Abstract
Neutralizing antibody (nAb) response is sporadic and has limited potency and breadth during infection with human immunodeficiency virus type 1 (HIV-1). In rare cases, broad and potent nAbs are actually induced in vivo. Identifying specific epitopes targeted by such broad and potent nAb response is valuable in guiding the design of a prophylactic vaccine aimed to induce nAb. In this study, we have defined neutralizing epitope usage in 7 out of 17 subjects with broad and potent nAbs by using targeted mutagenesis in known neutralizing epitopes of HIV-1 glycoproteins and by using in vitro depletion of serum neutralizing activity by various recombinant HIV-1 glycoproteins. Consistent with recent reports, the CD4 binding site (CD4BS) is targeted by nAbs in vivo (4 of the 7 subjects with defined neutralizing epitopes). The new finding from this study is that epitopes in the gp120 outer domain are also targeted by nAbs in vivo (5 of the 7 subjects). The outer domain epitopes include glycan-dependent epitopes (2 subjects), conserved non-linear epitope in the V3 region (2 subjects), and a CD4BS epitope composed mainly of the elements in the outer domain (1 subject). Importantly, we found indication for epitope poly-specificity, a dual usage of the V3 and CD4BS epitopes, in only one subject. This study provides a more complete profile of epitope usage for broad and potent nAb responses during HIV-1 infection.
Introduction
Human immunodeficiency virus type 1 (HIV-1) infection only in rare cases induces potent neutralizing antibody (nAb) responses that can effectively neutralize diverse primary strains of HIV-1 (Deeks et al., 2006; Dhillon et al., 2007; Li et al., 2007; Li et al., 2009; Sather et al., 2009; Shen et al., 2009). Even in such cases, serum neutralizing titers are often low, with IC50 values in the dilution range of 1:10 to 1:100. The nature of such broad and potent nAbs to HIV-1 envelope glycoproteins (Envs) is of interest for vaccine development, as passive immunization with broad and potent nAbs can prevent virus infection in the rhesus macaque model of SHIV infection (Mascola et al., 1999; Parren et al., 2001; Shibata et al., 1999; Veazey et al., 2003). The existence of nAbs with high neutralizing potency and breadth in vivo has been demonstrated by the characterization of monoclonal antibodies (mAbs) 2G12, 2F5 and 4E10, generated by hybridoma formation or EBV transformation of B lymphocytes (Muster et al., 1993; Stiegler et al., 2001; Trkola et al., 1996; Zwick et al., 2001). Of note, the broad and potent neutralizing mAb - b12 was produced using recombinant phage technology while mAbs PG9 and PG16 were produced using recombinant techniques based on near-clonal B-cell cultures (Burton et al., 1994; Walker et al., 2009). Many mAbs either have limited potency but relatively good breadth or have high potency but are strain-specific in neutralization [reviewed in (Wyatt and Sodroski, 1998; Zolla-Pazner, 2005)]. Hypothetically, broad and potent nAb responses in vivo may also be composed of a high concentration of antibodies with limited potency/good breadth or a large number of antibodies with limited breadth/high potency, or some combination of these extremes.
HIV-1 Envs exist on the virion surface as a trimer of gp120/gp41 heterodimers, representing the only viral target for Env-specific nAbs. Primary sequences of HIV-1 gp120 have five conserved (C1–C5) and five variable (V1–V5) regions; and the gp41 ectodomain is well-conserved among HIV-1 variants (Gaschen et al., 2001; Kuiken, Korber, and Shafer, 2003; Modrow et al., 1987; Myers et al., 1992; Starcich et al., 1986; Willey et al., 1986). The gp120 variable regions are exposed on the mature Env trimer and are heavily glycosylated, protecting the conserved gp120 structures from nAbs (Modrow et al., 1987; Starcich et al., 1986; Wyatt et al., 1998). Most known broad nAbs to HIV-1 Envs target epitopes in five structural regions. 1: The CD4BS is the binding target for b12, a broad and potent nAb, as well as some broad but less-potent nAbs, such as F105, F91 and 1.5e (Burton et al., 1994; Moore et al., 1994; Posner et al., 1993; Robinson et al., 1990). Typical CD4BS epitopes lie between the inner and outer domains of gp120, thus comprising elements of both domains. Both recombinant gp120 monomer and gp140 trimer contain such epitopes (Yang et al., 2000b; Yang et al., 2002). Interestingly, the b12 epitope is largely made of elements from gp120 outer domain and is capable of binding the recombinant form of gp120 outer domain known as OD1, in addition to gp120 and gp140 (Yang et al., 2004; Zhou et al., 2007). 2: The CD4-induced (CD4i) site is the gp120 core structure binding to the co-receptor, either CCR5 or CXCR4 (Moore and Sodroski, 1996; Thali et al., 1993; Xiang et al., 2003). It is poorly exposed prior to CD4-engagement and virus attachment, thus is a poor target for nAbs (Decker et al., 2005; Li et al., 2006). The V3 loop is adjacent to this site and contributes significantly to co-receptor binding. V3-specific nAbs are mostly strain-specific, but a few anti-V3 antibodies have also been described to have somewhat greater neutralizing breadth (Gorny et al., 1997; Gorny et al., 2002; Stanfield et al., 2004). The V3 region is a part of the gp120 outer domain. The OD1 protein, but not its V3-deleted version (ODΔV3), can bind V3-specific antibodies that are conformation-dependent and neutralizing but not those that target the linear V3 sequences and are non-neutralizing (Yang et al., 2004). 3: 2G12 is the only nAb targeting the glycan mass on the surface of gp120 outer domain, known as the “silent face” (Trkola et al., 1996). 2G12 efficiently binds to recombinant HIV-1 glycoproteins including gp140 trimer, gp120 monomer, and OD1. 4: The V2 loop is recently identified as the target for mAbs PG9 and PG16 which neutralize almost all group M HIV-1 strains with great strength (Walker et al., 2009). The epitope for PG16 also involves the V3 structure. 5: The membrane proximal external region (MPER) of the gp41 ectodomain contains two well-characterized neutralizing epitopes - 2F5 and 4E10 (Muster et al., 1993; Zwick et al., 2001). The gp140 trimer contains the sequences covering these epitopes, but not the gp120 monomer (Yang et al., 2002). Interestingly, the gp120 outer domain contributes major binding surfaces for the first 3 groups of nAbs, making it a high density target for nAb responses (Kwong et al., 1998; Yang et al., 2004; Zhou et al., 2007).
Recent efforts have demonstrated the use of CD4BS in a few individuals in vivo by employing recombinant HIV-1 gp120 and its epitope-specific mutant derivatives to bind and deplete nAbs from antisera containing broad and potent nAbs (Binley et al., 2008; Dhillon et al., 2007; Gray et al., 2009b; Li et al., 2007; Li et al., 2009; Sather et al., 2009). The gp41 MPER epitopes have also been reported as targets of nAbs with various neutralizing breadth and potency (Gray et al., 2009a; Gray et al., 2009b; Shen et al., 2009). In this report, we have characterized nAb responses in chronic HIV-1 infections using a panel of HIV-1 Env mutants to detect the change of sensitivity to neutralization due to alterations in known immunoepitopes. Complemented by the commonly used technique of depleting neutralizing activities from antiserum using recombinant HIV-1 Env proteins, we found that in addition to the CD4BS site, epitopes located in the gp120 outer domain, including glycan-dependent epitopes and conserved elements of the V3 region, are also targeted by broad and potent nAbs developed in chronic HIV-1 infection.
Materials and Methods
Patient Populations
The subjects came from three independent groups of patients chronically infected with HIV-1. The first group was from clinical centers located in Africa and the United States, managed by the Center for HIV/AIDS Vaccine Immunology (CHAVI) as the CHAVI001 study cohort. This group included adults based on standard clinical diagnosis of HIV-1 infection with no past or current usage of HAART. The second group included patients cared for by the King’s College Hospital, London, UK and the MRC Laboratories, Fajara, Gambia. Patients from the King’s College Hospital comprised a large proportion of immigrants from Africa. These patients were diagnosed with standard clinical tests for at least 6 months and had no history of HAART or had stopped all antiretroviral therapy for at least 3 months. The third group included patients at the University of Alabama Medical Center, Birmingham, Alabama. These patients were diagnosed by standard clinical tests with confirmation by the detection of HIV-1 vRNA. These subjects were selected to (1) have documented HIV-1 infection for more than 1 year, (2) have no history of HAART or to have stopped all antiretroviral therapy for at least 3 months, and (3) have lower viral load (<11,000 vRNA/ml plasma) without therapy. In summary, our study subjects were of diverse background and were chronically infected with HIV-1 but not treated with HAART at the time of sampling.
Titration of HIV-1-neutralizing Activity in Plasma or Serum Samples
All plasma and serum samples were first incubated at 56°C for 30 min to inactivate complements. Neutralization titers were determined against a standard panel of tier-2 reference strains of HIV-1 including 6 each from clades B and C, performed according to standard operating procedure in the laboratory of Dr. D. Montefiori (Duke Human Vaccine Institute, Duke University Medical Center) (Li et al., 2005). The pseudotyped HIV-1 reporter viruses were studied in a single-round entry format using the TZM-bl target cells. A specificity control of HIV-1 reporter virus carrying the glycoprotein of amphotropic murine leukemia virus (SVA-MLV) was included in each experiment. The IC50 value for each combination of serum/plasma sample and target strain of HIV-1 was generated using standard methods. For subjects whose samples were used for epitope mapping, neutralization titers were reconfirmed by independent experiments using another form of neutralization assay, as described below.
Reporter Virus Stocks, Single-round Infection Assay and Neutralization Assay
Wild-type and mutant HIV-1JR-FL gp160 glycoproteins were expressed from the pSVIIIenv vector (Sullivan et al., 1998). Mutants were created by the PCR-based QuikChange protocol (Stratagene). The integrity of construction was confirmed by DNA sequencing of the entire env reading frame. The names of the mutants designate the wild-type amino acid residue in single-letter code, the residue number, and the substituted amino acid. Residue numbering is based on that of the prototypic HIV-1HXBc2 gp160, according to current convention (Korber, 1998).
Recombinant HIV-1 encoding firefly luciferase and pseudotyped with the wild-type or mutant Envs was produced as previously described (Yang, Wyatt, and Sodroski, 2001). Briefly, 293T cells in 100-mm tissue culture dishes were co-transfected using the Effectene reagent (Qiagen) with 1 μg pSVIIIenv plasmid expressing the Env variants and 4 μg of the pNL4-3 Δenv.luc vector. The pNL4-3 Δenv.luc vector contains a luciferase reporter gene in the HIV-1 nef locus and all HIV-1 genes except env and nef (Connor et al., 1995). The viral stocks were harvested two days later, aliquoted and stored at −80°C. All viral stocks to be compared directly were prepared as a set.
The infectivity of recombinant viruses was measured by incubation of the viruses with the Cf2Th-CD4/CCR5 target cells in a 96-well format. Viral stocks were serially diluted in a total volume of 400 μl. After thoroughly removing the media from the target cells, 100 μl of the diluted virus suspension were added to each well in triplicates. After 48 hours, viral infectivity was quantified as the luciferase activity measured by a luciferase detection kit (Pharmingen) and an automated luminometer (Berthod). The mean value and range of variation of luciferase activity from the triplicate wells were used for further analysis.
For the neutralization assays, serial dilutions of the neutralizing agent, i.e. anti-serum/plasma or mAb, were made in medium in such a volume as to produce the designated final concentration after the target virus was added. The virus-antibody mix was incubated at 37°C for 2 hours, and its residual infectivity was determined using the single-round infection assay described above. The residual infectivity (%) was defined as the infectivity measured at a given concentration of the neutralizing agent divided by the infectivity of the same virus stock mocked-treated with cell culture medium. All experiments were performed at least twice. Comparable results were achieved and a typical set of results was reported. In rare cases that data variation was noticed between experimental sessions, the average and range of variation from 4-repeat experiments were presented, as noted in the figure legends.
Antibody Depletion by Recombinant HIV-1 Env Proteins
In vitro binding/depletion assays were conducted using various forms of recombinant HIV-1 Env proteins. Codon-optimized expression vectors were used to express the wild-type and D368R gp120 monomers as well as the gp140 trimer of HIV- 1JR-FL Env. The final products of gp120 monomers contain amino acids 31-508 with a - GSG- linker and a 6xHis tag. The soluble gp140 trimer is identical to the gp140(−/FT) construct, as reported (Yang et al., 2002) and the final product contains (1) a.a. 31-683, (2) R508/511S changes to block gp120/gp41 cleavage, (3) a –Gly-Ser-Gly- linker and the trimerization motif of T4 bacteriophage fibritin, and (4) a C-terminal 6xHis tag. The gp120 monomer and gp140 trimer were produced in 293F cells and affinity-purified using the C-terminal 6xHis tag. The OD1 protein contains amino acids 252 – 482 of HIV-1YU2 Env. A V3-deleted version (OD1ΔV3) was created by deleting the following V3 sequence: PNNNTRKSINIGPGRALYTTGEIIGDIRQ. The OD1 and OD1ΔV3 proteins were expressed in S2 Drosophila cells and affinity-purified via the C-terminal 6xHis tags, as previously reported (Yang et al., 2004).
To deplete neutralizing activities from anti-HIV-1 sera, equimolar amounts of recombinant Env proteins (45 ug of gp140, 13.3 ug of gp120, or 5 ug of OD1) were bound to 25 ul of Ni-NTA Superflow beads (Qiagen) in 1x PBS. After 3 washes, antiserum at 2x the volume of the quantity that caused 90% neutralization, as titrated in preliminary experiments, was incubated with the Env/Ni-NTA complex in 400 ul of 1x PBS at 4°C for 1.5 hours with shaking. The unbound fraction was harvested and the depletion was repeated two more times. The final product after 3 depletions was concentrated using Centricon-100 (Amicon) and resuspended in 1x PBS to 10 volume of the original serum. After sterilization by passing through a 0.22-um microfilter, the residual neutralizing activity that was not depleted by the designated Env proteins was titrated using the standard single-round entry/neutralization assay as described above. A mock depletion with empty Ni-NTA beads was always conducted in parallel.
V3 Peptide Competition Assay
A peptide corresponding to the HIV-1JR-FL gp120 V3 loop, T297RPNNNTRKSIHIGPGRAFYTTGEIIGDIRQAH330, was custom synthesized (New England Peptides, Inc). To perform a binding/competition assay, 10 ug/ml of the V3 peptide was mixed with an amount of antiserum that caused 90% neutralization, as titrated by preliminary experiments, in 1x PBS at a final volume of 10x the original serum. The serum/V3 peptide mixture was incubated at 4°C for 1.5 hours, and the residual neutralizing activity in this mixture was titrated using the single-round entry assay described above. Mock treatments without peptide and with the peptide only were always used in parallel to control for specificity.
Results
Neutralizing Antibody Responses in People with Chronic HIV-1 Infection
We collected serum or plasma samples from 244 subjects with chronic HIV-1 infection in 3 separate cohorts with diverse genetic backgrounds. Neutralizing activity in samples was titrated against a panel of tier-2 HIV-1 strains (6 each from Clades B & C) by a neutralization assay based on the single-round entry of pseudotyped viruses into the TZM-bl luciferase reporter cells as described (Li et al., 2006). Samples from 14 of 244 subjects (≈6%) neutralized 10 of the 12 tier-2 HIV-1 strains with an IC50 of 1:100 dilution or higher (Table 1). For our study, this level of neutralizing activity was defined as “broad and potent nAb responses in vivo”. These 14 samples did not inhibit the entry of SVA-MLV, indicating that the virus-inhibiting activity was specific for HIV-1 Env and that the samples are free of anti-HIV-1 drugs as consistent with the subjects’ self- reporting. The protein-A-binding immunoglobulins from these samples exhibited similar levels of neutralization against a subgroup of the 12-virus panel, indicating that the neutralizing activity was truly attributable to neutralizing antibodies (data not shown). The samples from these 14 patients were further investigated to dissect the neutralizing epitope usage in vivo during chronic HIV-1 infection.
Table 1.
Neutralization Titers of Sera from Selected Subjects Against Tier-2 HIV-1 Strains#
| Clade B, Tier-2$ |
Clade C, Tier 2 |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Subjects | B1 | B2 | B3 | B4 | B5 | B6 | C1 | C2 | C3 | C4 | C5 | C6 | SVA- MLV |
| C1-211$ | 251 | 518 | 58 | 679 | 396 | 748 | 1059 | 288 | 244 | 1175 | <20 | 228 | <20 |
| C1-210 | 245 | 315 | 157 | 114 | 257 | 34 | 2802 | 378 | 102 | 1692 | 971 | 138 | <20 |
| C1-343 | 1827 | 245 | 287 | 1000 | 156 | 593 | 206 | 238 | 362 | 195 | 48 | 54 | <20 |
| C1-540 | 151 | 58 | 184 | 1158 | 22 | 514 | 220 | 163 | 178 | 122 | 305 | 1262 | <20 |
| C1-581 | 263 | 39 | 284 | 1690 | 154 | 735 | 349 | 1349 | 250 | 136 | 704 | 173 | <20 |
| C1-090 | 235 | 44 | 34 | 120 | 224 | 397 | 192 | 209 | 3414 | 136 | 150 | 2522 | <20 |
| C1-111 | 124 | 109 | 1776 | 299 | 458 | 64 | 119 | 120 | 150 | 191 | 132 | 676 | <20 |
| C1-141 | 216 | 188 | 544 | 141 | 77 | 94 | 250 | 167 | 86 | 234 | 152 | 521 | <20 |
| C1-259 | 718 | 254 | 161 | 315 | 147 | 83 | 882 | 486 | 184 | 314 | 249 | 938 | <20 |
| C1-440 | 441 | 288 | 759 | 399 | 155 | 230 | 1480 | 282 | 190 | 312 | 191 | 369 | <20 |
| C8-117 | 44 | 190 | 508 | 97 | 118 | 103 | 662 | 163 | 209 | 327 | <20 | 295 | <20 |
| C8-258 | 262 | 196 | 371 | 242 | 57 | 180 | 289 | 123 | 356 | 567 | 625 | 640 | <20 |
| C1-328* | 68 | 42 | <20 | 134 | 68 | 72 | 143 | 88 | <20 | 371 | <20 | 82 | <20 |
| UAB-B | 721 | 157 | 224 | 123 | 315 | 160 | 106 | 208 | 183 | 44 | 116 | 51 | 22 |
| UAB-M | 525 | 255 | 756 | 1568 | 93 | 367 | 169 | 68 | 77 | 355 | 248 | 99 | <20 |
| UAB-D* | 83 | 105 | 349 | 845 | 69 | 121 | 372 | 35 | 95 | 206 | 362 | 330 | <20 |
| UAB-S* | 163 | 195 | 106 | 263 | 30 | 79 | 50 | 64 | 96 | 87 | 56 | 29 | <20 |
Note: All IC50s were obtained once and reported as it is.
HIV-1 strains: B1-B6 are 6535, QH0692.42, SO422661.8, RHPA4259.7, AC10.0.29, and PVO.4, and C1-C6 are Du156.12, Du172.17, Du422.1, ZM197M.PB7, ZM214M.PL15, and CAP45.2.x.
Subjects’ name with “C1-xxx”, “C8-XXX” and “UAB-x” were from cohorts CHAVI001, CHAVI008 and UAB, respectively.
Subjects not fitting the selection standards, i.e. with more than two IC50 values below 1:100.
Three additional subjects, marked with ‘*’ in Table 1, had neutralizing activities that did not fulfill our definition of “broad and potent nAb responses’ described above, but were also included in the analysis of neutralizing epitope usage in vivo. The samples of subjects UAB-S and UAB-D exhibited rather potent neutralization of HIV-1 strains of JR-FL, KB9, ADA, YU2 and HXBc2 in our initial screening (data not shown), thus being of interest for further study. The C1-328 sample exhibited potent inhibition of a few target viruses with especially high titers due to an experimental error; the neutralizing titers were subsequently re-measured in repeated experiments and reported in Table 1. Although the neutralizing titers of C1-328 did not meet the selection standards set for this study, the neutralizing epitope targeted in this sample was easily identified and thus is included in this report.
Usage of the Glycan-dependent Epitopes in the gp120 Outer Domain
The gp120 outer domain is heavily glycosylated. Multiple N-linked glycans in the gp120 “silent face” form a neutralizing epitope recognized by mAb 2G12 (Sanders et al., 2002; Scanlan et al., 2002; Trkola et al., 1996). Removal of a glycan by the N332S mutation results in neutralization escape from 2G12 in several strains of HIV-1 Envs (Yang et al., 2005). As expected, the N332S mutant of HIV-1JR-FL gp160 was neutralized by 2G12 with low efficiency compared to the parental Envs (Fig 1. A). The N332S mutant escaped neutralization by the C1-343 and C1-259 sera for more than two 1:2 dilutions compared with the parental Envs (Fig 1. B/C). For this study, a reduction of neutralization titer by two 1:2 dilutions was defined as a significant difference, indicating that the N332S mutant had a demonstratable escape from the neutralizing activity in the sample. Experimentally, this level of change in neutralizing titer is used as an indication that the majority of the neutralizing activity in that sample is targeted to the specific epitope. Moreover, the levels of neutralization escape by the N332S mutation from samples C1-343 and C1-259 were comparable to that of purified 2G12, suggesting that the nAbs targeting the epitopes involving in the N332 residue accounted for most of the neutralizing activity in these antisera. The neutralizing activities of C1-343 and C1-259 were dependent on the N332 residue which is glycosylated and is an integral part of the glycan-dependent epitope of 2G12 (Trkola et al., 1996). Samples from other subjects did not neutralize the wild-type and N332S Envs differently (data not shown). The recombinant OD1 protein contains a functional epitope of 2G12 and can be precipitated by 2G12 (Yang et al., 2004). Surprisingly, the neutralizing activities in C1-343 and C1-259 were not depleted by the OD1 protein (Fig 1. D/E). Finally, the neutralizing activity in C1-343 but not C1-259 was effectively depleted by recombinant gp120 monomer and gp140 trimer (Fig 1. D/E). Together, these data indicate that glycan-dependent epitopes in the gp120 outer domain are targeted by antisera C1-343 and C1-259 and that the internal compositions of the two epitopes differ from each other and also from the glycan composition of the 2G12 epitope. Additionally, to test whether the N332 glycan is the direct and main binding target by nAbs in the C1-343 and C1-259 antisera, we attempted to remove the glycan from virion-associated glycoproteins using glycosidases. However, even using a large panel of glycosidases, we could not achieve a level of de-glycosylation that selectively modifies neutralizing efficiency by mAb 2G12 and anti-sera C1-343 and C1-259, without inactivating the viral infectivity, (data not shown).
Figure 1.
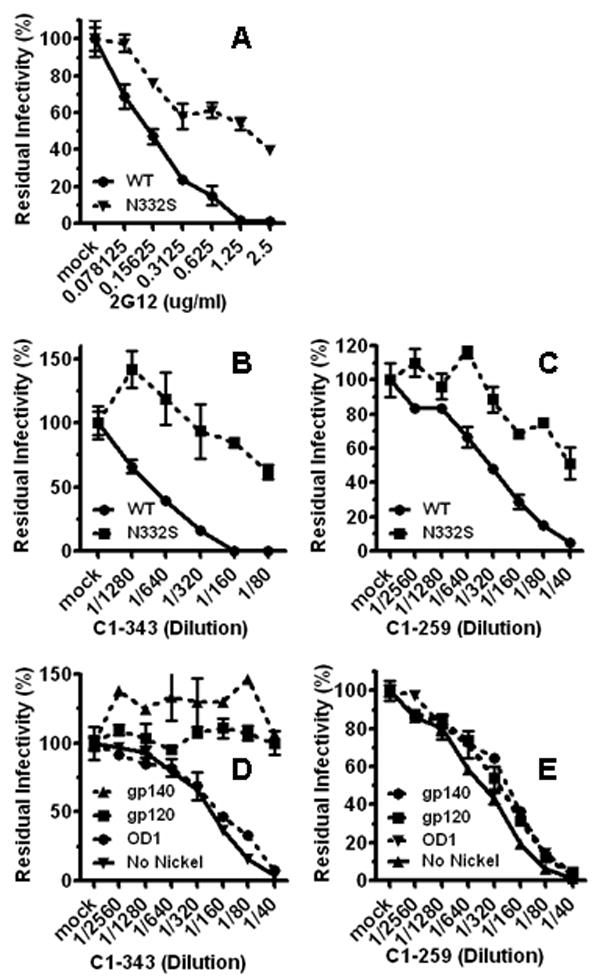
Use of the glycan-dependent epitopes on the gp120 outer domain. To quantify and compare the neutralization sensitivity of the wild-type and N332S HIV-1JR-FL gp160s to mAb 2G12 and subject antisera, the residual infectivity was measured using the single-round entry/neutralization assay. The luciferase activity of the mock-treated virus was used as the index of total infectivity, set as 100%, to which the infectivity of antibody-treated viruses was standardized as a percentage. The wild-type and N332S mutant Envs were neutralized by mAb 2G12 (A), antiserum C1-343 (B), and antiserum C1-259 (C). The same molar amounts of gp140 trimer (40 ug), gp120 monomer (13.3 ug), and OD1 (5 ug) were used to deplete neutralizing activity from antisera C1-343 (D) and C1-259 (E). The same amount of bare Ni-NTA Superflow beads (Qiagen), designated as “No Nickel”, was used as negative control.
Usage of Other Epitopes in the gp120 Outer Domain
The outer domain of gp120 contains most of the neutralizing epitopes against HIV-1 gp120 (Kwong et al., 1998). OD1 is a recombinant form of the gp120 outer domain, containing amino acids 252 to 482 (Yang et al., 2004; Zhou et al., 2007). OD1 binds mAb 2G12 as well as many conformation-dependent anti-V3 mAbs efficiently but not to those antibodies targeting linear sequences (Yang et al., 2004). OD1 binds mAb b12 weakly with an affinity detectable only by surface plasmon resonance but not by immnoprecipitation (Yang et al., 2004; Zhou et al., 2007). The protein was expressed in insect cells and purified to homogeneity, as reported previously (Yang et al., 2004). Using a vigorous depletion protocol, OD1 depleted neutralizing activity from anti-sera C1-211, C1-540, and UAB-D, indicating that the epitopes present on OD1 are targeted by nAbs in these subjects (Fig 2. A).
Figure 2.
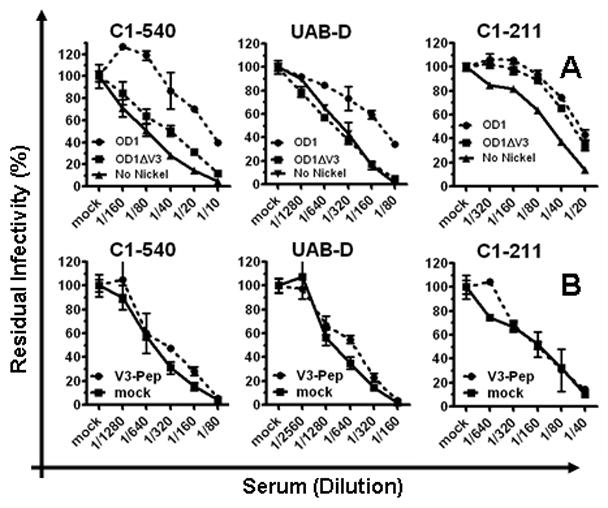
Use of other neutralizing epitopes on the gp120 outer domain. A. 5 ug of either OD1 or OD1ΔV3 proteins were used to deplete nAbs from subject antisera. Due to variation in different experimental sessions, the data on UAB-D represent averages of 4 independent experiments. B. The neutralizing activity that survived the competition with 10 ug/ml of the HIV-1JR-FL V3 peptide was measured using the standard neutralization assay, compared with same antiserum mock-treated with culture medium. The same quantity of peptide was also used to treat the target virus in parallel, and demonstrated no detectable inhibition of virus infectivity (data not shown).
A V3-deleted version of OD1 (OD1ΔV3) was also generated by deleting amino acids 299 to 328 (Yang et al., 2004). The OD1ΔV3 protein did not deplete neutralizing activity from the C1-540 serum. Also, the C1-540 neutralizing activity was not blocked by a V3 peptide in a binding/competition assay (Fig 2. B). Together, these data suggest that a conserved V3 structure, but not a linear V3 sequence, was targeted for neutralization by antiserum C1-540. The neutralizing activity in UAB-D was not depleted by OD1ΔV3 but by OD1 at a level of one to two 1:2 dilutions (Fig 2. A). The level of depletion by OD1 was modest but reproducible, consistent with the presence of nAbs with specificity for a conserved V3 structure in this antiserum; further studies revealed the presence of nAbs in this antiserum with additional specificity (see below in the CD4BS section). The neutralizing activity of UAB-D was not blocked by a V3 peptide in a competition assay (Fig 2. B). Additionally, the V3 peptide did not competitively block neutralizing activity in any of the 17 samples, indicating that linear epitopes within the gp120 V3 loop do not serve as targets for broad and potent nAb responses in these subjects (data not shown).
The neutralization activity in C1-211 was depleted by both recombinant OD1 and OD1ΔV3, indicating that the targeted epitope is located within the main body of the gp120 outer domain (Fig 2. A). Data from additional experiments suggest that the epitope targeted by this antiserum involves the CD4BS region (see below in the CD4BS section).
Detection of Epitope Use in the CD4BS Region by the T257A Mutant gp160
CD4 binds to the gp120 surface formed by elements of the outer domain and the bridging sheet (Kwong et al., 1998). This region, known as CD4BS, contains many neutralizing eptiopes, targeted by the recombinant mAb b12 that is broadly and potently neutralizing (Burton et al., 1994). We investigated the potential usage of a b12-like epitope by the subjects’ antisera with a large panel of mutant gp160 vairants, including P369L/D167N/D185N, N276A, A281V, S365A, D368Q, P369M, I371V, V372T, M373R, & K432S. Due to the heavy overlap between the CD4-binding surface and the b12 epitope, most of these mutants had significant defects in entry function and/or no escape from b12 neutralization; and additionally, neutralizing activity in none of the subject samples neutralized the wild type gp160 and these mutant vairants with discernible difference in efficiency (data not shown). The T257 residue has direct contact via its main peptide chain and side chain with b12 and it has no direct contact with the CD4 molecule, although it abuts the Phe43 cavity (Zhou et al., 2007). The T257A mutant exhibited no detectable escape from b12 neutralization (Fig 3. A). When tested against the 17 samples, the T257A virus was neutralized by serum C1-328 with a titer reduction of over two 1:2 dilutions compared to the wild-type virus, suggesting that the T257 residue was involved in the neutralizing epitope targeted by this antiserum (Fig 3. B). The T257A and wild-type viruses were neutralized equally by serum C1-343 and the other subject antisera (data not shown). The S375W change introduces an indole ring in the Phe43 cavity (Kwong et al., 1998; Xiang et al., 2002; Zhou et al., 2007). This mutation reduces gp120 binding to CD4BS antibodies, including b12, F105 and 1.5e (Xiang et al., 2002). The S375W virus was indistinguishable from the wild-type virus when subjected to antiserum C1-328 (Fig 3. B). Strikingly, the neutralizing activity in C1-328 was not depleted by the gp120 monomer or the gp140 trimer (see Fig 4. B), although both of these recombinant HIV-1 Env preparations are known to bind mAb b12 and other CD4BS mAbs (Yang et al., 2000a; Yang et al., 2000b). Together, these data support the notion that antiserum C1-328 contains a CD4BS-specific nAb(s) that is distinguishable from other known anti-CD4BS nAbs, including b12.
Figure 3.
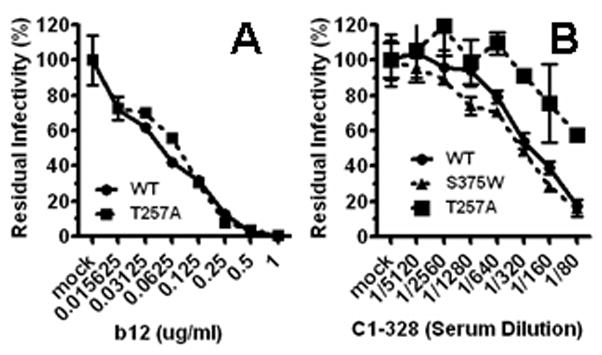
Serum neutralizing activity from subject C1-328 is sensitive to a change in the CD4BS region. Luciferase reporter viruses carrying the wild-type or T257A HIV-1JR-FL gp160s were subjected to neutralization by b12, (A) or antiserum C1-328 (B).
Figure 4.
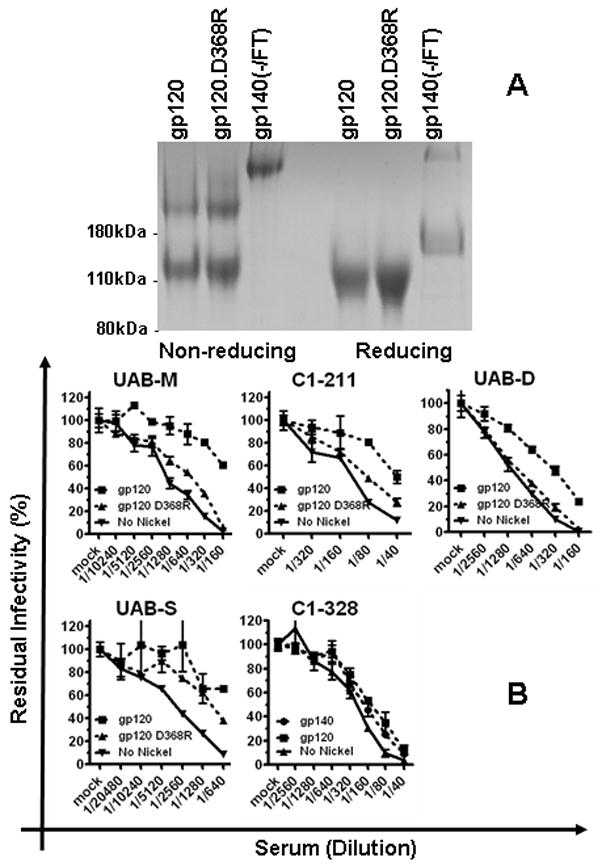
Detection of CD4BS nAbs using depletion by recombinant Env proteins. A. The wild-type and D368R mutant HIV-1JR-FL gp120 monomer and the gp140(−/FT) trimer were expressed in 293F cells and purified by Ni-NTA affinity column. Ten ug of the final products were analyzed on an 8% SDS-PAGE gel under reducing and non-reducing conditions and visualized by Coomassie blue staining. B. Same molar amounts of the wild-type or D368R gp120 monomer, or the gp140 trimer were used to deplete neutralizing activity from subject antisera. The neutralizing activity in the final flow-through was measured using the standard neutralization assay, in parallel to the same antiserum mock-depleted by empty Ni-NTA beads. Due to variations in different experimental sessions, the data on UAB-D represent averages of 4 independent experiments.
Detection of Epitope Use in the CD4BS Region by Depletion with Recombinant Env Proteins
The D368R mutation in recombinant gp120 results in a loss of the ability to bind and deplete antibodies directed against the CD4BS site (Li et al., 2007; Thali et al., 1992). This phenotype change has been utilized to characterize the presence of the CD4BS nAbs in patient sera by several groups (Binley et al., 2008; Dhillon et al., 2007; Gray et al., 2009b; Li et al., 2007; Li et al., 2009; Sather et al., 2009). We adopted this assay to complement the experiments described above. The recombinant gp120 monomers of wild-type and D368R HIV-1JR-FL Envs were expressed in 293F suspension cells and purified to homogeneity (Fig 4. A). Using very vigorous depletion conditions, wild-type gp120 depleted significant levels of neutralizing activity from UAB-M, C1-211, UAB-D, and UAB-S (Fig 4. B). In contrast, wild-type gp120 did not appreciably deplete neutralizing activity in 11 other samples (see Table 2), notably antiserum C1-328 which has nAb(s) to an epitope involving residue T257 (see above) (Fig 4. B and data not shown). The neutralizing activity was depleted by both the wild-type and D368R gp120 monomers, suggesting that the targeted neutralizing epitope is located outside the CD4BS region (Fig 4. B). The neutralizing activity in UAB-M was depleted by the wild- type gp120 but not by the D368R variant, similar to many known CD4BS nAbs (Fig 4. B). The neutralizing activity of C1-211 was depleted efficiently by the wild-type but not D368R gp120, suggesting that the CD4BS site was targeted by this antiserum (Fig 4. B). As shown in Fig 2, the neutralizing epitope targeted by antiserum C1-211 is located in the main body of the gp120 outer domain. Taken together, these results support the notion that the nAb(s) in antiserum C1-211 targets a CD4BS epitope that is mainly, if not exclusively, composed of elements from the gp120 outer domain.
Table 2.
Summary of Epitopes for Broad & Potent nAbs in HIV-1-infected Persons
| Subject ID | Specific escape | Depletion |
Epitope projection | ||||
|---|---|---|---|---|---|---|---|
| OD1 | OD1ΔV3 | gp120 | gp120.D368R | gp140 | |||
| C1-343 | N332S | no | n/d | yes | n/d | yes | OD glycan |
| C1-259 | N332S | no | n/d | no | n/d | no | OD glycan |
| C1-540 | None | yes | no | yes | n/d | yes | OD V3 |
| UAB-D | None | yes | no | yes | no | yes | OD V3/CD4BS |
| C1-211 | None | yes | yes | yes | no | yes | OD CD4BS |
| UAB-M | None | no | n/d | yes | no | yes | CD4BS |
| C1-328 | T257A | no | n/d | no | n/d | no | CD4BS |
| UAB-S | None | no | n/d | yes | yes | yes | Not-identified |
| C1-210 | None | no | n/d | no | no | no | Not-identified |
| C1-111 | None | no | n/d | no | no | no | Not-identified |
| C1-141 | None | no | n/d | no | no | no | Not-identified |
| C8-117 | None | no | n/d | no | no | no | Not-identified |
| UAB-B | None | no | n/d | no | n/d | no | Not-identified |
| C8-258 | None | no | n/d | no | no | no | Not-identified |
| C1-440 | None | no | n/d | no | no | no | Not-identified |
| C1-090 | None | no | n/d | no | no | no | Not-identified |
| C1-581 | None | no | n/d | no | no | no | Not-identified |
The neutralizing activity in antiserum UAB-D was depleted by the wild-type gp120 at a level between one to two 1:2 dilutions in repeated experiments, and this neutralizing activity was not depleted by the D368R gp120 (Fig 4. B). These data indicated the presence of a CD4BS nAb in this serum, even though this level of depletion is below what we define as a significant difference in this study. As shown in Fig 2, this antiserum also contains neutralization activity specific for a conserved V3 structure (see above). The simplest rational explanation for the combined data in Fig 4. B and Fig 2 is that the neutralizing activity in UAB-D comprises two specificities, one for CD4BS and one for the conserved V3 structure.
Summary of the Neutralizing Epitope Use during Chronic HIV-1 Infection
Table 2 summarizes the neutralizing epitope usage for the 17 subjects with chronic HIV-1 infection from diverse backgrounds. The most frequently used epitopes (4/17) is in the CD4BS region. There are three epitopes in the gp120 outer domain targeted for neutralization in 5 subjects. Among the epitopes residing in the gp120 outer domain, two are the N332S-sensitive, glycan-dependent epitopes on the silent face of gp120, two are composed of conserved V3 structural elements, and one is a CD4BS epitope. Neutralizing epitope usage in 10 of the 17 subjects is refractory to characterization by the two methods used in this study. Among these 10 subjects, 9 have neutralizing activity that is not sensitive to depletion by recombinant HIV-1 Env proteins, including OD1, gp120, gp140 trimer, and their derivatives. The failure to deplete neutralizing activity from most of the HIV-1-neutralizing antisera by similar recombinant HIV-1 proteins has been reported by multiple investigators, indicating significant differences in the structure of these recombinant proteins from that of a native, functional Env trimer presented on virion surface (Table 2) (Binley et al., 2008; Dhillon et al., 2007; Gray et al., 2009b; Li et al., 2007; Li et al., 2009; Sather et al., 2009).
Discussion
We studied 244 subjects with chronic HIV-1 infection from 3 cohorts with diverse backgrounds. Approximately 6% of the subjects (14 out of 244 subjects) has broad and potent nAb responses that can neutralize 80% of tier-2 HIV-1 strains with a serum-dilution titer of 1:100 or higher. This frequency of responses of broad and potent neutralizing antibodies during HIV-1 infection is consistent with other findings (Binley et al., 2008; Li et al., 2009; Simek et al., 2009). In samples from the 17 subjects, the CD4-binding site is targeted for nAb responses in vivo in 4 cases, consistent with recent findings (Binley et al., 2008; Dhillon et al., 2007; Gray et al., 2009b; Li et al., 2007; Li et al., 2009; Sather et al., 2009). The novel result from our study is that the gp120 outer domain is also frequently targeted (5 of the 17 subjects) by naturally arising nAbs in vivo. The neutralizing epitopes in the gp120 outer domain can be classified into three groups: 1) the CD4BS epitopes (1); 2) glycan-dependent epitopes (2); and 3) the conserved V3 structures (2). Using a synthetic peptide homologous to the target HIV-1JR-FL V3 loop to compete/block for V3-specific antibodies (see Material and Methods), we detected no neutralizing activity targeting the gp120 V3 linear peptide sequences in this group of 17 subjects. Using a K665N/W672 mutant to screen for neutralizing activity to the 2F5 and 4E10 epitopes (data not shown), no neutralizing activity targeting the 2F5/4E10 epitopes of MPER was found in the 17 subjects. However, limited experiments in this study cannot rule out nAbs to these epitopes may contribute to the neutralizing activity in some of the subjects. We have not yet attempted to detect the potential use of the V2/V3 epitopes targeted by mAbs PG9 and PG16 in this study.
In our experiments, the identification of targeted neutralizing epitopes is based on the detection of a change of neutralization efficiency by two or more 1:2 dilutions of an antiserum. In most cases (6 of the 7 subjects with defined epitope usage), the neutralizing epitope identified by this experiment standard is responsible for the majority of the neutralizing activity in the antiserum. The neutralizing activity in the antiserum UAB-D is sensitive to deletion of the V3 loop (depletion by the OD1 and OD1ΔV3 proteins) and also to the D368R mutation (in the context of depletion by the gp120 and gp120.D368R monomers). It is not clear why the V3 loop in the gp120.D368R monomer does not deplete the putative V3-specific nAb that is sensitive to depletion by OD1 but not OD1ΔV3; it may be that the V3 loop is simply exposed better in OD1 than in gp120 monomer, thus leading to better binding of OD1 and the targeted nAbs. Notwithstanding this possibility, poly-specificity of nAbs in subject UAB-D for both V3 and CD4BS epitopes is the most logical explanation for the differential depletion of neutralizing activity by OD1/OD1ΔV3 and gp120/gp120.D368R and for the observation that both gp120 and OD1 can only achieve a partial depletion of neutralizing activity, relative to that achieved for other antisera with apparently mono-specific neutralizing epitope usage (see Fig 2. A and 4. B). It is possible that the D368R mutation can affect antibody binding by the V3 loop in the context of monomeric gp120; and the UAB-D antiserum is specific for the V3 epitope only. However, based on current understanding of the structure of HIV-1 gp120, such an effect of the D368R change would not be expected (Chen et al., 2005; Kwong et al., 2000; Kwong et al., 1998; Liu et al., 2008; Zhu et al., 2003).
Our data indicate that glycan-dependent epitopes in the gp120 outer domain, related to the 2G12 epitope, are targeted in 2 of the 17 subjects, although binding of the glycan itself has not been proven directly. This adds to the evidence that the glycans concentrated on the gp120 outer domain can induce nAb responses in vivo, as indicated by the isolation of mAb 2G12 using the technology of EBV-transformation of B-lymphocytes from an HIV-1-infected individual (Trkola et al., 1996). In our study, we used a single glyan-deleting mutant, thus potentially underestimating the frequency at which this type of nAb is induced during HIV-1 infection. The composition of such glycan-dependent epitopes might be diverse. For example, both the gp120 and the gp140 trimer can deplete the neutralizing activity from the C1-343 antiserum but not from the C1-259 antiserum. Additionally, OD1 does not deplete the neutralizing activities from antisera C1-259 and C1-343, despite OD1 being able to efficiently bind to 2G12 (Yang et al., 2004). The diversity in glycan compositions of this epitope group may make the design of immunogens to induce similar nAb responses challenging, consistent with the outcome of a recent study (Luallen et al., 2008).
Two of the 17 subjects, C1-540 and C1-211, have nAbs targeting the conserved structural elements of V3, while none have nAbs to the linear peptide epitopes in V3. The V3 tip, especially those with the -GPGR- sequence, has been reported to bind potent and broad nAb in vitro (Gorny et al., 1993; Stanfield et al., 2004). However, while the multi-specificities of the nAbs in the C1-211 antiserum complicates the assessment of the neutralization breadth of the V3-specific nAbs in this antiserum, the neutralization breadth of the anti-V3 nAb in C1-540 is significantly wider than that of the anti-V3 antibodies reported previously (Gorny et al., 1993; Stanfield et al., 2004). The V3 base is also conserved structurally as shown in several X-ray crystal structures and in primary sequences (Kwong et al., 2000; Kwong et al., 1998). Although the exact nature of these V3 epitopes is not clear, they represent novel neutralizing targets worthy of further investigation.
Among the known neutralizing epitopes, the CD4BS region are targeted frequently by broad and potent nAbs in vivo while the frequency is very low among general population of HIV-1-infected people, consistent with recent reports (Dhillon et al., 2007; Gray et al., 2009b; Li et al., 2007; Li et al., 2009; Sather et al., 2009). This conclusion is also generally consistent with early reports of serum nAb specificities based on studying TCLA strains of HIV-1 (Haigwood et al., 1992; Kang et al., 1991; Moore and Ho, 1993). The CD4BS-specific nAbs in subjects UAB-D and UAB-M exhibit known characteristics commonly associated with CD4BS antibodies, such as not binding to gp120.D268R (Table 2) and sensitive to some mutations that affect the structural integrity of HIV-1 glycoprotein (data not shown). The CD4BS-specific nAbs in subjects C1-211 and C1-328 have distinct characteristics. The C1-211 epitope resides largely, if not totally, in the gp120 outer domain, as the neutralizing activity of this antiserum was efficiently depleted by the OD1 protein. It has been suggested that the propensity to bind larger part of the outer domain as its epitope contributes to the neutralization effectiveness of mAb b12, compared to other CD4BS antibodies that have less neutralizing capacity (Pantophlet et al., 2003; Saphire et al., 2001; Zhou et al., 2007). Thus, fine dissection of the antibody and the epitope in C1-211 might provide important clues for redesigning the OD1 protein to improve its immunogenicity (Wu et al., 2009; Yang et al., 2004). The epitope targeted in C1-328 is located within the CD4BS region as indicated by its sensitivity to the T275A mutation. Distinct from known CD4BS mAbs such as b12, F107, F91, this neutralizing activity is not sensitive to the S375W mutation and is resistant to depletion by recombinant gp120 and gp140 trimers (Fig 3. B & 4. B). Thus, the nAb(s) in this antiserum appears to have a distinct ability to recognize the native, infectious HIV-1 Env trimers but not the recombinant Env proteins. The inability of recombinant gp120 and gp140 trimer to deplete neutralizing activities from antisera in this study as well as in many other reports is indicative of the structural defect in these recombinant HIV-1 Env proteins when compared to a native HIV-1 Env trimer. The structure of a native, infectious HIV-1 Env trimer is still an illusive entity in the field, and clear understanding of it is critical for modification of recombinant Env proteins as vaccine candidates. Thus, characterizing the nAb(s) in antiserum C1-328 and defining the neutralizing epitope clearly will be helpful to define the properties of an HIV-1 Env trimer in its native state.
Finally, we showed that 11 of 17 subjects have nAbs which cannot be depleted by any of the recombinant Env proteins. Differential depletion using the wild-type and D368R mutant gp120s only contributed to the characterization of 2 antisera in this study, similar to several recent studies (Binley et al., 2008; Dhillon et al., 2007; Li et al., 2007; Li et al., 2009; Sather et al., 2009). In this study, we have developed some new approaches to identify the epitope usage in vivo by studying serum neutralizing activity. However, it is clearly that there is still an urgent need to further develop new and more effective methods to define the neutralizing epitope usage in HIV-1 infection.
Acknowledgments
We think Dr. Connie Gee for help in preparing this manuscript. We acknowledge the critical contributions of identifying and collecting samples from study subjects by the CHAVI 001 and 008 clinical working teams and the associates of Dr. Goepfert at UAB. We thank R.G. Overman for technical assistance. This work is supported by the NIAID Center for HIV/AIDS Vaccine Immunology grant AI067854 and a grant from NIH to X.Y. - RO1 AI073133.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Binley JM, Lybarger EA, Crooks ET, Seaman MS, Gray E, Davis KL, Decker JM, Wycuff D, Harris L, Hawkins N, Wood B, Nathe C, Richman D, Tomaras GD, Bibollet-Ruche F, Robinson JE, Morris L, Shaw GM, Montefiori DC, Mascola JR. Profiling the specificity of neutralizing antibodies in a large panel of plasmas from patients chronically infected with human immunodeficiency virus type 1 subtypes B and C. J Virol. 2008;82 (23):11651–68. doi: 10.1128/JVI.01762-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, Pyati J, Koduri R, Sharp SJ, Thornton GB, Parren PW, Sawyer LS, Hendry RM, Dunlop N, Nara PL, et al. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266(5187):1024–7. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- Chen B, Vogan EM, Gong H, Skehel JJ, Wiley DC, Harrison SC. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature. 2005;433(7028):834–41. doi: 10.1038/nature03327. [DOI] [PubMed] [Google Scholar]
- Connor RI, Chen BK, Choe S, Landau NR. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206 (2):935–44. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- Decker JM, Bibollet-Ruche F, Wei X, Wang S, Levy DN, Wang W, Delaporte E, Peeters M, Derdeyn CA, Allen S, Hunter E, Saag MS, Hoxie JA, Hahn BH, Kwong PD, Robinson JE, Shaw GM. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J Exp Med. 2005;201 (9):1407–19. doi: 10.1084/jem.20042510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks SG, Schweighardt B, Wrin T, Galovich J, Hoh R, Sinclair E, Hunt P, McCune JM, Martin JN, Petropoulos CJ, Hecht FM. Neutralizing antibody responses against autologous and heterologous viruses in acute versus chronic human immunodeficiency virus (HIV) infection: evidence for a constraint on the ability of HIV to completely evade neutralizing antibody responses. J Virol. 2006;80 (12):6155–64. doi: 10.1128/JVI.00093-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon AK, Donners H, Pantophlet R, Johnson WE, Decker JM, Shaw GM, Lee FH, Richman DD, Doms RW, Vanham G, Burton DR. Dissecting the neutralizing antibody specificities of broadly neutralizing sera from human immunodeficiency virus type 1-infected donors. J Virol. 2007;81 (12):6548–62. doi: 10.1128/JVI.02749-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaschen B, Kuiken C, Korber B, Foley B. Retrieval and on-the-fly alignment of sequence fragments from the HIV database. Bioinformatics. 2001;17 (5):415–8. doi: 10.1093/bioinformatics/17.5.415. [DOI] [PubMed] [Google Scholar]
- Gorny MK, VanCott TC, Hioe C, Israel ZR, Michael NL, Conley AJ, Williams C, Kessler JA, 2nd, Chigurupati P, Burda S, Zolla-Pazner S. Human monoclonal antibodies to the V3 loop of HIV-1 with intra- and interclade cross-reactivity. J Immunol. 1997;159 (10):5114–22. [PubMed] [Google Scholar]
- Gorny MK, Williams C, Volsky B, Revesz K, Cohen S, Polonis VR, Honnen WJ, Kayman SC, Krachmarov C, Pinter A, Zolla-Pazner S. Human monoclonal antibodies specific for conformation-sensitive epitopes of V3 neutralize human immunodeficiency virus type 1 primary isolates from various clades. J Virol. 2002;76 (18):9035–45. doi: 10.1128/JVI.76.18.9035-9045.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, Xu JY, Karwowska S, Buchbinder A, Zolla-Pazner S. Repertoire of neutralizing human monoclonal antibodies specific for the V3 domain of HIV-1 gp120. J Immunol. 1993;150 (2):635–43. [PubMed] [Google Scholar]
- Gray ES, Madiga MC, Moore PL, Mlisana K, Karim SS, Binley JM, Shaw GM, Mascola JR, Morris L. Broad HIV-1 neutralization mediated by plasma antibodies against the gp41 membrane proximal external region. J Virol. 2009a doi: 10.1128/JVI.01359-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray ES, Taylor N, Wycuff D, Moore PL, Tomaras GD, Wibmer CK, Puren A, Decamp A, Gilbert PB, Wood B, Montefiori DC, Binley JM, Shaw GM, Haynes BF, Mascola JR, Morris L. Antibody specificities associated with neutralization breadth in plasma from HIV-1 subtype C infected blood donors. J Virol. 2009b doi: 10.1128/JVI.00758-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigwood NL, Nara PL, Brooks E, Van Nest GA, Ott G, Higgins KW, Dunlop N, Scandella CJ, Eichberg JW, Steimer KS. Native but not denatured recombinant human immunodeficiency virus type 1 gp120 generates broad-spectrum neutralizing antibodies in baboons. J Virol. 1992;66(1):172–82. doi: 10.1128/jvi.66.1.172-182.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang CY, Nara P, Chamat S, Caralli V, Ryskamp T, Haigwood N, Newman R, Kohler H. Evidence for non-V3-specific neutralizing antibodies that interfere with gp120/CD4 binding in human immunodeficiency virus 1-infected humans. Proc Natl Acad Sci U S A. 1991;88 (14):6171–5. doi: 10.1073/pnas.88.14.6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber BF, Kuiken F, Pillai C, Sodroksi JS. Numbering positions in HIV relative to HXBc2. In: Korber BK, Foley C, Hahn F, McCutchan B, Mellor F, Sodroski J, editors. Human retroviruses and AIDS. Los Alamos National Laboratories; LOS Alamos: 1998. [Google Scholar]
- Kuiken C, Korber B, Shafer RW. HIV sequence databases. AIDS Rev. 2003;5(1):52–61. [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, Wyatt R, Majeed S, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Structure Fold Des. 2000;8(12):1329–39. doi: 10.1016/s0969-2126(00)00547-5. [DOI] [PubMed] [Google Scholar]
- Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393 (6686):648–59. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2005;79(16):10108–25. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Salazar-Gonzalez JF, Derdeyn CA, Morris L, Williamson C, Robinson JE, Decker JM, Li Y, Salazar MG, Polonis VR, Mlisana K, Karim SA, Hong K, Greene KM, Bilska M, Zhou J, Allen S, Chomba E, Mulenga J, Vwalika C, Gao F, Zhang M, Korber BT, Hunter E, Hahn BH, Montefiori DC. Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in Southern Africa. J Virol. 2006;80 (23):11776–90. doi: 10.1128/JVI.01730-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Migueles SA, Welcher B, Svehla K, Phogat A, Louder MK, Wu X, Shaw GM, Connors M, Wyatt RT, Mascola JR. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat Med. 2007;13 (9):1032–4. doi: 10.1038/nm1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Svehla K, Louder MK, Wycuff D, Phogat S, Tang M, Migueles SA, Wu X, Phogat A, Shaw GM, Connors M, Hoxie J, Mascola JR, Wyatt R. Analysis of neutralization specificities in polyclonal sera derived from human immunodeficiency virus type 1-infected individuals. J Virol. 2009;83 (2):1045–59. doi: 10.1128/JVI.01992-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455(7209):109–13. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luallen RJ, Lin J, Fu H, Cai KK, Agrawal C, Mboudjeka I, Lee FH, Montefiori D, Smith DF, Doms RW, Geng Y. An engineered Saccharomyces cerevisiae strain binds the broadly neutralizing human immunodeficiency virus type 1 antibody 2G12 and elicits mannose-specific gp120-binding antibodies. J Virol. 2008;82(13):6447–57. doi: 10.1128/JVI.00412-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascola JR, Lewis MG, Stiegler G, Harris D, VanCott TC, Hayes D, Louder MK, Brown CR, Sapan CV, Frankel SS, Lu Y, Robb ML, Katinger H, Birx DL. Protection of Macaques against pathogenic simian/human immunodeficiency virus 89.6PD by passive transfer of neutralizing antibodies. J Virol. 1999;73 (5):4009–18. doi: 10.1128/jvi.73.5.4009-4018.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrow S, Hahn BH, Shaw GM, Gallo RC, Wong-Staal F, Wolf H. Computer-assisted analysis of envelope protein sequences of seven human immunodeficiency virus isolates: prediction of antigenic epitopes in conserved and variable regions. J Virol. 1987;61 (2):570–8. doi: 10.1128/jvi.61.2.570-578.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JP, Ho DD. Antibodies to discontinuous or conformationally sensitive epitopes on the gp120 glycoprotein of human immunodeficiency virus type 1 are highly prevalent in sera of infected humans. J Virol. 1993;67 (2):863–75. doi: 10.1128/jvi.67.2.863-875.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JP, McCutchan FE, Poon SW, Mascola J, Liu J, Cao Y, Ho DD. Exploration of antigenic variation in gp120 from clades A through F of human immunodeficiency virus type 1 by using monoclonal antibodies. J Virol. 1994;68 (12):8350–64. doi: 10.1128/jvi.68.12.8350-8364.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JP, Sodroski J. Antibody cross-competition analysis of the human immunodeficiency virus type 1 gp120 exterior envelope glycoprotein. J Virol. 1996;70(3):1863–72. doi: 10.1128/jvi.70.3.1863-1872.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muster T, Steindl F, Purtscher M, Trkola A, Klima A, Himmler G, Ruker F, Katinger H. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J Virol. 1993;67 (11):6642–7. doi: 10.1128/jvi.67.11.6642-6647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers G, Berzofsky B, Korber BT, Smith R, Pavlakis G. A compilation and analysis of nucleic and amino acid sequecnes. Los alamos, NM: 1992. Human retroviruses and AIDS. [Google Scholar]
- Pantophlet R, Ollmann Saphire E, Poignard P, Parren PW, Wilson IA, Burton DR. Fine mapping of the interaction of neutralizing and nonneutralizing monoclonal antibodies with the CD4 binding site of human immunodeficiency virus type 1 gp120. J Virol. 2003;77 (1):642–58. doi: 10.1128/JVI.77.1.642-658.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parren PW, Marx PA, Hessell AJ, Luckay A, Harouse J, Cheng-Mayer C, Moore JP, Burton DR. Antibody protects macaques against vaginal challenge with a pathogenic R5 simian/human immunodeficiency virus at serum levels giving complete neutralization in vitro. J Virol. 2001;75 (17):8340–7. doi: 10.1128/JVI.75.17.8340-8347.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posner MR, Cavacini LA, Emes CL, Power J, Byrn R. Neutralization of HIV-1 by F105, a human monoclonal antibody to the CD4 binding site of gp120. J Acquir Immune Defic Syndr. 1993;6 (1):7–14. [PubMed] [Google Scholar]
- Robinson JE, Holton D, Pacheco-Morell S, Liu J, McMurdo H. Identification of conserved and variant epitopes of human immunodeficiency virus type 1 (HIV-1) gp120 by human monoclonal antibodies produced by EBV-transformed cell lines. AIDS Res Hum Retroviruses. 1990;6 (5):567–79. doi: 10.1089/aid.1990.6.567. [DOI] [PubMed] [Google Scholar]
- Sanders RW, Venturi M, Schiffner L, Kalyanaraman R, Katinger H, Lloyd KO, Kwong PD, Moore JP. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J Virol. 2002;76 (14):7293–305. doi: 10.1128/JVI.76.14.7293-7305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saphire EO, Parren PW, Pantophlet R, Zwick MB, Morris GM, Rudd PM, Dwek RA, Stanfield RL, Burton DR, Wilson IA. Crystal structure of a neutralizing human IGG against HIV-1: a template for vaccine design. Science. 2001;293(5532):1155–9. doi: 10.1126/science.1061692. [DOI] [PubMed] [Google Scholar]
- Sather DN, Armann J, Ching LK, Mavrantoni A, Sellhorn G, Caldwell Z, Yu X, Wood B, Self S, Kalams S, Stamatatos L. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J Virol. 2009;83 (2):757–69. doi: 10.1128/JVI.02036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlan CN, Pantophlet R, Wormald MR, Ollmann Saphire E, Stanfield R, Wilson IA, Katinger H, Dwek RA, Rudd PM, Burton DR. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of alpha1-->2 mannose residues on the outer face of gp120. J Virol. 2002;76(14):7306–21. doi: 10.1128/JVI.76.14.7306-7321.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Parks RJ, Montefiori DC, Kirchherr JL, Keele BF, Decker JM, Blattner WA, Gao F, Weinhold KJ, Hicks CB, Greenberg ML, Hahn BH, Shaw GM, Haynes BF, Tomaras GD. In vivo gp41 antibodies targeting the 2F5 monoclonal antibody epitope mediate human immunodeficiency virus type 1 neutralization breadth. J Virol. 2009;83 (8):3617–25. doi: 10.1128/JVI.02631-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata R, Igarashi T, Haigwood N, Buckler-White A, Ogert R, Ross W, Willey R, Cho MW, Martin MA. Neutralizing antibody directed against the HIV-1 envelope glycoprotein can completely block HIV-1/SIV chimeric virus infections of macaque monkeys. Nat Med. 1999;5 (2):204–10. doi: 10.1038/5568. [DOI] [PubMed] [Google Scholar]
- Simek MD, Rida W, Priddy FH, Pung P, Carrow E, Laufer DS, Lehrman JK, Boaz M, Tarragona-Fiol T, Miiro G, Birungi J, Pozniak A, McPhee DA, Manigart O, Karita E, Inwoley A, Jaoko W, Dehovitz J, Bekker LG, Pitisuttithum P, Paris R, Walker LM, Poignard P, Wrin T, Fast PE, Burton DR, Koff WC. Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J Virol. 2009;83 (14):7337–48. doi: 10.1128/JVI.00110-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanfield RL, Gorny MK, Williams C, Zolla-Pazner S, Wilson IA. Structural rationale for the broad neutralization of HIV-1 by human monoclonal antibody 447–52D. Structure (Camb) 2004;12 (2):193–204. doi: 10.1016/j.str.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Starcich BR, Hahn BH, Shaw GM, McNeely PD, Modrow S, Wolf H, Parks ES, Parks WP, Josephs SF, Gallo RC, et al. Identification and characterization of conserved and variable regions in the envelope gene of HTLV-III/LAV, the retrovirus of AIDS. Cell. 1986;45(5):637–48. doi: 10.1016/0092-8674(86)90778-6. [DOI] [PubMed] [Google Scholar]
- Stiegler G, Kunert R, Purtscher M, Wolbank S, Voglauer R, Steindl F, Katinger H. A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 2001;17 (18):1757–65. doi: 10.1089/08892220152741450. [DOI] [PubMed] [Google Scholar]
- Sullivan N, Sun Y, Binley J, Lee J, Barbas CF, 3rd, Parren PW, Burton DR, Sodroski J. Determinants of human immunodeficiency virus type 1 envelope glycoprotein activation by soluble CD4 and monoclonal antibodies. J Virol. 1998;72(8):6332–8. doi: 10.1128/jvi.72.8.6332-6338.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thali M, Furman C, Ho DD, Robinson J, Tilley S, Pinter A, Sodroski J. Discontinuous, conserved neutralization epitopes overlapping the CD4-binding region of human immunodeficiency virus type 1 gp120 envelope glycoprotein. J Virol. 1992;66(9):5635–41. doi: 10.1128/jvi.66.9.5635-5641.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thali M, Moore JP, Furman C, Charles M, Ho DD, Robinson J, Sodroski J. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol. 1993;67 (7):3978–88. doi: 10.1128/jvi.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore JP, Katinger H. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol. 1996;70 (2):1100–8. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veazey RS, Shattock RJ, Pope M, Kirijan JC, Jones J, Hu Q, Ketas T, Marx PA, Klasse PJ, Burton DR, Moore JP. Prevention of virus transmission to macaque monkeys by a vaginally applied monoclonal antibody to HIV-1 gp120. Nat Med. 2003;9 (3):343–6. doi: 10.1038/nm833. [DOI] [PubMed] [Google Scholar]
- Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Miiro G, Serwanga J, Pozniak A, McPhee D, Manigart O, Mwananyanda L, Karita E, Inwoley A, Jaoko W, Dehovitz J, Bekker LG, Pitisuttithum P, Paris R, Allen S, Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR. Broad and Potent Neutralizing Antibodies from an African Donor Reveal a New HIV-1 Vaccine Target. Science. 2009 doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willey RL, Rutledge RA, Dias S, Folks T, Theodore T, Buckler CE, Martin MA. Identification of conserved and divergent domains within the envelope gene of the acquired immunodeficiency syndrome retrovirus. Proc Natl Acad Sci U S A. 1986;83(14):5038–42. doi: 10.1073/pnas.83.14.5038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Zhou T, Yang ZY, Svehla K, O’Dell S, Louder MK, Xu L, Mascola JR, Burton DR, Hoxie JA, Doms RW, Kwong PD, Nabel GJ. Enhanced exposure of the CD4-binding site to neutralizing antibodies by structural design of a membrane-anchored human immunodeficiency virus type 1 gp120 domain. J Virol. 2009;83 (10):5077–86. doi: 10.1128/JVI.02600-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt R, Kwong PD, Desjardins E, Sweet RW, Robinson J, Hendrickson WA, Sodroski JG. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393(6686):705–11. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280 (5371):1884–8. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- Xiang SH, Kwong PD, Gupta R, Rizzuto CD, Casper DJ, Wyatt R, Wang L, Hendrickson WA, Doyle ML, Sodroski J. Mutagenic stabilization and/or disruption of a CD4-bound state reveals distinct conformations of the human immunodeficiency virus type 1 gp120 envelope glycoprotein. J Virol. 2002;76 (19):9888–99. doi: 10.1128/JVI.76.19.9888-9899.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang SH, Wang L, Abreu M, Huang CC, Kwong PD, Rosenberg E, Robinson JE, Sodroski J. Epitope mapping and characterization of a novel CD4-induced human monoclonal antibody capable of neutralizing primary HIV-1 strains. Virology. 2003;315(1):124–34. doi: 10.1016/s0042-6822(03)00521-x. [DOI] [PubMed] [Google Scholar]
- Yang X, Farzan M, Wyatt R, Sodroski J. Characterization of stable, soluble trimers containing complete ectodomains of human immunodeficiency virus type 1 envelope glycoproteins. J Virol. 2000a;74 (12):5716–25. doi: 10.1128/jvi.74.12.5716-5725.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Florin L, Farzan M, Kolchinsky P, Kwong PD, Sodroski J, Wyatt R. Modifications that stabilize human immunodeficiency virus envelope glycoprotein trimers in solution. J Virol. 2000b;74 (10):4746–54. doi: 10.1128/jvi.74.10.4746-4754.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Kurteva S, Lee S, Sodroski J. Stoichiometry of antibody neutralization of human immunodeficiency virus type 1. J Virol. 2005;79(6):3500–8. doi: 10.1128/JVI.79.6.3500-3508.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Lee J, Mahony EM, Kwong PD, Wyatt R, Sodroski J. Highly stable trimers formed by human immunodeficiency virus type 1 envelope glycoproteins fused with the trimeric motif of T4 bacteriophage fibritin. J Virol. 2002;76 (9):4634–42. doi: 10.1128/JVI.76.9.4634-4642.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Tomov V, Kurteva S, Wang L, Ren X, Gorny MK, Zolla-Pazner S, Sodroski J. Characterization of the outer domain of the gp120 glycoprotein from human immunodeficiency virus type 1. J Virol. 2004;78 (23):12975–86. doi: 10.1128/JVI.78.23.12975-12986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Wyatt R, Sodroski J. Improved elicitation of neutralizing antibodies against primary human immunodeficiency viruses by soluble stabilized envelope glycoprotein trimers. J Virol. 2001;75 (3):1165–71. doi: 10.1128/JVI.75.3.1165-1171.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, Yang X, Zhang MY, Zwick MB, Arthos J, Burton DR, Dimitrov DS, Sodroski J, Wyatt R, Nabel GJ, Kwong PD. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445(7129):732–7. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P, Chertova E, Bess J, Jr, Lifson JD, Arthur LO, Liu J, Taylor KA, Roux KH. Electron tomography analysis of envelope glycoprotein trimers on HIV and simian immunodeficiency virus virions. Proc Natl Acad Sci U S A. 2003;100 (26):15812–7. doi: 10.1073/pnas.2634931100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolla-Pazner S. Improving on nature: focusing the immune response on the V3 loop. Hum Antibodies. 2005;14(3–4):69–72. [PubMed] [Google Scholar]
- Zwick MB, Labrijn AF, Wang M, Spenlehauer C, Saphire EO, Binley JM, Moore JP, Stiegler G, Katinger H, Burton DR, Parren PW. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J Virol. 2001;75 (22):10892–905. doi: 10.1128/JVI.75.22.10892-10905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]


