Abstract
The major core promoter-binding factor in polymerase II transcription machinery is TFIID, a complex consisting of TBP, the TATA box-binding protein, and 13 to 14 TBP-associated factors (TAFs). Previously we found that the histone H2A-like TAF paralogs TAF4 and TAF4b possess DNA-binding activity. Whether TAF4/TAF4b DNA binding directs TFIID to a specific core promoter element or facilitates TFIID binding to established core promoter elements is not known. Here we analyzed the mode of TAF4b·TAF12 DNA binding and show that this complex binds DNA with high affinity. The DNA length required for optimal binding is ∼70 bp. Although the complex displays a weak sequence preference, the nucleotide composition is less important than the length of the DNA for high affinity binding. Comparative expression profiling of wild-type and a DNA-binding mutant of TAF4 revealed common core promoter features in the down-regulated genes that include a TATA-box and an Initiator. Further examination of the PEL98 gene from this group showed diminished Initiator activity and TFIID occupancy in TAF4 DNA-binding mutant cells. These findings suggest that DNA binding by TAF4/4b-TAF12 facilitates the association of TFIID with the core promoter of a subset of genes.
Two types of DNA elements regulate transcription of protein-encoding genes in eukaryotes. Enhancer elements, which may be localized proximally or distally relative to the transcription initiation site, are the binding sites for gene-specific transcription factors. A core promoter, situated close to the transcription start site (TSS),2 serves as the site on which RNA polymerase II and the general transcription factors bind and assemble into a pre-initiation complex (1, 2). Enhancer-bound transcription factors activate transcription by modulating chromatin structure or by recruiting the transcription machinery to the core promoter.
The major core promoter-binding factor within the general transcription apparatus is TFIID, a large complex composed of the TATA-binding protein (TBP) and about 14 TBP-associated factors (TAFs) (for recent reviews see Refs. 3, 4). Within TFIID TBP is responsible for recognition and binding of TATA-containing promoters. The TAFs are also important for core promoter recognition, and they bind primarily to non-TATA-box elements, interacting with sequences upstream and downstream to the TATA box (5–13). In addition certain TAF sub-complexes have been reported to specifically bind different core promoter elements. The TAF1·TAF2 complex binds to the Initiator element (14) and Drosophila TAF6 and TAF9 cross-linked to the downstream promoter element in the context of TFIID (15), and, as a reconstituted complex, these were shown to associate with a downstream promoter element-containing promoter (16).
A feature common to 9 of the 14 TAFs is the histone-fold domain (HFD) (17–20). The presence of histone-fold TAFs within TFIID led to the proposal that there is a nucleosomal-like interaction between HFD TAFs and DNA (21). Recently we reported that the H4-H3-like TAF6 and TAF9 have intrinsic DNA-binding activity that lies outside the HFD. However, when complexed through their HFDs, they show enhanced DNA-binding activity to the core promoter motif downstream promoter element (16). We also found that human, Drosophila, and yeast TAF4 is capable of DNA binding, which we mapped to its H2A-like histone-fold motif and a unique spacer domain that is not present in histone H2A. The interaction of the H2A-like TAF4b with the H2B-like TAF12 increased the stability of the DNA-bound complex (16). The ability of many TAFs to bind DNA suggests that they facilitate core promoter binding by TFIID. How TAF4/TAF4b·TAF12 accomplishes this is unknown.
In the present study we report on the unique biochemical and molecular features of the interaction of the histone-like pair TAF4b·TAF12 with DNA. We show that it binds DNA with high affinity most likely through one TAF4b·TAF12 heterodimer forming several contacts with one DNA molecule. For optimal binding the complex requires the DNA to have a length of ∼70 bp. The complex displays a weak sequence preference for the adenovirus major late (AdML) promoter, but the nucleotide composition of the DNA is less important than its length for high affinity binding. Expression profiling revealed a gene set that is down-regulated when TAF4 DNA binding is impaired. The vast majority of genes in this group have an Initiator core promoter element around the TSS and a high prevalence of the TATA-box. Examination of the Pel98 promoter from this group demonstrated that TAF4 DNA binding was critical for the core promoter function. Our findings suggest that, in this subset of genes, TAF4 can facilitate the binding of TFIID to the core promoter by providing additional contacts with DNA.
EXPERIMENTAL PROCEDURES
Protein Preparation and DNA Binding Assays
TAF4b and TAF12 were expressed in Escherichia coli BL-21 DE3 strain, refolded either alone or as complexes, and then purified as previously described (16) and further purified on a Sephadex 200 column. TAF4CRII and TAF4CRIImDB were expressed and refolded as previously described (16). For EMSA DNA was end-labeled using [γ-32P]ATP (Amersham Biosciences) and polynucleotide kinase. The DNA probe (4 ng) was incubated with the TAF4b·TAF12 complex in a 20-μl reaction volume in DNA binding buffer containing 10 mm Tris, pH 8.0, 75 mm KCl, 2.5 mm dithiothreitol, 10% glycerol, and 0.05% Nonidet P-40 for 20 min at 25 °C. The samples were loaded onto a 5% native polyacrylamide gel containing 0.5× TBE buffer (89 mm Tris-HCl, 89 mm boric acid, 2 mm EDTA) and run at 4 °C for 2 h. The gel was dried and visualized using a phosphorimaging device (Fuji BAS 2500). For DNA cellulose binding assays the proteins were incubated with either empty cellulose beads or DNA-containing cellulose beads (0.25 μg of double-stranded calf thymus DNA (Sigma) per reaction) for 45 min, at room temperature, in binding buffer composed of 10 mm Tris, pH 8.0, 50 mm KCl, 2.5 mm dithiothreitol, 0.1 mg/ml bovine serum albumin, 15% glycerol, and 0.2% Nonidet P-40. The beads were washed three times with binding buffer, and the proteins were eluted with 30 μl of binding buffer containing 1 m NaCl. 20% of the eluted bound proteins were analyzed by SDS-PAGE and visualized by silver staining.
Mass Spectrometric Analysis of TAF4b·TAF12
Mass spectrometry was performed under non-denaturing conditions on a QToF Q-Star XL (MDS Sciex, Concord, Ontario, Canada) mass spectrometer, modified for improved transmission of large, non-covalent complexes. The instrument was fitted with a high m/z quadrupole. In addition, the pressure regime in the early vacuum stage of the instrument was modulated to improve large ion transmission, by a flow-restricting sleeve surrounding part of the first quadrupole ion guide of the Sciex instrument (Chernushevich IV, Thomson BA; collisional cooling of large ions in electrospray mass spectrometry).
Plasmid Constructions
Construction of pGEX-TAF4CRII was previously described (Shao et al. 16). To construct the pGEX-TAF4CRIImDB two DNA fragments corresponding to TAF4 amino acids 828–1010 and 1052–1083 (end) were generated by PCR and sequentially inserted into pGEX-2TK. To generate expression plasmids for HA-tagged TAF4CRII and TAF4CRIImDB, DNAs encoding TAF4CRII and TAF4CRIImDB were amplified from the pGEX-TAF4CRII and pGEX-TAF4CRIImDB plasmids, respectively, by PCR with the oligonucleotides 5′-CCCCCCTCTAGAGACGATGATGACATTAATGA and 5′-GGGGGGGGATCCCCGGGAGCTGCATGTGTCAGAGG, and the fragments were first cloned into the pCGN expression vector downstream to the HA-epitope. The pCGN was then cut with SnaBI and EcoRI to generate DNA fragments that included TAF4CRII and TAF4CRIImDB with the HA tag in their N-terminal, and these fragments were cloned into pEIRES-P at NheI that was blunted by Klenow and EcoRI.
The PEL98 promoter from −488 to +10 was amplified by PCR from mouse genomic DNA using primers 5′-CCCCCCGGTACCCCAAGGTCCCTCCTGACTTG and 5′-CCCCCCCTCGAGAGAGAGGTTTGGGGAGAGCC and cloned into the promoter-less reporter gene pGL3-basic (Promega, Madison, WI) at the KpnI and XhoI sites of the multiple cloning site. The Initiator mutant was generated by PCR using the same forward primer and the reverse primer 5′-CCCCCCCTCGAGAGAGAGCACACCGGAGAGCC.
Isolation of Stable TAF4CRII and TAF4CRIImDB Fibroblasts
TAF4−/− cells, and their derivatives were grown in Dulbecco's minimal essential media supplemented with 10% fetal calf serum. TAF4CRII and TAF4CRIImDB expression plasmids were transfected into the TAF4−/− fibroblasts, and stable clones were picked out following puromycin selection.
Microarray Expression Profiling and Data Analysis
Poly-l-lysine-coated glass microarrays containing >23,000 different probes (mouse oligonucleotide set, Compugen) were purchased from the Center for Applied Genomics, New Jersey. The microarrays were probed with a mixture of cyanine 3- or cyanine 5-labeled cDNAs, generated from total RNA (100 μg) that was prepared from TAF4CRII and TAF4CRIImDB cell lines. The cDNA was synthesized using Moloney murine leukemia virus reverse transcriptase (Promega) with aminoallyl-modified dUTP nucleotide (Ambion) at a 4:1 aminoallyl-modified dUTP-to-dTTP ratio and labeled with an N-hydroxysuccinimide-activated cyanine 3 or cyanine 5 fluorescent probe (Amersham Biosciences) through aminoallyl-modified dUTP. These labeled cDNAs from TAF4CRII and TAF4CRIImDB cells were mixed with equivalent amounts of fluorescent dye (100 pmol each) in 2× SSC (1× SSC is 0.15 m NaCl plus 0.015 m sodium citrate), 0.08% SDS, 6 μl of blocking solution (Amersham Biosciences), and water to 100 μl. This target mixture was denatured at 95 °C for 3 min, chilled, and applied between a raised coverslip (LifterSlip, Erie Scientific Co.) and the array. The slide was then sealed in a microarray hybridization chamber and submerged in a darkened water bath set at 55 °C for hybridization. After 12 h, the slide was washed for 5 min in 2× SSC-0.5% SDS at 55 °C, 5 min in 0.5× SSC at room temperature, and 5 min in 0.05× SSC at room temperature. It was then quickly dried by centrifuging for 3 min at 1000 rpm and stored in the dark until scanned. Each cell line was represented by dye-swap microarray replicates.
To correct for dye bias Lowess normalization was performed. Bad spots were flagged out before normalization. Average log intensities were calculated using the R package Limma (22). Linear models and empirical Bayes methods were used for assessing differential expression in microarray experiments. All genes with <1.9-fold changes were excluded from the list.
RNA Preparation and Quantitative Reverse Transcription-PCR Analysis
Total RNA was prepared using the RNeasy Mini kit (Qiagen), according to the manufacturer's instructions. RNA preparations were treated with RQ1 DNase I (Promega) to avoid contamination by genomic DNA. First strand cDNA was synthesized from 1 μg of total RNA using an oligo(dT)15 primer and SuperScript II reverse transcriptase (Invitrogen). 1 μl of a 1/50 cDNA dilution was used for PCR. The real-time PCR was performed in 20-μl tubes using a SYBR Green PCR master mix (Applied Biosystems), according to the manufacturer's instructions in a 7300 real-time PCR system and was analyzed using 7300 system software. The oligonucleotides used for real-time PCR were as follows: β-actin, 5′-CCCTAAGGCCAACCGTGAA and 5′-TTGAAGGTCTCAAACATGATCTGG; mouse THBS1, 5′-CCATGAAGAGTTCCTTGGGTTT and 5′-TCTGGCTCTGTGAGTAAGGCAG; CYR61, 5′-TCAGGGACTAAGTGCCTCCAG and 5′-GCAAGGCACCATTCATCCTC; FGF7, 5′-AGGTCATGCTTCCACCTCGT and 5′-GGGCTGGAACAGTTCACACTC; SFRP2, 5′-TCCCAGTGGGTGGCTTCTC and 5′-TAGCTTTCCCGGACTGTGCTT; PEL98, 5′-AGCTGCATTCCAGAAGGTGA and 5′-ACATCATGGCAATGCAGGAC; OMD, 5′-TTCAGACACTCCAGAAGAGGGAG and 5′-CGACTGCTCTTCCGAAGGTC; MSLN, 5′-GAATGGCTGCAACACATCTCC and 5′-GTCGGAACCTTGGGTGTATGA; and IL1RL1, 5′-GAATGGGACTTTGGGCTTTG and 5′-CAGGACGATTTACTGCCCTCC.
Transient Transfection Assays and Chromatin Immunoprecipitation
Transfections into TAF4CRII and TAF4CRIImDB cell lines were performed using the jetPEI transfection reagent (PolyPlus Transfection) according to the manufacturer's instructions. For reporter assays, subconfluent cells were transfected in a 6-well plate using 1000 ng of the firefly luciferase reporter vector, 50 ng of Rous sarcoma virus-Renilla control reporter vector (containing Renilla luciferase), and 50 ng of cytomegalovirus-green fluorescent protein. 4 h after transfection the medium was replaced. 24 h after transfection luciferase and Renilla activities were measured.
ChIP assays were carried out as described (23). Equivalent amounts of cross-linked chromatin extract were used for immunoprecipitation. The input and the ChIP data were quantified by densitometric analysis using Quantity One one-dimensional analysis software (Bio-Rad), and the ChIP results were normalized to the input and the enrichment relative to the control antibodies was calculated. The ChIP primers were: PMM2 core promoter (forward, 5′-ACCGGTGTTCTGTGAACCAT; reverse, 5′-CCATCCATGTCGAAGAGACA), PEL98 promoter (forward, 5′-GCAAGAGCACAGTATCCATG; reverse, 5′-AGCAGTGCTATCAGACCAAC), and PEL98–1000 bp promoter upstream region (forward, 5′-CCTTTATGCTCCTTACTACTG; reverse, 5′-GTCTCATCTGATAGGACACG).
Additional Oligonucleotides Used in This Study
Different promoter fragments were generated by PCR using the primers as follows: IκB (forward, 5′-TCTGGTCTGACTGGCTTGG; reverse, 5′-GGACTGCTGTGGGCTCTG) and A20 (forward, 5-GAAATCCCCGGGCCTACAAC; reverse, 5-CAAGCTCGCTTGGCCCGCC). The template for the control fragment was the pTZ57R plasmid (forward, 5′-GACTCACTATAGGGAAAGCTTGC; reverse, 5-CGACGTTGTAAAACGACGGC).
For the AdML promoter the primers were: forward, 5′-GTGACCGGGTGTTCCTGAAGGGGGGC; reverse, 5′-CCATGATTACGCCAAGCTTGCATG; the AdML promoter was generated by PCR and then digested with ApaI restriction enzyme to get the 74-bp fragment containing the core promoter. In other experiments the AdML core promoter was generated by annealing two oligonucleotides and filling in with Klenow. Primers were forward, 5′-GTGACCGGGTGTTCCTGAAGGGGGGCTATAAAAGGGGGTGGGGGC; reverse, 5-CGGAAGAGAGTGAGGACGAACGCGCCCCCACCCCCTTTTATAGCC. The AdML promoter derivatives were generated by annealing synthetic oligonucleotides with the following sequences: AdML Δ5′ (forward, 5′-GTTCCTGAAGGGGGGCTATAAAAGGGGGTGGGGGCGCGTT; reverse, 5-CGGAAGAGAGTGAGGACGAACGCGCCCCCACCCCCTTTTATAGCC), AdML Δ3′ (forward, 5′-GTGACCGGGTGTTCCTGAAGGGGGGCTATAAAAGGGGGTGGGGGC; reverse, 5′-AACGCGCCCCCACCCCCTTTTATAGCCCCCCTTCAGGAAC), AdML Δ5′+Δ3′ (forward, 5-GTTCCTGAAGGGGGGCTATAAAAGGGGGTGGGGGCGCGTT; reverse, 5-AACGCGCCCCCACCCCCTTTTATAGCCCCCCTTCAGGAAC), AdML 5′ mutant (forward, 5′-TGTCAATTTGGTTCCTGAAGGGGGGCTATAAAAGGGGGTGGGGGC; reverse, 5′-CGGAAGAGAGTGAGGACGAACGCGCCCCCACCCCCTTTTATAGCC), AdML 3′-1 mutant (forward, 5′-GTGACCGGGTGTTCCTGAAGGGGGGCTATAAAAGGGGGTGGGGGC; reverse, 5′-ATTCCTCTAGTGAGGACGAACGCGCCCCCACCCCCTTTTATAGCC), and AdML 3′-2 mutant (forward, 5′-GTGACCGGGTGTTCCTGAAGGGGGGCTATAAAAGGGGGTGGGGGC; reverse, 5′-CGGAAGAGCTGTCTTCCGAACGCGCCCCCACCCCCTTTTATAGCC).
RESULTS
Stable DNA Binding by TAF4b·TAF12 Heterodimer
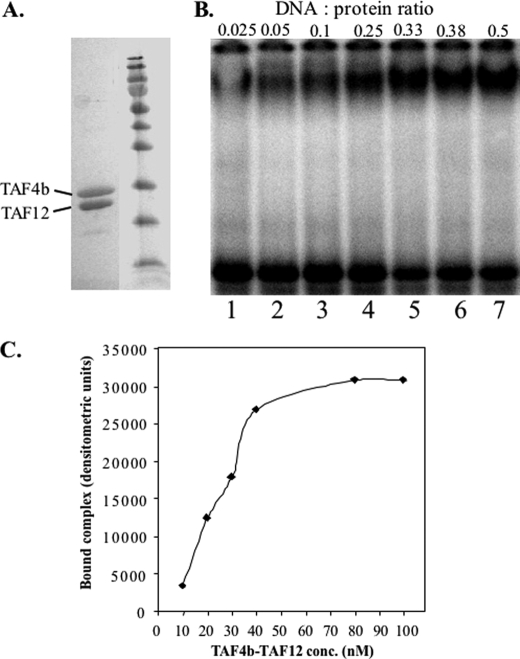
The H2A-like TAF4 and TAF4b interact with the H2B-like TAF12 in vitro and in native TFIID. Both TAF4/4b and TAF12 possess intrinsic DNA-binding activity, and their interaction through the HFD is important for stable DNA binding (16). To characterize TAF4b·TAF12 DNA binding further we first determined the composition of the TAF4b·TAF12 complex. When the TAF4b C-terminal DNA-binding domain (amino acids 561–769) and His6-TAF12 are either co-expressed in E. coli or expressed separately and then combined and purified on nickel beads, they co-purify, indicating complex formation (Fig. 1A). The TAF4b·TAF12 complex was loaded onto a Sephadex 200 gel filtration column, and it eluted in a single peak. The presence of TAF4b and TAF12 in the peak fraction was verified by mass spectrometry (data not shown). We performed a special mass spectrometric analysis to elucidate the oligomeric state of the TAF4b·TAF12 complex (24). This technique preserves non-covalent interactions between proteins, allowing determination of the molecular mass of the complex and the subunits ratio. The measured mass of the TAF4b·TAF12 complex was 41,233 ± 11 Da corresponding to a 1:1 stoichiometry between TAF4b and TAF12 (supplemental Fig. S1). To confirm the composition of the complex, tandem mass spectrometry experiments were conducted. The complex dissociates into two subunits with measured masses of 23,367 ± 2 and 17,794 ± 1 Da, which is in close agreement with the calculated masses of TAF4b and TAF12, respectively, without their first methionine (supplemental Fig. S1). This experiment confirms that TAF4b and TAF12 form a stable complex and suggests that the complex consists of a heterodimer.
FIGURE 1.
TAF4b and TAF12 bind DNA as a heterodimer. A, TAF4b (amino acids 561–769) and His6-TAF12 (full-length) were expressed in E. coli, refolded together as described under “Experimental Procedures,” purified on nickel beads, and run on an SDS-PAGE. The locations of TAF4b and TAF12 are indicated. B, EMSA analysis of the gel-filtration purified TAF4b·TAF12 using the AdML core promoter (−52 to +18) as a probe. Two pmol of TAF4b·TAF12 complex was incubated with 50 fmol of DNA probe (lane 1), and with increasing amounts of non-labeled DNA (lanes 2–7). The molar ratio between the protein complex and DNA is indicated at the top of each lane. C, determination of the apparent Kd of TAF4b·TAF12·DNA. DNA binding analysis by EMSA of increasing amounts of purified TAF4b·TAF12 complex in the presence of excess of DNA (AdML promoter). The graph shows the densitometric measurements of the bound DNA as a function of protein concentration (nanomolar). The apparent Kd is the concentration of the complex required to achieve 50% of maximal binding.
To examine the DNA-binding activity of this complex we employed the EMSA in which the ratio of DNA to TAF4b·TAF12 dimer was gradually increased up to 1:2 (Fig. 1B). The DNA used in this experiment is the AdML promoter that binds TAF4b·TAF12 preferentially (see below). Under conditions of excess protein, formation of the DNA·protein complex is inefficient and unstable, because it tends to dissociate during electrophoresis (lane 1). However, the complex becomes increasingly stable with increased DNA levels (lanes 2–7). When the DNA is in excess the complex remains stable and competition is observed (see Figs. 2B and 3A). The observation that the TAF4b·TAF12-DNA complex becomes more efficient and stable at a high DNA:protein ratio (compare lanes 4 to 5, Fig. 1B) raises the possibility that there are several points of contacts between TAF4b·TAF12 and the DNA. In a large excess of TAF4b·TAF12 some of these contacts are competed out and the complex becomes less stable.
FIGURE 2.
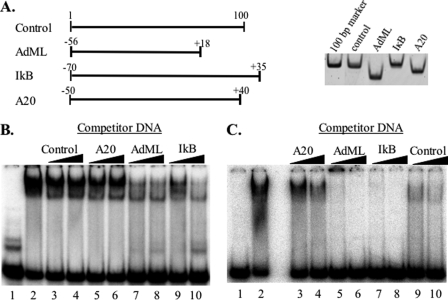
TAF4b·TAF12 displays sequence preference. A, graphic presentation of the different DNA fragments used for binding reactions with the purified TAF4b·TAF12 complex. The right panel shows an ethidium bromide-stained polyacrylamide gel with equal amounts of the DNA fragments. B and C, EMSAs were performed with purified TAF4b·TAF12 and radiolabeled AdML (B) or A20 (C) promoter fragments. Competition was by 2 and 5 m excess of unlabeled AdML, IkB, A20, and control fragments as indicated at the top. Lane 1 is the probe in the absence of protein, and lane 2 is a binding reaction in the absence of competitor.
FIGURE 3.
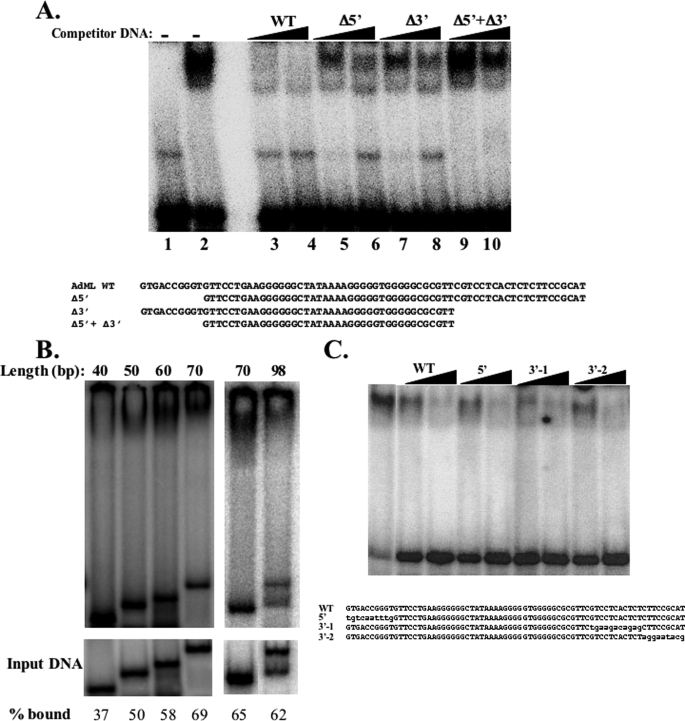
The optimal length of DNA for TAF4b·TAF12 binding is half the size of nucleosomal DNA. A, the TAF4b·TAF12 complex was incubated with a radiolabeled oligonucleotide containing a 70-bp AdML promoter (WT) in the absence (lane 2) or the presence (lanes 3–10) of cold DNA competitors as indicated at the top of the lanes. The sequences of the DNA used as competitors are shown below. B, EMSA experiment as in A using equivalent molar amounts of different length DNAs whose sequences are shown in A. The relative amount of bound DNA is indicated at the bottom. C, competition experiment as in A. Sequences of competitor DNA are shown at the bottom. Mutated nucleotides are in lowercase letters.
We determined the apparent dissociation constant (Kd) of the TAF4b·TAF12·DNA complex, in which the concentration was calculated according to its heterodimer composition, to be ∼30 nm (Fig. 1C), which is within the physiological range.
TAF4b·TAF12 Requires Long DNA for High Affinity DNA Binding and Has Weak Sequence Preference
To examine whether TAF4b·TAF12 binds DNA in a sequence-specific manner we examined the binding of the purified recombinant TAF4b·TAF12 complex to four different DNA fragments of 74- to 105-bp length (Fig. 2A). The first fragment, A20, is the core promoter of a gene regulated by TAF4b (25). Two others are the AdML and the IκB gene core promoters, and the last is a control DNA derived from a plasmid with no promoter sequences. We tested the binding of the TAF4b·TAF12 complex to these four DNA fragments with EMSA. Using labeled AdML DNA we examined the affinity of TAF4b·TAF12 to each of the DNA fragments by competition with an excess of unlabeled fragments (Fig. 2B). The experiment revealed differences in the ability of the fragments to compete with AdML suggesting that TAF4b·TAF12 discriminates between the different sequences. The AdML promoter had the highest affinity to the complex followed by IκB. The results were verified by reciprocal experiments in which either the A20 (Fig. 2C) or the IκB (data not shown) promoters were labeled. Enzymatic and chemical footprinting assays of the AdML promoter failed to reveal a specific DNA sequence bound by the complex indicating that the sequence preference is weak. The observation that the affinity for the A20 core promoter derived from a gene regulated by TAF4b was the lowest is surprising. In activating A20 transcription TAF4b serves as coactivator for NF-κB, an activity that requires direct interaction between TAF4b and NF-κB (25, 26). Thus, it seems that the DNA binding and coactivation functions of TAF4b are independent of each other.
We extended the characterization of TAF4b·TAF12 binding to the AdML promoter with a competition experiment between the AdML core promoter probe and an excess of cold DNAs, either the wild-type AdML core promoter or mutants in which the upstream or downstream segments or both were deleted (Fig. 3A). In this experiment we observed differences between the mutants for competition with the labeled 70-bp AdML promoter. The central 40-bp region lacking both the upstream and the downstream ends was the least effective competitor (compare lanes 3 and 4 to lanes 9 and 10), whereas mutants lacking either the upstream or the downstream region competed more efficiently than the central 40-bp but less than the full-length 70-bp AdML promoter (compare lanes 3 and 4 to lanes 5–8). The results indicate that areas both upstream and downstream to the central 40 bp area are important for binding by TAF4b·TAF12. To gain support for this finding, the full-length 70-bp AdML promoter, the mutants lacking either the upstream (60 bp) or the downstream (50 bp) or both (40 bp) and an extended promoter (98 bp) were each labeled and used for binding to the TAF4b·TAF12 complex by EMSA using equimolar amounts. The results show that the level and the stability of the TAF4b·TAF12·DNA complex increase gradually with the addition of the upstream and downstream sequences (Fig. 3B) up to 70 bp. Beyond this length the binding efficiency is similar. To examine further whether DNA length or the sequence surrounding the central AdML promoter contribute to increased DNA binding, the nucleotide sequence of the upstream and downstream regions were changed in the context of the full-length 70-bp AdML promoter and used for competition assay. As shown in Fig. 3C these substitutions did not reduce the binding affinity of TAF4b·TAF12, suggesting that DNA sequence may be less important than its length. To examine further the length requirement for binding we performed similar binding assays with the PEL98 promoter, which is distinct in its sequence from the AdML promoter (supplemental Fig. 2A). This promoter was chosen because of its dependency on TAF4 DNA binding (see below). With this sequence we observed a clear DNA length preference as was found for the AdML promoter as the affinity to 70 bp > 55 bp > 40 bp. In this promoter context we also examined whether the spacing between the TATA-box and the Initiator is important for high affinity binding. The 20 nucleotides between TATA and Initiator were either relocated to the 5′-end or deleted shortening the DNA to 50 bp (supplemental Fig. 2B). The results revealed that changing the spacing but retaining the length does not significantly affect binding affinity, whereas binding is reduced with the shorter 50-bp fragment. To test further the sequence preference we compared the binding between the PEL98 and the non-target promoter PMM2 and found that the complex binds preferentially the PEL98 DNA (supplemental Fig. 2C). These findings together confirm that high affinity DNA binding by TAF4b·TAF12 is achieved through contacts with DNA spanning 70 bp combined with weak sequence preference. A possible explanation that integrates these findings is that TAF4b·TAF12 DNA binding has several points of contact spanning the length of the DNA, some specific and some non-specific, that all contribute to stable binding.
TAF4 Family Members Bind Similarly to DNA
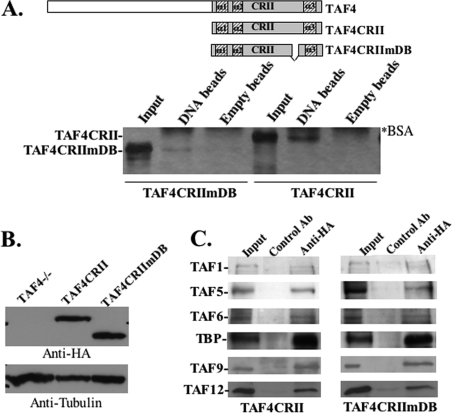
The TAF4b and TAF4 C termini (designated CRII for conserved region II) are highly homologous, and CRII is the most conserved domain between species. CRII mediates interactions with other TAFs (16) and includes within it the DNA-binding activity that is also conserved in human, Drosophila, and yeast TAF4 orthologs (16). To determine whether the DNA-binding properties of a TAF4 family member, described above, are functionally relevant in vivo, we decided to use a genetic approach. A set up that was available to us for this purpose was the TAF4−/− embryonic fibroblasts. Given the high degree of homology between TAF4 and TAF4b in the DNA binding region we considered that the mode by which TAF4 family members bind DNA should be generally similar, reminiscent of the resemblance between transcription factors that share a homologous DNA-binding domain. The CRII of the TAF4 family consists of an atypical H2A-like domain with a unique long spacer between the second and the third α-helices of the histone fold (Fig. 4A). Our previous analysis of hTAF4b and yeast TAF4 indicated that part of the unique spacer domain is necessary (but not sufficient) for their DNA-binding activity (16). To verify the similarity between TAF4 and TAF4b DNA binding we generated a mutation in TAF4 CRII by deleting the same part of the spacer domain that had impaired DNA binding in TAF4b (16). Wild-type and mutant recombinant TAF4 proteins were analyzed for binding to either empty or DNA-containing cellulose beads as previously described (16). The results show that DNA-binding activity of TAF4 is significantly weakened by partial deletion of the spacer region (Fig. 4A) confirming the similarity in DNA-binding characteristics between TAF4 and TAF4b. This finding prompted us to use TAF4 and TAF4−/− cells for functional studies.
FIGURE 4.
TAF4 DNA binding is not required for TAF4-mediated growth suppression. In A: top panel, schematic representation TAF4CRII TAF4CRIImDB relative to the full-length TAF4. The mutation in TAF4CRIImDB corresponds to amino acids 1011–1051 of the spacer domain. Lower panel, TAF4CRII and TAF4CRIImDB were fused to glutathione S-transferase, expressed in E. coli, and analyzed for binding to DNA-cellulose beads (DNA lanes). Binding to empty cellulose beads (Empty beads lanes) served as a control. The input represents 10% of the protein used for binding. 20% of the eluted proteins were analyzed by SDS-PAGE and silver staining. Positions of the protein are marked on the left, and the proteins fused in binding assays are indicated at the bottom. The asterisk indicates the bovine serum albumin that is added to the binding and elution buffers. B, TAF4−/− fibroblasts were transfected with HA-TAF4CRII and HA-TAF4CRIImDB, and an empty expression vector as a control. Stable clones were analyzed by immunoblot using anti-HA and anti-tubulin monoclonal antibodies. C, total cell extracts from TAF4CRII and TAF4CRIImDB cell lines were immunoprecipitated and assayed with non-relevant control and anti-HA antibodies as indicated at the top. The immunoprecipitated complexes were then subjected to immunoblot analysis with antibodies against a subset of TAFs and TBP antibodies as indicated.
TAF4 DNA Binding Is Not Required for the Growth Inhibitory Activity of TAF4
To determine the function of DNA-binding activity by the TAF4 family we examined the effect of a mutation in DNA binding using TAF4-deficient embryonic fibroblasts (27). Previous characterization of these cells established that expression of the CRII domain alone was as effective as the full-length TAF4 in complementing the TAF4 deficiency (27). We constructed plasmids for expression of TAF4 CRII (TAF4CRII) and its DNA-binding mutant derivative (TAF4CRIImDB), each carrying an N-terminal HA tag. These plasmids and a control parental plasmid were transfected into the TAF4−/− embryonic fibroblasts to generate stable clones. The control cells represent a pool of clones carrying the empty vector. An immunoblot with anti-HA antibody shows equivalent amounts of TAF4CRII and TAF4CRIImDB expression in the respective clones (Fig. 4B). The mutation in the spacer domain does not affect the ability of the CRII domain to interact with TAF12, TAF1, and TFIIA in vitro (Ref. 16 and data not shown). To verify the association of TFIID with the DNA-binding mutant of TAF4 we immunoprecipitated TAF4CRII and TAF4CRIImDB from the respective cell lines with the HA antibody and analyzed the immune complexes for the presence of a subset of TFIID subunits. As shown in Fig. 4C TBP and TAFs efficiently co-precipitated with both TAF4CRII and TAF4CRIImDB, confirming that the TAF4 DNA-binding mutant does not affect TFIID integrity.
Morphologically, TAF4−/− cells are elongated, and this elongation was reduced by expression of TAF4 CRII (supplemental Fig. 3A), as previously reported (27). Expression of TAF4CRIImDB had the same effect on cell morphology as TAF4CRII (supplemental Fig. 3A), indicating that loss of TAF4 DNA-binding activity does not influence cell morphology.
TAF4−/− cells grow faster and to a higher density than their wild-type counterpart or than cells expressing full-length TAF4 or TAF4CRII (27). We therefore examined whether DNA binding is important for the growth-repressing effect of TAF4 CRII by comparing the growth rate of TAF4CRII and TAF4CRIImDB to that of TAF4−/− cells. Cells were seeded at low density and counted 4, 6, 8, and 10 days after seeding. The results show that cells expressing TAF4 CRII display a slower growth rate and do not reach high density (supplemental Fig. 3B), as previously shown (27). Interestingly, TAF4CRIImDB displays almost identical growth features as TAF4CRII (supplemental Fig. 3B). Other independent clones of TAF4CRII and TAF4CRIImDB gave the same results (data not shown). Similarly, both TAF4CRII and TAF4CRIImDB fail to grow in low serum unlike the parental TAF4−/− (data not shown). Thus the DNA-binding activity is dispensable for the growth-inhibitory effect of TAF4. The ability of the DNA-binding mutant to preserve most of the known functional features of TAF4CRII confirms that the DNA-binding mutation has no gross effect on TFIID integrity. We also compared the activity of the full-length TAF4 with the CRII in reporter assays and found them to be similar (supplemental Fig. 4).
Identification of Genes Affected by Loss of TAF4 DNA Binding
To assess the impact of TAF4 DNA binding on gene expression, RNA was prepared from exponentially growing TAF4CRII or TAF4CRIImDB clones and used for microarray gene profiling with a gene chip containing more than 23,000 mouse cDNAs. Genes whose expression significantly and reproducibly differed between the TAF4CRII and TAF4CRIImDB clones are those affected by the mutation. The microarray expression analysis indicated that the mutation that diminished TAF4 DNA-binding activity resulted in a down-regulation >1.9-fold of 69 genes and up-regulation of 63 genes (supplemental Table S1).
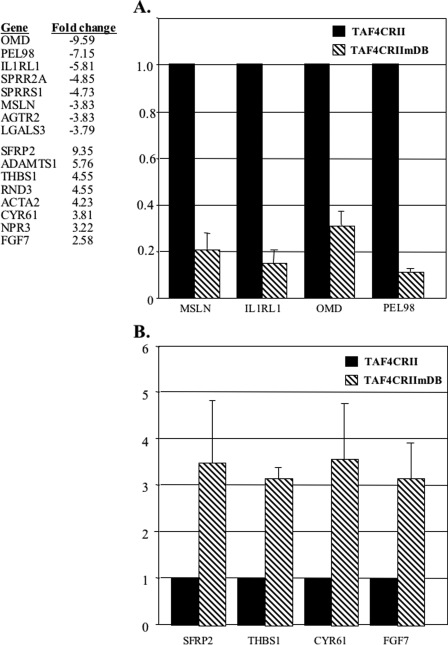
To confirm the microarray results the expression of four down-regulated and four up-regulated genes was also analyzed by reverse transcription real-time PCR in three independent samples. All the selected genes showed the expected down or up-regulation (Fig. 5).
FIGURE 5.
Gene expression differences between TAF4CRII and TAF4CRIImDB cell lines. A list of some of some differentially expressed genes is shown in the left. The full list is shown in supplemental file 2. A and B, changes in down-regulated (A) and up-regulated (B) selected genes are shown. Reverse transcription-real-time PCR was performed on the indicated genes using RNA from the TAF4CRII and TAF4CRIImDB. Results of three independent RNA preparations are shown.
TAF4 DNA Binding Is Required for Core Promoter Function of a Subset of Initiator-containing Genes
Considering that a central role of TFIID is core promoter recognition and binding, it is reasonable to expect that the DNA-binding activity of TAF4 would be linked to these functions. Therefore we set out to examine whether the core promoter of genes affected by the mutation in TAF4 DNA binding have features in common. We used bioinformatics tools to analyze the down and the up-regulated gene sets for the presence of specific DNA elements. The proximal promoter region, from −100 to +50, of genes differentially expressed in the TAF4CRIImDB cells was searched for common sequence elements using two distinct motif-identifying programs (MEME, AlignACE). Although certain motifs were identified, none were shared by most of the genes in either the down- or up-regulated groups (data not shown).
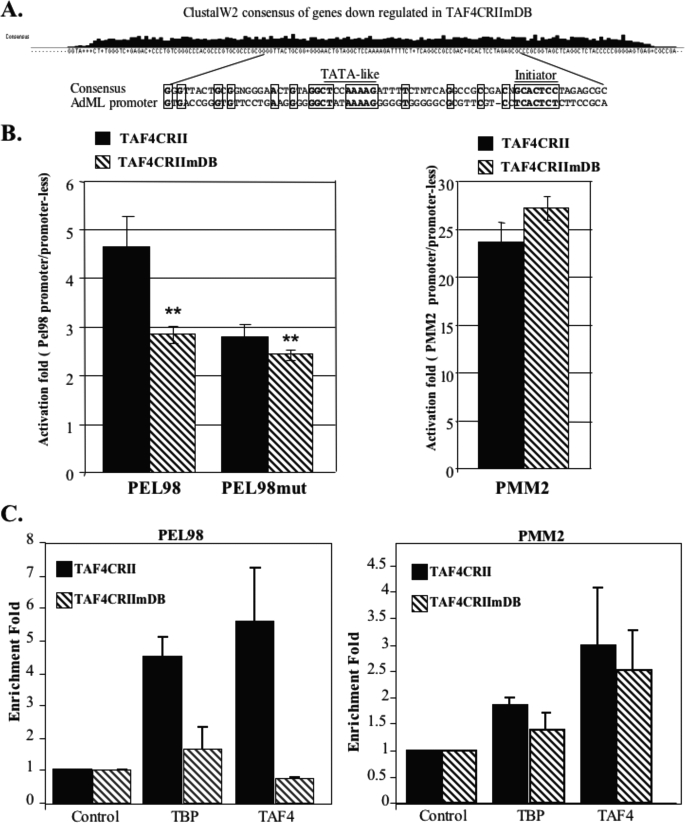
Taking into account the finding that stable DNA binding by the TAF4b·TAF12 complex requires contacts with the DNA over a length of at least 70 bp, we reasoned that it would be more appropriate to search for common features throughout the length of the proximal promoter. For this purpose sequences of the proximal promoter region (from −100 to +50) of the differentially expressed genes were subjected to ClustalW2 analysis, a multiple sequence alignment program, which calculates the best match for the selected sequences, lines them up and determines a consensus. Analysis of the up-regulated gene set by this program did not reveal common motifs or any other interesting features in their proximal promoters (data not shown). However, alignment of the proximal promoters of the down-regulated gene set resulted in a consensus sequence that has a TATA-like element and an Initiator with the expected spacing between them (Fig. 6A). Remarkably, these sequence motifs and additional flanking sequences match the TATA and the Initiator core promoter elements present in the AdML promoter (Fig. 6A, bottom panel), which we had found binds preferentially to TAF4b·TAF12 (see Fig. 2 above).
FIGURE 6.
A, proximal promoter sequences (from −100 to +50) of genes down-regulated in TAF4CRIImDB cells were retrieved from the Database of Transcription Start Sites and analyzed by the ClustalW2 program, which compares the sequences and provides a consensus sequence (top panel). Alignment of the AdML proximal promoter to the consensus sequence derived from the ClustalW2 analysis of the down-regulated genes is shown in the lower panel. Locations of the TATA-like and Initiator sequences are underlined. Homologous sequences are boxed. B, TAF4CRII and TAF4CRIImDB cells were transfected with luciferase reporter plasmids directed by the wild-type or initiator mutant Pel98 promoter. For control, the PMM2 promoter was also transfected. Rous sarcoma virus promoter-driven Renilla reporter plasmids were cotransfected with each plasmid to normalize for transfection efficiency. Luciferase activities were measured 24 h after transfection, and the relative luciferase activity is presented. The data represent the mean ± S.D. of three independent experiments each with independent duplicates. C, TAF4CRII and TAF4CRIImDB cells were subjected to ChIP with antibodies, indicated at the bottom, against TAF4 (anti-HA tag), TBP, and a non-relevant protein as control. The immunoprecipitated chromatin was analyzed by semi-quantitative PCR using primers for pel98 and PMM2 promoters. Quantified results, normalized to the input, were derived from three independent experiments and are presented as enrichment fold relative to the control antibodies.
To confirm these findings, we analyzed each promoter of the down-regulated genes for the presence of a minimal TATA box (TATAWA) and an Initiator (YYANA/TYY) at the expected location relative to the transcription start site, allowing up to two mismatches. The results show that TATA and Initiator, respectively, are present in 53 and 92% of these genes (Table 1 and supplemental file 2). For each of these elements these frequencies are significantly higher (p = 0.007 and 4.6E−10, respectively) than those in promoters in general (also with up to two mismatches) (28, 29). Because genes driven by the Initiator are TAF-dependent (14, 30, 31), we can deduce that the core promoter features of the down-regulated genes are TAF-dependent. These characteristics are specific to the down-regulated set, because enrichment of TATA and Initiator motifs was not found in the up-regulated genes.
TABLE 1.
The difference in distribution of TATA and Initiator core promoter elements in genes down-regulated in TAF4CRIImDB cells and genes in general, according to Moshonov et al. (29) and Gershenzon and Ioshikhes (28)
| TAF4CRIImDB | General (DBTSS)a | p value | |
|---|---|---|---|
| % | χ2 | ||
| TATA | 53 | 35.1 | 0.007 |
| Initiator | 92 | 48.4 | 4.6E-10 |
a DBTSS, Database of Transcription Start Sites.
Considering that 92% of the down-regulated genes share the Initiator core promoter element, we reasoned that if TAF4 DNA binding is important for core promoter function, down-regulation of a gene in TAF4CRIImDB cells should be dependent on the Initiator. To obtain evidence for this idea we analyzed further the PEL98 gene from the down-regulated set. This gene bears a TATA-box and an Initiator of the PyPyANA/TPyPy type at the functional locations. The PEL98 promoter was amplified from mouse genomic DNA by PCR and cloned upstream to a promoter-less luciferase reporter gene. The Initiator element of the promoter was then mutated by nucleotide substitutions. Wild-type and mutated promoters were transfected into the TAF4CRII and TAF4CRIImDB stable cell lines. As a control these cell lines were also transfected with a luciferase reporter driven by the promoter of PMM2, a gene whose mRNA production was not affected by the mutation in TAF4 DNA-binding domain. Consistent with the mRNA analysis, the luciferase activity of the PEL98 promoter was lower in TAF4CRIImDB than TAFCRII cells, whereas the activity of the PMM2 promoter was not significantly changed (Fig. 6B). Interestingly, in the absence of the Initiator element (PEL98mut) the luciferase activity in TAF4CRII cells is down-regulated to the same level as that of the wild-type PEL98 promoter in TAF4CRIImDB cells, and the difference in the activity of the wild-type and mutated promoters between the cell lines is substantially reduced. These findings confirm that TAF4 DNA binding is required for core promoter activity.
To determine whether the effect of TAF4 mutation is direct, we analyzed the occupancy of the PEL98 and PMM2 promoters by TFIID in TAFCRII and TAF4CRIImDB cell lines by chromatin immunoprecipitation assays using antibodies against TAF4 (anti-HA), TBP, and a non-relevant antibody as a control. After reverse cross-linking semi-quantitative PCR reactions were performed with primers corresponding to the core promoter regions of PEL98 and PMM2 genes and with primers for a region located 1000 bp upstream the PEL98 TSS. As shown in Fig. 6C, TBP and TAF4 are highly enriched on the PEL98 promoter in TAFCRII cells, but their occupancy is markedly reduced in TAF4CRIImDB cells, consistent with the down-regulation of PEL98 in these cells. TAF4 and TBP association with the core promoter is specific as no enrichment by these factors was detected 1000 bp upstream of TSS (data not shown). The enrichment of TAF4 and TBP on the PMM2 promoter is less pronounced and the DNA-binding mutation had much less effect on core promoter occupancy. Because the PMM2 promoter lacks TATA and Initiator this reduced enrichment may reflect the fact that neither TBP nor TAF4 are in direct contact with DNA. Together these results support the notion that TAF4 contributes to Initiator core promoter binding and function in a subset of genes.
DISCUSSION
The biochemical and functional analysis of TAF4b/TAF4·TAF12 DNA binding has revealed several interesting features. First, this complex binds DNA with high affinity as a heterodimer. Second, the homology of TAF4/TAF4b·TAF12 to histones H2A-H2B (19), and the observation that the optimal length for DNA binding is 70 bp, which is half the length of nucleosomal DNA (146 bp) are consistent with the notion that the TFIID interaction with DNA through the TAFs resembles in some way a nucleosome (20). Another feature of the TAF4b·TAF12 complex is that it can form a stable complex with DNA at a high DNA:protein ratio suggesting that it requires several contacts with the DNA. On the basis of these properties we propose that the TAF4b·TAF12 complex forms contacts with the DNA that are dispersed over a length of ∼70 bp. This could result in a DNA that wraps the protein complex in an analogous manner to the DNA in the nucleosome. Taking into consideration the weak sequence preference displayed by the complex the binding may occur in two steps: first, formation of specific contacts with the DNA and then additional nonspecific contacts that further stabilize the DNA-protein complex (the order of binding may be reversed). Resolving the mode by which this complex binds DNA awaits additional structural studies.
An intriguing observation from this study is that the binding preference for the AdML promoter revealed in the biochemical analysis matched the core promoter features shared by genes dependent upon TAF4 DNA binding. Almost all of these genes have an Initiator at a functional location and a significantly higher frequency of the TATA box. These findings received strong support from the observation that a mutation in the Initiator element in the PEL98 promoter down-regulated transcription to the same extent as the mutation in TAF4 DNA binding and rendered the promoter less sensitive to TAF4 DNA binding. However, we cannot rule out the possibility that changes observed in expression profiling may be the consequence of another unknown activity of TAF4. We propose that TAF4 DNA binding may facilitate the function of a subset of Initiator or TATA+Initiator-containing promoters, most likely by cooperating with TBP and/or TAF1·TAF2 subunits in their specific interaction with the TATA-box and Initiator by contacting other nucleotides throughout the length of the core promoter. It is also possible TAF4·TAF12 in the TFIID complex serves to compete with nucleosome for binding in the vicinity of the core promoter. Additional studies are required to determine the role of DNA length dependence in transcription.
Supplementary Material
Acknowledgments
We thank Dr. Shira Albeck for the gel-filtration analysis of TAF4b·TAF12, Dr. Diego Jaitin for his assistance with the microarray experiments, and Dr. Mali Slamon for her help in microarray data analysis and Dr. Laszio Tora (CNRS/INSERM/ULP, France) for the TAFs antibodies.
This work was supported by grants from the Israel Science Foundation and the Helen and Martin Kimmel Stem Cell Research Institute.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4 and Table 1.
- TSS
- transcription start site
- TBP
- TATA-binding protein
- TAF
- TBP-associated factor
- HFD
- histone-fold domain
- AdML
- adenovirus major late
- EMSA
- electrophoretic mobility shift assay
- HA
- hemagglutinin
- ChIP
- chromatin immunoprecipitation.
REFERENCES
- 1.Juven-Gershon T., Hsu J. Y., Kadonaga J. T. (2006) Biochem. Soc. Trans. 34, 1047–1050 [DOI] [PubMed] [Google Scholar]
- 2.Smale S. T. (2001) Genes Dev. 15, 2503–2508 [DOI] [PubMed] [Google Scholar]
- 3.Smale S. T., Kadonaga J. T. (2003) Annu. Rev. Biochem. 72, 449–479 [DOI] [PubMed] [Google Scholar]
- 4.Matangkasombut O., Auty R., Buratowski S. (2004) Adv. Protein Chem. 67, 67–92 [DOI] [PubMed] [Google Scholar]
- 5.Sawadogo M., Roeder R. G. (1985) Cell 43, 165–175 [DOI] [PubMed] [Google Scholar]
- 6.Nakatani Y., Horikoshi M., Brenner M., Yamamoto T., Besnard F., Roeder R. G., Freese E. (1990) Nature 348, 86–88 [DOI] [PubMed] [Google Scholar]
- 7.Kaufmann J., Smale S. T. (1994) Genes Dev. 8, 821–829 [DOI] [PubMed] [Google Scholar]
- 8.Oelgeschlager T., Chiang C. M., Roeder R. G. (1996) Nature 382, 735–738 [DOI] [PubMed] [Google Scholar]
- 9.Verrijzer C. P., Tjian R. (1996) Trends Biochem. Sci. 21, 338–342 [PubMed] [Google Scholar]
- 10.Emanuel P. A., Gilmour D. S. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 8449–8453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J. C., Van Dyke M. W. (1993) Biochim. Biophys. Acta 1216, 73–80 [DOI] [PubMed] [Google Scholar]
- 12.Purnell B. A., Emanuel P. A., Gilmour D. S. (1994) Genes Dev. 8, 830–842 [DOI] [PubMed] [Google Scholar]
- 13.Zhou Q., Lieberman P. M., Boyer T. G., Berk A. J. (1992) Genes Dev. 6, 1964–1974 [DOI] [PubMed] [Google Scholar]
- 14.Chalkley G. E., Verrijzer C. P. (1999) EMBO J. 18, 4835–4845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burke T. W., Kadonaga J. T. (1997) Genes Dev. 11, 3020–3031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shao H., Revach M., Moshonov S., Tzuman Y., Gazit K., Albeck S., Unger T., Dikstein R. (2005) Mol. Cell. Biol. 25, 206–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffmann A., Chiang C. M., Oelgeschlager T., Xie X., Burley S. K., Nakatani Y., Roeder R. G. (1996) Nature 380, 356–359 [DOI] [PubMed] [Google Scholar]
- 18.Selleck W., Howley R., Fang Q., Podolny V., Fried M. G., Buratowski S., Tan S. (2001) Nat. Struct. Biol. 8, 695–700 [DOI] [PubMed] [Google Scholar]
- 19.Werten S., Mitschler A., Romier C., Gangloff Y. G., Thuault S., Davidson I., Moras D. (2002) J. Biol. Chem. 277, 45502–45509 [DOI] [PubMed] [Google Scholar]
- 20.Xie X., Kokubo T., Cohen S. L., Mirza U. A., Hoffmann A., Chait B. T., Roeder R. G., Nakatani Y., Burley S. K. (1996) Nature 380, 316–322 [DOI] [PubMed] [Google Scholar]
- 21.Hoffmann A., Oelgeschlager T., Roeder R. G. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 8928–8935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smyth G. K. (2004) Stat. Appl. Genet. Mol. Biol. 3, Article3 [DOI] [PubMed] [Google Scholar]
- 23.Ainbinder E., Amir-Zilberstein L., Yamaguchi Y., Handa H., Dikstein R. (2004) Mol. Cell. Biol. 24, 2444–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharon M., Robinson C. V. (2007) Annu. Rev. Biochem. 76, 167–193 [DOI] [PubMed] [Google Scholar]
- 25.Yamit-Hezi A., Nir S., Wolstein O., Dikstein R. (2000) J. Biol. Chem. 275, 18180–18187 [DOI] [PubMed] [Google Scholar]
- 26.Silkov A., Wolstein O., Shachar I., Dikstein R. (2002) J. Biol. Chem. 277, 17821–17829 [DOI] [PubMed] [Google Scholar]
- 27.Mengus G., Fadloun A., Kobi D., Thibault C., Perletti L., Michel I., Davidson I. (2005) EMBO J. 24, 2753–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gershenzon N. I., Ioshikhes I. P. (2005) Bioinformatics 21, 1295–1300 [DOI] [PubMed] [Google Scholar]
- 29.Moshonov S., Elfakess R., Golan-Mashiach M., Sinvani H., Dikstein R. (2008) BMC Genomics 9, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amir-Zilberstein L., Ainbinder E., Toube L., Yamaguchi Y., Handa H., Dikstein R. (2007) Mol. Cell. Biol. 27, 5246–5259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Basehoar A. D., Zanton S. J., Pugh B. F. (2004) Cell 116, 699–709 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.