Abstract
In Saccharomyces cerevisiae, the Rho-type GTPase Cdc42 regulates polarized growth through its effectors, including the p21-activated kinases (PAKs) Ste20, Cla4, and Skm1. Previously, we demonstrated that Ste20 interacts with several proteins involved in sterol synthesis that are crucial for cell polarization. Under anaerobic conditions, sterols cannot be synthesized and need to be imported into cells. Here, we show that Ste20, Cla4, and Skm1 form a complex with Sut1, a transcriptional regulator that promotes sterol uptake. All three PAKs can translocate into the nucleus and down-regulate the expression of genes involved in sterol uptake, including the Sut1 targets AUS1 and DAN1 by a novel mechanism. Consistently, deletion of either STE20, CLA4, or SKM1 results in an increased sterol influx and PAK overexpression inhibits sterol uptake. For Ste20, we demonstrate that the down-regulation of gene expression requires nuclear localization and kinase activity of Ste20. Furthermore, the Ste20-mediated control of expression of sterol uptake genes depends on SUT1 but is independent of a mitogen-activated protein kinase signaling cascade. Together, these observations suggest that PAKs translocate into the nucleus, where they modulate expression of sterol uptake genes via Sut1, thereby controlling sterol homeostasis.
INTRODUCTION
The small Rho GTPase Cdc42 plays a central role in the regulation of cellular polarity in eukaryotic cells (Etienne-Manneville, 2004; Jaffe and Hall, 2005). In the budding yeast Saccharomyces cerevisiae, Cdc42 regulates different types of polarized growth during various phases of its life cycle, including budding during vegetative growth, mating between haploid cells of opposite mating types, and filamentous growth upon nutrient limitation (Park and Bi, 2007). Cdc42 promotes polarized growth through multiple pathways, including polarization of the actin cytoskeleton and directed vesicle trafficking. Among the Cdc42 effectors that trigger these pathways are Ste20, Cla4 and Skm1, members of the p21-activated kinase (PAK) family of serine/threonine protein kinases (Hofmann et al., 2004; Park and Bi, 2007).
Cdc42 recruits Ste20 and Cla4 from the cytoplasm and activates them at the plasma membrane at sites of polarized growth. Therefore, Ste20 and Cla4 are enriched at tips of buds and mating projections (Peter et al., 1996; Leberer et al., 1997; Holly and Blumer, 1999). The recruitment of Ste20 and Cla4 to these sites is governed by protein–protein as well as protein–lipid interactions. PAKs carry a conserved Cdc42/Rac-interactive binding (CRIB) domain that mediates binding to Cdc42 and regulates their activity (Cvrckova et al., 1995; Peter et al., 1996; Leberer et al., 1997; Lamson et al., 2002; Ash et al., 2003). Ste20 and Cla4 also bind to Bem1, a scaffold protein that brings Cdc42, its activator Cdc24 and either Ste20 or Cla4, into proximity (Leeuw et al., 1998; Gulli et al., 2000; Bose et al., 2001; Winters and Pryciak, 2005; Yamaguchi et al., 2007). In addition, phosphoinositide-binding domains promote the association with membrane lipids. Cla4 carries a pleckstrin homology (PH) domain and Ste20 contains a short basic-rich (BR) region domain, both of them binding to membrane lipids.
Ste20 regulates multiple mitogen-activated protein kinase (MAPK) pathways that control mating, filamentous growth, and osmotic stress response, and it is also involved in exit from mitosis and hydrogen peroxide-induced apoptosis (Leberer et al., 1992; Ramer and Davis, 1993; Liu et al., 1993; Roberts and Fink, 1994; O'Rourke and Herskowitz, 1998; Raitt et al., 2000; Höfken and Schiebel, 2002; Ahn et al., 2005). Probably the best characterized function of Cla4 is the assembly of the septin ring, which plays a fundamental role in cytokinesis and cell compartmentalization (Weiss et al., 2000; Schmidt et al., 2003; Kadota et al., 2004; Versele and Thorner, 2004). In addition, Cla4 regulates mitotic entry and exit (Höfken and Schiebel, 2002; Seshan et al., 2002; Sakchaisri et al., 2004). Very little is known about Skm1, and no clear function has been attributed to this PAK (Martín et al., 1997). Interestingly, Ste20 and Cla4 interact with Erg4, Cbr1 and Ncp1, which are all involved in sterol biosynthesis, and the deletion of the corresponding genes results in various polarity defects, suggesting that sterol biosynthesis is crucial for cell polarization (Ni and Snyder, 2001; Tiedje et al., 2007).
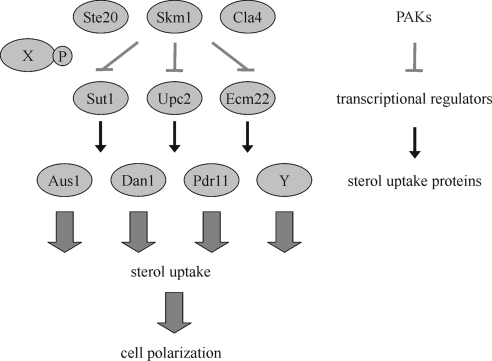
Many aspects of sterol homeostasis are conserved between yeast and human, and ergosterol, the predominant sterol of yeast, is structurally and functionally related to sterols of higher eukaryotes (Sturley, 2000). Ergosterol biosynthesis can occur only when oxygen is available. Although sterol synthesis is an energy-consuming process, cells do not take up significant amounts of exogenous sterol under aerobiosis. The physiological significance of this phenomenon, termed aerobic sterol exclusion, is poorly understood (Lewis et al., 1985). It may be a way for cells to ensure that only the best fitting sterols accumulate in its membranes (Parks and Casey, 1995). In contrast, under anaerobic conditions, when sterol biosynthesis is compromised, cells become capable of importing sterols whose presence in the medium is then required for growth. Because completely anaerobic conditions are difficult to maintain, most studies used sterol auxotrophs or mutants in heme synthesis. Heme acts as an intermediary in regulating the expression of oxygen-responsive genes. Therefore, deficiency in heme biosynthesis, e.g., in a hem1Δ background, mimics anaerobic conditions in the presence of oxygen, and as a consequence these cells accumulate sterol from the medium (Gollub et al., 1977). The molecular mechanisms of sterol import are poorly understood. Sterol uptake is controlled by the transcriptional regulators Sut1, Upc2, and Ecm22, which are members of the Zn(II)2Cys6 family of DNA-binding proteins (Schjerling and Holmberg, 1996). Under anaerobic conditions, Upc2 and Sut1 up-regulate the expression of the ATP-binding cassette transporter AUS1 and PDR11, and the putative cell wall mannoprotein DAN1 (Figure 1) (Régnacq et al., 2001; Wilcox et al., 2002; Alimardani et al., 2004). These proteins then mediate sterol influx. Presumably, other proteins contribute to this process as well (Wilcox et al., 2002; Alimardani et al., 2004).
Figure 1.
Transcriptional regulation of genes mediating sterol uptake. In the presence of oxygen, yeast cells synthesize sterols and are unable to take up sterols from the extracellular medium. In contrast, under anaerobiosis cells rely on sterol uptake, because sterol synthesis requires oxygen. Under these conditions, the transcriptional regulators Sut1, Upc2, and Ecm22 promote the expression of AUS1, DAN1, and PDR11 and less well characterized genes (Y), which mediate sterol import. The underlying mechanisms of this transcriptional regulation are largely unknown. Here, we show that the PAKs Ste20, Cla4, and Skm1 can translocate into the nucleus, where they bind to the transcriptional regulator Sut1. The fact that the PAKs negatively regulate expression of AUS1, DAN1, and PDR11, suggests that PAKs inhibit Sut1 and/or Upc2 and Ecm22 by a novel mechanism. As demonstrated for Ste20, the down-regulation of gene expression requires its kinase activity. However, since there is no evidence for Sut1 phosphorylation by PAKs, an unknown protein (X), phosphorylated by PAKs, may be involved in the control of transcriptional regulators. Because sterol synthesis is crucial for cell polarization under aerobic conditions, it seems likely that sterol uptake plays an equally important role in the absence of oxygen.
Here, we characterize the link between proteins regulating sterol uptake and the PAKs Ste20, Cla4 and Skm1. Our data suggest a model (Figure 1), according to which PAKs shuttle into the nucleus, where they interact with Sut1 and possibly other transcriptional regulators and down-regulate the expression of AUS1, DAN1, and PDR11. As a consequence, sterol influx is also negatively regulated. Thus, PAKs seem to play an important role in the control of sterol uptake.
MATERIALS AND METHODS
Yeast Strains, Plasmids, and Growth Conditions
All yeast strains are listed in Supplemental Table S1. The strains used in this study were in the YPH499 background with the exception of strains used for filamentous growth and sterol uptake. For filamentous growth, the Σ1278b background was used (MLY48, MLY49, PPY966, and THY696). Cholesterol uptake experiments were performed with the BY4742 background (Winzeler et al., 1999) (MLY180, MLY181, MLY185, THY725, THY734, THY737, YRS1707, and YRS1962). Yeast strains were grown in yeast extract, peptone, dextrose (YPD) or synthetic complete (SC) medium. For induction of the GAL1 promoter, yeast cells were grown in yeast extract, peptone (YP) or SC medium with 3% raffinose instead of glucose. Galactose (final concentration 2%) was added to induce the GAL1 promoter. hem1Δ cells were grown in medium supplemented either with 40 μg/ml δ-aminolevulinic acid or with 80 μg/ml ergosterol solubilized in Tergitol NP-40/ethanol (1:1) and 1% Tween 80. Yeast strains were constructed using polymerase chain reaction (PCR)-amplified cassettes (Longtine et al., 1998; Knop et al., 1999; Janke et al., 2004). All constructs used in this work are listed in Supplemental Table S2.
Split-Ubiquitin Technique
The split-ubiquitin screen using STE20 as bait is described in Tiedje et al. (2007). For the interaction assays, 105 ste20Δ cells carrying the split-ubiquitin plasmids were spotted on SC-His/Leu and SC-His/Leu/Ura plates and were grown for 2 d at 30°C.
Pull-Down Assays and Antibodies
Glutathione transferase (GST), GST-Sut1, His6-Sec6, and His6-Sut1 were expressed in Escherichia coli BL21 (DE3) and purified using glutathione-Sepharose (GE Healthcare, Chalfont St. Giles, Buckinghamshire, United Kingdom) and nickel-nitrilotriacetic acid (Ni-NTA) agarose (QIAGEN, Valencia, CA), respectively. These immobilized recombinant proteins were presented to yeast lysates of 3HA-STE20, 3HA-CLA4, and 3HA-SKM1, respectively, for 90 min at 4°C in immunoprecipitation (IP) buffer (20 mM Tris, pH 7.5, 100 mM NaCl, 10 mM EDTA, 1 mM EGTA, 5% glycerol, 1% NP-40, and 1% bovine serum albumin). After five washes with IP buffer, the associated proteins were eluted with sample buffer and analyzed by immunoblotting.
Monoclonal mouse anti-hemagglutinin (HA) (12CA5) was obtained from Roche Diagnostics (Indianapolis, IN). Rabbit polyclonal anti-Cdc11 and goat polyclonal anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA), and rabbit polyclonal anti-green fluorescent protein (GFP) was from Fitzgerald Industries (Acton, MA). Secondary antibodies were from Jackson ImmunoResearch Laboratories (West Grove, PA).
Microscopy
For fluorescence microscopy, cells were fixed with 4% formaldehyde for 10 min, washed twice with phosphate-buffered saline (PBS), and resuspended in water or in 0.25 μg/ml 4,6-diamidino-2-phenylindole (DAPI) in 50% glycerol. Cells were examined with an Axiovert 200M fluorescence microscope (Carl Zeiss, Jena, Germany) equipped with a 100× Plan oil-immersion objective, and images were captured using an AxioCam MRm charge-coupled device camera (Carl Zeiss).
β-Galactosidase Assays
Densities of cell cultures were measured by OD600. One to 10 ml of cells was harvested by centrifugation and resuspended in 1 ml of Z buffer (100 mM sodium phosphate, pH 7.0, 10 mM KCl, 1 mM MgSO4, and 50 mM β-mercaptoethanol). Cells were permeabilized by addition of 20 μl of chloroform and 20 μl of 0.1% SDS. After 15-min incubation at 30°C, the reaction was started by addition of 160 μl of o-nitrophenyl-β-d-galactopyranoside (4 mg/ml in 100 mM sodium phosphate, pH 7.0) and incubated at 30°C until the solution became yellow, and then the reaction was stopped by addition of 400 μl of 1 M Na2CO3. Samples were centrifuged, and the OD420 and OD550 of the supernatant was determined. β-Galactosidase activity was calculated in Miller units as 1000 × [OD420 − (1.75 × OD550)]/reaction time (min) × culture volume (ml) × OD600.
Invasive Growth and Pheromone Response Assays
For agar invasion assays, 105 cells of an overnight culture were spotted on YPD and grown for 2 d at 30°C. Plates were photographed before and after being rinsed under a gentle stream of deionized water.
For examination of the formation of a mating projection, logarithmically growing cells were incubated with 1 μg/ml α-factor for 3 h. These cells were fixed with formaldehyde for morphological examination.
Sterol Uptake
Sterol uptake was essentially analyzed as described previously (Reiner et al., 2006). Hem1Δ mutant cells were cultured in δ-aminolevulinic acid-containing media, washed, and diluted in minimal media supplemented with Tween 80 (5 mg/ml), cholesterol (20 μg/ml), and 0.025 μCi/ml [14C]cholesterol (American Radiolabeled Chemicals, St. Louis, MO). After 2, 4, 8, and 24 h, equal OD units of cells were collected and washed with 0.5% Tergitol. [3H]Palmitic acid was added to the cell pellet as internal standard. Cells were disrupted with glass beads in the presence of chloroform/methanol (1:1), and lipids were extracted into the organic phase. Lipids were separated by thin layer chromatography (TLC) (Merck, Darmstadt, Germany), with the solvent system petroleum ether:diethylether:acetic acid (70:30:2, per vol.), and free cholesterol and cholesteryl esters were quantified by scanning with a Tracemaster 40 Automatic TLC-Linear Analyzer (Berthold Technologies, Bad Wildbad, Germany). TLC plates were then exposed to a phosphorimager screen and visualized using a phosphorimager (Bio-Rad Laboratories, Hercules, CA).
RESULTS
PAKs Interact with the Transcriptional Regulator Sut1
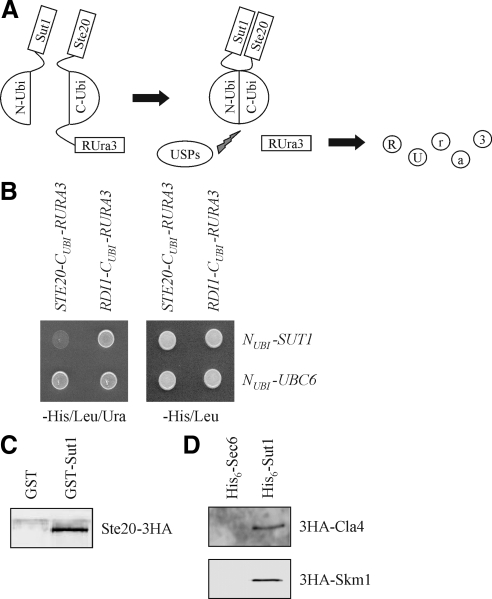
Previously, we described a screen to identify novel interactors of Ste20 by using the split-ubiquitin technique (Tiedje et al., 2007). This method is based on the ability of N- and C-terminal halves of ubiquitin to assemble into a quasi-native ubiquitin (Johnsson and Varshavsky, 1994; Wittke et al., 1999). If two proteins, which are attached to the N- and C-terminal halves, respectively, interact, the ubiquitin peptides may be forced into proximity and a ubiquitin-like molecule is reconstituted (Figure 2A). Ubiquitin-specific proteases recognize this molecule, but not its halves, and cleave off the enzyme Ura3, that carries an additional arginine at the extreme N terminus (RUra3). The freed RUra3 reporter is rapidly degraded by proteases of the N-end rule, resulting in uracil auxotrophy. Conversely, growth on 5-fluoroorotic acid (5-FOA) indicates protein interaction as well, because 5-FOA is converted by Ura3 into 5-fluorouracil, which is toxic for the cell. By using this technique, we identified Sut1 as a putative Ste20-interacting protein (Figure 2B). SUT1 encodes a transcriptional regulator involved in sterol uptake under anaerobic conditions (Bourot and Karst, 1995; Ness et al., 2001). To confirm the interaction between Ste20 and Sut1 by an independent approach, recombinant GST and GST-Sut1 purified from E. coli were bound to glutathione-Sepharose beads, which were then incubated with a yeast extract of STE20-3HA cells. Ste20-3HA interacted with GST-Sut1 but not with GST alone (Figure 2C). Next, it was tested whether Cla4 and Skm1, which are related to Ste20, also interact with Sut1 using a pull-down assay. 3HA-Cla4 and 3HA-Skm1, both expressed from yeast, bind specifically to recombinant His6-Sut1 but not to the unrelated His6-Sec6, which was used as a negative control (Figure 2D).
Figure 2.
PAKs form a complex with Sut1. (A) The split-ubiquitin technique. See text for details. USPs, ubiquitin-specific proteases. (B) Ste20 interacts with Sut1 using the split-ubiquitin system. Cells (105) of the indicated plasmid combinations in a ste20Δ background were spotted onto media either lacking histidine and leucine to select for the plasmids or lacking histidine, leucine, and uracil to monitor protein interactions. The unrelated proteins Rdi1 and Ubc6 served as negative controls. (C) Ste20 interacts with Sut1 in vitro. Purified GST and GST-Sut1 bound to glutathione-Sepharose beads were incubated with an extract from cells expressing STE20-3HA from the plasmid pKA86. Eluted proteins were analyzed by immunoblotting using anti-HA antibodies. (D) Cla4 and Skm1 bind to Sut1. Recombinant His6-Sut1 and His6-Sec6 bound to Ni-NTA beads were incubated with lysates from cells expressing 3HA-CLA4 and 3HA-SKM1, respectively. Eluted proteins were analyzed by immunoblotting using anti-HA antibodies.
Nuclear Localization of PAKs
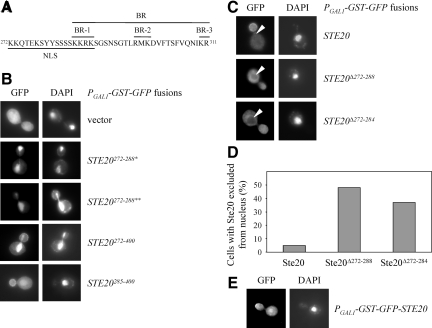
Sut1 localizes to the nucleus (Ness et al., 2001), whereas Ste20 can be found in the cytosol and at the plasma membrane at sites of polarized growth (Peter et al., 1996; Leberer et al., 1997) (Figure 3B). Therefore, it is not clear where the interaction between Sut1 and Ste20 takes place. Analysis of the Ste20 sequence revealed a putative bipartite NLS between amino acids 272-288 with two clusters of positively charged amino acids separated by a spacer of 11 amino acids (Figure 3A). Notably, this putative nuclear localization signal (NLS) overlaps with the BR domain, which targets Ste20 to the plasma membrane (Takahashi and Pryciak, 2007). In fact, the first of three basic clusters within the BR domain (BR-1) is identical to the second cluster of the NLS (Figure 3A). To test whether the identified NLS is functional, it was fused with green fluorescent protein (GFP) and expressed in yeast cells. This GFP fusion also contained a homodimerizing GST moiety, which can help to reveal weak localization determinants by increasing binding avidity (Winters et al., 2005). However, the GFP-NLS fusion protein was found in the cytoplasm (data not shown). We reasoned that the NLS might not be recognized by the nuclear import machinery, possibly due to steric hindrance. Therefore, we added a short stretch of amino acids (SGAGAGAGAGAIL), which is used as a spacer for GFP fused to a protein of interest (Miller and Lindow, 1997) at the C terminus of the NLS. This protein (termed GFP-Ste20272-288*) exclusively localized to the nucleus as demonstrated by DAPI costaining (Figure 3B). To rule out the possibility that the attached spacer sequence was responsible for the nuclear localization, another unrelated random sequence (LGYFLFFEGGPGTQFAL) was C-terminally fused to the NLS. This fusion protein (termed GFP-Ste20272-288**) was restricted to the nucleus as well (Figure 3B). From these data we conclude that the NLS is functional but requires a short nonspecific stretch of amino acids at the C terminus.
Figure 3.
Ste20 localizes to the nucleus. (A) Sequence of the NLS and BR domain. The NLS is composed of two clusters of basic residues, whereas the BR domain consists of three clusters (BR-1, -2, and -3) (Takahashi and Pryciak, 2007). The second cluster of the NLS is identical to BR-1. (B) The Ste20 NLS is functional. Exponentially growing wild-type cells carrying the indicated Ste20 fragments fused to GST-GFP were incubated with galactose for 1 h. Cells were subsequently fixed with formaldehyde and stained with DAPI. One asterisk denotes the fusion with the peptide SGAGAGAGAGAIL. Two asterisks indicate addition of the sequence LGYFLFFEGGPGTQFAL. (C) Ste20 lacking the NLS is excluded from the nucleus. Logarithmically growing wild-type strains carrying the indicated GFP-STE20 alleles under control of a galactose-inducible promoter on a centromeric plasmid were cultivated in galactose-containing media for 1 h. Arrowheads indicate the nucleus. (D) Quantification of C. n > 100. (E) In few cells, Ste20 was enriched in the nucleus. Exponentially growing wild-type cells harboring GFP-Ste20 under control of a galactose-inducible promoter on a centromeric plasmid were incubated with galactose for 1 h.
As mentioned above, the NLS overlaps with the BR domain (Figure 3A) (Takahashi and Pryciak, 2007). To find out whether the BR domain alone is sufficient for nuclear localization, we compared the localization of GFP fused with Ste20 residues 272-400 (comprising the entire NLS and BR domain plus some C-terminal amino acids) and Ste20 residues 285-400 (comprising only the second cluster of positively charged amino acids of the NLS, the complete BR domain and some C-terminal amino acids). Notably, GFP-Ste20272-400 was strongly enriched in the nucleus and present in the cytoplasm and at the plasma membrane, whereas GFP-Ste20285-400 was associated with the bud cortex and only a weak nuclear signal was observed (Figure 3B). Thus, the entire NLS is required for efficient nuclear targeting of Ste20, whereas the BR domain alone, which includes the second basic cluster of the NLS, plays only a minor role in this process. Importantly, in contrast to wild-type Ste20 fused to GFP, GFP-Ste20 without the complete NLS (GFP-Ste20Δ272-288) was excluded from the nucleus (Figure 3, C and D). Similarly, much of GFP-Ste20 lacking the first basic cluster but with an intact BR domain (GFP-Ste20Δ272-284) was no longer present in the nucleus (Figure 3, C and D). However, the effect was less pronounced compared with GFP-Ste20Δ272-288 (Figure 3D). Together, these data show that the BR-1 region contributes only to a minor extent to nuclear targeting of Ste20. Furthermore, our observations suggest that during normal vegetative growth at least some Ste20 localizes to the nucleus. Consistently, in few cells we observed nuclear localization of wild-type Ste20 (Figure 3E). However, since Ste20 was only weakly enriched in the nucleus compared with the surrounding cytoplasm, the number of cells with a nuclear Ste20 signal was difficult to quantify.
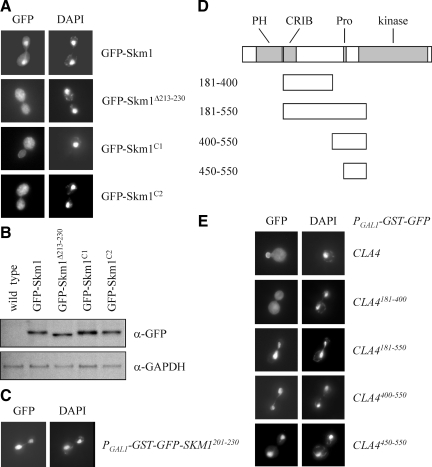
Very little is known about Skm1, and its localization has not been reported previously (Martín et al., 1997). Therefore, we first analyzed the localization of full-length Skm1 fused to GFP. SKM1-GFP under control of its endogenous promoter could be detected by immunoblotting but was to weak to be visualized by fluorescence microscopy (data not shown). Therefore, we overexpressed GFP-SKM1 from the inducible GAL1 promoter. Unexpectedly, full-length Skm1 was strongly enriched in the nucleus with only a faint signal in the cytoplasm and at the plasma membrane (Figure 4A). Between amino acids 213-230 we identified a putative bipartite NLS (KRTNSIKRSVSRTLRKGK; positively charged amino acids of the two basic clusters are in italics). Skm1 lacking this putative NLS was no longer enriched in the nucleus (Figure 4A). We also made point mutations in either the first or the second cluster of positively charged amino acids. The mutant protein Skm1C1, in which Lys213 and Arg214 of the first basic cluster were changed to Ala, mostly localized to the cytoplasm (Figure 4A). In a second mutant, Arg227, Lys228, and Lys230 of the second positively charged cluster were changed to Ala. The corresponding protein, termed SkmC2, was not or only faintly enriched in the nucleus (Figure 4A). By immunoblotting it was confirmed that Skm1 wild-type and mutant proteins were expressed at comparable levels (Figure 4B). Thus, both clusters play an important role in nuclear targeting of Skm1. We also fused Skm1 residues 213-230 to GFP. This fusion protein was present in the cytoplasm (data not shown), but a slightly larger Skm1 fragment (residues 201-230) exclusively localized to the nucleus (Figure 4C). Together, it seems that, similar to Ste20, the predicted Skm1 NLS is functional but requires a few more amino acids to target GFP to the nucleus.
Figure 4.
Nuclear localization of Skm1 and Cla4. (A) Skm1 is strongly enriched in the nucleus. Exponentially growing cells carrying the indicated GFP-SKM1 alleles were induced for 1 h by the addition of galactose. Cells were fixed with formaldehyde and stained with DAPI. (B) Wild-type SKM1 and its derivatives are expressed at comparable levels. The indicated strains were incubated with galactose for 1 h. Cells were lysed and equal amounts of protein extract were analyzed by immunoblotting using antibodies against GFP and GAPDH (loading control). (C) Residues 201-230 are sufficient for nuclear targeting of Skm1. The Skm1 fragment was expressed as galactose-inducible GST-GFP fusion. (D) Fragments used to map the sequence that targets Cla4 to the nucleus. The kinase domain and regions that are required for membrane association are highlighted. The CRIB domain is the minimal Cdc42-binding motif (Cvrckova et al., 1995), a proline-rich region (Pro) mediates binding to the scaffold protein Bem1 (Winters and Pryciak, 2005), and the PH domain is required for the association with membrane phosphoinositides (Wild et al., 2004). (E) Cla4 fragments localize to the nucleus. Cla4 fragments were expressed as GST-GFP fusions from a galactose-inducible promoter in wild-type cells.
Like Ste20, Cla4 associates with the bud cortex (Holly and Blumer, 1999) (Figure 4E) and analysis of the Cla4 protein sequence did not reveal an NLS. To determine whether Cla4 could translocate into the nucleus, a series of Cla4 fragments fused to GFP were constructed (Figure 4D). In line with a previous report (Wild et al., 2004), Cla4 fragments lacking the N-terminal 180 residues, which contain a PH domain that binds to membrane lipids, did not associate with the plasma membrane (Figure 4E). A fragment containing residues 181-550 exclusively localized to the nucleus, whereas a smaller portion of Cla4 (residues 181-400) was cytoplasmic (Figure 4E). Therefore, we reasoned that a nuclear targeting sequence may be present between residues 400-550. Indeed, a fragment comprising this sequence and also a slightly smaller fragment (amino acids 450-550) were only found in the nucleus (Figure 4E). Further truncations resulted in nuclear enrichment but in these cells we also observed cytoplasmic staining (data not shown). Thus, residues 450-550 are sufficient for proper nuclear targeting of Cla4. Together, we demonstrated that all three PAKs can localize to the nucleus, where they could interact with the transcriptional regulator Sut1.
PAKs Specifically Down-Regulate the Expression of AUS1, DAN1, and PDR11
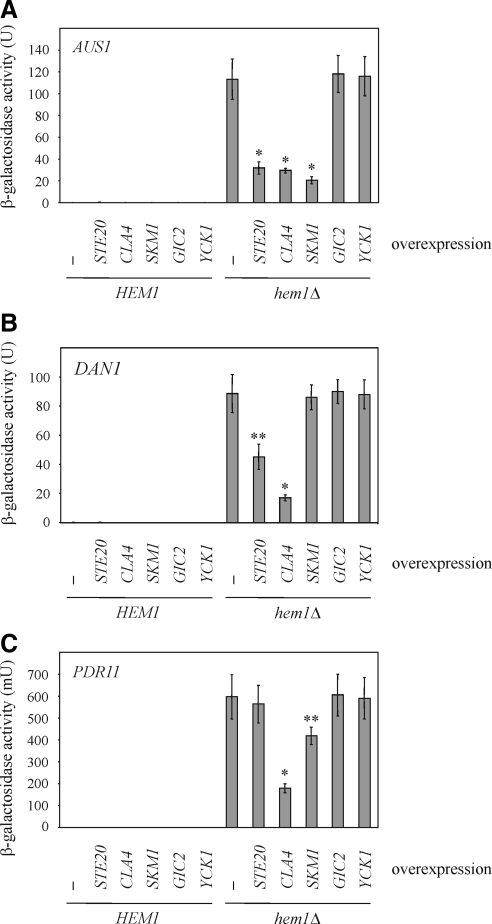
Sut1 promotes sterol uptake under anaerobic conditions by enhancing the transcription of AUS1 and DAN1 (Régnacq et al., 2001; Alimardani et al., 2004). Therefore, we wanted to know whether the expression of these genes also depends on Ste20, Cla4, and Skm1. To this end, the promoter regions of AUS1, DAN1, and PDR11, were fused to the reporter gene lacZ. PDR11 was included because it also mediates sterol influx but is not regulated by Sut1 (Wilcox et al., 2002; Alimardani et al., 2004). Using these constructs, we confirmed that AUS1, DAN1, and PDR11 are not expressed in wild-type cells under aerobic conditions (Figure 4, A–C). In contrast, in a hem1Δ background mimicking anaerobic conditions these genes were strongly induced (Figure 4, A–C). Furthermore, SUT1 overexpression under aerobic conditions led to expression of AUS1 and DAN1 but not of PDR11 (Supplemental Figure S1). This is in line with previous reports (Régnacq et al., 2001; Alimardani et al., 2004) and indicates that AUS1 and DAN1 are both target genes of Sut1. Interestingly, AUS1 expression levels were markedly reduced in hem1Δ cells overexpressing either STE20, CLA4, or SKM1 (Figure 5A), whereas high levels of either GIC2, which, like the PAKs, is a Cdc42 effector (Brown et al., 1997; Chen et al., 1997), or YCK1, a kinase that is involved in cell polarity (Robinson et al., 1993), did not affect AUS1 expression (Figure 5A). Thus, the effects of PAKs are specific. Overexpression of STE20 and CLA4, respectively, but not of SKM1 lowered DAN1 levels (Figure 5B). Finally, high levels of CLA4 and SKM1, but not of STE20 decreased PDR11 expression (Figure 5C). In summary, all three PAKs down-regulate the expression of genes involved in sterol uptake, but with very different specificities. High levels of GIC2 and YCK1 did not affect DAN1 and PDR11 expression levels (Figure 5, B and C).
Figure 5.
PAKs down-regulates the expression of DAN1, AUS1, and PDR11. (A) AUS1 expression is regulated by Ste20, Cla4, and Skm1. The indicated strains harboring a plasmid on which the lacZ was fused to the promoter region of AUS1 were grown in selective raffinose medium in the presence of ergosterol and Tween 80. The indicated genes were overexpressed for 18 h by the addition of galactose. Shown is the mean of β-galactosidase activity of at least six independent experiments with SD, *p < 0.0001, compared with hem1Δ cells without any overexpression. (B) Ste20 and Cla4 control the expression of DAN1. The indicated strains carrying a DAN1-lacZ plasmid were treated as described in A. Shown is the mean of β-galactosidase activity of at least six independent experiments. *p < 0.0001 and **p < 0.001, compared with hem1Δ cells without any overexpression. (C) PDR11 expression is negatively regulated by Cla4 and Skm1. β-Galactosidase activity of the indicated strains harboring a PDR11-lacZ plasmid was determined in at least 6 independent experiments. *p < 0.0001 and **p < 0.005, compared with hem1Δ cells without any overexpression.
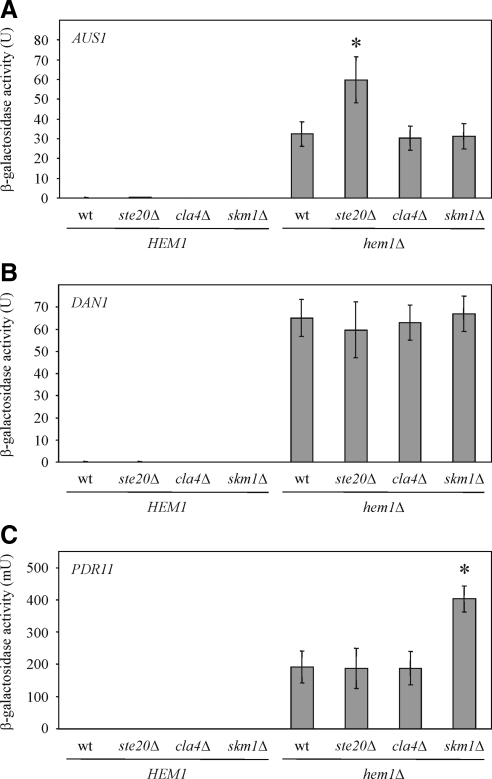
Next, it was tested whether deletion of PAKs has an effect on the expression of AUS1, DAN1 and PDR11. We observed higher AUS1 levels in cells lacking STE20 (Figure 6A), and SKM1 deletion led to increased PDR11 expression (Figure 6C). Notably, these results are consistent with the data obtained following PAK overexpression (Figure 5, A and C).
Figure 6.
Expression of AUS1, DAN1, and PDR11 in PAK deletion strains. (A) STE20 deletion results in higher AUS1 expression. The indicated strains carrying an AUS1-lacZ plasmid were grown in selective medium supplemented with ergosterol and Tween 80, and β-galactosidase activity was determined in at least six independent experiments. wt, wild type; *p < 0.001, compared with hem1Δ cells. (B) PAK deletion does not affect DAN1 levels. Shown is the mean β-galactosidase activity of at least six independent experiments with SD of the indicated strains harboring DAN1-lacZ on a plasmid. (C) Cells lacking SKM1 express higher PDR11 levels. β-Galactosidase activity of the indicated strains harboring a PDR11-lacZ plasmid was determined in at least 6 independent experiments. *p < 0.0005, compared with hem1Δ cells.
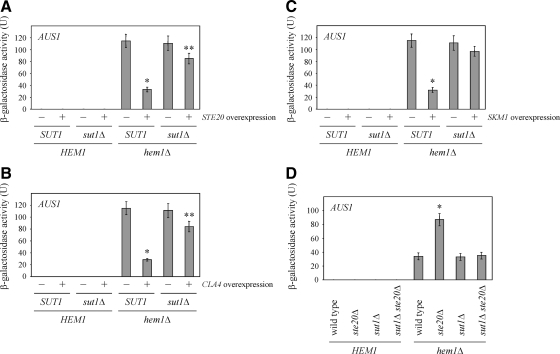
To test whether the observed effects of PAKs on gene expression are mediated by Sut1, expression studies were performed in cells lacking SUT1 using AUS1 as a reporter. The deletion of SUT1 did not affect AUS1 expression levels (Figure 7A), which is not surprising since AUS1 expression is also under control of Upc2 and possibly other transcriptional regulators, which could compensate for the loss of SUT1 (Wilcox et al., 2002). High levels of either STE20, CLA4, or SKM1 in the absence of SUT1 led to no (Figure 7B) or only a minor reduction of AUS1 levels (Figure 7, A and C). Thus, the PAK-mediated down-regulation of AUS1 depends mostly on SUT1. Possibly, PAKs to some extent also regulate Upc2 and maybe other transcriptional regulators of AUS1. This would explain the slight decrease of AUS1 levels observed after overexpression of STE20 and CLA4 in cells lacking SUT1.
Figure 7.
Down-regulation of AUS1 expression by PAKs depends on SUT1. (A) Effect of SUT1 deletion on AUS1 levels following STE20 overexpression. The β-galactosidase activity of the indicated strains was determined in six independent experiments as described in Figure 5A. *p < 0.0001 and **p < 0.05, compared with hem1Δ cells without STE20 overexpression. (B) Regulation of AUS1 expression by Cla4 depends on SUT1. Shown is the mean β-galactosidase activity of six independent experiments with SD of the indicated strains. *p < 0.0001 and **p < 0.05, compared with hem1Δ cells without CLA4 overexpression. (C) The down-regulation of AUS1 expression by Skm1 requires SUT1. β-Galactosidase activity of the indicated strains was determined in at least six independent experiments. *p < 0.0001, compared with hem1Δ cells without SKM1 overexpression. (D) Effect of SUT1 deletion on AUS1 expression in cells lacking STE20. The β-galactosidase activity of the indicated strains was determined in at least six independent experiments as described in Figure 6A. *p < 0.0005, compared with hem1Δ cells.
The role of Sut1 was also examined in cells deleted for STE20. The additional deletion of STE20 in the absence of SUT1 had no effect on AUS1 expression levels (Figure 7D). This is consistent with the notion that specific inhibition of Sut1 by Ste20 results in a down-regulation of AUS1 expression. In ste20Δ cells, Sut1 seems to have a higher activity, which results in stronger AUS1 expression, possibly due to the missing inhibition by Ste20. As mentioned above, the sut1Δ strain probably expresses AUS1 at levels comparable to the wild type because of proteins such as Upc2, which are functionally redundant with Sut1. The fact that AUS1 expression is unaltered in sut1Δ ste20Δ cells suggests that Ste20 negatively regulates for the most part Sut1 and not or only to a minor extent other transcriptional regulators like Upc2. Whereas the experiment in Figure 7D suggests that Sut1 activates AUS1 expression in a manner that is antagonized by Ste20, the experiments in Figure 7, A–C, shows that overexpression of PAKs inhibits AUS1 expression in a manner that requires SUT1. It seems that Sut1 negatively regulates AUS1 expression, a process that is promoted by high PAK levels. Importantly, in both models the effect on AUS1 expression would be the same but it is not clear how these two different modes of action can be reconciled. Unfortunately, it is not known how Sut1 controls the expression of genes such AUS1. Thus, it is even less clear how PAKs could regulate Sut1 activity.
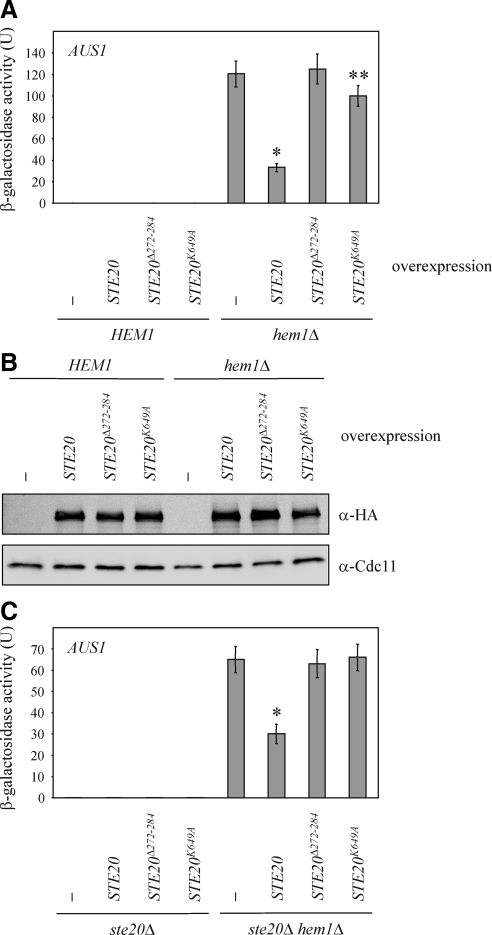
Next, the mechanisms by which PAKs contribute to the regulation of gene expression were further characterized. For these experiments we focused on the regulation of AUS1 by Ste20. Whereas overexpression of wild-type STE20 decreased AUS1 levels, overexpression of a mutated STE20 lacking a major part of the NLS but leaving the BR domain intact (STE20Δ272-284) did not affect AUS1 (Figure 8A). Thus, the nuclear localization of Ste20 is essential for its role in the control of gene expression. High levels of STE20K649A, an allele that lacks kinase activity (Wu et al., 1995), resulted in a minimal decrease of AUS1 expression, but this effect, which is possibly due to minute residual kinase activity, was much weaker than after overexpression of wild-type STE20 (Figure 8A). Thus, Ste20 kinase activity is required for the regulation of AUS1 expression. Notably, wild-type STE20 and the described mutant alleles were expressed at comparable levels as confirmed by immunoblotting (Figure 8B). Therefore, the observed effects are not due to different amounts of Ste20 protein. We also examined phenotypes of STE20 mutant alleles when expressed from the native STE20 promoter. Deletion of STE20 led to an increased AUS1 expression in the hem1Δ background (Figure 6A). Therefore, we brought back wild-type and mutated versions of STE20 under control of its own promoter into the ste20Δ hem1Δ strain. As expected STE20 reduced the expression of AUS1 (Figure 8C). In contrast, the kinase dead derivative STE20K649A and the NLS mutant STE20Δ272-284 had no effect on AUS1 levels (Figure 8C), suggesting that Ste20 kinase activity and nuclear localization are required for the inhibition of AUS1 expression.
Figure 8.
The regulation of AUS1 expression requires kinase activity and nuclear localization of Ste20. (A) Wild-type STE20 and mutant versions were overexpressed in the indicated strains as described in Figure 5A. Shown is the mean β-galactosidase activity of at least six independent experiments with SD, *p < 0.0001 and **p < 0.05, compared with hem1Δcells without STE20 overexpression. (B) Wild-type STE20 and its mutant alleles are expressed at comparable levels. The indicated strains were incubated with galactose for 18 h. Cells were lysed and equal amounts of protein extract were analyzed by immunoblotting. An N-terminal 3HA tag allowed the detection of Ste20. Cdc11 was used as loading control. (C) Wild-type and mutant alleles of STE20 expressed from the native STE20 promoter were integrated into ste20Δ and ste20Δ hem1Δ cells. AUS1 expression was determined in at least six independent experiments as described in Figure 6A. *p < 0.0005, compared with ste20Δ hem1Δ cells.
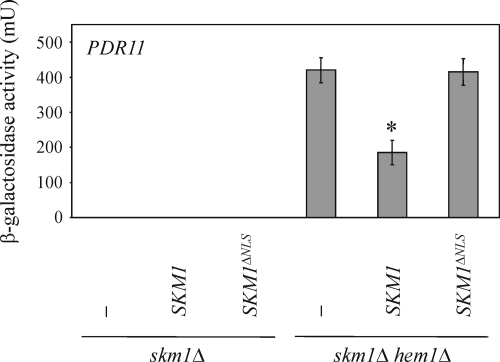
In a similar experiment, we tested whether the NLS of Skm1 plays a role in the down-regulation of sterol uptake genes. As shown in Figure 6C, SKM1 deletion results in an up-regulation of PDR11. Integrating wild-type SKM1 into the skm1Δ hem1Δ double mutant reduced PDR11 expression to normal levels, whereas SKM1ΔNLS had no effect (Figure 9). Thus, nuclear localization of Skm1 seems to be important for the down-regulation of PDR11.
Figure 9.
The down-regulation of PDR11 expression requires nuclear localization of Skm1. SKM1 and SKM1ΔNLS, respectively, were integrated into skm1Δ and skm1Δ hem1Δ cells. PDR11 expression was determined in at least six independent experiments as described in Figure 6C. *p < 0.0005, compared with skm1Δ hem1Δ cells.
The regulation of AUS1 Expression Is Independent of Ste20 MAPK Signaling
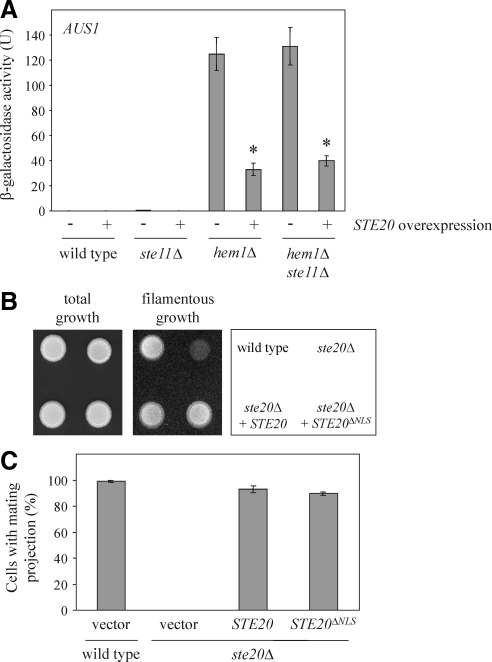
Ste20 controls the expression of numerous genes by the activation of MAPK cascades regulating mating, filamentous growth, and the hyperosmotic response (Roberts and Fink, 1994; O'Rourke and Herskowitz, 1998; Raitt et al., 2000). All three MAPK pathways share the MAPK kinase kinase Ste11, which is a target of Ste20 and essential for Ste20-dependent activation of MAPK cascades (Roberts and Fink, 1994; Wu et al., 1995). To examine whether the Ste20-mediated transcriptional changes observed by us depend on MAPK signaling, we determined AUS1 expression in cells lacking STE11. The decrease of AUS1 levels after STE20 overexpression was independent of STE11 (Figure 10A). Thus, Ste20 seems to regulate gene expression by two distinct mechanisms: the activation of MAPK cascades and a Sut1-dependent pathway. Because the down-regulation of AUS1 required nuclear Ste20 (Figure 8, A and C), we tested whether the nuclear localization of Ste20 was also necessary for its role in filamentous growth and mating. As described previously, Ste20 is essential for haploid filamentous growth and the formation of a mating projection in response to pheromone (Leberer et al., 1992; Roberts and Fink, 1994) (Figure 10, B and C). Importantly, we observed normal filamentous growth for cells with Ste20 lacking the NLS (Figure 10B). These cells also responded normally to mating pheromone by arresting in G1 phase and formation of a mating projection (Figure 10C).
Figure 10.
Ste20 regulates gene expression by two distinct mechanisms. (A) The down-regulation of AUS1 expression is independent of the MAPK kinase kinase Ste11. The β-galactosidase activity was determined in eight independent experiments as described in Figure 5A. *p < 0.0001, compared with hem1Δ cells without STE20 overexpression. (B) Haploid invasive growth does not require nuclear localization of Ste20. Cells (105) of the indicated strains were spotted on a YPD plate and incubated for 2 d at 30°C. Pictures were taken before and after gentle rinsing with water. (C) The NLS of Ste20 is not necessary for the formation of a mating projection. Exponentially growing cells of the indicated strains were incubated in selective medium with 1 μg/ml α-factor for 3 h. Cells were then fixed with formaldehyde and the percentage of cells with a mating projection was determined. Given is the mean of three independent experiments with SD bars (n > 100 for each experiment).
Together, Ste20 controls gene expression by two different mechanisms. MAPK cascades depend on Ste11, but do not require nuclear Ste20. In contrast, the Sut1-mediated pathway relies on nuclear Ste20, but is independent of Ste11.
Ste20, Cla4, and Skm1 Negatively Regulate Sterol Uptake
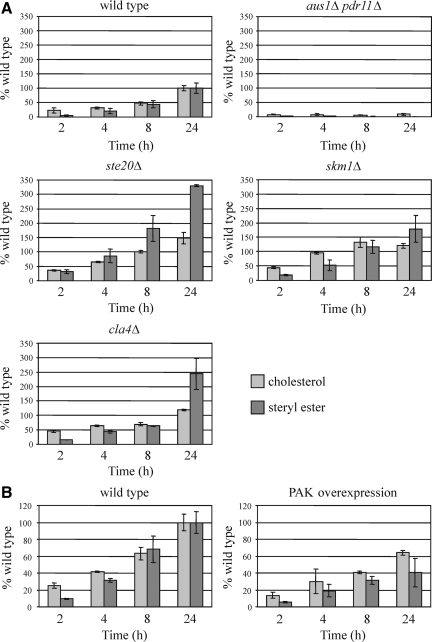
Because Ste20, Cla4, and Skm1 down-regulate the expression of genes involved in sterol influx, it was tested whether individual deletion of either STE20, CLA4, or SKM1 affects sterol uptake. To this end, heme-deficient wild-type and deletion strains were grown in the presence of [14C]cholesterol, and uptake and esterification were quantified by TLC analysis of radiolabeled lipids. Consistent with the gene expression data, increased levels of free and esterified sterol were observed for ste20Δ, cla4Δ, and skm1Δ (Figure 11A). Sterol uptake was also analyzed in a strain that simultaneously overexpresses STE20, CLA4, and SKM1. In these cells, sterol import was markedly reduced compared with the wild type (Figure 11B). Thus, PAKs have a negative effect on sterol uptake. Finally, it was tested whether the inhibition of sterol import by Ste20 depends on nuclear localization of this PAK. Sterol uptake of cells expressing STE20ΔNLS from its own promoter was indistinguishable from the wild type (Supplemental Figure S2), suggesting that Ste20 may also have functions in sterol uptake that do not require nuclear translocation (see Discussion).
Figure 11.
Ste20, Cla4, and Skm1 negatively regulate sterol uptake. (A) Ste20, Skm1, and Cla4 overaccumulate free sterols. Heme-deficient cells of the indicated genotype were cultured in media containing [14C]cholesterol for the indicated period of time. Cells were collected, and lipids were isolated, separated by TLC, and quantified by radioscanning. Data are normalized to free sterols and steryl ester levels in wild-type cells after 24 h of uptake. Values are means and SD of two independent experiments. The aus1Δ pdr11Δ mutant was used as negative control. (B) Overexpression of PAKs results in a decreased sterol import. For PAK overexpression, SKM1 and CLA4 were placed under control of the GAL1 promoter by genomic integration. These cells also harbored STE20 expressed from the GAL1 promoter on a plasmid. Wild-type cells carried the corresponding empty vector. Both strains were grown in SC-Ura medium supplemented with galactose and [14C]cholesterol. Sterols and steryl ester were analyzed as described in A.
DISCUSSION
Kinases of the PAK-family have a well established role in cell polarization (Hofmann et al., 2004). Budding yeast expresses three PAKs: Ste20, Cla4, and Skm1. To gain insight into molecular mechanisms of Ste20 action, we screened for interactors of Ste20 using the split-ubiquitin technique (Tiedje et al., 2007). Here, we show that Sut1, a transcriptional regulator that controls sterol uptake (Bourot and Karst, 1995; Ness et al., 2001), forms a complex with Ste20. By using pull-down assays, we demonstrate that not only Ste20 but also Cla4 and Skm1 bind to Sut1. Sut1 localizes exclusively to the nucleus, even after strong overexpression (data not shown) (Ness et al., 2001). Thus far, Ste20 and Cla4 have been shown to localize mainly to the plasma membrane at sites of polarized growth and to the cytoplasm, whereas the localization of Skm1 has not been reported previously (Peter et al., 1996; Leberer et al., 1997; Holly and Blumer 1999). In this work, we show strong nuclear enrichment of Skm1 and we reveal an NLS. Furthermore, we identify an NLS for Ste20. The fact that a mutated Ste20, which lacks the entire NLS or a major part of it, is excluded from the nucleus under normal growth conditions, suggests that at least a small amount of Ste20 is present in the nucleus at any time. Consistently, in few cells, we observed a nuclear enrichment of wild-type Ste20. Cla4 does not contain an obvious NLS and the full-length protein is not enriched in the nucleus. Nevertheless, we observed that several Cla4 fragments that lacked the PH domain translocated into the nucleus. The PH domain is necessary for membrane association of Cla4 by binding to phosphoinositides (Wild et al., 2004). We suggest that similar to Ste20, Cla4 that is not associated with membranes can localize to the cytoplasm and the nucleus. Therefore, it seems likely that the interaction between Sut1 and PAKs takes place in the nucleus. This notion is confirmed by our observation that the down-regulation of the Sut1 target AUS1 by Ste20 and the inhibition of PDR11 expression by Skm1 depends on the nuclear localization of these PAKs.
The expression of AUS1, DAN1, PDR11, and of other genes contributing to sterol uptake is repressed under aerobic conditions. In anaerobiosis, transcriptional regulators such as Sut1 and Upc2 allow expression of these genes, which is required for sterol uptake under these conditions (Régnacq et al., 2001; Wilcox et al., 2002; Alimardani et al., 2004). Here, we demonstrate that PAKs negatively regulate the expression of genes involved in sterol influx. Interestingly, we observed very different expression profiles for Ste20, Cla4, and Skm1. Ste20 regulates the expression of AUS1 and DAN1, Skm1 controls AUS1 and PDR11, whereas Cla4 inhibits the expression of AUS1, DAN1, and PDR11. The reason for these different specificities is not clear.
Importantly, we demonstrate for all PAKs that the down-regulation of gene expression depends for the most part on SUT1. Because AUS1 and DAN1 are target genes of Sut1 and PAKs form a complex with Sut1, it seems very likely that PAKs control gene expression via Sut1. Nevertheless, it cannot be excluded that PAKs mediate their effects on transcription through other factors such as Upc2 or the less well characterized Ecm22 (Shianna et al., 2001) as well. PDR11 is also regulated by Cla4 and Skm1, but its transcription is controlled by Upc2 and there is no evidence that PDR11 is a target of Sut1. Further, we observed a minor reduction of AUS1 levels in the absence of SUT1 after overexpression of either STE20 or CLA4. Regulation of Upc2 or other factors by Ste20 and Cla4 could account for these weak effects on AUS1 expression. We observed that sterol uptake in contrast to the regulation of AUS1 expression is independent of the Ste20 NLS. The reason for this discrepancy is not clear. However, sterol uptake is a more complex process than the transcription of an individual gene. Possibly, Ste20 contributes also to sterol uptake by other mechanisms. Ste20 could control the activity of transcriptional regulators such as Upc2 and Ecm22, which localize to the cytoplasm as well (Marie et al., 2008).
Thus far, the mechanisms of sterol uptake are only poorly understood. It seems that the transcriptional regulators involved in this process act redundantly, but very little is known about Ecm22 and it is not clear how Sut1 promotes the expression of AUS1 and DAN1. Therefore, it is difficult to establish the mechanisms of inhibition of Sut1 and possibly other transcriptional regulators by PAKs. The down-regulation of AUS1 expression requires Ste20 kinase activity. Nevertheless, we did not observe Sut1 phosphorylation by PAKs using in vivo and in vitro kinase assays (data not shown). Thus, it is conceivable that PAKs control transcriptional regulators via an unknown protein.
The control of gene expression by Ste20, Cla4, and Skm1 described here is clearly distinct from the regulation of transcription by Ste20 via MAPK modules as reported previously. During mating, filamentous growth and in response to hyperosmolarity Ste20 activates the MAPK kinase kinase Ste11 by phosphorylation, which eventually results in a change of the transcriptional pattern of numerous genes (Roberts and Fink, 1994; Wu et al., 1995; O'Rourke and Herskowitz, 1998; Raitt et al., 2000). As demonstrated for AUS1, the Sut1-mediated regulation of expression is independent of Ste11. Furthermore, it requires nuclear localization of Ste20. In contrast, the MAPK pathways promoting filamentation and the formation of a mating projection are independent of nuclear Ste20, but Ste11 is essential for these processes. Thus, the two mechanisms seem to be independent.
It has been reported that Ste20 translocates into the nucleus during hydrogen peroxide-induced cell death and directly phosphorylates histone H2B (Ahn et al., 2005). This presumably results in chromatin condensation. However, it has not been examined whether Ste20 alters gene expression under these conditions. Whether the inhibition of a transcriptional regulator involved in sterol uptake by Ste20 also involves chromatin modification as suggested for hydrogen peroxide-induced cell death remains to be tested.
Importantly, we demonstrated that the control of genes involved in sterol import mediated by Ste20, Cla4, and Skm1 affects sterol uptake. As expected from their role as negative regulators, the deletion of either STE20, CLA4 or SKM1 results in an increased sterol influx and subsequent esterification. Furthermore, sterol uptake was markedly reduced in cells overexpressing all three PAKs.
Our data raise the question why Ste20, Cla4, and Skm1 regulate sterol uptake. The fact that all three PAKs are involved in this process, suggests that the control of sterol influx is crucial for the cell and possibly for cell polarization. Previously, we could show that Ste20 binds to Erg4, Cbr1, and Ncp1, which all catalyze important steps in sterol biosynthesis (Tiedje et al., 2007). Interestingly, these proteins are also involved in bud site selection, apical bud growth, mating, filamentous growth, and exit from mitosis (Ni and Snyder, 2001; Keniry et al., 2004; Tiedje et al., 2007). These observations highlight the importance of sterol synthesis for cell polarization. Because Ste20 plays a crucial role in all these processes as well, it seems likely that Ste20 and sterol biosynthetic proteins act in the same pathway(s). In the absence of oxygen, sterols cannot be synthesized and cells completely depend on import of sterols from the extracellular medium. Under these conditions, control of sterol uptake may be as important for cell polarization as sterol biosynthesis in aerobiosis. Therefore, it is not surprising that PAKs regulate sterol uptake. Importantly, Cla4 not only has a negative effect on sterol uptake but also down-regulates sterol biosynthesis and storage (our unpublished data). Thus, it seems that Cla4 modulates sterol homeostasis under aerobic conditions and that all PAKs are involved in sterol import under anaerobiosis. In addition, homologues of oxysterol-binding proteins, a family of proteins, which regulate synthesis and transport of sterols, were found to participate in Cdc42-dependant cell polarity (Kozminski et al., 2006). How sterols contribute to cell polarization has been discussed controversially (Pichler and Riezman, 2004; Alvarez et al., 2007), and the elucidation of underlying molecular mechanisms will require further work.
In summary, we describe here a novel function for all three PAKs. They control transcription in the nucleus, which results in a change of sterol influx rate.
Supplementary Material
ACKNOWLEDGMENTS
We thank Nils Johnsson (University of Ulm, Ulm, Germany) for providing the split-ubiquitin library. We thank Thierry Berges (University of Poitiers, Poitiers Cedex, France) for plasmids, Peter Pryciak (University of Massachusetts Medical School, Worcester, MA) for constructs and communicating unpublished data, and Silke Horn for excellent technical support. The project was supported by the Deutsche Forschungsgemeinschaft (HO 2098/2 and HO 2098/3 (to T. H.) and the Swiss National Science Foundation (PP00A-110450 and 3100-120650 (to R. S).
Abbreviations used:
- BR
basic-rich
- CRIB
Cdc42/Rac-interactive binding
- PAK
p21-activated kinase
- PH
pleckstrin homology
- TLC
thin layer chromatography.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-01-0034) on September 30, 2009.
REFERENCES
- Ahn S. H., Cheung W. L., Hsu J. Y., Diaz R. L., Smith M. M., Allis C. D. Sterile 20 kinase phosphorylates histone H2B at serine 10 during hydrogen peroxide-induced apoptosis in S. cerevisiae. Cell. 2005;120:25–36. doi: 10.1016/j.cell.2004.11.016. [DOI] [PubMed] [Google Scholar]
- Alimardani P., Régnacq M., Moreau-Vauzelle C., Ferreira T., Rossignol T., Blondin B., Bergès T. SUT1-promoted sterol uptake involves the ABC transporter Aus1 and the mannoprotein Dan1 whose synergistic action is sufficient for this process. Biochem. J. 2004;381:195–202. doi: 10.1042/BJ20040297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez F. J., Douglas L. M., Konopka J. B. Sterol-rich plasma membrane domains in fungi. Eukaryot. Cell. 2007;6:755–763. doi: 10.1128/EC.00008-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ash J., Wu C., Larocque R., Jamal M., Stevens W., Osborne M., Thomas D. Y., Whiteway M. Genetic analysis of the interface between Cdc42p and the CRIB domain of Ste20p in Saccharomyces cerevisiae. Genetics. 2003;63:9–20. doi: 10.1093/genetics/163.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourot S., Karst F. Isolation and characterization of the Saccharomyces cerevisiae SUT1 gene involved in sterol uptake. Gene. 1995;165:97–102. doi: 10.1016/0378-1119(95)00478-o. [DOI] [PubMed] [Google Scholar]
- Bose I., Irazoqui J. E., Moskow J. J., Bardes E. S., Zyla T. R., Lew D. J. Assembly of scaffold-mediated complexes containing Cdc42p, the exchange factor Cdc24p, and the effector Cla4p required for cell cycle-regulated phosphorylation of Cdc24p. J. Biol. Chem. 2001;276:7176–7186. doi: 10.1074/jbc.M010546200. [DOI] [PubMed] [Google Scholar]
- Brown J. L., Jaquenoud M., Gulli M. P., Chant J., Peter M. Novel Cdc42-binding proteins Gic1 and Gic2 control cell polarity in yeast. Genes Dev. 1997;11:2972–2982. doi: 10.1101/gad.11.22.2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G. C., Kim Y. J., Chan C. S. The Cdc42 GTPase-associated proteins Gic1 and Gic2 are required for polarized cell growth in Saccharomyces cerevisiae. Genes Dev. 1997;11:2958–2971. doi: 10.1101/gad.11.22.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvrckova F., De Virgilio C., Manser E., Pringle J. R., Nasmyth K. Ste20-like protein kinases are required for normal localization of cell growth and for cytokinesis in budding yeast. Genes Dev. 1995;9:1817–1830. doi: 10.1101/gad.9.15.1817. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S. Cdc42-the centre of polarity. J. Cell Sci. 2004;117:1291–1300. doi: 10.1242/jcs.01115. [DOI] [PubMed] [Google Scholar]
- Gollub E. G., Liu K. P., Dayan J., Adlersberg M., Sprinson D. B. Yeast mutants deficient in heme biosynthesis and a heme mutant additionally blocked in cyclization of 2,3-oxidosqualene. J. Biol. Chem. 1977;252:2846–2854. [PubMed] [Google Scholar]
- Gulli M. P., Jaquenoud M., Shimada Y., Niederhäuser G., Wiget P., Peter M. Phosphorylation of the Cdc42 exchange factor Cdc24 by the PAK-like kinase Cla4 may regulate polarized growth in yeast. Mol. Cell. 2000;6:1155–1167. doi: 10.1016/s1097-2765(00)00113-1. [DOI] [PubMed] [Google Scholar]
- Höfken T., Schiebel E. A role for cell polarity proteins in mitotic exit. EMBO J. 2002;21:4851–4862. doi: 10.1093/emboj/cdf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann C., Shepelev M., Chernoff J. The genetics of Pak. J. Cell Sci. 2004;117:4343–4354. doi: 10.1242/jcs.01392. [DOI] [PubMed] [Google Scholar]
- Holly S. P., Blumer K. J. PAK-family kinases regulate cell and actin polarization throughout the cell cycle of Saccharomyces cerevisiae. J. Cell Biol. 1999;147:845–856. doi: 10.1083/jcb.147.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe A. B., Hall A. Rho GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- Janke C. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- Johnsson N., Varshavsky A. Split ubiquitin as a sensor of protein interactions in vivo. Proc. Natl. Acad. Sci. USA. 1994;91:10340–10344. doi: 10.1073/pnas.91.22.10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota J., Yamamoto T., Yoshiuchi S., Bi E., Tanaka K. Septin ring assembly requires concerted action of polarisome components, a PAK kinase Cla4p, and the actin cytoskeleton in Saccharomyces cerevisiae. Mol. Biol. Cell. 2004;15:5329–5345. doi: 10.1091/mbc.E04-03-0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keniry M. E., Kemp H. A., Rivers D. M., Sprague G. F. The identification of Pcl1-interacting proteins that genetically interact with Cla4 may indicate a link between G1 progression and mitotic exit. Genetics. 2004;166:1177–1186. doi: 10.1534/genetics.166.3.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M., Siegers K., Pereira G., Zachariae W., Winsor B., Nasmyth K., Schiebel E. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Kozminski K. G., Alfaro G., Dighe S., Beh C. T. Homologues of oxysterol-binding proteins affect Cdc42p- and Rho1p-mediated cell polarization in Saccharomyces cerevisiae. Traffic. 2006;7:1224–1242. doi: 10.1111/j.1600-0854.2006.00467.x. [DOI] [PubMed] [Google Scholar]
- Lamson R. E., Winters M. J., Pryciak P. M. Cdc42 regulation of kinase activity and signaling by the yeast p21-activated kinase Ste20. Mol. Cell Biol. 2002;22:2939–2951. doi: 10.1128/MCB.22.9.2939-2951.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leberer E., Dignard D., Harcus D., Thomas D. Y., Whiteway M. The protein kinase homologue Ste20p is required to link the yeast pheromone response G-protein beta gamma subunits to downstream signalling components. EMBO J. 1992;11:4815–4824. doi: 10.1002/j.1460-2075.1992.tb05587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leberer E., Wu C., Leeuw T., Fourest-Lieuvin A., Segall J. E., Thomas D. Y. Functional characterization of the Cdc42p binding domain of yeast Ste20p protein kinase. EMBO J. 1997;16:83–97. doi: 10.1093/emboj/16.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeuw T., Wu C., Schrag J. D., Whiteway M., Thomas D. Y., Leberer E. Interaction of a G-protein beta-subunit with a conserved sequence in Ste20/PAK family protein kinases. Nature. 1998;391:191–195. doi: 10.1038/34448. [DOI] [PubMed] [Google Scholar]
- Lewis T. A., Taylor F. R., Parks L. W. Involvement of heme biosynthesis in control of sterol uptake by Saccharomyces cerevisiae. J. Bacteriol. 1985;163:199–207. doi: 10.1128/jb.163.1.199-207.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Styles C. A., Fink G. R. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science. 1993;262:1741–1744. doi: 10.1126/science.8259520. [DOI] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Marie C., Leyde S., White T. C. Cytoplasmic localization of sterol transcription factors Upc2p and Ecm22p in S. cerevisiae. Fungal Genet. Biol. 2008;45:1430–1438. doi: 10.1016/j.fgb.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín H., Mendoza A., Rodríguez-Pachón J. M., Molina M., Nombela C. Characterization of SKM1, a Saccharomyces cerevisiae gene encoding a novel Ste20/PAK-like protein kinase. Mol. Microbiol. 1997;23:431–444. doi: 10.1046/j.1365-2958.1997.d01-1870.x. [DOI] [PubMed] [Google Scholar]
- Miller W. G., Lindow S. E. An improved GFP cloning cassette designed for prokaryotic transcriptional fusions. Gene. 1997;191:149–153. doi: 10.1016/s0378-1119(97)00051-6. [DOI] [PubMed] [Google Scholar]
- Myers A. M., Tzagoloff A., Kinney D. M., Lusty C. J. Yeast shuttle and integrative vectors with multiple cloning sites suitable for construction of lacZ fusions. Gene. 1986;45:299–310. doi: 10.1016/0378-1119(86)90028-4. [DOI] [PubMed] [Google Scholar]
- Ness F., Bourot S., Régnacq M., Spagnoli R., Bergès T., Karst F. SUT1 is a putative Zn[II]2Cys6-transcription factor whose upregulation enhances both sterol uptake and synthesis in aerobically growing Saccharomyces cerevisiae cells. Eur. J. Biochem. 2001;268:1585–1595. [PubMed] [Google Scholar]
- Ni L., Snyder M. A genomic study of the bipolar bud site selection pattern in Saccharomyces cerevisiae. Mol. Biol. Cell. 2001;12:2147–2170. doi: 10.1091/mbc.12.7.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke S. M., Herskowitz I. The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev. 1998;12:2874–2886. doi: 10.1101/gad.12.18.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H. O., Bi E. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol. Mol. Biol. Rev. 2007;71:48–96. doi: 10.1128/MMBR.00028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks L. W., Casey W. M. Physiological implications of sterol biosynthesis in yeast. Annu. Rev. Microbiol. 1995;49:95–116. doi: 10.1146/annurev.mi.49.100195.000523. [DOI] [PubMed] [Google Scholar]
- Peter M., Neiman A. M., Park H. O., van Lohuizen M., Herskowitz I. Functional analysis of the interaction between the small GTP binding protein Cdc42 and the Ste20 protein kinase in yeast. EMBO J. 1996;15:7046–7059. [PMC free article] [PubMed] [Google Scholar]
- Pichler H., Riezman H. Where sterols are required for endocytosis. Biochim. Biophys. Acta. 2004;1666:51–61. doi: 10.1016/j.bbamem.2004.05.011. [DOI] [PubMed] [Google Scholar]
- Raitt D. C., Posas F., Saito H. Yeast Cdc42 GTPase and Ste20 PAK-like kinase regulate Sho1-dependent activation of the Hog1 MAPK pathway. EMBO J. 2000;19:4623–4631. doi: 10.1093/emboj/19.17.4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramer S. W., Davis R. W. A dominant truncation allele identifies a gene, STE20, that encodes a putative protein kinase necessary for mating in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 1993;90:452–456. doi: 10.1073/pnas.90.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Régnacq M., Alimardani P., El Moudni B., Bergès T. SUT1p interaction with Cyc8p(Ssn6p) relieves hypoxic genes from Cyc8p-Tup1p repression in Saccharomyces cerevisiae. Mol. Microbiol. 2001;40:1085–1096. doi: 10.1046/j.1365-2958.2001.02450.x. [DOI] [PubMed] [Google Scholar]
- Reiner S., Micolod D., Zellnig G., Schneiter R. A genomewide screen reveals a role of mitochondria in anaerobic uptake of sterols in yeast. Mol. Biol. Cell. 2006;17:90–103. doi: 10.1091/mbc.E05-06-0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. L., Fink G. R. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 1994;8:2974–2985. doi: 10.1101/gad.8.24.2974. [DOI] [PubMed] [Google Scholar]
- Robinson L. C., Menold M. M., Garrett S., Culbertson M. R. Casein kinase I-like protein kinases encoded by YCK1 and YCK2 are required for yeast morphogenesis. Mol. Cell Biol. 1993;13:2870–2881. doi: 10.1128/mcb.13.5.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakchaisri K., Asano S., Yu L. R., Shulewitz M. J., Park C. J., Park J. E., Cho Y. W., Veenstra T. D., Thorner J., Lee K. S. Coupling morphogenesis to mitotic entry. Proc. Natl. Acad. Sci. USA. 2004;101:4124–4129. doi: 10.1073/pnas.0400641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer N., Stolz J. SUC1 and SUC 2, two sucrose transporters from Arabidopsis thaliana; expression and characterization in baker's yeast and identification of the histidine-tagged protein. Plant J. 1994;6:67–77. doi: 10.1046/j.1365-313x.1994.6010067.x. [DOI] [PubMed] [Google Scholar]
- Schjerling P., Holmberg S. Comparative amino acid sequence analysis of the C6 zinc cluster family of transcriptional regulators. Nucleic Acids Res. 1996;24:4599–4607. doi: 10.1093/nar/24.23.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M., Varma A., Drgon T., Bowers B., Cabib E. Septins, under Cla4p regulation, and the chitin ring are required for neck integrity in budding yeast. Mol. Biol. Cell. 2003;14:2128–2141. doi: 10.1091/mbc.E02-08-0547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshan A., Bardin A. J., Amon A. Control of Ltel localization by cell polarity determinants and Cdc14. Curr. Biol. 2002;12:2098–2110. doi: 10.1016/s0960-9822(02)01388-x. [DOI] [PubMed] [Google Scholar]
- Shianna K. V., Dotson W. D., Tove S., Parks L. W. Identification of a UPC2 homolog in Saccharomyces cerevisiae and its involvement in aerobic sterol uptake. J. Bacteriol. 2001;183:830–834. doi: 10.1128/JB.183.3.830-834.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturley S. L. Conservation of eukaryotic sterol homeostasis: new insights from studies in budding yeast. Biochim. Biophys. Acta. 2000;1529:155–163. doi: 10.1016/s1388-1981(00)00145-1. [DOI] [PubMed] [Google Scholar]
- Takahashi S., Pryciak P. M. Identification of novel membrane-binding domains in multiple yeast Cdc42 effectors. Mol. Biol. Cell. 2007;18:4945–4956. doi: 10.1091/mbc.E07-07-0676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiedje C., Holland D. G., Just U., Höfken T. Proteins involved in sterol synthesis interact with Ste20 and regulate cell polarity. J. Cell Sci. 2007;120:3613–3624. doi: 10.1242/jcs.009860. [DOI] [PubMed] [Google Scholar]
- Versele M., Thorner J. Septin collar formation in budding yeast requires GTP binding and direct phosphorylation by the PAK, Cla4. J. Cell Biol. 2004;164:701–715. doi: 10.1083/jcb.200312070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss E. L., Bishop A. C., Shokat K. M., Drubin D. G. Chemical genetic analysis of the budding-yeast p21-activated kinase Cla4p. Nat. Cell Biol. 2000;2:677–685. doi: 10.1038/35036300. [DOI] [PubMed] [Google Scholar]
- Wilcox L. J., Balderes D. A., Wharton B., Tinkelenberg A. H., Rao G., Sturley S. L. Transcriptional profiling identifies two members of the ATP-binding cassette transporter superfamily required for sterol uptake in yeast. J. Biol. Chem. 2002;277:32466–32472. doi: 10.1074/jbc.M204707200. [DOI] [PubMed] [Google Scholar]
- Wild A. C., Yu J. W., Lemmon M. A., Blumer K. J. The p21-activated protein kinase-related kinase Cla4 is a coincidence detector of signaling by Cdc42 and phosphatidylinositol 4-phosphate. J. Biol. Chem. 2004;279:17101–17110. doi: 10.1074/jbc.M314035200. [DOI] [PubMed] [Google Scholar]
- Winters M. J., Lamson R. E., Nakanishi H., Neiman A. M., Pryciak P. M. A membrane binding domain in the ste5 scaffold synergizes with gbetagamma binding to control localization and signaling in pheromone response. Mol. Cell. 2005;20:21–32. doi: 10.1016/j.molcel.2005.08.020. [DOI] [PubMed] [Google Scholar]
- Winters M. J., Pryciak P. M. Interaction with the SH3 domain protein Bem1 regulates signaling by the Saccharomyces cerevisiae p21-activated kinase Ste20. Mol. Cell Biol. 2005;25:2177–2190. doi: 10.1128/MCB.25.6.2177-2190.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler E. A., et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Wittke S., Lewke N., Müller S., Johnsson N. Probing the molecular environment of membrane proteins in vivo. Mol. Biol. Cell. 1999;10:2519–2530. doi: 10.1091/mbc.10.8.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Whiteway, Thomas M., D. Y., Leberer E. Molecular characterization of Ste20p, a potential mitogen-activated protein or extracellular signal-regulated kinase kinase (MEK) kinase kinase from Saccharomyces cerevisiae. J. Biol. Chem. 1995;270:15984–15992. doi: 10.1074/jbc.270.27.15984. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y., Ota K., Ito T. A novel Cdc42-interacting domain of the yeast polarity establishment protein Bem1. Implications for modulation of mating pheromone signaling. J. Biol. Chem. 2007;282:29–38. doi: 10.1074/jbc.M609308200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.