Abstract
The DIE NEUTRALIS (DNE) locus in garden pea (Pisum sativum) was previously shown to inhibit flowering under noninductive short-day conditions and to affect a graft-transmissible flowering signal. In this study, we establish that DNE has a role in diurnal and/or circadian regulation of several clock genes, including the pea GIGANTEA (GI) ortholog LATE BLOOMER 1 (LATE1) and orthologs of the Arabidopsis thaliana genes LATE ELONGATED HYPOCOTYL and TIMING OF CHLOROPHYLL A/B BINDING PROTEIN EXPRESSION 1. We also confirm that LATE1 participates in the clock and provide evidence that DNE is the ortholog of Arabidopsis EARLY FLOWERING4 (ELF4). Circadian rhythms of clock gene expression in wild-type plants under constant light were weaker in pea than in Arabidopsis, and a number of differences were also seen in the effects of both DNE/ELF4 and LATE1/GI on clock gene expression. Grafting studies suggest that DNE controls flowering at least in part through a LATE1-dependent mobile stimulus, and dne mutants show elevated expression of a FLOWERING LOCUS T homolog under short-day conditions. However, the early flowering of the dne mutant is not associated with altered expression of a previously described CONSTANS-like gene. Collectively, our results characterize the clock system and reveal its importance for photoperiod responsiveness in a model legume.
INTRODUCTION
In many species, photoperiod is an important environmental signal influencing the onset of flowering, and rapid advances have recently been made in understanding how plants sense and respond to photoperiod. Most of this progress has come from studies in Arabidopsis thaliana, but more recent work has expanded to several other species, including rice (Oryza sativa) and the temperate cereals wheat (Triticum aestivum) and barley (Hordeum vulgare; Hayama and Coupland, 2004; Trevaskis et al., 2007). At the most general level, photoperiodic flowering results from photoperiod-specific expression of genes in the FLOWERING LOCUS T (FT) family. The biochemical function of FT proteins is unclear, but they have been shown to move from leaf to apex and interact with bZIP transcription factors to regulate inflorescence identity genes (Kobayashi and Weigel, 2007; Turck et al., 2008). While several mechanisms contribute to the photoperiod-specific expression of FT genes in the leaf, all appear to involve interactions between light and the circadian clock (Doi et al., 2004; Imaizumi and Kay, 2006; Hayama et al., 2007; Jung et al., 2007; Fujiwara et al., 2008).
Circadian clocks are molecular oscillators that generate output rhythms of ∼24 h under constant conditions, which can be entrained to a cycle of exactly 24 h by diurnal variations in light or temperature. The molecular nature of the plant circadian clock is best understood in Arabidopsis and is thought to consist of three interlocking negative feedback loops in which myb transcription factors COLD CLOCK ASSOCIATED 1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY) reciprocally regulate the expression of the pseudo-response regulator TIMING OF CHLOROPHYLL A/B BINDING PROTEIN EXPRESSION 1 (TOC1) and several other related proteins (Gardner et al., 2006; McClung, 2008). A number of other genes whose biochemical function is less well understood also have an important influence on the clock, including EARLY FLOWERING 4 (ELF4), GIGANTEA (GI), and LUX ARRHYTHMO (LUX), which are proposed to be core clock components (Hazen et al., 2005; Locke et al., 2005; McWatters et al., 2007), and EARLY FLOWERING 3 (ELF3), TIME FOR COFFEE (TIC), and FAR-RED ELONGATED HYPOCOTYLS 3 (FHY3), which are thought to function in gating of light signals to the clock (McWatters et al., 2000; Allen et al., 2006; Ding et al., 2007).
In Arabidopsis, up to 15% of genes show rhythmic cycling of transcript abundance under constant conditions, including genes acting in a wide variety of different metabolic processes, emphasizing the importance of circadian regulation for adaptation to the daily light/dark cycle (Gardner et al., 2006; McClung, 2008). The specific importance of the clock for photoperiodic flowering is demonstrated by the circadian regulation of many Arabidopsis flowering genes and the fact that many Arabidopsis mutants with a primary effect on clock also show altered photoperiod responses. In Arabidopsis, the main output mechanism by which the clock controls flowering is through the rhythmic regulation of the B-box transcription factor CONSTANS (CO), such that CO expression occurs during the light period under long days (LDs) but not short days (SDs) (Suárez-López et al., 2001; Yanovsky and Kay, 2002). More recently, other mechanisms for clock regulation of FT have been proposed to act independently of CO, through ELF3 (Kim et al., 2005), miRNA172 (Jung et al., 2007), and the MADS protein SHORT VEGETATIVE PHASE (Fujiwara et al., 2008).
The nature of the circadian clock in other species is much less well understood than in Arabidopsis. One recent study in Lemna gibba used RNA interference to address the conservation of the core clock mechanism (Serikawa et al., 2008) and demonstrated important roles for LHY, GI, and ELF3 homologs in regulation of Arabidopsis CCA1 and TOC1 reporters in a transient expression system. Although expression studies have been conducted in various other species, functional analyses have otherwise been limited to overexpression studies in rice (Murakami et al., 2003, 2007). The identification of flowering time genes Heading date 6 in rice and Photoperiod -H1 in barley as homologs of Arabidopsis genes implicated in clock function (Takahashi et al., 2001; Turner et al., 2005) does suggest that photoperiod response in these species also depends on normal function of the circadian system. Comparative studies in rice and potato (Solanum tuberosum) have identified CO-like genes as clock outputs important for regulation of FT expression and photoperiod response (Kojima et al., 2002; Rodríguez-Falcón et al., 2006), and involvement of CO in photoperiod-dependent FT expression has been inferred from expression studies in poplar (Populus spp; Böhlenius et al., 2006). However, CO-independent clock regulation of FT genes has also been demonstrated in both rice and Pharbitis (Doi et al., 2004; Hayama et al., 2007). suggesting that as in Arabidopsis, CO-like genes may not be the only clock output necessary for photoperiod-responsive growth and flowering.
Another model system prominent in early genetic studies of flowering time control is garden pea (Pisum sativum). Pea is the best-studied legume model for control of flowering, and more than a dozen major flowering loci have been identified, including several that affect photoperiod responsiveness and graft-transmissible signals (Weller et al., 1997; Weller, 2007). We recently identified LATE BLOOMER 1 (LATE1) as the pea ortholog of Arabidopsis GI and showed it to be necessary for promotion of flowering, the production of a mobile flowering stimulus, and induction of a FT homolog under LD conditions (Hecht et al., 2007). We also described the isolation of pea orthologs of Arabidopsis clock genes LHY (previously called MYB1), TOC1, and ELF4 (Hecht et al., 2005) and showed that LATE1 influences diurnal expression rhythms of several of these genes (Hecht et al., 2007).
In contrast with LATE1, other pea photoperiod response loci inhibit flowering under SD conditions. Mutants at the STERILE NODES (SN), DIE NEUTRALIS (DNE), and PHOTOPERIOD (PPD) loci all flower earlier than the wild type in SDs and show characteristics of LD-grown wild-type plants, such as reduced branching and rapid termination of apical growth (Murfet, 1971; King and Murfet, 1985; Taylor and Murfet, 1996). Little is known about the molecular nature of these loci, but the fact that many early-flowering photoperiod response mutants in Arabidopsis affect the circadian clock has suggested that these pea genes may also affect the circadian system (Weller, 2005). We have been characterizing the effect of these mutants on the circadian system and pursuing a candidate gene approach to identify the SN, DNE, and PPD genes (Weller, 2007). In this study, we show that the DNE and LATE1 genes function in the pea circadian clock. We also provide evidence that DNE is an ortholog of Arabidopsis ELF4 and examine its interactions with LATE1 in the control of flowering and photoperiod responsiveness. Our results make a significant contribution to comparative genetics of the plant circadian clock and identify both similarities and differences with the Arabidopsis model. They also argue against the long-standing hypothesis that the photoperiod response in pea is primarily determined through the action of a mobile flowering inhibitor.
RESULTS
The dne Mutant Shows Early, Photoperiod-Insensitive Flowering
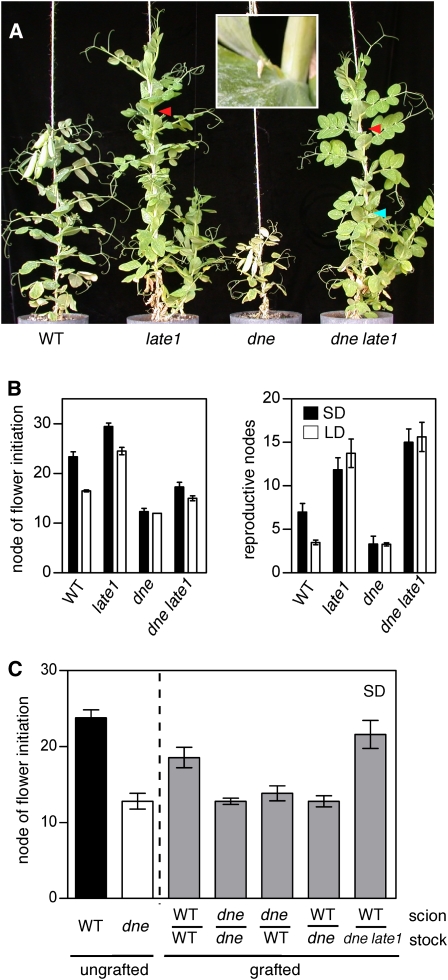
The DNE locus is known from a single mutant allele, dne-1, which was isolated in the cv Torsdag genetic background (King and Murfet, 1985). The dne mutant flowers early in SDs and shows other traits characteristic of a wild-type plant grown in LDs, including reduced branching, rapid termination of apical growth after flowering, and rapid senescence (Figure 1). The dne mutant thus appears to show constitutive activation of a LD developmental program and is essentially unresponsive to photoperiod differences.
Figure 1.
The dne Mutant Is Early Flowering and Insensitive to Photoperiod.
(A) Representative 6-week-old wild-type line NGB5839 and isogenic dne-1 mutant plants.
(B) Node of flower initiation (left) and final number of reproductive nodes. Data are mean ± se for n = 6 to 8 plants.
All plants were grown in the phytotron under standard SD or LD conditions.
This is true regardless of the genotype at the LE locus, which governs the synthesis of the active gibberellin, GA1 (Lester et al., 1997; see Supplemental Figure 1 online). Under LDs, the dne mutant does typically flower slightly earlier than the wild type, but the wild type and dne are otherwise similar in phenotype (King and Murfet, 1985).
The dne Mutant Shows Altered Rhythms of Gene Expression under Light/Dark Cycles
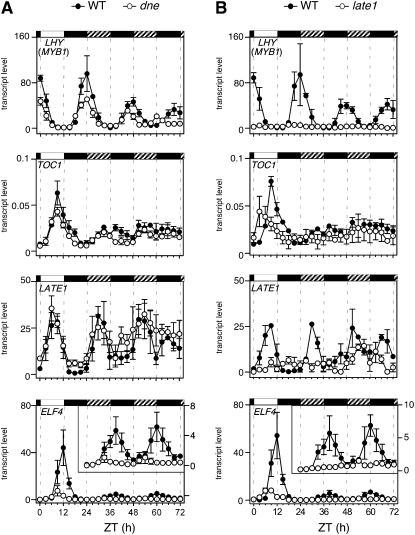
The early-flowering phenotype of the dne mutant is similar to that of Arabidopsis circadian clock mutants elf3, elf4, lux, and the cca1 lhy double mutant (Hicks et al., 1996; Doyle et al., 2002; Mizoguchi et al., 2002; Hazen et al., 2005), and we therefore considered that dne might have a defect in rhythmic expression of clock gene homologs. We previously found that the pea genes LHY, TOC1, ELF4, and LATE1 show LD expression rhythms that are similar to Arabidopsis and that these are altered in a late1 mutant (Hecht et al., 2007).
Figure 2 shows that dne had no clear effect on expression of LHY under either SDs or LDs, nor on TOC1 under LDs (Figures 2A and 2B). However, under SDs, the expression rhythm of TOC1 in the wild type showed a relatively sharp peak at dusk (ZT8) and dropped significantly by ZT12, whereas in the dne mutant, TOC1 expression continued into the night, remaining high at ZT12 (Figure 2A), suggesting a small phase delay. ELF4 expression in the wild type under SDs showed a sharp peak early in the night (ZT12), whereas in the dne mutant, the peak occurred at dusk (ZT8) (Figure 2A). The earlier rise of ELF4 expression during the day and the earlier drop during the night are consistent with a phase advance in dne. Under LDs, the shape of the wild-type ELF4 rhythm differed from SD, with a broader peak from ZT12 to ZT16 (Figure 2B). In LDs, the ELF4 rhythm in dne peaked late in the day (ZT12) and also showed a small phase advance relative to the wild type. The dne mutation also affected LATE1 expression under both SDs and LDs (Figures 2A and 2B). There was no clear indication of a phase shift under either photoperiod, but LATE1 transcript levels were higher in dne than in the wild type throughout the night, similar to the effect of the sn mutant on LATE1 (Hecht et al., 2007). Expression of the TOC1-related genes PSEUDO-RESPONSE REGULATOR 37 (PRR37) and PRR59 in SDs was apparently unaffected by dne (Figures 2A and 2B), whereas in LDs, expression of PRR59 in dne was elevated relative to the wild type during the night (Figure 2B). In summary, the dne mutant affects the diurnal expression of TOC1, ELF4, LATE1, and PRR59 under SD and/or LD photoperiods but had no apparent effect on LHY or PRR37 expression.
Figure 2.
DNE Affects Rhythmic Expression of Clock Gene Homologs under Light/Dark Cycles.
(A) SD conditions (8-h photoperiod).
(B) LD conditions (16-h photoperiod).
All plants were grown for 21 d from sowing at 20°C before harvesting commenced. Data are mean ± se for n = 3 biological replicates, each consisting of pooled material from two plants. Day and night periods are indicated by open and closed bars, respectively, above the graph.
DNE and LATE1 Affect Rhythms of Clock Gene Expression in Constant Light
We were next interested to examine whether DNE might affect circadian rhythms. The circadian clock has not been directly examined in pea, but in Arabidopsis, most circadian analyses have been performed under constant white light (LL), and robust rhythms for leaf movement and gene expression generally persist over several circadian cycles. We initially found that the strong expression rhythms seen for LHY, TOC1, LATE1, and ELF4 in wild-type pea seedlings under diurnal cycles were significantly damped during the first subjective night after transfer to LL of moderate irradiance, resulting in lower peak levels for LHY and ELF4 and higher trough levels for TOC1 and LATE1 (see Supplemental Figure 2 online). However, under LL of lower irradiance, clear rhythmic expression was maintained for all four genes through at least one circadian cycle (Figure 3). For ELF4, the rhythm was maintained for at least 48 h with a strong amplitude similar to the LD rhythm, whereas rhythms for LHY, LATE1, and TOC1 showed some damping by the second circadian cycle, toward trough levels for LHY and intermediate levels for TOC1 and LATE1 (Figure 3). Although rhythms were only followed for two full circadian cycles, all four genes gave some indication of a shorter period, with peaks 18 h apart for ELF4 and 21 h apart for LHY and TOC1.
Figure 3.
DNE and LATE1 Affect Circadian Rhythms of Clock Gene Homologs in LL.
Plants were grown in growth cabinets under a light/dark cycle (12L:12D) at 20°C for 21 d before transfer to continuous white light at 25 μmol m−2 s−1 at ZT36. Data are mean ± se for n = 2 biological replicates, each consisting of pooled material from two plants. Zeitgeber time (ZT) refers to the time since lights-on of the last full entraining cycle. Bars above the graph refer to periods of light (open or stippled bars) or darkness (closed bars). The heavy and light stippled bars refer to periods of subjective night and subjective day, respectively, during the period of continuous light.
(A) Expression of clock genes in the wild type and dne.
(B) Expression of clock genes in the wild type and late1.
We also examined the effect of the dne and late1 mutations in the same experiment (Figures 3A and 3B). The clearest effects were seen for late1, which showed substantial reduction in the peak expression level for all four genes and little evidence of any residual rhythm (Figure 3B). By contrast, dne had more subtle effects, including an apparent small phase advance of TOC1 and LATE1 expression in the second circadian cycle, suggestive of a shorter period. Interestingly, there is also a suggestion that the phase difference for ELF4 expression between wild type and dne under light/dark cycles may diminish after transfer to LL.
DNE and LATE1 Also Affect Rhythms of Clock Gene Expression in Constant Darkness
Rhythmic expression of clock genes also persisted after transfer of wild-type plants from entraining conditions to constant darkness (DD) but differed from LL in several respects (Figure 4). The strongest rhythm in DD was seen for LHY expression, which, as in LL, showed only moderate damping over the two circadian cycles (Figure 4A). In contrast with LL, the ELF4 rhythm was strongly damped in DD, although still clearly rhythmic. Both ELF4 and LHY rhythms showed periods of close to 24 h. TOC1 expression under DD was not clearly rhythmic and, in contrast with LL, damped to a low rather than intermediate level, appearing to lose the induction during the subjective day, instead of the repression phase during the subjective night in LL (Figure 3). Finally, the rhythm of LATE1 under DD was very similar to LL both in amplitude and apparent period shortening.
Figure 4.
DNE and LATE1 Affect Circadian Rhythms of Clock Gene Homologs in DD.
Plants were grown in growth cabinets under a light/dark cycle (12L:12D) at 20°C for 3 weeks before transfer to continuous darkness. Data are mean ± se for n = 2 to 3 biological replicates, each consisting of pooled material from two plants. Zeitgeber time (ZT) refers to the time since lights-on of the last full entraining cycle. Bars above the graph refer to periods of light (open bars) or darkness (closed or hatched bars). The hatched bars indicate the periods of subjective day during the period of continuous darkness.
(A) Expression of clock genes in the wild type and dne.
(B) Expression of clock genes in the wild type and late1.
As in LL, late1 had clear effects on expression rhythms of LHY and ELF4 under DD, essentially eliminating the expression of both genes. The late1 mutation also appeared to eliminate rhythmic expression of LATE1 itself, which although expressed, showed no discernable rhythm in late1 (Figure 4B). TOC1 expression was already low and essentially arrhythmic in DD in wild-type seedlings and was not significantly affected by late1. In the dne mutant, LHY expression continued to cycle in DD, but with an apparently shorter period, with peaks at ZT45 and ZT60 compared with ZT48 and ZT69 in the wild type. No clear effect of dne on TOC1 or LATE1 expression was detected, but the residual rhythm of ELF4 itself was also altered in the dne mutant under DD, with a lower peak expression level and a phase advance of ∼8 h compared with the wild type (Figure 4A).
Genetic and Physiological Interaction of DNE and LATE1 in the Control of Flowering
As both DNE and LATE1 appear to have a primary influence on the circadian clock, it seemed possible that both genes might affect flowering through the same pathway, and to test this, we constructed a dne late1 double mutant. Figure 5A shows that, under LD, the dne late1 mutant is similar in overall appearance to the late1 single mutant, with delayed senescence, increased branching, and an increased number of reproductive nodes compared with the wild type. Despite these similarities, the double mutant initiated the formation of its first flower at a much lower node than in the single late1 mutant (Figure 5B). Interestingly, however, the growth of flowers at the first few reproductive nodes of dne late1 plants was arrested at an early stage (Figure 5A, inset), and fully developed, open flowers were not produced until approximately the node at which flowering commenced in the late1 single mutant (Figure 5A). The dne mutation was thus clearly able to promote the initiation of flowering in the absence of LATE1, but LATE1 clearly influenced the subsequent development of these early-initiated flower primordia and was epistatic to DNE in other respects.
Figure 5.
Interaction of LATE1 and DNE in the Control of Flowering.
(A) Representative 8-week-old plants grown under LDs. Inset shows the first initiated flower primordium in the dne late1 mutant, which is also indicated by the blue arrowhead in the main panel. The red arrowheads indicate the node of first open flower in late1 and dne late1.
(B) Node of flower initiation (left) and final number of reproductive nodes (right). Data are mean ± se for n = 6 to 8 plants.
(C) Node of flower initiation in ungrafted controls, self-grafts, and reciprocal grafts between the wild type, dne, and dne late1. Data are mean ± se for n = 12 plants.
All plants were grown in the phytotron under standard SD or LD conditions.
Previous studies suggested that LATE1 is necessary for the production of a mobile stimulus of flowering (Hecht et al., 2007), and the LATE1–DNE interaction raised the possibility that DNE might act, in part, through the same mobile signal. Figure 5C shows that under SDs, dne graft stocks possessing three or four true foliage leaves strongly promoted flowering of wild-type scions relative to wild type–on–wild type self-grafts. By contrast, flowering of dne scions grafted to wild-type stocks was not significantly delayed compared with dne self-grafts (P = 0.74). This implies that the early flowering of the dne mutant in SDs is associated with increased production of a mobile stimulus, rather than reduced production of an inhibitor as previously suggested (King and Murfet, 1985). Moreover, in dne late1 double mutant stocks, the ability of dne to promote flowering of wild-type scions was completely blocked by the late1 mutation (Figure 5C). This suggests that LATE1 not only controls a mobile flowering signal in LD, but also acts downstream of DNE in the regulation of a similar signal in SDs.
Effects of DNE on Expression of CO and FT-Like Genes
In Arabidopsis, one of the main ways the circadian clock influences flowering is through control of the expression rhythm of the CO gene. Pea and Medicago both possess a single group Ia CO-like gene (COLa) that is orthologous to the CO/COL1/COL2 clade in Arabidopsis and shares the diurnal expression pattern of COL1 and COL2 but not CO (Hecht et al., 2007; P. Diwadkar, R.E. Laurie, and R.C. Macknight, unpublished data). In a previous study, we showed that although late1 affected the diurnal regulation of several clock-related genes and impaired the induction of an FT homolog, FTL, there was no clear effect of late1 on the expression rhythm of COLa (Hecht et al., 2007). Figure 6A shows that there was also no significant difference in the expression rhythm of COLa under SDs between the wild type and dne.
Figure 6.
Effect of dne on Expression of CO and FT Homologs.
(A) Diurnal rhythms of COLa transcript accumulation in 21-d-old plants grown in SDs.
(B) Relative transcript levels of FTL in LDs (20-d-old plants) and SDs (35-d-old plants).
All plants were grown in growth cabinets at 20°C under either SD (8 h) or LD (16 h) conditions. Data are mean ± se for n = 3 biological replicates, each consisting of pooled material from two plants.
We also examined whether the dne mutation might also affect expression of FTL and how dne and late1 might interact in this respect. Figure 6B shows that in SDs, where the early-flowering phenotype of the dne mutant is most evident, FTL expression in leaf tissue was significantly higher in dne than in the wild type. In LDs, FTL expression was significantly lower in late1 than in the wild type, consistent with our previous report (Hecht et al., 2007), whereas the expression level in dne was not significantly different from wild type (P = 0.19). However, FTL expression in the dne late1 double mutant was low like the late1 single mutant (Figure 6), despite the fact that it initiated flowering much earlier (Figure 5). Taken together, these results suggest (1) that DNE and LATE1 interact to control flowering and other photoperiodic traits through a mobile signal; (2) that regulation of FTL expression correlates with the response to photoperiod; and (3) the promotion of flower initiation by DNE in LDs can occur independently of LATE1 and transcriptional regulation of the FTL gene.
DNE Is the Likely Pea Ortholog of Arabidopsis ELF4
The results from expression analyses demonstrate that the dne mutant affects the rhythmic expression of clock genes, suggesting that DNE might itself be a homolog of a known clock-related gene. We therefore investigated whether any homologs of known clock-related genes were located in the DNE genomic region, making use of the genomic resources of the related legume Medicago truncatula. Database searches identified a Medicago ELF4 homolog that mapped to a region of chromosome 3 syntenic to the region of pea linkage group III containing the DNE locus (see Supplemental Figure 3 online), suggesting that Ps ELF4 (monitored in the expression experiments in Figures 2 to 4 above) could be a candidate for DNE. We therefore extended the previously reported partial sequence of Ps ELF4 (Hecht et al., 2005) to obtain a full-length cDNA. The Mt ELF4 and Ps ELF4 genes cluster with other legume ELF4-like genes and At ELF4 in a well-supported clade of apparent ELF4 orthologs (see Supplemental Figure 4 online). Three other ELF4-like (ELF4-L) sequences from Medicago cluster in another well-supported clade with previously described Arabidopsis ELF4-like genes ELF4-L2, ELF4-L3, and ELF4-L4 (Khanna et al., 2003; see Supplemental Figure 4 online). We mapped Ps ELF4 close to marker R12_320, previously shown to be closely linked to DNE (Rameau et al., 1998) (see Supplemental Figure 3 online), confirming the conserved genomic location of these genes in pea and Medicago. Sequencing of Ps ELF4 from dne-1 revealed a mutation predicted to replace Gln-64 (CAG) with a stop codon (TAG) (Figure 7A), which cosegregated perfectly with the early-flowering phenotype in >500 progeny from segregating families. This shows that DNE is tightly linked to the ELF4 gene at a distance of <0.2 centimorgans.
Figure 7.
DNE Is the Likely Pea Ortholog of Arabidopsis ELF4.
(A) Alignment of ELF4 protein sequences. Conserved amino acids are shaded in black, and the location of the Gln-64 residue altered by the dne-1 mutation is indicated by an asterisk. Ps, Pisum sativum; At, Arabidopsis thaliana; Le, Lycopersicon esculentum; Vv, Vitis vinifera; He, Helianthus exilis.
(B) to (E) Complementation of the Arabidopsis elf4 mutant. All plants were grown in growth cabinets at 20°C in either LD (16 h) or SD (8 h) conditions.
(B) Total leaf number at flowering of Arabidopsis plants in LDs and SDs. Data are mean ± se for n = 3.
(C) Representative plants grown in LDs. Flower induction has occurred in elf4-1 and elf4-1 expressing mutated Ps ELF4 (dne-1), whereas other plants have not yet flowered.
(D) Representative plants grown in SDs, showing elongated petioles in elf4-1 and elf4-1 expressing mutated Ps ELF4 (dne-1), whereas other plants have normal petiole length.
(E) Representative plants grown in SD, showing flower induction in elf4-1 and elf4-1 complemented with mutated Ps ELF4 (dne-1), whereas other plants have not yet flowered.
Alignment of ELF4 sequences revealed a highly conserved central domain, but little sequence similarity in the short C- and N-terminal extensions (Figure 7A). As the truncated ELF4 protein in the dne mutant would lack most of the conserved central domain, it is likely to be largely functionally inactive, and we tested this by complementation in Arabidopsis. Arabidopsis elf4 mutants are unable to sense daylength and flower early under both LDs and SDs, with elongated hypocotyls and petioles (Doyle et al., 2002; McWatters et al., 2007). Figures 7B to 7E show that Ps ELF4 expressed under the control of the 35S promoter complemented the Arabidopsis elf4-1 mutation under both LDs and SDs, strongly delaying flowering in a manner similar to 35S:At ELF4. By contrast, overexpression of Ps ELF4 carrying the dne-1 mutation had much less of an effect than wild-type Ps ELF4 under both photoperiods despite comparable expression levels (see Supplemental Figure 5 online), and plants continued to produce elongated petioles, suggesting the Ps ELF4 activity had been mostly eliminated by the dne-1 mutation. Under SDs, however, flowering time was significantly later than elf4-1, suggesting some residual function of the truncated dne-1 protein (Figure 7). Nevertheless, the strong impairment of Ps ELF4 function caused by this mutation and the tight cosegregation of the dne mutant phenotype with the mutation strongly support a conclusion that DNE is Ps ELF4.
DNE Also Regulates Stem Elongation
In addition to effects on flowering, the circadian clock regulates other traits, including rhythmic regulation of hypocotyl elongation and leaf expansion (Dowson-Day and Millar, 1999). As Arabidopsis ELF4 is also proposed to function in phyB-mediated deetiolation (Khanna et al., 2003), we also examined deetiolation in the dne mutant. Mutant dne seedlings were indistinguishable from the wild type in both darkness and white light but showed shorter internodes under red, blue, and far-red light, with the proportionately strongest effect seen under blue (Figure 8A). This is in clear contrast with the elongated hypocotyl phenotype seen in the elf4 mutant (Khanna et al., 2003). By contrast, the dne mutant and the wild type did not differ in leaf expansion in darkness or under any light condition (Figure 8A).
Figure 8.
Effects of DNE on Stem Elongation.
(A) Effect of the dne mutation on spectral sensitivity for deetiolation responses. Seedlings were grown for 14 d from sowing under continuous light or darkness. Internode elongation was quantified as the length between nodes 1 and 3, and leaf area was estimated as the product of the length and width of a single leaflet from leaf 3.
(B) Hypocotyl length of Arabidopsis plants showing elongated hypocotyl in elf4-1 and elf4-1 complemented with mutated Ps ELF4 (dne-1) compared with normal hypocotyl lengths in wild-type plants and in elf4-1 mutant plants complemented with wild-type Ps ELF4 (DNE).
All plants were grown in growth cabinets under continuous far-red, red, or blue light at 15 μmol m−2 s−1 (A) or white light at 100 μmol m−2 s−1 ([A] and [B]). Data are mean ± se for n = 8 to 12 (A) or n = 12 to 20 (B).
We also examined whether Ps ELF4 could complement the hypocotyl elongation phenotype of the Arabidopsis elf4 mutant. Figure 8B shows that overexpression of Ps ELF4 in the Arabidopsis elf4-1 mutant resulted in shortened hypocotyls that were comparable in length to the wild type (Wassilewskija [Ws]). No change in hypocotyl length was observed in seedlings overexpressing the dne-1 mutant ELF4 protein, supporting the conclusion from the flowering-time experiment (Figure 7) that the dne-1 mutation severely impairs Ps ELF4 protein function. This result also suggests that the difference in elongation phenotype of the dne and elf4 mutants is due to a species-specific context for DNE/ELF4 protein function, rather than being inherent to the two proteins.
DISCUSSION
Early studies of photoperiod-responsive flowering in pea centered on the physiological and genetic analysis of three loci necessary for inhibition of flowering under noninductive SD photoperiods: SN, DNE, and PPD (Murfet, 1971; King and Murfet, 1985; Taylor and Murfet, 1996). More recent studies have identified genes necessary for promotion of flowering under inductive LD photoperiods, including PHYA (Weller et al., 2004) and LATE1, the ortholog of Arabidopsis GI (Hecht et al., 2007), but the primary physiological role and molecular nature of the SN, DNE, and PPD loci have remained unclear. Here, we show that DNE is necessary for the normal rhythmic regulation of circadian clock genes and identify DNE as the pea ortholog of Arabidopsis ELF4.
ELF4 is thought to be a core component Arabidopsis circadian clock and functions in the CCA1/LHY-TOC1 feedback loop of the central oscillator. ELF4 also plays a role in the entrainment of the clock, functioning as part of the light input pathway. Despite these important roles, little is known about the function of ELF4-like genes outside of Arabidopsis, and true orthologs of ELF4 may not exist in monocots (Khanna et al., 2003; Murakami et al., 2007). The identification of DNE thus provides the first opportunity to examine the function of this gene in another species.
Rhythmic Expression of Pea Clock Genes
Diurnal expression rhythms described here for LHY, TOC1, ELF4, and LATE1 are consistent with a previous report (Hecht et al., 2007) and are similar to those of the corresponding Arabidopsis genes (Fowler et al., 1999; Matsushika et al., 2000; Doyle et al., 2002; Mizoguchi et al., 2002) with peak expression of LHY in the morning and peak expression of TOC1, ELF4, and LATE1 in the evening. Clearly rhythmic expression is also seen for pea genes under low irradiance LL, but these rhythms are strongly damped at higher irradiances (Figure 3; see Supplemental Figure 2 online). A direct comparison with Arabidopsis data is difficult due to incomplete reporting of growth conditions in many published studies, but it is clear that strong rhythms do persist for all four genes under LL irradiances above 60 μmol m−2 s−1 (e.g., Park et al., 1999; Alabadi et al., 2001; Doyle et al., 2002; Mizoguchi et al., 2002; Hazen et al., 2005). In addition, even under low-irradiance LL, pea genes show signs of damping by the second circadian cycle. These apparent differences deserve further study but in general do suggest that pea and Arabidopsis differ in their regulation by light.
In Arabidopsis, selective impairment of circadian rhythms under LL is reported for mutants in ELF3, TIC, LUX, and FHY3, genes that are all thought to have a role in input of light signals to the clock (Hicks et al., 1996; McWatters et al., 2000; Covington et al., 2001; Hall et al., 2003; Hazen et al., 2005; Allen et al., 2006). One explanation for the suppressed rhythmicity seen for several pea genes under higher irradiances of LL could be that our standard wild-type line NGB5839 is a natural mutant with reduced function of one of these genes or in another gene needed for light input to the clock. In the future, this could presumably be evaluated using standard release-from-light and phase-response assays. It will also be important to determine whether this unusual circadian phenotype is common to other cultivars or, indeed, to the entire species. In this respect, it is notable that most garden pea cultivars (including NGB5839) and many spring-sown field pea cultivars carry recessive alleles at the HIGH RESPONSE (HR) locus (Murfet, 1973; Lejeune-Hénaut et al., 2008), which confer earlier flowering under SDs and a reduction in the flowering response to photoperiod. The light input mutants elf3, lux, and tic are also early flowering in short days (Zagotta et al., 1996; Hall et al., 2003; Hazen et al., 2005), and it will therefore be of interest to test if the weaker LL rhythms we observe reflect loss of HR function.
In addition to the unexpected damping or loss of rhythms under LL, we also observed differences in the expression rhythms of pea clock genes in DD in comparison to their Arabidopsis counterparts. The patterns of ELF4 and TOC1 expression in DD are similar in pea and Arabidopsis, with both rhythms damping rapidly to trough and median levels, respectively (Strayer et al., 2000; Doyle et al., 2002), and rhythmic GI/LATE1 expression persists in both species (Park et al., 1999; Figure 4). However, the persistence of rhythmic Ps LHY expression in DD (Figure 4) differs from expression patterns of CCA1 and At LHY, which both damp rapidly to near trough levels (Wang and Tobin, 1998), and the residual low amplitude rhythm we observed for pea ELF4 is not apparent in the Arabidopsis data (Doyle et al., 2002). In addition, whereas in Arabidopsis the period of expression for several clock genes in DD is generally longer than 24 h (Hicks et al., 1996; Wang and Tobin, 1998), the rhythms of Ps LHY and LATE1 expression rhythms under DD appeared significantly shorter than 24 h.
In summary, despite the fundamental importance of the circadian clock, significant differences in the expression of clock genes are evident between Arabidopsis and pea. This suggests that the function of clock components and mechanisms for clock entrainment may differ between plant species. Interestingly, a similar conclusion has been drawn from work in another legume, Phaseolus (Kaldis and Prombona, 2006).
Roles of DNE and LATE1 in Diurnal and Circadian Rhythms
To assess the effect of the dne mutation on the pea circadian clock, we analyzed the expression of pea clock genes under both LL and DD. We found that in contrast with Arabidopsis elf4 mutations, which severely impair rhythmic expression of LHY, CCA1, TOC1, and ELF4 under LL (Doyle et al., 2002; McWatters et al., 2007), dne has only relatively minor effects on phase of TOC1, LATE1, and ELF4 in LL, and all four genes cycle with amplitude similar to the wild type. In DD, the dne mutation also causes a reduction in amplitude and a phase advance in the DNE rhythm and an apparent period shortening of the LHY rhythm in DD. Less is known about the effects of Arabidopsis elf4 in DD except that, as in LL, it severely impairs the CCA1 expression rhythm (Doyle et al., 2002). Overall, these results suggest that DNE may have a more subtle role in clock regulation than Arabidopsis ELF4, that there may be a greater degree of redundancy within the family of ELF4-like genes in pea, or that there may be some residual DNE activity in the dne-1 mutant, as suggested from the Arabidopsis complementation experiments. However, regardless of which of these explanations may be true, it is evident that a strong effect of the dne-1 mutation on photoperiodic flowering is associated with only relatively minor effects on circadian rhythms of clock gene expression.
Under LD cycles, Arabidopsis elf4 had no effect on CCA1 expression, while under SDs the CCA1 rhythm in elf4 showed a reduced ability to anticipate dawn and an increased sensitivity to light immediately after dawn (McWatters et al., 2007). By contrast, the pea LHY rhythm was not significantly affected by dne under SDs or LDs, and we found no clear evidence for impaired anticipation of dawn in dne, although this could in part reflect the lower resolution of our measurements and in future should be examined in more detail. It will also be interesting to examine whether DNE, like Arabidopsis ELF4, has a role in the light induction of LHY and in the gating of light signals to the clock. It is notable that in SDs the dne mutation causes a phase delay in TOC1 expression but a phase advance in expression of DNE itself, despite both genes being expressed in the evening. This is difficult to reconcile with a primary effect of DNE on the core clock mechanism and may instead reflect a role in light input. Such a role may also be suggested by the fact that the timing of peak TOC1 expression is less sensitive to photoperiod in dne than in the wild type.
Other comparisons also suggest that DNE does not have a simple interaction with the putative core clock components LHY and TOC1. For example, in DD, where dne clearly affected the LHY rhythm, it had little effect on expression of the evening genes TOC1 and LATE1 (Figure 4A), but under SD cycles, where the rhythm of LHY was not affected, dne did affect both TOC1 and LATE1 (Figure 2A). This could imply either that dne mutation may independently affect light signaling to LHY and TOC1 and/or that it may affect the coupling of LHY and TOC1 expression. In Arabidopsis, most analyses of the core clock mechanism have been conducted in constant light, where coupling of antiphased CCA1/LHY and TOC1 expression rhythms are normally observed. However, it has been shown in deetiolating Arabidopsis seedlings that ELF4 can act independently of TOC1 to regulate CCA1/LHY expression and that rhythmic TOC1 expression does not completely depend on the regulation of CCA1/LHY (Kikis et al., 2005). The presence of an additional factor X necessary for the coupling of the TOC1-GI loop to CCA1/LHY has been predicted from computational modeling, and ELF4 has been proposed as one candidate for X (Locke et al., 2005; Zeilinger et al., 2006). Given the complexity of the circadian clock, more detailed comparisons between pea and Arabidopsis will require the application of similar modeling approaches in both species.
We previously showed that LATE1 has a role in regulating diurnal rhythms in expression of several clock genes under LD cycles (Hecht et al., 2007), implying that LATE1 might function in the circadian clock, and this has now been confirmed by the finding that late1 eliminates rhythmic expression of LHY and DNE after transfer to LL or DD (Figures 3B and 4B). Thus, without LATE1 function, several putative core components of the pea circadian clock are significantly misregulated under both constant and photoperiodic conditions. The effects of late1 on LHY and TOC1 expression in LL are similar to those reported for gi mutants in Arabidopsis (Fowler et al., 1999; Park et al., 1999; Mizoguchi et al., 2005; Gould et al., 2006), whereas effects of gi on ELF4 expression have not been reported. A major role of GI in the Arabidopsis clock is suggested to be the light-dependent regulation of TOC1 protein stability (Kim et al., 2007), but computational modeling also suggests that GI may contribute to the function of a hypothetical clock component Y necessary for regulation of TOC1 expression (Locke et al., 2006). Under LD cycles, gi mutations (in contrast with late1) reduce the amplitude but do not affect the phase of LHY expression. This suggests that either the mechanisms by which GI and LATE1 affect LHY expression may be fundamentally different or that the additional effect of late1 on LHY phase may be a combined effect of the late1 and hr mutations.
Coupling of DNE and LATE1 to Flowering Output Pathways
Previous models for flowering in pea proposed that the dne mutation blocks production of a mobile inhibitor of flowering in SDs (King and Murfet, 1985), a suggestion difficult to reconcile with current understanding of photoperiodic flowering in Arabidopsis, where the primary target of clock regulation (FT) acts as a mobile stimulus (Turck et al., 2008). However, we show here that in grafts with leafy donor tissue, the major effect of DNE in SDs is to inhibit a graft-transmissible flowering stimulus (Figure 5C) and that increased ability of the dne mutant to promote flowering across a graft union is associated with elevated expression of the FT-like gene FTL. The correlation between flowering time, effect on a mobile stimulus, and FTL expression is also seen for the late-flowering photoperiod response mutant late1 under LDs (Hecht et al., 2007).
Together, these results are superficially similar to those from Arabidopsis showing that the induction of FT expression is necessary for the LD response and that ELF4 and GI act in opposite ways to regulate expression of FT (Suárez-López et al., 2001; Doyle et al., 2002). In Arabidopsis, this regulation occurs at least in part through CO (Doyle et al., 2002; Mizoguchi et al., 2005). However, as reported previously for the late1 mutant (Hecht et al., 2007), dne also had no effect on the COLa expression rhythm under conditions where its flowering phenotype is strongest (Figure 6A). It is possible that other COL genes may have assumed the function of Arabidopsis CO, and this can now be addressed using reverse genetics (Hecht et al., 2005; Dalmais et al., 2008; Tadege et al., 2008).
We also used a dne late1 double mutant to examine the interaction between DNE and LATE1 in control of flowering. In most respects late1 and dne show a straightforward interaction in which late1 is epistatic to dne, with respect to overall phenotype in both SDs and LDs (Figures 5A and 5B) and graft-transmissible effects on flowering in SDs (Figure 5C), suggesting that LATE1 is necessary for DNE effects on both a mobile flowering stimulus and on general photoperiod responsiveness. However, a more complex interaction between dne and late1 in the control of flower initiation is suggested by the early flower initiation of flower primordia in the dne late1 double mutant. This distinct phenotype of the dne late1 double suggests that dne can affect flowering independently of late1, a conclusion that is superficially contradictory to the observation that late1 completely blocked the effect of dne on flowering in graft stocks (Figure 5C). However, these two experiments differ in that the intact dne late1 plants carried the dne and late1 mutations in all tissues, whereas the grafted plants carried the mutations in the graft stock only, which might mean that the overall effect of the dne mutation was less in these plants. Alternatively, the difference could reflect the existence of a heterogenous mobile flowering signal and differential effects of dne and late1 on components of such a signal. In this respect, an interesting feature of the early initiation in dne late1 is that it occurs in the apparent absence of any increase in FTL expression level in leaf tissue. This indicates that DNE can act independently of both LATE1 and FTL specifically to regulate the induction of flowering. One possible interpretation for these results is that the role of FT in pea may not be performed by FTL alone but by one or more additional FT-like genes.
In Arabidopsis, the FT family has two members, FT and TSF, which have similar regulatory characteristics and functions (Yamaguchi et al., 2005), but several recent studies have shown that the FT family in other species is expanded relative to Arabidopsis, with individual members showing distinct patterns of regulation with respect to daylength, season, and tissue specificity (Izawa et al., 2002; Faure et al., 2007; Danilevskaya et al., 2008; Igasaki et al., 2008) and interactions with different downstream partners (Li and Dubcovsky, 2008). In M. truncatula, the FT gene family consists of five genes, and the pea FTL gene described here is the apparent ortholog of Mt FTLe (Hecht et al., 2005; see Supplemental Figure 6 online). If the early initiation of flowering in the dne late1 double mutant is due to expression of another FTL gene, this would imply the existence of at least two pea FTL genes induced under LD: one associated with general photoperiod responsiveness and one with a narrower role in initiation of flowering. We have recently found that in Medicago both FTLe and FTLa are upregulated under LD and that FTLa has a significant role in regulation of flower induction under LD (R.E. Laurie, M. Tadege, K. Mysore, J.L. Weller, and R.C. Macknight, unpublished data). It will be of interest to determine whether this is also the case in pea, whether these genes are differentially regulated by DNE and LATE1, and whether they have distinct functions. Unraveling the roles of the different legume FTL genes and understanding how they are regulated will be the focus of future work.
METHODS
Plant Material, Growth Conditions, and Grafting
The origins of the le-3, dne-1, and late1-2 mutants have been described previously (King and Murfet, 1985; Hecht et al., 2007). Seedling deetiolation experiments (Figure 8) gene expression studies (Figures 2 to 4 and 6) and Arabidopsis thaliana flowering experiments (Figure 7) were conducted in growth cabinets at 20°C, whereas photoperiod and grafting experiments (Figures 1 and 5) were conducted in the Hobart phytotron, using previously described growth media, light sources, phytotron conditions, and grafting protocols (Hecht et al., 2007). Standard phytotron SD conditions consisted of an 8-h photoperiod of natural light, which was extended for 8 h with white light from fluorescent tubes at an irradiance of 10 μmol m−2 s−1 to give a 16-h LD. Spectral scans for all artificial light sources used can be viewed at http://www.utas.edu.au/glasshouse/gh_facilities.html.
Gene Isolation, Mapping, and Molecular Genotyping
The full-length Ps ELF4/DNE cDNA was obtained by rapid amplification of cDNA ends-PCR using the BD-SMART RACE cDNA amplification kit (Clontech) and gene-specific primers (ELF4-GSP2 and ELF4-2R for the 5′ region). All PCR fragments were cloned in pGEM-T easy (Promega) and sequenced at the Australian Genome Research Facility. The dne-1 mutation was detected as a cleaved amplified polymorphic sequence (marker, and cosegregation with the dne phenotype was confirmed in segregating progenies from several different crosses. For mapping of Ps ELF4/DNE, a polymorphism was identified and scored as a derived cleaved amplified polymorphic sequence marker in the JI281 × JI399 recombinant inbred line population (Hall et al., 1997). All primer details are given in Supplemental Table 1 online. For phylogenetic trees shown in Supplemental Figures 4 and 6 online, amino acid sequences of proteins related to ELF4 and FT were aligned using ClustalX (Thompson et al., 1997) Distance and parsimony-based methods were used for phylogenetic analyses in PAUP*4.0b10 (http://paup.csit.fsu.edu/) using the alignments shown. The tree in Supplemental Figure 4 online is rooted with a putative ELF4 ortholog from the chlorophyte Chlamydomonas reinhardtii as the outgroup, whereas the tree of FT-related proteins in Supplemental Figure 6 online is rooted at the midpoint between the FT clade and the other clades.
Complementation Studies
The Arabidopsis elf4-1 mutation in the Ws background has been previously described (Doyle et al., 2002). Full-length cDNA fragments for ELF4 were generated by PCR from pea (Pisum sativum) wild-type line NGB5839 and the isogenic dne-1 mutant, and from Arabidopsis accession Ws, using primers listed in Supplemental Table 1 online. The cDNA fragments were recombined into the binary vector pB2GW7 (Invitrogen) using Gateway cloning (Karimi et al., 2002) and confirmed by sequencing. To measure hypocotyl length, seeds were surface sterilized and plated on 4 g/L Murashige and Skoog without sucrose and 8 g/L agar. Plates were stored at 4°C in the dark for 48 h and transferred into growth chambers with the appropriate light regimes.
Gene Expression Studies
Harvested tissue consisted of both leaflets from the uppermost fully expanded leaf. Samples were frozen in liquid nitrogen and total RNA extracted using the Promega SV Total RNA isolation system (Promega). RNA concentrations were determined using Ribogreen RNA quantification reagent (Molecular Probes) in a Picofluor fluorometer (Turner Biosystem). Reverse transcription was conducted in 20 μL with 1 μg of total RNA using the ImProm-II reverse transcription system (Promega) according to the manufacturer's instructions. RT-negative (no enzyme) controls were routinely performed to monitor for contamination with genomic DNA. First-strand cDNA was diluted five times, and 2 μL was used in each real-time PCR reaction. Real-time PCR reactions using SYBR green chemistry (Quantace Sensimix) were set up with a CAS-1200N robotic liquid handling system (Corbett Research) and run for 50 cycles in a Rotor-Gene RG3000 (Corbett Research). Two technical replicates and two to three biological replicates were performed for each sample. Transcript levels for experimental genes were evaluated against the constitutive gene ACTIN (ACT) as previously described (Weller et al., 2009). Primer sequences are given in Supplemental Table 1 online.
Accession Numbers
Genomic and cDNA sequences are deposited in GenBank/EMBL under the following accession numbers: AY830926 (Ps ELF4 genomic/cDNA), FJ609177 (PRR37 cDNA), FJ609178 (PRR37 genomic), FJ609179 (PRR59 cDNA), and FJ609180 (PRR59 genomic). Accession numbers for other sequences used are as follows: P. sativum ACTIN (X68649), LHY (AY826730), TOC1 (AY830927), LATE1 (EF185297), COLa (AY830921), and FTL (AAX47174), and Arabidopsis ELF4 (NM_129566, At2G40080).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Effect of dne Mutation on Flower Initiation in the Tall (LE) Genetic Background.
Supplemental Figure 2. Circadian Regulation of Pea Clock Gene Expression in LL at 150 μmol m−2 s−1.
Supplemental Figure 3. Comparative Map of Pea Linkage Group III and Medicago Chromosome 3.
Supplemental Figure 4. Phylogram for ELF4-Like Protein Sequences Aligned with ClustalX and Rooted to Cs ELF4.
Supplemental Figure 5. 35S:Ps ELF4 and 35S:Ps ELF4(dne-1) Are Expressed at Similar Levels in Arabidopsis elf4-1 Plants.
Supplemental Figure 6. Phylogram for FTL Protein Sequences Aligned with ClustalX.
Supplemental Table 1. Primers Sequences Used in Gene Isolation, Mapping, and Mutation Detection and Real-Time PCR.
Supplementary Material
Acknowledgments
We thank Ian Cummings, Tracey Winterbottom, Chris Blackman, and Scott Taylor for plant maintenance, Ian Cummings for assistance with controlled environments, and Noel Ellis for use of the JI281 × JI399 mapping population. This work was supported by the Australian Research Council (J.L.W.), a University of Tasmania International Postgraduate Research Scholarship (L.C.L.), an AGMARDT Postdoctoral Fellowship (R.E.L.), and the New Zealand Foundation for Research, Science, and Technology (R.C.M.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: James L. Weller (jim.weller@utas.edu.au).
Online version contains Web-only data.
References
- Alabadi, D., Oyama, T., Yanovsky, M.J., Harmon, F.G., Mas, P., and Kay, S.A. (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293 880–883. [DOI] [PubMed] [Google Scholar]
- Allen, T., Koustenis, A., Theodorou, G., Somers, D.E., Kay, S.A., Whitelam, G.C., and Devlin, P.F. (2006). Arabidopsis FHY3 specifically gates phytochrome signaling to the circadian clock. Plant Cell 18 2506–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhlenius, H., Huang, T., Charbonnel-Campaa, L., Brunner, A.M., Jansson, S., Strauss, S.H., and Nilsson, O. (2006). CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 312 1040–1043. [DOI] [PubMed] [Google Scholar]
- Covington, M.F., Panda, S., Liu, X.L., Strayer, C.A., Wagner, D.R., and Kay, S.A. (2001). ELF3 modulates resetting of the circadian clock in Arabidopsis. Plant Cell 13 1305–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmais, M., Schmidt, J., Le Signor, C., Moussy, F., Burstin, J., Savois, V., Aubert, G., Brunaud, V., de Oliveira, Y., Guichard, C., Thompson, R., and Bendahmane, A. (2008). UTILLdb, a Pisum sativum in silico forward and reverse genetics tool. Genome Biol. 9 R43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilevskaya, O.N., Meng, X., Hou, Z., Ananiev, E.V., and Simmons, C.R. (2008). A genomic and expression compendium of the expanded PEBP gene family from maize. Plant Physiol. 146 250–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, Z., Millar, A.J., Davis, A.M., and Davis, S.J. (2007). TIME FOR COFFEE encodes a nuclear regulator in the Arabidopsis thaliana circadian clock. Plant Cell 19 1522–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi, K., Izawa, T., Fuse, T., Yamanouchi, U., Kubo, T., Shimatani, Z., Yano, M., and Yoshimura, A. (2004). Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 18 926–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowson-Day, M.J., and Millar, A.J. (1999). Circadian dysfunction causes aberrant hypocotyl elongation patterns in Arabidopsis. Plant J. 17 63–71. [DOI] [PubMed] [Google Scholar]
- Doyle, M.R., Davis, S.J., Bastow, R.M., McWatters, H.G., Kozma-Bognar, L., Nagy, F., Millar, A.J., and Amasino, R.M. (2002). The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419 74–77. [DOI] [PubMed] [Google Scholar]
- Faure, S., Higgins, J., Turner, A., and Laurie, D.A. (2007). The FLOWERING LOCUS T-Like gene family in barley (Hordeum vulgare). Genetics 176 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler, S., Lee, K., Onouchi, H., Samach, A., Richardson, K., Morris, B., Coupland, G., and Putterill, J. (1999). GIGANTEA: A circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 18 4679–4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara, S., Oda, A., Yoshida, R., Niinuma, K., Miyata, K., Tomozoe, Y., Tajima, T., Nakagawa, M., Hayashi, K., Coupland, G., and Mizoguchi, T. (2008). Circadian clock proteins LHY and CCA1 regulate SVP protein accumulation to control flowering in Arabidopsis. Plant Cell 20 2960–2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner, M.J., Hubbard, K.E., Hotta, C.T., Dodd, A.N., and Webb, A.A. (2006). How plants tell the time. Biochem. J. 397 15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould, P.D., Locke, J.C.W., Larue, C., Southern, M.M., Davis, S.J., Hanano, S., Moyle, R., Milich, R., Putterill, J., Millar, A.J., and Hall, A. (2006). The molecular basis of temperature compensation in the Arabidopsis circadian clock. Plant Cell 18 1177–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, A., Bastow, R.M., Davis, S.J., Hanano, S., McWatters, H.G., Hibberd, V., Doyle, M.R., Sung, S., Halliday, K.J., Amasino, R.M., and Millar, A.J. (2003). The TIME FOR COFFEE gene maintains the amplitude and timing of Arabidopsis circadian clocks. Plant Cell 15 2719–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, K.J., Parker, J.S., Ellis, T.H., Turner, L., Knox, M.R., Hofer, J.M., Lu, J., Ferrandiz, C., Hunter, P.J., Taylor, J.D., and Baird, K. (1997). The relationship between genetic and cytogenetic maps of pea. II. Physical maps of linkage mapping populations. Genome 40 755–769. [DOI] [PubMed] [Google Scholar]
- Hayama, R., Agashe, B., Luley, E., King, R., and Coupland, G. (2007). A circadian rhythm set by dusk determines the expression of FT homologs and the short-day photoperiodic flowering response in Pharbitis. Plant Cell 19 2988–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama, R., and Coupland, G. (2004). The molecular basis of diversity in the photoperiodic flowering responses of Arabidopsis and rice. Plant Physiol. 135 677–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen, S.P., Schultz, T.F., Pruneda-Paz, J.L., Borevitz, J.O., Ecker, J.R., and Kay, S.A. (2005). LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc. Natl. Acad. Sci. USA 102 10387–10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht, V., Foucher, F., Ferrandiz, C., Macknight, R., Navarro, C., Morin, J., Vardy, M.E., Ellis, N., Beltran, J.P., Rameau, C., and Weller, J.L. (2005). Conservation of Arabidopsis flowering genes in model legumes. Plant Physiol. 137 1420–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht, V., Knowles, C.L., Vander Schoor, J.K., Liew, L.C., Jones, S.E., Lambert, M.J.M., and Weller, J.L. (2007). Pea LATE BLOOMER1 is a GIGANTEA ortholog with roles in photoperiodic flowering, deetiolation, and transcriptional regulation of circadian clock gene homologs. Plant Physiol. 144 648–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks, K.A., Millar, A.J., Carre, I.A., Somers, D.E., Straume, M., Meeks-Wagner, D.R., and Kay, S.A. (1996). Conditional circadian dysfunction of the Arabidopsis early-flowering 3 mutant. Science 274 790–792. [DOI] [PubMed] [Google Scholar]
- Igasaki, T., Watanabe, Y., Nishiguchi, M., and Kotoda, N. (2008). The FLOWERING LOCUS T/TERMINAL FLOWER 1 family in Lombardy Poplar. Plant Cell Physiol. 49 291–300. [DOI] [PubMed] [Google Scholar]
- Imaizumi, T., and Kay, S.A. (2006). Photoperiodic control of flowering: Not only by coincidence. Trends Plant Sci. 11 550–558. [DOI] [PubMed] [Google Scholar]
- Izawa, T., Oikawa, T., Sugiyama, N., Tanisaka, T., Yano, M., and Shimamoto, K. (2002). Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Genes Dev. 16 2006–2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, J.-H., Seo, Y.-H., Seo, P.J., Reyes, J.L., Yun, J., Chua, N.-H., and Park, C.-M. (2007). The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 19 2736–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaldis, A.D., and Prombona, A. (2006). Synergy between the light-induced acute response and the circadian cycle: A new mechanism for the synchronization of the Phaseolus vulgaris clock to light. Plant Mol. Biol. 61 883–895. [DOI] [PubMed] [Google Scholar]
- Karimi, M., Inzé, D., and Depicker, A. (2002). GATEWAYTM vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7 193–195. [DOI] [PubMed] [Google Scholar]
- Khanna, R., Kikis, E.A., and Quail, P.H. (2003). EARLY FLOWERING 4 functions in phytochrome B-regulated seedling de-etiolation. Plant Physiol. 133 1530–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikis, E.A., Khanna, R., and Quail, P.H. (2005). ELF4 is a phytochrome-regulated component of a negative-feedback loop involving the central oscillator components CCA1 and LHY. Plant J. 44 300–313. [DOI] [PubMed] [Google Scholar]
- Kim, W.-Y., Fujiwara, S., Suh, S.-S., Kim, J., Kim, Y., Han, L., David, K., Putterill, J., Nam, H.G., and Somers, D.E. (2007). ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449 356–362. [DOI] [PubMed] [Google Scholar]
- Kim, W.-Y., Hicks, K.A., and Somers, D.E. (2005). Independent roles for EARLY FLOWERING 3 and ZEITLUPE in the control of circadian timing, hypocotyl length, and flowering time. Plant Physiol. 139 1557–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, W.M., and Murfet, I.C. (1985). Flowering in Pisum: A sixth locus, Dne. Ann. Bot. (Lond.) 56 835–846. [Google Scholar]
- Kobayashi, Y., and Weigel, D. (2007). Move on up, it's time for change—Mobile signals controlling photoperiod-dependent flowering. Genes Dev. 21 2371–2384. [DOI] [PubMed] [Google Scholar]
- Kojima, S., Takahashi, Y., Kobayashi, Y., Monna, L., Sasaki, T., Araki, T., and Yano, M. (2002). Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 43 1096–1105. [DOI] [PubMed] [Google Scholar]
- Lejeune-Hénaut, I., et al. (2008). The flowering locus Hr colocalizes with a major QTL affecting winter frost tolerance in Pisum sativum L. Theor. Appl. Genet. 116 1105–1116. [DOI] [PubMed] [Google Scholar]
- Lester, D.R., Ross, J.J., Davies, P.J., and Reid, J.B. (1997). Mendels stem length gene (Le) encodes a gibberellin 3B-hydroxylase. Plant Cell 9 1435–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C., and Dubcovsky, J. (2008). Wheat FT protein regulates VRN1 transcription through interactions with FDL2. Plant J. 55 543–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke, J.C., Kozma-Bognar, L., Gould, P.D., Feher, B., Kevei, E., Nagy, F., Turner, M.S., Hall, A., and Millar, A.J. (2006). Experimental validation of a predicted feedback loop in the multi-oscillator clock of Arabidopsis thaliana. Mol. Syst. Biol. 2 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke, J.C., Southern, M.M., Kozma-Bognar, L., Hibberd, V., Brown, P.E., Turner, M.S., and Millar, A.J. (2005). Extension of a genetic network model by iterative experimentation and mathematical analysis. Mol. Syst. Biol. 1: 2005.0013. [DOI] [PMC free article] [PubMed]
- Matsushika, A., Makino, S., Kojima, M., and Mizuno, T. (2000). Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: Insight into the plant circadian clock. Plant Cell Physiol. 41 1002–1012. [DOI] [PubMed] [Google Scholar]
- McClung, C.R. (2008). Comes a time. Curr. Opin. Plant Biol. 11 514–520. [DOI] [PubMed] [Google Scholar]
- McWatters, H.G., Bastow, R.M., Hall, A., and Millar, A.J. (2000). The ELF3 zeitnehmer regulates light signalling to the circadian clock. Nature 408 716–720. [DOI] [PubMed] [Google Scholar]
- McWatters, H.G., Kolmos, E., Hall, A., Doyle, M.R., Amasino, R.M., Gyula, P., Nagy, F., Millar, A.J., and Davis, S.J. (2007). ELF4 is required for oscillatory properties of the circadian clock. Plant Physiol. 144 391–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi, T., Wheatley, K., Hanzawa, Y., Wright, L., Mizoguchi, M., Song, H.R., Carre, I.A., and Coupland, G. (2002). LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev. Cell 2 629–641. [DOI] [PubMed] [Google Scholar]
- Mizoguchi, T., Wright, L., Fujiwara, S., Cremer, F., Lee, K., Onouchi, H., Mouradov, A., Fowler, S., Kamada, H., Putterill, J., and Coupland, G. (2005). Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell 17 2255–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami, M., Ashikari, M., Miura, K., Yamashino, T., and Mizuno, T. (2003). The evolutionarily conserved OsPRR quintet: Rice pseudo-response regulators implicated in circadian rhythm. Plant Cell Physiol. 44 1229–1236. [DOI] [PubMed] [Google Scholar]
- Murakami, M., Tago, Y., Yamashino, T., and Mizuno, T. (2007). Comparative overviews of clock-associated genes of Arabidopsis thaliana and Oryza sativa. Plant Cell Physiol. 48 110–121. [DOI] [PubMed] [Google Scholar]
- Murfet, I.C. (1971). Flowering in Pisum. A three-gene system. Heredity 27 93–110. [Google Scholar]
- Murfet, I.C. (1973). Flowering in Pisum. Hr, a gene for high response to photoperiod. Heredity 31 157–164. [Google Scholar]
- Park, D.H., Somers, D.E., Kim, Y.S., Choy, Y.H., Lim, H.K., Soh, M.S., Kim, H.J., Kay, S.A., and Nam, H.G. (1999). Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285 1579–1582. [DOI] [PubMed] [Google Scholar]
- Rameau, C., Denoue, D., Fraval, F., Haurogne, K., Josserand, J., Laucou, V., Batge, S., and Murfet, I.C. (1998). Genetic mapping in pea. 2. Identification of RAPD and SCAR markers linked to genes affecting plant architecture. Theor. Appl. Genet. 97 916–928. [Google Scholar]
- Rodríguez-Falcón, M., Bou, J., and Prat, S. (2006). Seasonal control of tuberization in potato: conserved elements with the flowering response. Annu. Rev. Plant Biol. 57 151–180. [DOI] [PubMed] [Google Scholar]
- Serikawa, M., Miwa, K., Kondo, T., and Oyama, T. (2008). Functional conservation of clock-related genes in flowering plants: Overexpression and RNA interference analyses of the circadian rhythm in the monocotyledon Lemna gibba. Plant Physiol. 146 1952–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strayer, C., Oyama, T., Schultz, T.F., Raman, R., Somers, D.E., Mas, P., Panda, S., Kreps, J.A., and Kay, S.A. (2000). Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289 768–771. [DOI] [PubMed] [Google Scholar]
- Suárez-López, P., Wheatley, K., Robson, F., Onouchi, H., Valverde, F., and Coupland, G. (2001). CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410 1116–1120. [DOI] [PubMed] [Google Scholar]
- Tadege, M., Wen, J., He, J., Tu, H., Kwak, Y., Eschstruth, A., Cayrel, A., Endre, G., Zhao, P.X., Chabaud, M., Ratet, P., and Mysore, K.S. (2008). Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J. 54 335–347. [DOI] [PubMed] [Google Scholar]
- Takahashi, Y., Shomura, A., Sasaki, T., and Yano, M. (2001). Hd6, a rice quantitative trait locus involved in photoperiod sensitivity, encodes the alpha subunit of protein kinase CK2. Proc. Natl. Acad. Sci. USA 98 7922–7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, S.A., and Murfet, I.C. (1996). Flowering in Pisum: Identification of a new ppd allele and its physiological action as revealed by grafting. Physiol. Plant. 97 719–723. [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. (1997). The ClustalX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevaskis, B., Tadege, M., Hemming, M.N., Peacock, W.J., Dennis, E.S., and Sheldon, C. (2007). Short vegetative phase-like MADS-box genes inhibit floral meristem identity in barley. Plant Physiol. 143 225–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck, F., Fornara, F., and Coupland, G. (2008). Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu. Rev. Plant Biol. 59 573–594. [DOI] [PubMed] [Google Scholar]
- Turner, A., Beales, J., Faure, S., Dunford, R.P., and Laurie, D.A. (2005). The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310 1031–1034. [DOI] [PubMed] [Google Scholar]
- Wang, Z.-Y., and Tobin, E.M. (1998). Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93 1207–1217. [DOI] [PubMed] [Google Scholar]
- Weller, J.L. (2005). Mobile flowering signals in pea. Flowering Newslett. 36 15–24. [Google Scholar]
- Weller, J.L. (2007). Update on the genetics of flowering. Pisum Genet. 39 1–8. [Google Scholar]
- Weller, J.L., Batge, S.L., Smith, J.J., Kerckhoffs, L.H.J., Sineshchekov, V.A., Murfet, I.C., and Reid, J.B. (2004). A dominant mutation in the pea PHYA gene confers enhanced responses to light and impairs the light-dependent degradation of phytochrome A. Plant Physiol. 135 2186–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller, J.L., Hecht, V., Vander Schoor, J.K., Davidson, S.E., and Ross, J.J. (2009). Light regulation of gibberellin biosynthesis in pea is mediated through the COP1/HY5 pathway. Plant Cell 21 800–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller, J.L., Reid, J.B., Taylor, S.A., and Murfet, I.C. (1997). The genetic control of flowering in pea. Trends Plant Sci. 2 412–418. [Google Scholar]
- Yamaguchi, A., Kobayashi, Y., Goto, K., Abe, M., and Araki, T. (2005). TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant Cell Physiol. 46 1175–1189. [DOI] [PubMed] [Google Scholar]
- Yanovsky, M.J., and Kay, S.A. (2002). Molecular basis of seasonal time measurement in Arabidopsis. Nature 419 308–312. [DOI] [PubMed] [Google Scholar]
- Zagotta, M.T., Hicks, K.A., Jacobs, C.I., Young, J.C., Hangarter, R.P., and Meeks-Wagner, D.R. (1996). The Arabidopsis ELF3 gene regulates vegetative photomorphogenesis and the photoperiodic induction of flowering. Plant J. 10 691–702. [DOI] [PubMed] [Google Scholar]
- Zeilinger, M.N., Farre, E.M., Taylor, S.R., Kay, S.A., and Doyle III, F.J. (2006). A novel computational model of the circadian clock in Arabidopsis that incorporates PRR7 and PRR9. Mol. Syst. Biol. 2 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.