WB. Mice lacking renal chloride channel, CLC-5, are a model for Dent's disease, a nephrolithiasis disorder associated with defective receptor-mediated endocytosis. Hum Mol Genet. 2000;9:2937–2945. doi: 10.1093/hmg/9.20.2937. [DOI] [PubMed] [Google Scholar]
- 9.Gunther W, Piwon N, Jentsch TJ. The ClC-5 chloride channel knock-out mouse - An animal model for Dent's disease. Pflugers Arch. 2003;445:456–462. doi: 10.1007/s00424-002-0950-6. [DOI] [PubMed] [Google Scholar]
- 10.Piwon N, Gunther W, Schwake M, Bosl MR, Jentsch TJ. ClC-5 Cl- -channel disruption impairs endocytosis in a mouse model for Dent's disease. Nature. 2000;408:369–373. doi: 10.1038/35042597. [DOI] [PubMed] [Google Scholar]
- 11.Jentsch TJ, Neagoe I, Scheel O. CLC chloride channels and transporters. Curr Opin Neurobiol. 2005;15:319–325. doi: 10.1016/j.conb.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 12.Waldegger S, Jentsch TJ. Functional and structural analysis of ClC-K chloride channels involved in renal disease. J Biol Chem. 2000;275:24527–24533. doi: 10.1074/jbc.M001987200. [DOI] [PubMed] [Google Scholar]
- 13.Christensen EI, Devuyst O, Dom G, Nielsen R, Van der Smissen P, Verroust P, Leruth M, Guggino WB, Courtoy PJ. Loss of chloride channel ClC-5 impairs endocytosis by defective trafficking of megalin and cubilin in kidney proximal tubules. Proc Natl Acad Sci USA. 2003;100:8472–8477. doi: 10.1073/pnas.1432873100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scheinman SJ, Guay-Woodford LM, Thakker RV, Warnock DG. Genetic disorders of renal electrolyte transport. N Engl J Med. 1999;340:1177–1187. doi: 10.1056/NEJM199904153401507. [DOI] [PubMed] [Google Scholar]
- 15.Lifton RP, Gharavi AG, Geller DS. Molecular mechanisms of human hypertension. Cell. 2001;104:545–556. doi: 10.1016/s0092-8674(01)00241-0. [DOI] [PubMed] [Google Scholar]
- 16.Schedl A, Hastie ND. Cross-talk in kidney development. Curr Opin Genet Dev. 2000;10:543–549. doi: 10.1016/s0959-437x(00)00125-8. [DOI] [PubMed] [Google Scholar]
- 17.Vainio S, Lin Y. Coordinating early kidney development: Lessons from gene targeting. Nat Rev Genet. 2002;3:533–543. doi: 10.1038/nrg842. [DOI] [PubMed] [Google Scholar]
- 18.Simon DB, Lu Y, Choate KA, Velazquez H, Al-Sabban E, Praga M, Casari G, Bettinelli A, Colussi G, Rodriguez-Soriano J, McCredie D, Milford D, Sanjad S, Lifton RP. Paracellin-1, a renal tight junction protein required for paracellular Mg2+ resorption. Science. 1999;285:103–106. doi: 10.1126/science.285.5424.103. [DOI] [PubMed] [Google Scholar]
- 19.Muller D, Kausalya PJ, Claverie-Martin F, Meij IC, Eggert P, Garcia-Nieto V, Hunziker W. A novel claudin 16 mutation associated with childhood hypercalciuria abolishes binding to ZO-1 and results in lysosomal mistargeting. Am J Hum Genet. 2003;73:1293–1301. doi: 10.1086/380418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharpe J, Ahlgren U, Perry P, Hill B, Ross A, Hecksher-Sorensen J, Baldock R, Davidson D. Optical projection tomography as a tool for 3D microscopy and gene expression studies. Science. 2002;296:541–545. doi: 10.1126/science.1068206. [DOI] [PubMed] [Google Scholar]
- 21.Kerwin J, Scott M, Sharpe J, Puelles L, Robson SC, Martinez-de-la-Torre M, Ferran JL, Feng G, Baldock R, Strachan T, Davidson D, Lindsay S. 3 dimensional modelling of early human brain development using optical projection tomography. BMC Neurosci. 2004;5:27. doi: 10.1186/1471-2202-5-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moore AW, Schedl A, McInnes L, Doyle M, Hecksher-Sorensen J, Hastie ND. YAC transgenic analysis reveals Wilms' tumour 1 gene activity in the proliferating coelomic epithelium, developing diaphragm and limb. Mech Dev. 1998;79:169–184. doi: 10.1016/s0925-4773(98)00188-9. [DOI] [PubMed] [Google Scholar]